Abstract
Background
We determined the burden of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in air and on surfaces in rooms of patients hospitalized with coronavirus disease 2019 (COVID-19) and investigated patient characteristics associated with SARS-CoV-2 environmental contamination.
Methods
Nasopharyngeal swabs, surface, and air samples were collected from the rooms of 78 inpatients with COVID-19 at 6 acute care hospitals in Toronto from March to May 2020. Samples were tested for SARS-CoV-2 ribonucleic acid (RNA), cultured to determine potential infectivity, and whole viral genomes were sequenced. Association between patient factors and detection of SARS-CoV-2 RNA in surface samples were investigated.
Results
Severe acute respiratory syndrome coronavirus 2 RNA was detected from surfaces (125 of 474 samples; 42 of 78 patients) and air (3 of 146 samples; 3 of 45 patients); 17% (6 of 36) of surface samples from 3 patients yielded viable virus. Viral sequences from nasopharyngeal and surface samples clustered by patient. Multivariable analysis indicated hypoxia at admission, polymerase chain reaction-positive nasopharyngeal swab (cycle threshold of ≤30) on or after surface sampling date, higher Charlson comorbidity score, and shorter time from onset of illness to sampling date were significantly associated with detection of SARS-CoV-2 RNA in surface samples.
Conclusions
The infrequent recovery of infectious SARS-CoV-2 virus from the environment suggests that the risk to healthcare workers from air and near-patient surfaces in acute care hospital wards is likely limited.
Keywords: aerosol, contamination, COVID-19, SARS-CoV-2, surface
The infrequent recovery of infectious SARS-CoV-2 virus from the environment suggests that the risk to healthcare workers from air and near-patient surfaces in acute care hospital wards is likely limited, particularly several days after admission.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 causing the coronavirus disease 2019 (COVID-19) pandemic [1] and many hospital outbreaks of COVID-19 [2]. Understanding the role of surface and air (environmental) contamination in the transmission of SARS-CoV-2 is essential to ensuring the prevention of transmission of SARS-CoV-2 between patients and to healthcare workers in acute care hospitals.
Severe acute respiratory syndrome coronavirus 2 ribonucleic acid (RNA) has been detected from surfaces and air in hospitals [3–12]. However, a minority of studies have attempted to culture virus [13, 14]. This limits our understanding of exposure and transmission risk.
This study aimed to determine the burden of SARS-CoV-2 in the air and on surfaces in hospital rooms of acutely ill inpatients with COVID-19 in Toronto, Ontario, Canada. We determined the association between patient factors and detection of SARS-CoV-2 from environmental samples. We also apply a genomics approach to environmental samples for SARS-CoV-2, thus linking environmental contamination of SARS-CoV-2 to the source [15–18].
METHODS
Study Population
The Toronto Invasive Bacterial Diseases Network (TIBDN) performs population-based surveillance for infectious diseases in metropolitan Toronto and the regional Municipality of Peel, south-central Ontario, Canada (population 4.2 million in 2016). The TIBDN clinical microbiology laboratories report clinical specimens yielding SARS-CoV-2 to TIBDN’s central study office. At 6 TIBDN hospitals, consecutive patients hospitalized on any ward with laboratory-confirmed COVID-19 between March and May 2020 were eligible for this study. Research ethics approval was granted by all participating Toronto Invasive Bacterial Diseases Network hospitals. (Sunnybrook’s Research Ethics Board, The Mount Sinai Hospital Research Ethics Board, Toronto East Health Network Research Ethics Board, William Osler Health System’s Research Ethics Board, and Scarborough Health Network’s Research Ethics Board). Informed consent from was obtained from all patients. Findings were reported in accordance with the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) guidelines for reporting observational studies [19].
Data and Specimen Collection
Demographic, clinical, and COVID-19 risk factor data were collected by participant interview and chart review. Study staff obtained nasopharyngeal (NP) swabs from patients at enrollment and every 3 days until refusal, hospital discharge, or death [20]. A set of surface samples was collected at enrollment and every 3 days, including (1) bathroom doorknob, (2) phone (all surfaces of the patient’s phone and room phone), (3) overbed table and chair (pooled), (4) bed (bed rail and pillow) and light switch or pullcord in patient’s bedspace (pooled), and (5) toilet and sink faucet handles (pooled) (Supplementary Figure 1). Surface samples were collected by thoroughly wiping each surface type using the rough side of a dry 6 cm × 6 cm Swiffer cloth (Swiffer; Procter & Gamble, Toronto, Canada). Nasopharyngeal swabs and Swiffer cloths were immediately placed into universal transport medium (UTM; Copan Diagnostics, Murrietta, CA).
