Abstract
Accurate detection of COVID-19 is one of the challenging research topics in today's healthcare sector to control the coronavirus pandemic. Automatic data-powered insights for COVID-19 localization from medical imaging modality like chest CT scan tremendously augment clinical care assistance. In this research, a Contour-aware Attention Decoder CNN has been proposed to precisely segment COVID-19 infected tissues in a very effective way. It introduces a novel attention scheme to extract boundary, shape cues from CT contours and leverage these features in refining the infected areas. For every decoded pixel, the attention module harvests contextual information in its spatial neighborhood from the contour feature maps. As a result of incorporating such rich structural details into decoding via dense attention, the CNN is able to capture even intricate morphological details. The decoder is also augmented with a Cross Context Attention Fusion Upsampling to robustly reconstruct deep semantic features back to high-resolution segmentation map. It employs a novel pixel-precise attention model that draws relevant encoder features to aid in effective upsampling. The proposed CNN was evaluated on 3D scans from MosMedData and Jun Ma benchmarked datasets. It achieved state-of-the-art performance with a high dice similarity coefficient of 85.43% and a recall of 88.10%.
Keywords: COVID-19; Segmentation; Deep learning; Attention; Decoder, CNN
1. Introduction
COVID-19 outbreak has posed an unprecedented global health crisis and has adversely impacted human life. COVID-19 is a viral infection that is transmitted via droplets from an infected person's cough, sneeze, or breath. It can show mild symptoms like fever, sore throat, and fatigue. In some cases, it can lead to severe respiratory illness and difficulty in breathing. According to WHO, over 118 M corona virus cases have been reported worldwide, of which 2.62 M were casualties as of March 2021. The United States is the most affected country with the maximum number of infected cases (over 6 M) and the highest death toll of 184 K.
Currently, Reverse-Transcription Polymerase Chain Reaction (RT-PCR) is the most widely used technique for testing COVID-19. RT-PCR test detects the presence of the COVID-19 viral RNA from respiratory samples. Though PCR testing is the current major clinical diagnostic test, it is faced with some limitations: it displays varying sensitivity with time (i.e. negative PCR patient can test positive in up to 5 days) [1], it has a long turnaround time from a few hours to 2 days [2]. More recently, Rapid Diagnostic Test (RPT) was used for detecting the corona virus antigens with artificial antibodies. They show sensitivity between 34–80% and take less than 30 min [4]. But cases inadequate antigen traces in the nasal samples can go undetected on RPT.
While biomedical tests demand intensive laboratory settings and patient testing requirements, medical imaging on the other hand, is relatively easier to obtain. When combined with Artificial Intelligence (AI) based techniques, the medical imaging modality can serve as an effective screening aid for automatic COVID-19 detection. Many recent clinical studies have suggested that chest Computed Tomography (CT) imaging can be a potential tool for COVID-19 diagnosis due to its high sensitivity and low infection miss rate. In a study with 1014 patients in Wuhan China, CT-based screening registered 97% sensitivity as confirmed by the PCR test [1]. CT had also shown a 75% detection rate in identifying false-negative cases missed by the PCR test. In another clinical experiment with 51 patients, CT was shown to have a recall of 96.07% with the typical findings being ground-glass opacities, consolidation, and septal thickening [3]. Some CT patterns were found to overlap as well as contrast with that of Severe Acute Respiratory Syndrome (SARS) and adenovirus infection. In a study with 34 subjects involving 4121 COVID-19 patients, CT scans for 91.4% of them had presented with bilateral lung involvement that is suggestive of COVID-19 [4]. Guan et al. observed abnormal CT findings of Ground Glass Opacity (GGO) and bilateral patchy shadowing in 86.2% COVID-19 cases [5]. For a study population in Italy, the use of CT for assessing COVID-19 on PCR positive patients gave a high detection accuracy of 97% [6]. The highest sensitivity recorded for chest CT is 98% on a test group of 81 patients in Shanghai, China [7].
The most common CT manifestations of COVID-19 include peripheral and posterior ground-glass opacities. In a few other cases (esp. with severe symptoms), interlobular septal thickening, air bronchogram are likely to occur [3]. Crazy paving patterns, reverse ‘halo’ signs are also reported as CT features for COVID-19 detection [4]. AI-based analysis can help localize regions on chest CT that can potentially be a COVID-19 infection. With supervised learning, AI can learn possible patterns that distinctly characterize COVID-19 on CT. Though detecting the presence of the virus is useful for confirmatory diagnosis, the segmentation of the infected regions from CT is more insightful and provides finer details for further clinical assistance. Some challenging elements in CT that affect AI modeling are as follows: 1) heterogeneity of the target COVID-19 infection that might hugely vary in size, shape, and location. 2) Unclear boundaries and limited contrast that is hard for pixel-precise delineation. 3) Interference from other manifestations that affect the class-discriminability of the target structure. Suitable AI modeling for addressing these issues would attempt to learn a robust attention model for accurately harnessing the nature of a feature pixel. In this way, AI for automated COVID-19 segmentation can cut down a huge amount of clinician's time and effort put into the delineation process.
In this work, we propose a Contour-aware Pixelwise Attention (CPA) decoder Convolutional Neural Network (CNN) for accurate COVID-19 infection segmentation from chest CT. In broad sense, the decoder explores additional contextual information from contour regions to guide in detecting the spread of infection. It employs dual convolution pathways to extract this relevant lower-order context (boundary, edge, shape, etc.) from the contour map. The attention logic used in the decoder learns to densely fuse these contour features with the incoming intermediate feature map. Also, to robustly upsample the resolution of the decoder map, a Cross-Context Attention Fusion upsampling module has been presented. It exploits the encoded feature map at the corresponding level as supplementary context for upsampling. This module interpolates high-resolution pixel features by computing the upsampled region as an attention function over the input low-resolution map and the supplementary context. By aggregating such variable receptive field information from diverse contextual maps, the upsampler achieves lossless transformation, suppressing any unwanted artefacts.
The following are the main contributions of this research.
-
1.
We propose a novel contour-enhanced pixel attention decoder in this work that can be leveraged to enhance discriminability of lesion from normal pixels. Though enhancing decoder with boundary/edge awareness has been investigated in prior works [24,25], the decoder model presented in this work explores a new fusion approach that combines information from multiple contour feature maps via attention.
-
2.
We also propose a novel upsampling module that takes advantage of structural details present in the encoder features for interpolating high-resolution pixels. Different from previous works, this upsampler employs a pixel-precise attention model to extract relevant information from the encoded maps through cross-correlations.
The rest of the manuscript is organized as follows. In Section 2, related works in AI-assisted COVID-19 CT segmentation are reviewed. Also, deep attention methods for feature upsampling and decoding are discussed. Section 3 and 4 describe the proposed work and discussion on the experimental results respectively. Section 5 concludes the key findings of the work.
2. Related works
This section reviews related works in automated COVID-19 segmentation and deep attention models for feature upsampling and decoding.
2.1. COVID-19 segmentation from CT
COVID-19 diagnosis from chest CT has been associated with the typical manifestations of the infection on the CT modality. AI techniques for inspecting COVID-19 mainly search for the ground-glass opacities, consolidations, and interstitial changes to characterize the infection. Several research works have explored different deep CNN architectures for the detection and segmentation of the COVID-19 infected regions from CT.
Semi-supervised approaches learn from active annotation feedback and can alleviate the problem of sparsity in manually delineated data. For instance, Fan et al. presented a semi-supervised CNN framework for segmentation of COVID-19 infection from CT slices [8]. The CNN aggregates high-level features from multiple levels using a parallel partial decoder. It employs reverse attention and edge attention modules to mine boundary and edge information from spatial CT. Multiple Instance Learning (MIL) enables training on limited data labeling via leveraging relationships between the instance features. In a joint COVID-19 segmentation cum severity assessment CNN, He et al. proposed hierarchical MIL to first learn embedding-level representation for each 2D patch instance in the 3D scan bag. Further bag-level MIL aggregation over the instance embeddings was used for final classification [9]. Loosely labeled CT data simulated by a time-series model can reveal trends in COVID-19 infection patterns across time. Zhou et al. proposed a dynamic simulation framework to map the distribution of progression in infection regions across different points in time [10]. The simulated 2.5D data is spread out along three planes (x-y, y-z, x-z) and it is jointly segmented on a three-way segmentation U-net model.
U-net efficiently combines encoded features with high-resolution layers to enable precise target region localization and pixel-accurate classification. Chaganti et al. utilized the Dense U-net model to derive COVID-19 related lung abnormalities segmentation from CT [11]. The lung segmented 3D chest CT regions are passed through Dense U-net feature extraction and classified into affected and unaffected voxels. A new form of the shared encoder with two bifurcated U-net decoders was proposed by Yazdekhasty et al. for COVID-19 segmentation [12].
Few works propose new optimization objectives that focus on a distinct criterion or enhance existing algorithms. Elaziz et al. proposed an improved version of the Marine Predators Algorithm (MPA) to generate multi-level thresholds for effective COVID-19 CT segmentation [13]. The approach optimizes the MPA search space exploration using the Moth Flame Optimization (MFO) technique. Clustering techniques learn from unannotated datasets and generate patterns that can be used to better explicate COVID-19 CT images. Chakraborty et al. applied a superpixel-based clustering approach to process spatial features in COVID-19 infected scans [14]. A modified flower pollination algorithm was designed to explore the search space for forming clusters.
Ensemble networks fuse diverse contextual information generated from various feature transformations. Ouyang et al. proposed an ensemble of 3D ResNet34 with uniform and size-balanced data sampling for COVID-19 detection from CT [15]. An online 3D attention mechanism generates attention maps that are explicitly trained against segmented infection regions. Learning from noisy labels can be adaptively tuned in response to self-ensemble network optimization. Wang et al. proposed a noise-robust SqueezeNet-based self-ensembling framework that trains for COVID-19 segmentation with noisy labels [16]. The student and teacher networks in the ensemble are updated adaptively through noise-robust dice and consistency losses.