During the study period, 4 bioaerosol samplers were used for sampling the first 45 patients enrolled that were not intubated. For each patient, 1 to 2 different bioaerosol samplers were used in each run. Using an air sampling pump (GilAir Plus Personal Air Sampling Pump, Sensidyne, St. Petersburg, FA), air samples were obtained using the 1-μm pore size, 37-mm polytetrafluoroethylene (PTFE) membrane filters (SKC Inc., Eighty Four, PA), the 37-mm 3-piece cassette with 0.8-μm polycarbonate (PC) filter (Zefon International, Ocala, FL), and 25-mm gelatin membrane filters (SKC Inc.). Before sampling, the pumps were calibrated to a flow rate of 3.5 L/minute using the corresponding filter used for sampling that day (Gilibrator 3, Standard Flow Dry Cell Calibrator; Sensidyne, St. Petersburg, FL). In the patient rooms, samplers were placed at 1 m and 2 m from the head of patient at the approximate level of and anterior to mouth and nose, with samples collected over a 2-hour period. All filters were placed in coolers at the end of the sampling period for transport and processed immediately. Air samples were also collected using the NIOSH 2-stage cyclone bioaerosol sampler (National Institute for Occupational Safety and Health, Morgantown, WV). The NIOSH cyclone bioaerosol sampler comprises stages collecting larger particles (>4 μm) in the first stage into 15-mL conical tubes, smaller particles (1–4 μm) in the second stage into 1.5-mL conical tubes, and particles <1 μm onto a PTFE filter. The NIOSH cyclone samplers were assembled in the laboratory in a biosafety cabinet and calibrated to a flow rate of 3.5 L/minute (BIOS DC-1 DryCal flow calibrator; SKC Inc.). In the patient rooms, the sampler was placed 1 m from the patient as described above and sampling occurred over a 2-hour period.
Laboratory Procedures
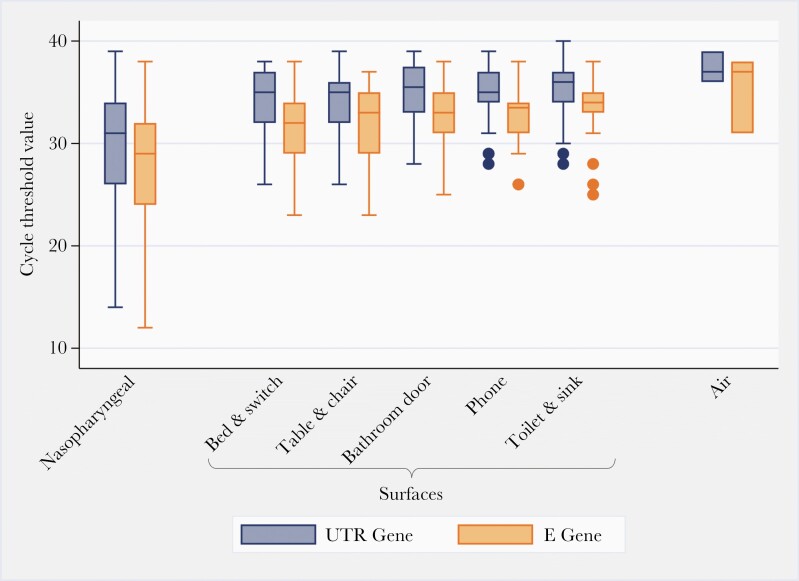
All samples were processed at Sunnybrook Research Institute on the day of collection. Nasopharyngeal swabs and environmental samples were vortexed for 20 seconds before aliquoting and storage at −80°C. The PTFE, PC, and gelatin membrane filters were placed in 3-mL transport media before being vortexed for 20 seconds, followed by aliquoting and storage at −80°C. For the NIOSH cyclone bioaerosol sampler, 1 mL transport media was added to the first stage, 500 μL was added to the second stage, and 3 mL transport media were aliquoted onto the PTFE filter. Samples were vortexed for 20 seconds before aliquoting and storage at −80°C. Ribonucleic acid extractions were performed using QIAmp viral RNA mini kit (QIAGEN, https://www.qiagen.com) according to manufacturer’s instructions; samples were eluted into 40 μL. Reverse-transcriptase polymerase chain reaction (RT-PCR) reactions were performed using the Luna Universal Probe One-Step RT-qPCR Kit (New England BioLabs Inc., https://www.neb.ca/). Two separate gene targets were used for detection of SARS-CoV-2, the 5’-untranslated region (UTR), and the envelope (E) gene, with human RNaseP as an internal control [21]. The cycling conditions were as follows: 1 cycle of denaturation at 60°C for 10 minutes then 95°C for 2 minutes followed by 44 amplification cycles of 95°C for 10 seconds and 60°C for 15 seconds. Rotor-Gene Q software (QIAGEN) was used to determine cycle thresholds (Cts), and samples with Cts <40 in both UTR and E genes were considered positive. Correlation analysis indicates almost perfect correlation between Ct values for the UTR and E gene (0.99). Therefore, we present Ct values for the UTR gene target within the text; the Ct value results for both gene targets are summarized in Figure 1. See Supplementary Methods for details on genome sequencing and analysis.
Figure 1.
Boxplot summary of the cycle threshold values for the untranslated region (UTR) gene (blue) and E gene (orange) targets from the severe acute respiratory syndrome coronavirus 2 polymerase chain reaction analysis for each sample type investigated for 78 coronavirus disease 2019-positive patients in Toronto, Canada. It is notable that air sampling pumps were calibrated to a flow rate of 3.5 L/minutes for 2 hours; each air sample represents 420 liters of air.
Virus Isolation
Virus isolation was attempted on (1) PCR-positive NP swabs and air samples and (2) PCR-positive environmental surface samples with a Ct of <34.0 in containment level 3 at the University of Toronto.
Vero E6 cells were seeded at a concentration of 3 × 105 cells/well in a 6-well plate. The next day, 500 μL sample containing 16 μg/mL TPCK-treated trypsin (New England BioLabs Inc.), 2× penicillin/streptomycin (Pen/Strep), and 2× antibiotic-antimycotic (Wisent, https://www.wisentbioproducts.com/en/) were used to inoculate cells. Plates were returned to a 37°C, 5% CO2 incubator for 1 hour and rocked every 15 minutes. After 1 hour, the inoculum was removed and replaced with Dulbecco’s modified Eagle’s medium containing 2% fetal bovine serum, 6 μg/mL TPCK-treated trypsin, 2× Pen/Strep, and 2× antibiotic-antimycotic. Cells were observed daily under a light microscope for cytopathic effect (CPE) for 5 days postinfection. Cell cultures not showing any CPE were blind passaged onto fresh Vero cells and observed for a further 5 days. The RT-PCR assay described above was used to confirm SARS-CoV-2 isolation from supernatant.
Statistical Analysis
All analyses were conducted using Stata/SE 15.1 (StataCorp, College Station, TX; http://www.stata.com). Descriptive statistics were used for patient characteristics, PCR results, and culture results. To explore putative associations between patient characteristics and SARS-CoV-2 PCR-positive environmental surface samples, we reviewed the literature and surveyed Canadian COVID-19 researchers to identify factors of interest that might be associated with environmental contamination. The following variables were investigated: age, sex, Charlson comorbidity index [22], smoking history, Clinical Frailty Score [23], presence or absence of symptoms from onset to 24 hours postadmission (cough, fever, diarrhea, delirium/confusion), hypoxia at admission (defined as oxygen saturation <92%), admission to intensive care unit at time of sample collection, use of exogenous oxygen during stay, prone position, receiving steroids for treatment on day of sampling, room type (regular private room or negative pressure room), and the presence or absence of a PCR-positive NP swab on or after environmental sampling date (PCR-positive NP swabs were further categorized to Ct >30 and Ct ≤30); the sampling date refers to the date the sample was taken. If use of exogenous oxygen during stay was significant, oxygen delivery methods (intubation, facemask/nasal prong, high flow) were included to investigate individual oxygen requirements. Because samples were taken serially from each patient over the course of this study, we included onset of illness to sample date as a fixed-effect control to account for temporal variability. See Supplementary Table 2 for further variable details. The outcome of interest was SARS-CoV-2 PCR-positive environmental surface samples.
A causal diagram was constructed to examine possible confounding and intervening relationships among exploratory variables relative to a SARS-CoV-2 PCR-positive environmental surface sample. Mixed-effects logistic regression models with a random intercept for unique patient identification to account for clustering were constructed using stepwise backwards elimination. Variables that were significant, potential confounders, part of a significant interaction term, or a control variable (ie, onset of illness to sample date) were included in the final model.
Pearson and deviance residuals were explored for outlying observations. Model fit was assessed by determining whether the best linear unbiased predictors (BLUPs) met the assumptions of normality and homogeneity of variance.
RESULTS
Study Population and Samples Collected
There were 78 inpatients with COVID-19 who consented to participate. All were confirmed to have COVID-19 with a positive nasal, mid-turbinate, or NP swab tested in a licensed diagnostic laboratory in Toronto before enrollment; diagnostic samples used for initial COVID-19 confirmation were not included in the present study. Patients were deidentified and randomly assigned a number from 1 to 78.
The median age of participants was 67 years (interquartile range [IQR], 53–79). The median duration between onset of illness and hospital admission was 5 days (IQR, 2–8). Patient characteristics are shown in Table 1. Numbers of additional NP swabs, surface samples, and air samples collected are shown in Table 2; a detailed breakdown of samples collected is shown in Supplementary Table 1.