2.2. Attention upsampling and decoder networks for segmentation
In the context of medical image analysis, feature interpolation from a fine-grained low-resolution map to the target high-resolution demands lossless property. Moreover, enhancing the location information and selection of the most salient low-level attention features can result in optimal segmentation performance. Several works have investigated ways to achieve effective feature upsampling that is sensitive to the target objects and suppresses irrelevant features. For instance, Yin et al. proposed a Total Generalized Variation (TGV) scheme for restoring the feature map from the loss of information due to upsampling with bilinear interpolation [17]. TGV is used to effectively reconstruct target maps from low-resolution features. The model exploits first and second-order derivatives for incorporating information from various channels in the feature map into the upsampled map.
Learning an attention gating function from the high-level feature map can serve as guidance for localizing areas from the low-level map. Huang et al. applied a global pooling upsample attention model at each decoder layer for semantic segmentation using Feature Pyramid Networks [18]. The global knowledge context from the high-level features was used to derive attention coefficients for the low-level feature maps. Attention-guided dense upsampling convolutions were exploited for learning precise feature information. In the work by Sun et al., dense attention upsampling convolution and bilinear upsampling were used for deriving upsampled feature maps from high-level features [19]. The densely upsampled map is summed with low-level features and concatenated with the bilinear upsampled map. The resultant feature map is attended with channel-wise spatial attention. Chen et al. used sub-pixel convolution with pixel shuffling as a dense upsampling convolution operation [20]. Multiple periodic low-level feature maps are merged into a high-resolution map in positions determined by the period shuffling operation. This technique overcomes the problem of block artifacts in the Fully Convolutional Network (FCN) due to pixel replication from the same feature map.
Decoder modules provide a mechanism to fuse low-level details like edges, boundaries, regions with high-level features to create accurate segmentation maps. Exploiting primitive features helps CNN decode precise boundaries, recover minor objects, and refine the feature activations from generating false positives. Spatial and cross-channel attention are the two major attention models for implicitly learning salient regions and key features. The Spatial Channel Attention U-Net (SCAU-net) employs spatial and channel attention models for feature decoding and recalibration [21]. The spatial attention model adaptively learns to generate weighted maps for the features that identify key areas and prune irrelevant features. The channel attention module selectively exploits global information to enhance useful features for the decoder. In the work by Karthik et al., an implicit spatial attention mask is derived for weighing the relative importance between pixels in a feature map [22]. Peng et al. applied a dual attention mechanism to the decoder that employs two key components: 1) global attention upsampling to derive channel attention mask for the low-level feature map 2) spatial attention mask generated from the low-level features to recover boundary details for the decoder [23].
Edge information from low-level features can serve as important cues for localizing objects. Zhang et al. proposed an edge guidance network that propagates edge attention features from the initial encoding layers onto the decoder [24]. A weighted aggregation of multi-scale decoder features and edge-attention representations are used to derive the final segmentation mask. The effectiveness of exploiting saliency information in deeper layers and use it to extract meaningful structural information in shallower layers was demonstrated in [25].
Decoders can be also set to capture specific aspects of the CNN, like factoring multi-class specific information, aggregating contexts, etc. Hong et al. proposed a decoder that reconstructs dense foreground segmentation masks from categorical adaptive saliency attention maps produced by an attention model [26]. Class-specific attention weights defined over all the channels in the feature map are processed by the decoder to render the final segmentation map. By fusing features across modalities, a mask-guided attention model can be applied to learn rich feature patterns from multi-modality data [27].
Learning to adaptively aggregate multi-scale pyramid feature maps via attention can result in enhanced pixel-wise delineation. In the work, the authors proposed an efficient decoder that highlights salient regions and suppresses noise through defining attentive spatial gating and feature interaction mechanisms over the FCN [28]. Zhang et al. proposed a multi-scale parallel decoder design that aggregates local and long-range contextual information from the branches of a HRNet [29]. These aggregated feature sets are a result of adaptive spatial pooling and spatial reasoning module which capture the large local receptive field and long-range spatial correlations respectively.
3. Proposed work
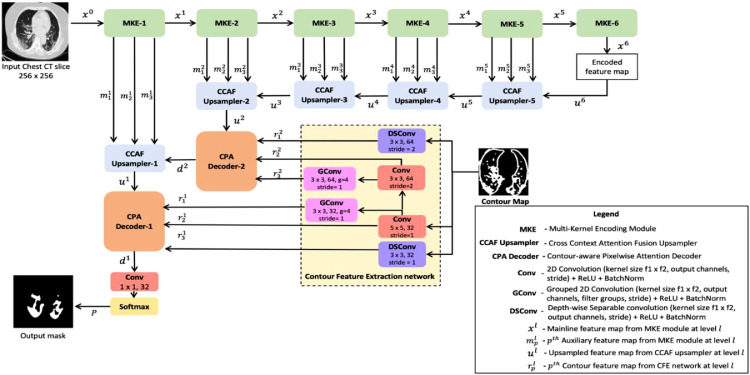
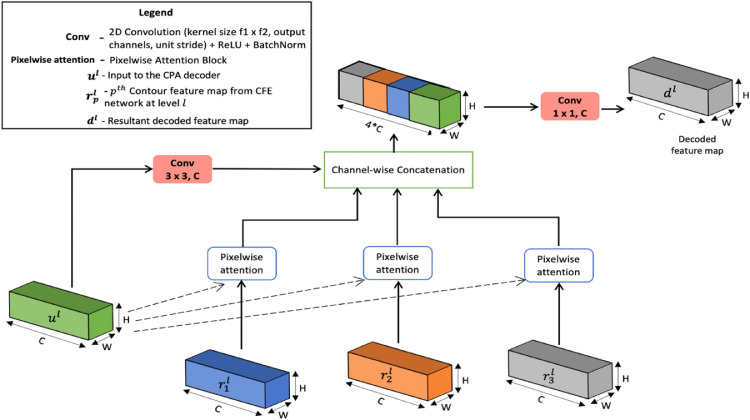
The architecture of the proposed contour-enhanced attention decoder CNN is presented in Fig. 1 . It is modeled as an encoder-decoder paradigm. It starts out with a series of encoding layers. Each encoder block is made up of three convolutional branches with different receptive field sizes. The encoder block emits one mainline output to the downstream encoder and three auxiliary outputs for use at the corresponding decoder.
Fig. 1.
Architectural diagram of the proposed CNN.
At every decoding step, the CNN uses a cross-attention model to merge this auxiliary encoder feature-set with the incoming feature map at that step. From Fig. 1, the upsampling module selectively fuses this encoder context with the input feature map to generate the decoded map. Through a series of such attention-aided decoding steps, the encoded map is transformed back to high resolution segmentation image.
In the final two steps of the decoding, the CNN exploits CT contour based features to guide in accurate tracing of the infected areas. A feature extraction network is employed to generate robust features from CT contour map. The decoder then integrate these structural features containing shape, boundary information with the deep semantic feature map through spatial-attention, to refine its prediction of the infection spread.
Being indirectly linked to the shallower encoder blocks, this contour attention decoder induces the encoding layers to generate effective representations that complement it. This feedback enables the encoder to produce salient features that augment the decoder in highlighting the infection pixels from normal pixels. This complementary learning not only enhances discriminability of the infection, but also suppresses noise from entering the decoder.
Section 3.1 presents the design of the encoder block. The upsampler module is described in Section 3.2. Finally, Section 3.3 elucidates the pixelwise attention decoder that uses contour features to refine the predicted infection regions. The output from the final layer is deeply supervised with categorical cross-entropy loss.
3.1. Multi-Kernel encoding (MKE) module
In semantic segmentation, the design of the encoder network directly affects the performance of the model to a large extent. As opposed to a linear convolutional chain, widening the number of parallel feature extraction branches at a single layer boosts the representational capability of the CNN. It enables the CNN to observe and correlate large variations in the spatial properties of the infection. The MKE module is inspired by the hybrid Inception-ResNet-V2 architecture, which factorizes the convolution into multiple separable steps, resulting in high computational efficiency and a richer feature-set.
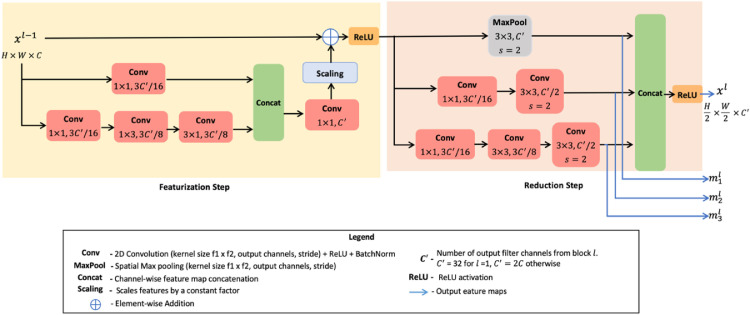
As shown in Fig. 2 , the MKE block consists of two steps, namely1) the featurization step and 2) reduction step. Let be the input feature map for the MKE module at network level . Here denote the spatial height, spatial width, and the number of channels respectively. First, the featurization layer is applied on . This step derives two auxiliary residual connections via linear and separable convolutions. The dimensionality of the stacked residues is scaled up to match the channel number of through 1 × 1 filter-expansion convolution. Residual links offer multiple paths for feature flow and prevent gradient signals from vanishing. When these residual connections are combined with the Inception the training is greatly speeded-up. The magnitude of the aggregated residues is scaled by a factor of 0.3 to stabilize learning. The scaling factor was chosen as per the design parameters reported in the Inception net [46].
Fig. 2.
Architectural overview of the multi-Kernel encoding module.
The ReLU activated feature map is fed down the reduction layer that results in 1) downsizing the spatial resolution by half, 2) doubling the number of feature channels. As shown in Fig. 2, outputs from the multi-branch reduction step are as follows: one mainline output feature map and three auxiliary feature maps . The mainline result is consumed by the downstream encoder layers, while the auxiliary/sideline features are exploited by the decoder.
As per Fig. 2, the number of output channels in the resulting mainline feature map is given by .The value of = 32 for level = 1. For subsequent levels, is set to twice the number of input channels . That is, for input passed into the MKE module, the resultant feature map is given by .
Similarly, resolution of the emitted three auxiliary feature maps is also reduced by half. These auxiliary features are denoted as and . Overall, introducing such multi-branched convolutions offers wider filter banks. It enables the encoder to efficiently process the spatial grid-size reduction in several hops. Furthermore, by forming a channel-wise dense concatenation of these feature maps, it integrates variably-sized receptive field information locally at each level in the processing hierarchy.
3.2. Cross context attention fusion (CCAF) upsampler
To learn an adaptive feature interpolation scheme that can recover spatial features from high-dimensional semantic representations, a dense attention upsampler has been proposed. Learning such a dense attention function to retrieve salient structural details can attribute more awareness to the semantic features learnt in these deep layers.
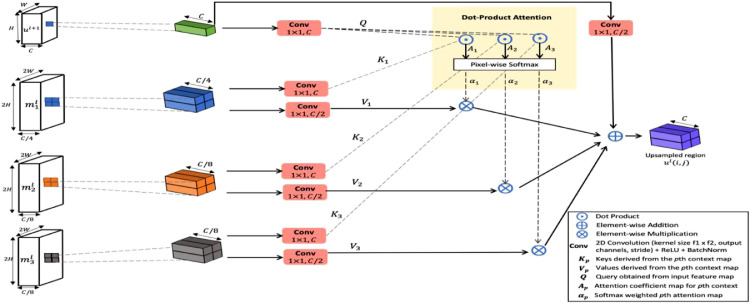
Especially, when reconstructing to higher resolution, retrieving the relevant lower-order context significantly enhances the localization information in the decoder layers. Considering these design aspects, the proposed attention upsampler draws a spatial cross-correlation amongst the three auxiliary encoded feature maps. The upsampler architecture is presented in Fig. 3 .
Fig. 3.
Schematic diagram of cross context attention fusion upsampler.
For CCAF upsampler at level , consider input feature map , where H, W, C denote the spatial height, spatial width, and a number of channels respectively. Let and be the three auxiliary lower-order feature maps from the encoder level . The output yielded by the upsampler block is denoted as , where the spatial resolution is doubled and the channel number is halved. Doubling the spatial resolution can be seen as projecting every pixel in the low-resolution map to a 2 × 2 region in the upsampled map. Specifically, for a pixel location in the low-resolution map , the upsampler learns an attention function over the aligning 2 × 2 sub-region in the contextual maps , . By weighing the contributions from these sub-regions along with the pixel feature, the pixel at location is densely interpolated to a 2 × 2 region in the upsampled map .
As shown in Fig. 3, for pixel at location in , the corresponding 2 × 2 area is sampled from the lower-order contextual maps , , . Let be used to identify the respective contextual map . Then from Fig. 3, three 1 × 1 convolutions are applied to transform every region of size 2 × 2 to different embeddings called the keys. In Fig. 3, denote the key embeddings for the corresponding contextual map. Here the channel number is in dimensions. Similarly, the pixel feature in is linearly transformed into . Then the attention coefficients map is generated via matrix multiplication between query and keys. Eq. (1) presents the relation for computing .
| (1) |
This form of attention weighing is defined as dot-product attention. Every point in the 2 × 2 gives the degree of correlation between the features and the corresponding pixel in . Further the attention weights are pixel-wise softmax normalized across (shown in Fig. 3). Therefore the weights assigned to every position in the 2 × 2 attention map are cross-correlated over the contextual maps. This step yields the new spatial attention map for each .
Similar to the calculation of keys , new embeddings denoted as are calculated from the contextual maps . Three more convolutional layers with 1 × 1 filters are applied on to create the values . It represents the contextual features in number of channels. Derivation of from is depicted in Fig. 3. The values mainly help with feature adaptation.
The attention fusion utilizes these normalized weights to aggregate the values . The final upsampled 2 × 2 sub-region for a pixel as per Eq. (2).
| (2) |
where is a 1 × 1 convolution that projects to channel number. denotes the upsampled area for the pixel at . The same mechanism is parallelly run overall spatial points to generate the upsampled map . From Eq. (2), it is seen that residual learning is placed on these attentive features against the feature flow . Due to this, the attention branch is seen as applying suitable infection segmentation refinement over the mainstream feature map . The spatial attention weights are adjusted to reflect the inclusion of lower-order context towards enhancing the semantic maps. Since these contextual maps receive active attention feedback via back propagation, the MKE feature extraction is auto-tuned to complement the refinement of semantic features. Thus highly effective upsampler learning is achieved via adaptive attention recalibration and active feedback.
3.3. Contour-aware pixelwise attention (CPA) decoder
To obtain precise high-resolution region segmentation with sharp boundaries, the final decoder layers are infused with explicit coarse contour area maps as presented in Fig. 1. Towards the end, the decoding layers mainly focus on attributing the semantic context to recover spatial resolution. As a result, they retain lesser feature channels compared to the inner layers which learn comprehensive representational features. When these layers learn to link class labeling with the pixels, the contour region maps serve as supplementary information channels that aid in boundary pixel discrimination. These contour region features on CT serve as an important cue to delineate the intricate morphologies of the target COVID-19 infection. Moreover, diffusing such lower-order context via an attention model enables the decoder to selectively integrate only the salient shape profile that boosts boundary awareness. It also automatically discards the noise inherent to the coarse contour regions. The contour region features do not induce semantic context, but they highlight homogeneous regions. This region information enriched with semantic features from the deeper layers is leveraged by the decoder to distinguish contrast variations between COVID-19 infected tissues and other lung manifestations.
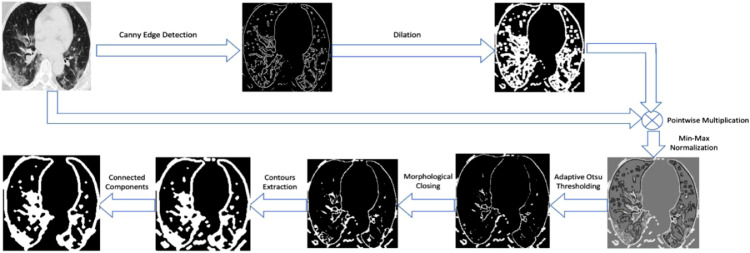
Fig. 4 presents the process flow diagram describing the various steps in the extraction of connected regions from the CT image. The input CT scan is processed through stages of edge detection, intensity thresholding, and contours extraction to reveal the prominent candidate-connected regions within the lung area. Firstly, the CT is subjected to Canny edge detection. The edges are dilated and masked against the base CT image, to obtain a sharpened image containing the regions enclosed within these edges. The resulting map is spatially min-max intensity normalized, to preserve the edge contrast details. The region fragments displaying high boundary contrast against the surrounding background are deduced using Otsu binary thresholding. This is followed by a morphological closing operation that connects discontinuities in the foreground objects. Contour lines are then inferred from the image.
Fig. 4.
Processing pipeline for connected contour regions extraction from chest CT.
The areas enclosed within the contours are filled as foreground. As a result, only the connected components that fall inside the lung segments are rendered as the contour regions map.
The Contour Feature Extraction (CFE) network shown in Fig. 1, extracts two levels of multi-type convolutional features from the contour region map described in Fig. 4. To minimize the size of the auxiliary network and create robust feature-sets, multi-branching of depth-wise separable and grouped convolutions is formed at the CFE block. There are two levels of convolutional features in the CFE network, i.e. and . Each level offers triple branching of different convolutional types, whose outputs , , connect to the mainstream decoder blocks at the respective level. The motivations behind designing the CFE network as an array of such convolution types are as follows: 1) they facilitate rich-location context at a much lower computation cost 2) each branch can be scaled separately and thus highly tuneable via the decoder attention, 3) bind information flow from variable spatial scale profiling, since convolutional branches in the same CFE-level observe different feature scales, 4) boost gradient flow between CFE levels due to aggregation of multi-scale features. Moreover, the depth-wise separable convolution efficiently draws channel-wise correlations and offers dense pixel connectivity. The grouped convolution acts as a regularizer.
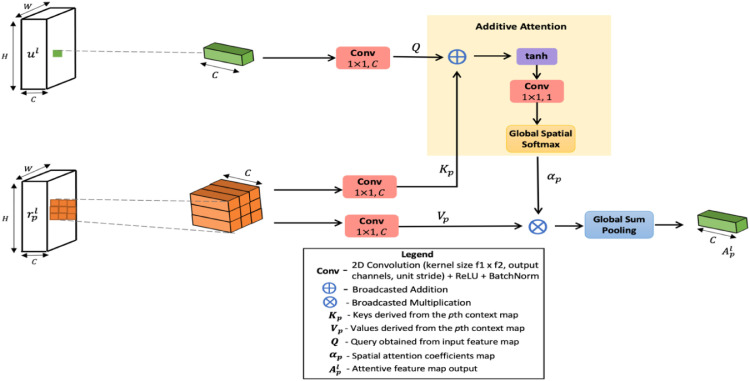
The proposed attention decoding module utilizes these , region contextual features from the CFE net to refine COVID-19 infection segments. The pixelwise attention decoder exploits the 8-point connectivity of a pixel to build an attention function over the corresponding position and its neighboring positions on the contour region map. The architectural diagram of the proposed decoder is shown in Fig. 5 .