Table 1.
Patient Demographics and Clinical Characteristics for 78 Hospitalized Patients With COVID-19 in Toronto, Canada
| Patient Characteristics | No. Patients (%) |
|---|---|
| (N = 78) | |
| Age: <65 years | 37 (47) |
| ≥65 years | 41 (53) |
| Sex (number, %, male) | 44 (56) |
| Charlson comorbidity index: 0 | 36 (46) |
| 1–2 | 29 (37) |
| ≥3 | 13 (17) |
| Underlying illness: diabetes mellitus | 24 (41) |
| Pulmonary | 16 (27) |
| Cardiac | 14 (23) |
| History of smoking | 17 (22) |
| Clinical Frailty Score (n = 77)a: not frail (1–4) | 57 (74) |
| Mild to moderate (5–6) | 12 (16) |
| Severe (7–8) | 8 (10) |
| Symptoms/signs: Cough | 64 (82) |
| Fever | 61 (78) |
| Diarrhea | 24 (31) |
| Delirium/confusion | 12 (15) |
| O2 saturation <92% at admission | 41 (53) |
| Oxygen Requirements During Admission | |
| No oxygen required | 21 (27) |
| Required oxygen by face mask or nasal prong only | 43 (55) |
| Required high-flow oxygen | 9 (12) |
| Required intubation | 15 (19) |
| Required oxygen by facemask/nasal prong | 54 (69) |
| Required high-flow oxygen, not intubated | 5 (6) |
| Management: prone positioning | 7 (9) |
| Received steroids | 14 (18) |
| Required ICU admission | 25 (32) |
| Accommodation: regular private room | 35 (45) |
| Negative pressure room | 43 (55) |
Abbreviations: COVID-19, coronavirus disease 2019; ICU, intensive care unit.
The clinical frailty was collapsed into 3 categories: nonfrail (1–4), mild-to-moderately frail (5–6), and severely frail (7–8).
Table 2.
Summary of Sample Types Collected and Results of PCR Testing and Cell Culture for SARS-CoV-2 in 78 Hospitalized Patients With COVID-19 in Toronto, Canada
| PCR | Culturea | |||
|---|---|---|---|---|
| Sample Type | No. Positive Samples/Total (%) | No. Patients With ≧1 Sample Positive/Total (%)b | No. Positive Samples/Total Cultured (%) | No. Patient With ≧1 Sample Positive/Total (%) |
| Nasopharyngeal swab | 172/219 (79) | 74/78 (95) | 30/110 (27) | 21/65 (32) |
| Environmental surface (all) | 125/474 (26) | 42/78 (54) | 6/36 (17) | 3/16 (19) |
| Bathroom door | 12/88 (14) | 12/69 (17) | 1/4 (25) | 1/4 (25) |
| Bed and switch (pooled) | 39/102 (38) | 33/78 (42) | 2/13 (15) | 2/11 (18) |
| Phone | 24/88 (27) | 21/70 (30) | 1/5 (20) | 1/4 (25) |
| Table and chair (pooled) | 29/95 (31) | 24/74 (32) | 1/10 (10) | 1/9 (11) |
| Toilet and sink (pooled) | 21/101 (21) | 20/74 (27) | 1/4 (25) | 1/3 (33) |
| Airc | 3/146 (2) | 3/45 (7) | 0/3 (0) | 0/3 (0) |
| 1 m from head of patient | 3/101 (3) | 3/45 (7) | 0/3 (0) | 0/3 (0) |
| 2 m from head of patient | 0/45 (0) | 0/45 (0) | .. | .. |
Abbreviations: COVID-19, coronavirus disease 2019; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
All PCR-positive nasopharyngeal swabs and air samples, and PCR-positive environmental surface samples with a cycle threshold of <34.0 were submitted for virus isolation.
Total number of patients for each category were different because samples were taken every 3 days postenrollment until refusal, hospital discharge, or death.
Air sampling pumps were calibrated to a flow rate of 3.5 L/minute for 2 hours; each air sample represents 420 liters of air.
Nasopharyngeal Swabs
A total of 219 follow-up NP swabs were collected. The median number of NP swabs collected per patient was 2 (IQR, 1–4). Overall, 172 (79%) NP swabs from 74 (95%) patients were positive for SARS-CoV-2 by PCR (Table 2). Among patients with at least 1 positive NP swab, the median number of positive swabs was 2 (IQR, 1–3).
The median Ct value among positive follow-up NP swabs was 31.4 (IQR, 26.8–34.5) (Figure 1). Overall, 30 (27%) of the 110 cultured NP swabs from 21 unique patients yielded viable virus; the highest Ct observed to yield viable virus was 27.2.