Fig. 5.
Schematic diagram of the proposed CPA decoder.
As shown in Fig. 1, the CPA decoder at level , feeds on the mainline feature map from the upstream CCAF upsampler. It learns a transformation of to through attentive aggregation over the diverse region contextual maps from level . These region context maps have the same dimensions as the incoming feature map , as shown in Fig. 5. Let denote the output feature map from the CPA decoder module.
The attention mechanism is formed at the pixel level, as shown in Fig. 6 . Every pixel in the output map is a result of attending to the aligning 3 × 3 spatial window from the region context maps . Let p = {1,2,3} be used to identify the corresponding region context features . Then, to decode a pixel at spatial position , salient region information around that position is drawn from each region context map, , via an attention model. Fig. 6 presents the internal processing of this attention strategy for . Specifically, for the input pixel features , the attention model decodes the matching 3 × 3 region from each to yield attentive feature vector for each . As shown in Fig. 5, such attention features derived for each are densely aggregated to render the decoded map .
Fig. 6.
Pixelwise attention module.
Fig. 6 describes the attention mechanism to create attention-weighed features from the region context map, i.e. . Firstly a 1 × 1 convolution is applied to pixel features at position to form the query vector . Similarly, a 1 × 1 convolution performed over the 3 × 3 corresponding region from , constructs the set of keys . Then, an additive attention scheme is defined over the query and keys . The query vector is first added to every spatial location in the 3 × 3 region . The resulting map is scaled by the hyperbolic tangent activation and linearly projected to a 3 × 3 matrix of attention weights. This linear transformation is parameterized in and shown as 1 × 1 convolution in Fig. 6. Upon spatial softmax normalization, the attention coefficient matrix is obtained. The positional index denotes that the attention coefficients are formed for a 3 × 3 window around that pixel. Eq. (3) presents the computation steps for deriving .
| (3) |
Here, denotes the pixel-wise sum between the query and keys. Softmax scaling is performed over the spatial locations. The hyperbolic tangent activation is employed because it includes both positive and negative values into the non-linearity and regulates gradient-flow. Identical to the keys derivation, a new set of embeddings are calculated for the 3 × 3 region corresponding to the position in . As shown in Fig. 6, is obtained via convolution with 1 × 1 filters. These values are combined with the attention weights as a weighted linear sum to render the final attended feature vector for the position . This is given in Eq. (4).
| (4) |
where denotes pixelwise multiplication and GSP denotes Global Sum Pooling. The overall feature map computed overall in the image is collected from region context map .
As shown in Fig. 5, the CPA decoder forms a dense convolution of stacked on top of these attentive feature maps. The dense stack is transformed by a 1 × 1 convolution layer to emit the decoded feature map from level . In effect, the contour-aware decoder inspects the region context feature maps through a precise attention model fit local to each pixel in the input. As a result of such a dense attention fusion, the decoder can efficiently discriminate the infection boundary pixels and produce accurate segmentation.
4. Results and discussions
In this section, the efficacy of the proposed CNN is evaluated via ablation experiments. We also present a quantitative performance comparison of the CNN with state-of-the-art semantic segmentation methods. In each performance analysis sub-section, the observations are substantiated with relevant discussions on the architectural elements.
4.1. Data collection
The two data sources used to train and evaluate the proposed CNN are given in Table 1 . The COVID-19 lung infection segmentation dataset by Jun Ma consists of 20 COVID-19 regions annotated 3D CT scans [30]. The infected regions were delineated by radiologists on 20 CT scans sourced from Corona cases Initiative and Radiopaedia. The average 2D spatial resolution was 550 × 550. The other data source, MosMed CT scans, was acquired from municipal hospitals in Moscow [31]. Out of 1110 studies, a subset of 50 samples has been labeled with COVID-19 affected areas and facilitates use in segmentation tasks. The corresponding COVID-19 patients were diagnosed with mild corona symptoms and typical chest CT manifestations were that of ground-glass opacities and consolidation. Under all experiments discussed in the subsequent sections, the results are reported for the model trained on these two data sources - Jun Ma dataset and MosMed data individually, as well as on the combined set. The3D CT scans were split in the ratio 70:10:20to create training, validation, and testing sets respectively. Then, the 3D scans under each partition were converted to 2D slices for training and testing the proposed CNN.
Table 1.
3D CT scan datasets curated from different sources.
| S.No | Source | Details | Number of COVID-19 labeled 3D scans | Average number of slices per scan | Average 2D resolution |
|---|---|---|---|---|---|
| 1 | Jun Ma benchmark dataset [30] | 3D scans from Corona cases Initiative | 10 | 258 | 550 × 550 |
| Annotated scans from Radiopaedia | 10 | 94 | 550 × 550 | ||
| 2 | MosMedData [31] | Municipal hospitals in Moscow | 50 | 41 |
|
4.2. Data augmentation
To solve the challenge of data insufficiency and improve generalizability of predictions to real-world CT scan, this work utilizes various data augmentation techniques to enable model fitting on a larger set of samples. The dataset was augmented by applying random rotation, translation, shearing, and horizontal flipping. The parameter range for these affine transformations was chosen such that the infection surface is not distorted on the binary mask. These matrix functions are parameterized as an angle of rotation in the range of to , translation change along x-y directions are within 10% of the image's height and width, horizontal and vertical shear factors are between and for the shear angle of , and randomized horizontal flipping with a 50% probability. The four transformations were serially applied to data batches sampled in the training phase. For validation and testing, raw CT samples were used.
4.3. System setup
The proposed CNN was trained on dual 12GB NVIDIA Tesla K80 GPUs in a VM instance rented on Google Cloud. The models were implemented in PyTorch. The upsampler and decoder logic was designed as torch network modules that can be plugged into the base CNN. The system specifications are as follows: Ubuntu 18.04, 4vCPUs, and 16GB RAM. For all experiments, the models were trained with Adam optimizer with an initial learning rate of 0.01. The learning rate decay was scheduled to drop by a factor of 0.1 when no improvement is seen in validation dice over 10 epochs. To balance the proportion of samples with small and large net infection surface area in a single batch, the stratified data sampling technique was used. Such a weighted-class sampling approach guarantees an equal number of small and large infection samples in a batch.
4.4. Ablation experiments
In this section, the effectiveness of the two building blocks of the proposed CNN, i.e. CCAF upsampler and CPA decoder are individually determined. Further, the performance of the two modules is compared against the state-of-the-art upsampling/decoding schemes. In the final sub-section, the training and validation results of the proposed CNN are discussed.
4.4.1. Effectiveness of the upsampler
To ensure fair comparison, the performance mainly due to inclusion of CCAF upsampler in the proposed CNN has to be measured. For that reason, a modified form of the proposed CNN (in Fig. 1) is instantiated with the following changes: 1) the MKE encoder blocks are retained in-place. 2) CPA decoder and contour region feature extraction are eliminated from the network. We term this modified CNN as the ‘CCAF upsampler CNN’. Thus the effectiveness of incorporating the CCAF upsampling logic can be estimated by evaluating this model.
These results are compared to similar upsampling models in the literature, in terms of two metrics- Dice Similarity Coefficient (DSC) and Intersection over Union (IoU). Table 2 compiles the performance obtained for different upsampling strategies on the individual Jun Ma, MosMed data source, also on the set formed by combining those two sources. All the compared methods were implemented and run on the prepared dataset at hand. The same training, validation, and testing partitions were used in all experiments. The metric values projected in Table 2 are recorded on the common test set. Of these methods reported in Table 2, the first two methods directly reconstruct the feature map to a higher resolution. The remaining methods combine lower-order feature maps from the respective encoder level.
Table 2.
Comparison of various upsampling schemes for semantic segmentation. The results are presented both on the individual MosMeddata and Jun Ma datasets as well as on the combined set. The same test set partition is used to test the model.
| S. No. | Method | Jun Ma dataset [30] | MosMedData [31] | Combined dataset | |||
|---|---|---|---|---|---|---|---|
| DSC | IoU | DSC | IoU | DSC | IoU | ||
| 1 | Bilinear upsampling | 70.49 | 58.80 | 63.54 | 52.17 | 66.27 | 53.15 |
| 2 | Sub-pixel shuffling dense upsample [20] | 73.52 | 59.69 | 67.83 | 55.47 | 69.12 | 57.77 |
| 3 | Global Attention Upsample [18] | 71.63 | 57.56 | 70.97 | 58.38 | 71.35 | 60.01 |
| 4 | Attention-guided dense-upsampling [19] | 78.17 | 66.32 | 73.02 | 58.97 | 74.86 | 61.88 |
| 5 | Data-dependent Upsampling [31] | 77.31 | 64.80 | 74.95 | 58.62 | 75.92 | 63.12 |
| 6 | Proposed CCAF upsampler CNN | 80.43 | 69.87 | 75.19 | 65.30 | 77.67 | 65.79 |
To ensure a fair comparison of these upsampling schemes with the CCAF upsampler CNN, an experimental procedure was established. As per this, for each upsampling method (listed in Table 2) a new model is created by modifying the proposed CNN as follows: 1) MKE module is unaltered, while the CFE network and CPA decoder are removed. 2) the specific upsampling module is embedded in place of the CCAF module in the CNN. For the compared upsampling techniques that access the lower-order feature map (methods 3,4,5 in Table 2), the respective encoded map is passed into that module.
Bilinear interpolation achieves a DSC of 66.27% on the combined dataset and it is established as a baseline for comparing enhancements from neural network-based upsampling schemes. Bilinear upsampling is faced with two limiting factors: 1) it does not exploit the structural properties and semantic context of the image, 2) places a tight upper limit for reconstruction and loses information. The sub-pixel dense upsampling proposed by Chen et al. yields 69.12% DSC. It is a special form of deconvolution that interweaves pixel features from different channels [20]. But it suffers from checkerboard/block artifacts when used without a corrective mechanism or appropriate kernel initialization. Especially in the multi-step reconstruction of medical images, it is prone to introduce artifacts due to gradient saturation.
In contrast, infusing salient low-level features enrich upsampler learning. Global Attention Upsampling (GAU) creates global channel descriptors for low-level feature maps and attentively combines this fine-grained information to reconstruct infected regions. It reaches a high DSC of 71.35% on the joint dataset due to the inclusion of low-level features and global context aggregation. The GAU conceptually covers design aspects of attention upsampling, but it offers large scope for architectural enhancements. The attention-guided dense upsampling proposed by Sun et al. enhance this model by applying dense convolution and Squeeze-Excitation-based channel-wise attention. Thus it improves GAU's DSC by 4.92% for COVID-19 segmentation on the combined set. Compared to global channel-attention, it is observed that spatial attention applied over neighboring pixels produces finer upsampled features. Also, compared to fusing low and high-level features via addition or dense convolution, the proposed Query-Key-Value-based cross attention model has yielded better representational capability.
A new form of Data-Dependent approach to upsampling was proposed by Tian et al. [31]. An inverse projection matrix technique was employed to recover the upsampled pixels from low-resolution data [31]. It obtains a DSC of 75.92% which is closest to the proposed work. While it is computationally efficient and uses sub-region aggregation, in the context of COVID-19 segmentation exploring ‘multiple’ fine-grained deep structural encoder representations has led to better performance. This is because the CCAF upsampler not only learns selective feature recovery but also imparts some level of class-awareness to the MKE encoder whose learning complements that of the upsampler. Compared to data-dependent upsampling, the CCAF upsampler exhibits better adaptive scaling of reconstruction parameters which account for the 2.30% increase in DSC.
4.4.2. Effectiveness of the decoder
In this section, the efficacy of the CPA decoder is analyzed by spawning an altered form of the proposed CNN.
In the modified network, the CCAF upsampler block is substituted with the following configuration: 1) the input is bilinearly upsampled and densely stacked with respective encoder features , 2) the result is convolved with 3-by-3 convolution to reduce channel depth by half. The new network is called the ‘CPA decoder CNN’. Given these changes, it is ensured that only enhancements offered by the decoder are reflected in the results.
Table 3 provides a comparison of the different decoder approaches in the literature. As discussed in Section 4.4.1, the experiments were orchestrated in a way that gains exclusively due to decoder are fairly compared across different works. In that view, the decoder strategies compared in Table 3 are incorporated into the proposed framework as follows: 1) the CCAF upsampler and CPA decoder blocks in Fig. 1 are entirely replaced with the decoder model considered for comparison, 2) multi-scale encoder feature maps are made available to the decoder logic when fusing low-level features.
Table 3.
Analysis of the proposed CPA decoder with existing decoder architectures used for semantic segmentation on datasets described in Section 4.1. To ensure fair comparison the models were trained and tested on the same data partitions.
| S.No. | Method | Jun Ma dataset [30] | MosMedData [31] | Combined dataset | |||
|---|---|---|---|---|---|---|---|
| DSC | IoU | DSC | IoU | DSC | IoU | ||
| 1 | Point-wise attention decoder [32] | 75.19 | 64.35 | 72.77 | 62.08 | 73.77 | 62.98 |
| 2 | Stride spatial pyramid pooling and dual attention decoder [23] | 79.33 | 67.29 | 74.14 | 65.24 | 76.25 | 67.85 |
| 3 | Cross-granular attention decoder [33] | 78.13 | 69.81 | 83.85 | 73.19 | 78.89 | 68.65 |
| 4 | Proposed CPA decoder | 82.63 | 72.20 | 83.49 | 72.78 | 80.12 | 70.42 |
The point-wise decoder aggregates features of different resolutions progressively via point-wise attention gating [32]. It achieved a DSC of 73.77% on the combined dataset. The point-wise operations apply dedicated attention weights to map every point in the feature map. But it is observed that learning dense attention weighing over local/global context for a pixel yields higher performance.
On the other hand, generating multi-scale semantic information from a high-level feature map was attempted by Peng et al. using stride spatial pyramid pooling [23]. Due to multi-scale feature fusion via dual attention, it achieves a high DSC of 76.25% on the combined set. But for COVID-19 segmentation, further region refinement through explicit edge context has improved finer boundary discrimination. The proposed CPA decoder has shown better sensitivity to exploit coarse contour region maps for segmentation refinement.
The cross-granularity attention decoder proposed by Zhu et al. substantiates this argument by learning a mesh network to propagate semantically and contour region features across layers [33]. Boundary awareness in the deeper layers is achieved by applying supervision to the contour attention branch with Sobel edges. The cross-granular decoder achieves a DSC of 78.89%, which is close to the proposed CPA decoder in the given architectural setting. The CPA decoder outperforms the cross-granular decoder by 1.56%, owing to the pixelwise attention-based refinement of semantic details. Further extracting multiple contour region feature maps in the same layer builds high scale adaptability than deep supervision.
4.4.3. CNN training and validation
This section presents the empirical results of the proposed Contour-enhanced Attention CNN. We investigate the two major design blocks of the proposed CNN, i.e. CCAF upsampler and CPA decoder modules. These modules were evaluated as standalone CNN models as per the experimental settings described in Sections 4.4.1 and 4.4.2 respectively.
These three models were evaluated on the common test set of samples using evaluation metrics such as Dice Similarity Coefficient (DSC), Precision, Sensitivity, Specificity and Area under Curve (AUC). Batch sizes of 128, 40, and 16 were used for training the three models on GPU respectively. The test set results of the three ablation experiments are tabulated in Table 4 .
Table 4.
Observations of the Ablation studies. The results are grouped dataset-wise. The proposed models were trained and tested under each dataset.
| S No | Dataset | Method | DSC | IoU | Precision | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|---|---|
| 1 | Jun Ma dataset [30] | CCAF upsampler CNN | 80.43 | 69.87 | 78.85 | 82.46 | 99.75 | 77.00 |
| CPA decoder CNN | 82.63 | 72.20 | 80.26 | 96.06 | 99.76 | 81.69 | ||
| Proposed CNN | 88.01 | 75.03 | 85.57 | 90.05 | 99.77 | 85.03 | ||
| 2 | MosMedData [31] | CCAF upsampler CNN | 75.19 | 65.30 | 73.11 | 77.32 | 99.70 | 77.34 |
| CPA decoder CNN | 83.49 | 72.78 | 84.66 | 82.19 | 99.75 | 83.26 | ||
| Proposed CNN | 83.71 | 71.51 | 82.43 | 84.58 | 99.75 | 82.21 | ||
| 3 | Combined dataset | CCAF upsampler CNN | 77.67 | 62.65 | 76.32 | 79.21 | 99.72 | 75.60 |
| CPA decoder CNN | 80.12 | 68.70 | 80.96 | 79.39 | 99.79 | 78.69 | ||
| Proposed CNN | 85.43 | 73.44 | 81.23 | 89.88 | 99.77 | 84.61 |
From Table 4, it is evident that the CPA decoder is highly precise in limiting false positives. This can be attributed to the attention-based boundary refinement strategy in the decoder. It delineates complicated infection morphologies. The proposed CNN displays the highest overall metrics in terms of DSC and IoU. The high recall score of 89.88% on the combined dataset resonates with the DSC. Furthermore, it can be inferred that when the CCAF upsampler is coupled with the CPA decoder, it significantly boosts the sensitivity factor of the CNN. The proposed CNN also inherits the precise region distinction offered by the CPA decoder. The specificity is high in all experiments, which reinforces the models’ ability to identify unaffected pixels. The Area Under the PR Curve (AUC) quantifies the extent of class separability, by drawing a trade-off between precision and sensitivity for pixel classification. The PR curve is a suitable choice of metric given the large imbalance in the proportion of infected to non-infected pixels. In the order of experiments listed in Table 5 , the AUC scores of 75.60%, 78.69%, and 84.61% testify progressive refinement in class discriminability displayed by the three models.
Table 5.
Learning curves showing epoch-wise trends in the decay of cross-entropy loss and evolution of DSC. Additionally, the PR curve recorded on the validation set is provided.
 |
Table 5 presents the training and validation trends in loss and DSC tracked through epochs. Moreover, the Precision-Recall curve for the infection class is measured on the validation set. When training the CCAF upsampler CNN, convergence was attained in 93 epochs. The increase in DSC values with training epochs was smooth and consistent. Besides, the model generalized excellently to the test set with a DSC of 77.67%. On the other hand, the CPA decoder learnt precise attention weighing over the contour region contextual maps in 96 epochs. It took extra time for the pixelwise attention model to converge on the optimal parameter set. Compared to the previous two experiments, the proposed CNN converged at 120 epochs. This is because of reduced training batch size on GPU due to additional parameterization. Also, the multi-level attention structures demanded a longer time to converge. The proposed CNN had the least validation loss and converged in a sustained manner. For all the models, the effective divergence between the training and validation dice was minimal in the second half of training. The mean pixel-wise cross-entropy training loss decayed consistently in all experiments.
Table 6 summarises the experimental results across training, validation, and testing over the dice and IoU metrics. It is evident that the training and validation dice scores matched closely for all models, while the test scores slightly differed from the validation scores. Additionally, the inference times and parameters were also recorded under each ablation experiment. The proposed CNN took ∼40 ms to generate segmentation for a 256 × 256 CT image on NVIDIA Tesla K80 GPU.
Table 6.