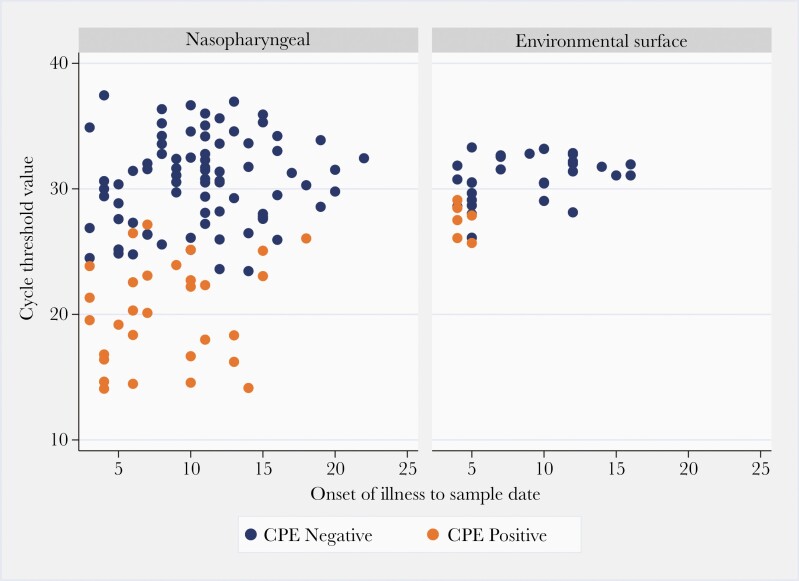
The median time between onset of illness and sampling date was 13 days (IQR, 9–20; range, 3–56 days) for all nasopharyngeal swabs, 12 days (IQR, 8–17; range, 3–52 days) for PCR-positive swabs, and 7 days (IQR, 5–11; range, 3–18 days) for culture positive swabs (Figure 2).
Figure 2.
Virus isolation results from 110 nasopharyngeal swabs and 36 surface samples in relation to polymerase chain reaction cycle threshold value and time since symptom onset. CPE, cytopathic effect.
Surface Samples
A total of 474 surface samples were collected. Sixty-one patients (78%) had at least 1 complete set of surface samples, 12 (15%) had 4 of 5 surface types, 2 (3%) had 3, and 3 (4%) had 2. Overall, 125 (25%) surface samples from 42 (54%) patient rooms yielded SARS-CoV-2 RNA; virus was most frequently detected from the bedrail/light switch pool and least frequently on the bathroom doorknob (Table 2).
The median Ct value of surface samples testing positive for SARS-CoV2-2 was 35.1 (IQR, 32.8–36.9). Cycle thresholds across surface types were similar (Figure 1). Thirty-six surface samples had Ct values <34.0 and 6 of these (17%), from 3 (4%) patients, yielded infectious virus; the highest Ct of a sample yielding virus by culture was 29.1 (Figure 2). Viable virus was recovered from each surface type investigated.
The median time between onset of illness and surface sample date was 10 days (IQR, 6–12) for all environmental surface swabs, 9 days (IQR, 5–12; range, 3–20 days) for PCR-positive surface swabs, and 4 days (range, 4–5) for culture-positive surface swabs (Figure 2).
Air Samples
A total of 146 air samples were collected; 101 samples (17 gelatin filters, 39 PC filters, 6 PTFE filters, and 13 of each NIOSH stage) at a distance of 1 m from the patient and 45 samples (39 PC filters and 6 PTFE filters) at a distance of 2 m from the patient. Three (2%) air samples taken from 3 (7%) different rooms at 1 m from the patient were positive for SARS-CoV-2 RNA by PCR; none yielded viable virus. The median time between onset of illness and air sampling was 11 days (IQR, 7–14); the time between onset of illness and sampling date for all 3 PCR-positive air samples was 4 days. Each of the 3 PCR-positive air samples were collected by a different air sampling method, including PTFE and PC filters and the NIOSH sampler where viral RNA was detected from stage 1.