Experimental observations of model training, validation, and testing evaluated on Dice and IoU scores. Additional runtime analysis including Inference times, Number of learnable parameters and number of floating-point operations (FLOPs) are also computed for the ablation models.
| S.No | Experiment | Dice-coefficient (%) |
Mean IoU (%) |
Inference Time (milliseconds/image) | Number of Parameters (in millions) | Giga FLOPs | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Training | Validation | Testing | Training | Validation | Testing | |||||
| 1 | CCAF upsampler CNN | 81.68 | 80.11 | 77.67 | 66.52 | 65.99 | 62.65 | 20.47 | 18.82 | 7.04 |
| 2 | CPA Decoder CNN | 85.42 | 83.76 | 80.12 | 73.19 | 71.61 | 66.70 | 26..95 | 22.45 | 8.55 |
| 3 | Proposed CNN | 88.01 | 87.54 | 85.43 | 77.57 | 76.73 | 73.44 | 38.24 | 30.51 | 13.78 |
Visualizations of the predicted segmentation maps are provided in Table 7 . The samples are chosen in a way that diversity and heterogeneity of infection spread are well captured for a holistic view. In many cases, the CCAF upsampler CNN predictions have lower recall than the other models. In some cases, the CCAF upsampler CNN predicted regions have dilated boundaries exceeding the actual boundary traced by the infection islands. These irregularities are overcome by the CPA decoder CNN, in which the shape boundaries are comparatively well delineated. In addition to lesser false positives, the CPA decoder CNN also detects minuscule infection spots. It exploits the contour features well to discriminate well between COVID and other artifacts. By far, the proposed CNN has the most accurate infection annotation over the previous models. The proposed CNN segmentations meticulously capture the COVID-19 morphologies on CT. Even subtler holes, cuts, spills within the infected area are precisely expressed. It shows excellent sensitivity to minor regions same as the CPA decoder CNN.
Table 7.
Visual comparison of COVID-19 segmentations results from different experiments.
 |
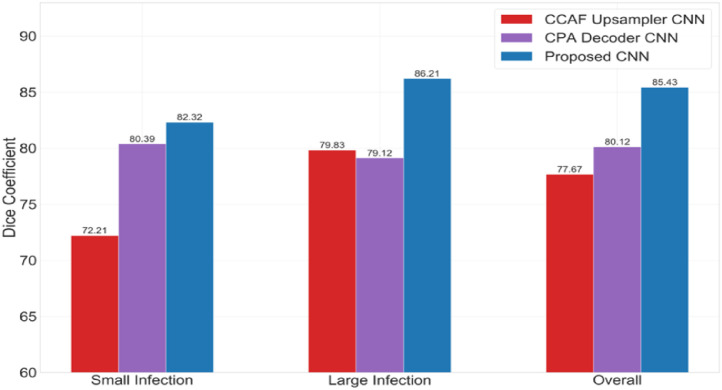
The CT infections can be broadly classified based on size. Splitting on the 50th percentile of the infection surface areas, they can be small or large infections. Fig. 7 shows the mean DSC registered by the three models in capturing small and large infection spread. The proposed CNN displays high sensitivity towards small infection localization, similar to the CPA decoder CNN. For large infections, it exhibits high detectability in the same manner as the CCAF sampler CNN.
Fig. 7.
Effectiveness of the proposed modules in improving segmentation performance for small and large infection regions (in terms of surface area).
4.5. Performance analysis
The proposed CNN is compared with the state-of-the-art attention-based segmentation models. The comparison is further extended to other supervised and semi-supervised CNN architectures for semantic segmentation. All the compared models were directly re-implemented for the COVID-19 CT dataset. The results were demonstrated on a test dataset common across all experiments. The metrics described in Section 4.4.3 were used for the evaluation.
4.5.1. Comparison with state of the art attention-based semantic segmentation models
This section compares the performance of the proposed CNN with state-of-the-art attention models on the COVID-19 infection segmentation dataset. The results of the models’ test-set performance are tabulated in Table 8 . All the compared methods were implemented and run over the datasets prepared in this work to generate these results. Under each dataset, the compared models were trained and evaluated on the same training and testing partitions. A runtime analysis of these models, including inference time, number of parameters, and FLOPs is presented in Table 9 . The inference time for single CT image prediction was calculated on NVIDIA Tesla K80 GPU.
Table 8.
Quantitative comparison of the proposed attention model with state-of-the-art attention models for semantic segmentation. The experiments are grouped by the dataset. Results are shown for both individual Jun Ma and Mosmed data, also on the set formed by combining these two sources.
| S No | Dataset | Method | DSC | IoU | Precision | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|---|---|
| 1 | Jun Ma dataset [30] | FocusNet [34] | 75.67 | 66.38 | 73.64 | 77.17 | 99.67 | 73.45 |
| Dual Attention Network [35] | 80.15 | 70.61 | 77.82 | 81.49 | 99.72 | 79.09 | ||
| Asymmetric Non-local networks [36] | 81.12 | 71.78 | 80.16 | 82.08 | 99.73 | 82.03 | ||
| Multi-scale self-guided attention [37] | 86.67 | 75.31 | 88.42 | 84.05 | 99.75 | 84.45 | ||
| Criss Cross Attention [38] | 85.58 | 74.60 | 82.84 | 88.12 | 99.75 | 83.21 | ||
| Semi Inf Net [8] | 88.45 | 76.07 | 90.47 | 85.11 | 99.78 | 86.55 | ||
| Proposed CNN | 88.01 | 75.03 | 85.57 | 90.05 | 99.77 | 86.74 | ||
| 2 | MosMedData [31] | FocusNet [34] | 73.49 | 63.23 | 71.22 | 75.88 | 99.70 | 71.54 |
| Dual Attention Network [35] | 75.02 | 61.00 | 74.82 | 75.70 | 99.71 | 72.10 | ||
| Asymmetric Non-local networks [36] | 82.17 | 69.19 | 83.25 | 80.67 | 99.74 | 81.67 | ||
| Multi-scale self-guided attention [37] | 80.97 | 68.78 | 80.24 | 81.33 | 99.72 | 77.34 | ||
| Criss Cross Attention [38] | 82.32 | 70.05 | 84.68 | 80.92 | 99.74 | 80.64 | ||
| Semi Inf Net [8] | 83.23 | 72.55 | 85.76 | 79.61 | 99.74 | 82.50 | ||
| Proposed CNN | 83.71 | 71.51 | 82.43 | 84.58 | 99.75 | 81.49 | ||
| 3 | Combined dataset | FocusNet [34] | 73.81 | 62.13 | 68.41 | 80.15 | 99.71 | 71.95 |
| Dual Attention Network [35] | 77.39 | 64.16 | 74.59 | 80.42 | 99.68 | 76.23 | ||
| Asymmetric Non-local networks [36] | 81.96 | 66.08 | 80.25 | 83.74 | 99.72 | 78.76 | ||
| Multi-scale self-guided attention [37] | 82.05 | 71.17 | 79.47 | 84.79 | 99.75 | 80.49 | ||
| Criss Cross Attention [38] | 83.85 | 72.54 | 79.68 | 88.47 | 99.73 | 82.75 | ||
| Semi Inf Net [8] | 84.56 | 72.32 | 80.50 | 89.05 | 99.74 | 83.71 | ||
| Proposed CNN | 85.43 | 73.44 | 81.23 | 89.88 | 99.74 | 84.57 |
Table 9.
Runtime analysis of the attention-based CNN models considered for comparison in Table 7. To maximum possible extent, in most experiments the backbone was uniformly chosen to be ResNet50 in order to enable comparison of different attention approaches on top of the same CNN.
| S. No. | Method | Backbone | Inference time (milliseconds/image) | Number of Parameters (in millions) | Giga FLOPs |
|---|---|---|---|---|---|
| 1 | FocusNet [34] | SE-Net50 | 12.38 | 26.82 | 2.74 |
| 2 | Dual Attention Network [35] | ResNet50 | 35.45 | 49.51 | 14.27 |
| 3 | Asymmetric Non-local networks [36] | ResNet50 | 52.78 | 44.04 | 12.57 |
| 4 | Multi-scale self-guided attention [37] | ResNet50 | 60.73 | 38.78 | 10.19 |
| 5 | Criss Cross Attention [38] | ResNet50 | 25.14 | 28.18 | 6.32 |
| 6 | Semi Inf Net [8] | Res2Net | 44.23 | 33.12 | 7.36 |
| 7 | Proposed CNN | Inception-ResNet-V2 based MKE module | 38.24 | 30.51 | 13.78 |
From Tables 8 and 9, the FocusNet CNN based on residual cum Squeeze net architecture, exhibited the least runtime complexity and a fair DSC of 73.81% on the combined dataset. It transfers selective semantic details to the encoder via a gated attention model, thus offers good recall for infection segmentation. The Dual Attention network (DAnet) leverages global self-attention in spatial and channel dimensions to capture long-range feature dependencies [35]. It offers the highest learning complexity, nevertheless run predictions at faster inference rate. It detects COVID-19 infection at a mean DSC of 77.39%on the merged dataset, whereas its DSC in tracing large lesions was only 69.24%. The DANet doesn't explore multi-scale feature fusion, therefore, it does not lend to medical imaging data, where the target structures diversely vary in size, contrast, morphology. The proposed CCAF upsampler overcomes this by attentively exploiting variable spatial scale information present in the multiple encoder maps.
The multi-scale self-guided attention model [37] is an enhanced version of the DANet that builds a global context from multi-scale feature maps. It also explores semantic-guided refinement of the attentive features and consequently improves DSC by 6.02% compared to DANet on the combined dataset. It displays highest inference time owing to the attention refinement. In contrast, since the proposed CPA decoder also draws explicit region awareness to contour region features, it exhibits an even higher sensitivity to even minuscule infection areas.
By using spatial pyramid pooling to sample fewer pixel locations as representative features, asymmetric non-local network captures long-range spatial dependencies [36]. At the same time, it fuses low-level features to enhance semantic details. This approach achieved a DSC of 81.96% in segmenting COVID-19, which was very close to [37]. Criss-Cross (CC) attention also accumulates global contextual information efficiently, as a two-step recurrent operation [38]. CC-attention on top of ResNet50 resulted in better DSC than the multi-scale attention approach [37]. It has the least attention model running complexity over the ResNet but yielded the highest performance. Although Asymmetric Non-local networks and CC-net accurately focus on relevant pixels, in some cases they predict smoother segmentations for large lesions and lose fine boundary information. Although global context is useful in creating robust representations, with respect to medical image segmentation, efficient multi-step local feature reconstruction at decoder enables recovery of finer structure details. The proposed CCAF upsampler and CPA decoder modules carry out this progressive refinement of the discriminative regions with explicit boundary awareness leading to sharp infection segmentation.
Semi-supervised attention-guided refinement of coarse localization maps was carried out by Fan et al. [8]. Recurrent reverse attention and edge-attention guidance techniques were applied to refine the rough encoder estimation. It obtains a DSC of 84.56% and optimal inference times which is close to that of the proposed CNN. It exhibited almost the same precision as the proposed CNN. However, the proposed model scores better recall mainly due to the pixelwise attention correlation amongst the region context maps.
4.5.2. Comparison with state of the art CNN architectures for semantic segmentation
Table 10 presents a comparison of different segmentation techniques that were fit to the COVID-19 segmentation task. Table 11 gives the runtime analysis of these compared techniques.
Table 10.
Performance comparison of the proposed work against state-of-the-art segmentation methods. All the models were freshly instantiated and run on the individual datasets listed in Table 1, also on the combined set. The results are grouped dataset-wise.
| S No | Dataset | Method | DSC | IoU | Precision | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|---|---|
| 1 | Jun Ma dataset [30] | U-Net [39] | 68.98 | 52.96 | 65.38 | 72.24 | 99.50 | 64.72 |
| Attention U-net [40] | 72.45 | 58.32 | 70.66 | 74.34 | 99.67 | 70.76 | ||
| R2U Net [41] | 77.14 | 64.91 | 79.12 | 74.78 | 99.76 | 76.13 | ||
| FCN8s (ResNet50 backbone) [42] | 75.56 | 63.65 | 75.32 | 74.81 | 99.70 | 73.49 | ||
| Wang et al. [47] | 78.98 | 65.81 | 74.45 | 83.71 | 99.77 | 77.01 | ||
| DeepLabV3 (ResNet50 backbone) [43] | 77.41 | 64.37 | 75.09 | 78.91 | 99.75 | 77.19 | ||
| Link Net [44] | 79.09 | 65.99 | 75.76 | 81.92 | 99.78 | 77.45 | ||
| PSPNet [45] | 84.56 | 72.38 | 85.90 | 83.31 | 99.82 | 80.33 | ||
| Proposed CNN | 88.01 | 75.03 | 85.57 | 90.05 | 99.77 | 86.17 | ||
| 2 | MosMedData [31] | U-Net [39] | 62.77 | 48.79 | 60.23 | 64.73 | 99.53 | 60.81 |
| Attention U-net [40] | 70.41 | 55.08 | 75.86 | 65.13 | 99.61 | 68.39 | ||
| R2U Net [41] | 70.11 | 57.34 | 69.51 | 70.47 | 99.69 | 69.63 | ||
| FCN8s (ResNet50 backbone) [42] | 71.36 | 58.47 | 78.44 | 65.33 | 99.65 | 69.23 | ||
| Wang et al. [47] | 73.99 | 60.00 | 75.57 | 72.33 | 99.70 | 72.72 | ||
| DeepLabV3 (ResNet50 backbone) [43] | 76.46 | 62.80 | 77.21 | 75.37 | 99.74 | 74.22 | ||
| Link Net [44] | 76.94 | 64.15 | 74.83 | 79.10 | 99.76 | 74.51 | ||
| PSPNet [45] | 79.29 | 64.80 | 76.78 | 81.44 | 99.78 | 78.10 | ||
| Proposed CNN | 83.71 | 71.51 | 82.43 | 84.58 | 99.82 | 81.18 | ||
| 3 | Combined dataset | U-Net [39] | 65.00 | 51.91 | 61.08 | 69.48 | 99.44 | 60.40 |
| Attention U-net [40] | 71.76 | 55.52 | 65.09 | 79.50 | 99.46 | 66.97 | ||
| R2U Net [41] | 72.18 | 57.83 | 77.27 | 67.79 | 99.70 | 71.34 | ||
| FCN8s (ResNet50 backbone) [42] | 73.24 | 60.28 | 77.13 | 69.95 | 99.89 | 73.10 | ||
| Wang et al. [47] | 75.31 | 65.82 | 73.60 | 77.10 | 99.69 | 76.15 | ||
| DeepLabV3 (ResNet50 backbone) [43] | 76.78 | 63.47 | 74.50 | 79.17 | 99.71 | 75.56 | ||
| Link Net [44] | 77.45 | 65.36 | 81.67 | 73.65 | 99.76 | 76.51 | ||
| PSPNet [45] | 81.32 | 67.38 | 81.12 | 81.50 | 99.75 | 79.97 | ||
| Proposed CNN | 85.43 | 73.44 | 81.23 | 89.88 | 99.74 | 84.57 |
Table 11.
Inference time analysis of the CNN models compared in Table 10 recorded on NVIDIA Tesla K80 GPUs. In all the compared methods, ResNet50 was used as the common backbone.
| S. No. | Method | Backbone | Inference time (milliseconds/image) | Parameters (in millions) | Giga FLOPs |
|---|---|---|---|---|---|
| 1 | U-Net [39] | ResNet50 encoder | 34.37 | 32.51 | 10.56 |
| 2 | Attention U-net [40] | ResNet50 encoder | 47.12 | 36.07 | 12.45 |
| 3 | R2U Net [41] | ResNet50 | 57.80 | 27.31 | 15.80 |
| 4 | FCN8s [42] | ResNet50 | 25.54 | 26.10 | 7.71 |
| 5 | DeCovNet [47] | ResNet50 | 20.25 | 22.12 | 6.36 |
| 6 | DeepLabV3 [43] | ResNet50 | 62.09 | 39.62 | 40.71 |
| 7 | Link Net [44] | ResNet50 | 27.25 | 31.17 | 10.63 |
| 8 | PSPNet [45] | ResNet50 | 11.05 | 24.29 | 2.83 |
| 9 | Proposed CNN | Inception-ResNet-V2 based MKE module | 38.24 | 30.51 | 13.78 |
From Table 11 it is clear that PSPNet has the lowest runtime complexity and better accuracy compared to other methods. DeepLabV3, Attention U-Net has a large learning complexity, while the R2U Net generates more inference time due to recurrent residual connection.
From Table 10, performance of the U-net CNN with DSC of 65.00% is taken to be the baseline for comparing proposed architecture. The Attention U-net refines the U-net design by introducing additive attention gates to process coarser features. Due to the attention gating, it improved the U-net DSC by 10.4% on the combined dataset. In contrast, the Residual Recurrent convolutional network (R2U Net) employs recurrent convolution logic over residual links leading to a high DSC of 72.18% dice and precision of 77.27%. On the other hand, FCN8s with ResNet50 backbone use large strides to generate pixel-wise semantic predictions from encoded features. It exhibits sharp precision in drawing object shapes, which is evident from precision of 77.13%. The weakly-supervised DeCovNet proposed by Wang et al. based on 3D residual network reached a DSC of 75.31%, which comes close to DeepLabV3.The DeepLabV3 applies multi-grid atrous convolutions and spatial pyramid pooling to learn from multi-scale features. It obtains a DSC of 76.78% on the COVID-19 test set, which is marginally better than FCN8s for the same ResNet50 backbone. In comparison to FCN8s, DeepLabV3 detects a larger share of COVID-19 infections. The Linknet CNN links the output from the encoder with a full convolution at the corresponding decoder block. It improved the DeepLab's DSC by 0.87%, owing to the high precision of 81.67%. In contrast, the PSP Net uses variably sized sub-region average pooling over the DeepLab encoded feature map, which results in a competitive DSC of 81.32%. The proposed CNN outperforms these models by a large DSC margin, which can be attributed to a high recall score of 89.88%. The highly accurate segmentations were a result of the coupled attention upsampling/decoding modules. They jointly learnt to focus on salient regions by attending to different contextual feature maps.
5. Conclusion
In this work, a novel attention-guided upsampler and decoder embedded CNN modelwas proposed for segmenting COVID-19 infected regions from chest CT. The key takeaways are the following: 1) Learning a cross-correlation of encoder feature maps via attention helps extract salient contextual details that directly improves upsampling accuracy at the decoder. Pixels on the upsampled map are a result of aggregating variably sized receptive fields from diverse low-level encoder feature maps. 2) Introducing boundary, shape awareness into the decoding scheme through a specialized pixel-locality attention model greatly improves infection segmentation results. The CT contour regions offer explicit low-level cues to focus on infected tissues. The attention decoder exploits these contour features by forming a dense fusion over a localized spatial window around the pixel locations. It complements upsampler learning in the final CNN layers, thereby achieves strong discriminability of COVID-19 and precise delineation of intricate COVID-19 morphology.
From the experimental results, it is evident that the proposed CNN has learned highly accurate COVID-19 segmentation that captures even subtler holes, minor distortions within the infected area. As future work, the proposed CNN can be experimented with to segment other types of interstitial lung abnormalities from CT. Global context aggregation and channel-wise attention techniques can be explored to further enhance the design. Architectural fusion with different encoder backbones can be tried to study the role of the encoder structure in enhancing the upsampler performance.