Genome Sequencing
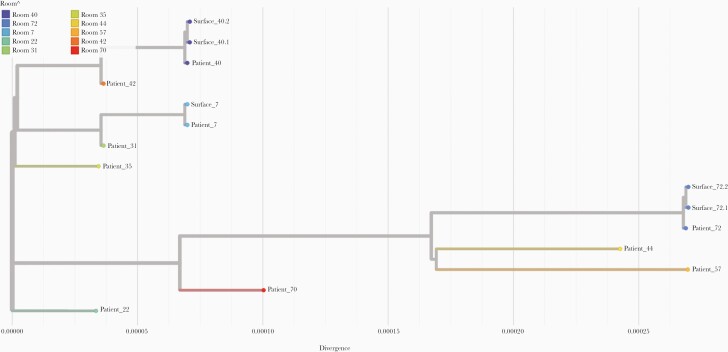
In total, 152 surface samples and NP swabs with Ct values ranging from 16.3 to 33.2 (UTR gene) underwent whole-genome sequencing for SARS-CoV-2. Fifty passed the Canadian COVID Genomics Network (CanCOGeN) quality control for public release of SARS-CoV-2 genomes and were submitted to GSAID [24]; 23 of these were from surface samples. Air samples were excluded from the analyses due to poor quality sequences. A phylogenetic analysis of NP and surface swabs is presented in Figure 3. For ease of visualization, we included up to 2 surface samples per room passing quality control in the phylogenetic analyses. All surface samples cluster with the corresponding NP swabs from patients occupying the same room.
Figure 3.
Phylogenetic tree of 15 severe acute respiratory syndrome coronavirus 2 genomes from inpatients’ nasopharyngeal swabs and environmental surface swabs from 10 patients’ rooms. Augur pipeline from Nextstain was used to build the phylogenetic tree based on the IQTREE method. The root of the tree is obtained with the first isolate from Wuhan-Hu-1 referenced MN908947.3 in National Center for Biotechnology Information. The tree is refined using RAxML.
Factors Associated With Positive Environmental Swabs
In the final multivariable mixed-effects model, the following were found to be associated with the detection of SARS-CoV-2 RNA in environmental samples: hypoxia on admission, PCR-positive NP swab with Ct ≤30 on or after the environmental sampling date, higher Charlson comorbidity index score, and shorter time from onset of illness to environmental sample date (Table 3).
Table 3.
Results From the Final Mixed-Effects Logistic Regression Analysis, With a Random Effect for Patient, Exploring the Association Between Patient Factors and Detection of SARS-CoV-2 RNA in Environmental Samples From 78 Hospitalized Patients With COVID-19 in Toronto, Canadaa
| Explanatory Variable | Category | Odds Ratio (95% CI) | P Valueb |
|---|---|---|---|
| Hypoxic on admission | No | Referent | |
| Yes | 7.25 (2.00–26.33) | .003 | |
| PCR-positive nasopharyngeal swab on or after sample date | No | Referent | <.001 |
| Ct >30 | 1.81 (0.30–10.91) | .515 | |
| Ct ≤30 | 15.56 (2.21–109.32) | .006 | |
| Charlson score | No comorbidities | Referent | .006 |
| Mild (1–2) | 4.48 (1.28–15.77) | .019 | |
| Moderate–severe (≥3) | 13.72 (2.39–78.80) | .003 | |
| Onset to sample date | ≤7 days | Referent | |
| >7 days | 0.27 (0.09–0.79) | .017 | |
| Variance | Patient | 3.66 (1.73–7.74) |
Abbreviations: CI, 95% confidence interval; COVID-19, coronavirus disease 2019; Ct, cycle threshold; PCR, polymerase chain reaction; RNA, ribonucleic acid; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Mixed-effects logistic regression model was constructed via stepwise backwards elimination and investigated the following variables: age, sex, Charlson comorbidity index, smoking history, Clinical Frailty Score, presence or absence of symptoms from onset to 24 hours postadmission (cough, fever, diarrhea, delirium/confusion), hypoxia at admission (defined as oxygen saturation <92%), admission to intensive care unit at time of sample collection, use of exogenous oxygen during stay, prone position, receiving steroids for treatment on day of sampling, room type (regular private room or negative pressure room), and the presence or absence of a PCR-positive nasopharyngeal (NP) swab on or after environmental sampling date (PCR-positive NP swabs were further categorized to Ct >30 and Ct ≤30).
Significance of variables with 3 or more categories were assessed via Wald’s χ2 test.
The intraclass correlation coefficient between observations at the patient level was 53% (95% confidence interval, 34%–70%). No outlying observations were identified. Graphical exploration of the BLUPs for patient identification seemed to meet the assumptions of homogeneity of variance and normality.
DISCUSSION
In this prospective cohort study in Ontario, Canada, SARS-CoV-2 RNA was detected from surfaces (25%) and air (2%) in the acute care setting. The genomic analyses of whole SARS-CoV-2 sequences in the present work confirmed that patients were the source of viral contamination of their immediate surroundings in this setting. Although direct comparison of our results to other studies is limited due to heterogeneity in sampling, processing, and detection methodologies, proportionally higher rates of recovery of viral RNA from surfaces compared with the air are broadly consistent with other studies investigating SARS-CoV-2 surface and air contamination [1, 5–7, 10–12].
A limited number of studies to date have recovered viable SARS-CoV-2 virus from environmental samples [14]. We attempted to recover SARS-CoV-2 virus from 36 environmental surface samples, 6 (17%) of which from 3 (4%) patients yielded viable virus; positive cultures were confirmed with SARS-CoV-2 RT-PCR. It is notable that all surface samples that yielded viable virus were collected from patients within 5 days of illness onset.
The PCR-positive air samples were collected from within 1 m of the patient in 3 cases. However, we were unable to culture viable virus from any of these air samples. To our knowledge, only 1 study has isolated SARS-CoV-2 from air samples in this setting [25]. However, it is important to note that the authors concentrated their samples before cell culture, potentially optimizing viable virus recovery from samples despite low concentrations of SARS-CoV-2 in the sample. In addition, although no CPE was observed, Santarpia et al [9] did observe increases in viral RNA in cell culture; Western blot and transmission election microscopy also showed evidence of viral proteins and intact virions. The difficulty in culturing virus from air samples likely relates to a combination of low viral concentrations, dilution effects, the effects of sampling itself on viral cell membrane, and surface protein integrity [3].
In the multivariable analysis, hypoxia on admission, a PCR-positive NP swab with a Ct of ≤30 on or after the environmental sampling date, higher Charlson comorbidity index score, and shorter time from symptom onset to environmental sampling were significantly associated with the detection of SARS-CoV-2 RNA in environmental surface samples (Table 3). Although, to our knowledge, no other study has investigated putative patient factors associated with environmental contamination using multivariable modeling, our findings are consistent with several observational studies that show that viral load peaks in the first week of illness in COVID-19 patients, with active viral replication in the upper respiratory tract in the first 5 days of illness [1, 10, 26, 27]. In addition, both hypoxia and a high Charlson comorbidity index have previously been found to be associated with higher SARS-CoV-2 viral loads in the nasopharynx [28, 29].
Our study has several limitations. First, although we had a large number of surface and air samples, the samples were recovered from only 78 patients, resulting in a relatively small effective sample size when accounting for clustering. Although this still facilitated an exploratory analysis, this limited the power of our multivariable analysis as indicated by the wide confidence intervals for some of the significant variables in our final model. The small effective sample size also prohibited us from investigating factors associated with viable virus in environmental surface samples, including time from symptom onset. Second, the present work focused only on acute care inpatients, excluded critically ill individuals, and had first samples obtained several days after onset of illness. Working in acute care allowed us ready access to patient areas for sampling and clinical data to garner a granular understanding of environmental contamination in hospital settings; however, the generalizability of our findings to other settings is limited, particularly where room ventilation is highly variable such as homes, schools, long-term care residences, other workplaces, and public spaces, or where patients may be presymptomatic or early in the course of their disease. Because of the pandemic, we were not able to access individual rooms to measure air exchanges. It is important to note that these data were collected before the emergence of the SARS-CoV-2 variants of concern (VOCs) in late 2020. Both the alpha and delta variants (B.1.617.2) are transmitted more efficiently [30], but the modes of transmission and the effect of the difference in the variants on surface and air contamination are unknown. As such, the results from the present work represents a baseline that can be used to understand the transmission dynamics of VOCs. There was no apparent difference in the yield of SARS-CoV-2 with different air samplers and filters, but our study was underpowered to detect such differences. Finally, we did not use a standard curve for our RT-PCR analysis and could not calculate the virus concentration per volume of air. Therefore, we were not able to estimate a limit of detection for our aerosol samples.
CONCLUSIONS
The findings of this study provide insights into surface and air contamination with SARS-CoV-2 in hospitalized COVID-19 patients. We found that SARS-CoV-2 RNA was detected on a minority of surfaces in COVID-19 patients’ rooms and rarely from air samples, and only early in the course of their hospitalization. These data suggest environmental sources are unlikely to pose a major exposure risk in hospitals with similar personal protective equipment, surface decontamination procedures, and ventilation in place. In addition, hypoxia on admission, PCR-positive NP swab on or after sampling date, and comorbidities were found to be important factors associated with the detection of SARS-CoV-2 RNA in the environment. These findings taken together, suggest that the overall likelihood transmission from the air and surfaces in hospitalized patients with COVID-19 is likely limited. Nonetheless, early detection and isolation of COVID-19 patients is important to ensure minimization of exposure risk to healthcare workers, particularly when patients are admitted shortly after onset of symptoms.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Dr. David Bulir at McMaster University for providing the primer and probe sequences for the UTR gene target for the severe acute respiratory syndrome coronavirus 2 polymerase chain reaction assay used in the present work. Computational support was provided by the McMaster Service Lab and Repository computing cluster, supplemented by hardware donations and loans from Cisco Systems Canada, Hewlett Packard Enterprise, and Pure Storage. Many thanks to the patients with coronavirus disease 2019 who agreed to participate in this research. We are grateful to the laboratory, infection prevention and control, public health, and research ethics staff who make the Toronto Invasive Bacterial Diseases Network possible.