Declaration of Competing Interest
None Declared.
Biographies
R. Karthik obtained his Master's degree from Anna University, India. He received his Doctoral degree from VIT University in the area of medical image processing. He is currently serving as Senior Assistant Professor in the Center for Cyber Physical Systems, Vellore Institute of Technology, Chennai. His research interest includes digital image processing, medical image analysis and deep learning. He has published several papers in peer reviewed journals and conferences.
R. Menaka completed her Masters in Applied Electronics from Anna University, Chennai, India. She received her Doctoral degree from Anna University. She is currently serving as Professor and Director in the Center for Cyber Physical Systems, Vellore Institute of Technology, Chennai. Her areas of interest are image processing, neural networks and fuzzy logic. She has published several papers in peer reviewed journals and conferences.
M Hariharan completed his Bachelor's degree in Computer Science and Engineering in Vellore Institute of Technology, Chennai. His major research interests include Deep learning, Computer Vision, Machine Learning etc.
Daehan Won received his Doctoral degree from University of Washington. He is currently serving as Assistant Professor in System Sciences and Industrial Engineering, Binghamton University. His research interest includes Healthcare Analytics, Machine learning etc. He has published several papers in peer reviewed journals and conferences.
References
- 1.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 Cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hope M.D., Raptis C.A., Shah A., Hammer M.M., Henry T.S. A role for CT in COVID-19? What data really tell us so far. Lancet. 2020;395(10231):1189–1190. doi: 10.1016/S0140-6736(20)30728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J., Zhong Z., Li H., Ji P., Pang J., Li B., Zhang J. CT imaging features of 4121 patients with COVID-19: a meta-analysis. J. Med. Virol. 2020;92(7):891–902. doi: 10.1002/jmv.25910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J.…Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caruso D., Zerunian M., Polici M., Pucciarelli F., Polidori T., Rucci C., Guido G., Bracci B., De Dominicis C., Laghi A. Chest CT Features of COVID-19 in Rome, Italy. Radiology, 2020;296(2):E79–E85. doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of Chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 9.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., Shen D. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. Pattern Recognit. 2021;113 doi: 10.1016/j.patcog.2021.107828. ... &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., Xie D., Zhao L., Fan M., Hashmi S., Abdelkareem F., Eiada R., Xiao X., Li L., Qiu Z., Gao X. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaganti S., Grenier P., Balachandran A., Chabin G., Cohen S., Flohr T., Comaniciu D. Automated quantification of CT patterns associated with COVID-19 from chest CT. Radiology. 2020;2(4) doi: 10.1148/ryai.2020200048. ... &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yazdekhasty P., Zindari A., Nabizadeh-ShahreBabak Z., Roshandel R., Khadivi P., Karimi N., Samavi S. Pattern Recognition. ICPR International Workshops and Challenges. Springer International Publishing; 2021. Bifurcated autoencoder for segmentation of COVID-19 infected regions in CT images; pp. 597–607. [Google Scholar]
- 13.Elaziz M.A., Ewees A.A., Yousri D., Alwerfali H.S.N., Awad Q.A., Lu S., Al-Qaness M.A.A. An improved marine predators algorithm with fuzzy entropy for multi-level thresholding: real world example of COVID-19 CT image segmentation. IEEE Access. 2020;8:125306–125330. doi: 10.1109/ACCESS.2020.3007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty S., Mali K. SuFMoFPA: a superpixel and meta-heuristic based fuzzy image segmentation approach to explicate COVID-19 radiological images. Expert Syst. Appl. 2021;167 doi: 10.1016/j.eswa.2020.114142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., Shi F., Yuan H., Wei Y., Cao X., Gao Y., Wu D., Wang Q., Shen D. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 16.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin X., Li Y., Shin B.-S. TGV upsampling: a making-up operation for semantic segmentation. Comput. Intell. Neurosci. 2019;2019:1–12. doi: 10.1155/2019/8527819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Z., Zhong Z., Sun L., Huo Q. 2019 IEEE Winter Conference on Applications of Computer Vision (WACV) 2019. Mask R-CNN with pyramid attention network for scene text detection. [Google Scholar]
- 19.Sun H., Li C., Liu B., Liu Z., Wang M., Zheng H., Wang S. AUNet: attention-guided dense-upsampling networks for breast mass segmentation in whole mammograms. Phys. Med. Biol. 2020;65(5) doi: 10.1088/1361-6560/ab5745. ... &. [DOI] [PubMed] [Google Scholar]
- 20.Chen X., Zhang R., Yan P. IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) 2019. Feature fusion encoder decoder network for automatic liver lesion segmentation. 2019. [Google Scholar]
- 21.Zhao P., Zhang J., Fang W., Deng S. SCAU-net: spatial-channel attention U-net for gland segmentation. Front. Bioeng. Biotechnol. 2020;8:1–9. doi: 10.3389/fbioe.2020.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karthik R., Hariharan M., Anand S., Mathikshara P., Johnson A., Menaka R. Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. 2020;86 [Google Scholar]
- 23.Peng C., Ma J. Semantic segmentation using stride spatial pyramid pooling and dual attention decoder. Pattern Recognit. 2020;107 [Google Scholar]
- 24.Zhang Z., Fu H., Dai H., Shen J., Pang Y., Shao L. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; Cham: 2019. Et-net: a generic edge-attention guidance network for medical image segmentation; pp. 442–450. [Google Scholar]
- 25.Zhang Q., Shi Y., Zhang X. Attention and boundary guided salient object detection. Pattern Recognit. 2020;107 [Google Scholar]
- 26.Hong S., Oh J., Lee H., Han B. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning transferrable knowledge for semantic segmentation with deep convolutional neural network; pp. 3204–3212. [Google Scholar]
- 27.Zhang D., Huang G., Zhang Q., Han J., Han J., Yu Y. Cross-modality deep feature learning for brain tumor segmentation. Pattern Recogni. 2021;110 [Google Scholar]
- 28.Zhang P., Liu W., Wang H., Lei Y., Lu H. Deep gated attention networks for large-scale street-level scene segmentation. Pattern Recognit. 2019;88:702–714. [Google Scholar]
- 29.Zhang J., Lin S., Ding L., Bruzzone L. Multi-scale context aggregation for semantic segmentation of remote sensing images. Remote Sens. 2020;12(4):701. [Google Scholar]
- 30.Morozov S., Andreychenko A., Pavlov N., Vladzymyrskyy A., Ledikhova N., Gombolevskiy V., Blokhin I., Gelezhe P., Gonchar A., Chernina V., Babkin V. Cold Spring Harbor Laboratory; 2020. MosMedData: Chest CT Scans with COVID-19 Related Findings. [Google Scholar]
- 31.Tian Z., He T., Shen C., Yan Y. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. Decoders matter for semantic segmentation: data-dependent decoding enables flexible feature aggregation; pp. 3126–3135. [Google Scholar]
- 32.Li G., Jiang S., Yun I., Kim J., Kim J. Depth-wise asymmetric bottleneck with point-wise aggregation decoder for real-time semantic segmentation in urban scenes. IEEE Access. 2020;8:27495–27506. [Google Scholar]
- 33.Zhu L., Wang T., Aksu E., Kamarainen J.K. Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops. 2019. Cross-granularity attention network for semantic segmentation. 0-0. [Google Scholar]
- 34.Kaul C., Manandhar S., Pears N. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) IEEE; 2019. Focusnet: an attention-based fully convolutional network for medical image segmentation; pp. 455–458. [Google Scholar]
- 35.Fu J., Liu J., Tian H., Li Y., Bao Y., Fang Z., Lu H. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. Dual attention network for scene segmentation; pp. 3146–3154. [Google Scholar]
- 36.Zhu Z., Xu M., Bai S., Huang T., Bai X. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. Asymmetric non-local neural networks for semantic segmentation; pp. 593–602. [Google Scholar]
- 37.Sinha A., Dolz J. Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. 2021;25(1):121–130. doi: 10.1109/JBHI.2020.2986926. [DOI] [PubMed] [Google Scholar]
- 38.Huang Z., Wang X., Wei Y., Huang L., Shi H., Liu W., Huang T.S. CCNet: criss-cross attention for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2020:1. doi: 10.1109/TPAMI.2020.3007032. –1. [DOI] [PubMed] [Google Scholar]
- 39.Ronneberger O., Fischer P., Brox T. International Conference on Medical image computing and computer-assisted intervention. Springer; Cham: 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 40.Schlemper J., Oktay O., Schaap M., Heinrich M., Kainz B., Glocker B., Rueckert D. Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 2019;53:197–207. doi: 10.1016/j.media.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alom M.Z., Yakopcic C., Taha T.M., Asari V.K. NAECON 2018 - IEEE National Aerospace and Electronics Conference. 2018. Nuclei segmentation with recurrent residual convolutional neural networks based U-net (R2U-net) [Google Scholar]
- 42.Long J., Shelhamer E., Darrell T. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 43.Chen L.C., Zhu Y., Papandreou G., Schroff F., Adam H. Proceedings of the European Conference on Computer Vision (ECCV) 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation; pp. 801–818. [Google Scholar]
- 44.Chaurasia A., Culurciello E. 2017 IEEE Visual Communications and Image Processing (VCIP) 2017. LinkNet: exploiting encoder representations for efficient semantic segmentation. [Google Scholar]
- 45.Zhao H., Shi J., Qi X., Wang X., Jia J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Pyramid scene parsing network; pp. 2881–2890. [Google Scholar]
- 46.Szegedy C., Ioffe S., Vanhoucke V., Alemi A. Proceedings of the AAAI Conference on Artificial Intelligence. Vol. 31. 2017. Inception-v4, inception-resnet and the impact of residual connections on learning. [Google Scholar]
- 47.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. ... &. [DOI] [PubMed] [Google Scholar]