Disclaimer. The funding body had no role the design of the study or in the collection, analysis, and interpretation of data nor in the process of writing the present manuscript.
Financial support. This study was funded by the Canadian Institutes of Health Research (CIHR No. RN419944-439999) as well as The Canadian COVID Genomics Network funding. J. D. K. is supported by an AMMI Canada/BioMérieux Fellowship in Microbial Diagnostics. A. J. J. was supported by the Vanier Canada Graduate Scholarship at the time of this work. H. M. is funded with a postdoctoral fellowship from le Fond de Recherche du Québec – Nature et Technologie and is the recipient of the Lab Exchange Visitor Program Award from the Canadian Society for Virology. C. D. is holder of Canada Research Chair on Bioaerosols. A. G. M. holds McMaster’s inaugural David Braley Chair in Computational Biology, generously supported by the family of the late David Braley.
Potential conflicts of interest. All authors report no conflicts relevant to this article. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Zhou J, Otter JA, Price JR, et al. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin Infect Dis 2020; 73:e1870–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richterman A, Meyerowitz EA, Cevik M.. Hospital-acquired SARS-CoV-2 infection: lessons for public health. JAMA 2020; 324:2155–6. [DOI] [PubMed] [Google Scholar]
- 3. Chia PY, Coleman KK, Tan YK, et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun 2020; 11:2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colaneri M, Seminari E, Novati S, et al. Severe acute respiratory syndrome coronavirus 2 RNA contamination of inanimate surfaces and virus viability in a health care emergency unit. Clin Microbiol Infect 2020; 26:1094.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo Z-D, Wang Z-Y, Zhang S-F, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis 2020; 26:1583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y, Ning Z, Chen Y, et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 2020; 582:557–60. [DOI] [PubMed] [Google Scholar]
- 7. Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA 2020; 323:1610–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santarpia JL, Rivera DN, Herrera VL, et al. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci Rep 2020; 10:12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santarpia JL, Herrera VL, Rivera DN, et al. The size and culturability of patient-generated SARS-CoV-2 aerosol. J Expo Sci Environ Epidemiol 2021. doi: 10.1038/s41370-021-00376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J, Feng H, Zhang S, et al. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the coronavirus disease 2019 outbreak in a Chinese hospital. Int J Infect Dis 2020; 94:103–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu S, Wang Y, Jin X, Tian J, Liu J, Mao Y.. Environmental contamination by SARS-CoV-2 in a designated hospital for coronavirus disease 2019. Am J Infect Control 2020; 48:910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye G, Lin H, Chen S, et al. Environmental contamination of SARS-CoV-2 in healthcare premises. J Infect 2020; 81:e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Birgand G, Peiffer-Smadja N, Fournier S, Kerneis S, Lescure F-X, Lucet J-C.. Assessment of air contamination by SARS-CoV-2 in hospital settings. JAMA Netw Open 2020; 3:e2033232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cherrie J, Cherrie M, Davis A, et al. Contamination of air and surfaces in workplaces with SARS-CoV-2 virus: a systematic review. Ann Work Expo Health 2021; 65:879–92. doi: 10.1101/2021.01.25.21250233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oude Munnink BB, Nieuwenhuijse DF, Stein M, et al. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med 2020; 26:1405–10. [DOI] [PubMed] [Google Scholar]
- 16. Gorzynski JE, De Jong HN, Amar D, et al. High-throughput SARS-CoV-2 and host genome sequencing from single nasopharyngeal swabs [preprint]. medRxiv 2021. [Google Scholar]
- 17. Handrick S, Bestehorn-Willmann M, Eckstein S, et al. Whole genome sequencing and phylogenetic classification of Tunisian SARS-CoV-2 strains from patients of the Military Hospital in Tunis. Virus Genes 2020; 56:767–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nasir JA, Kozak RA, Aftanas P, et al. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses 2020; 12:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Elm E, Altman DG, Egger M, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet Lond Engl 2007; 370:1453–7. [DOI] [PubMed] [Google Scholar]
- 20. Marty FM, Chen K, Verrill KA.. How to obtain a nasopharyngeal swab specimen. N Engl J Med 2020; 382:e76. [DOI] [PubMed] [Google Scholar]
- 21. LeBlanc JJ, Gubbay JB, Li Y, et al. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J Clin Virol 2020; 128:104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Charlson ME, Pompei P, Ales KL, MacKenzie CR.. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 23. Rockwood K, Song X, MacKnight C, et al. A global clinical measure of fitness and frailty in elderly people. CMAJ 2005; 173:489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shu Y, McCauley J.. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro Surveill 2017; 22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lednicky JA, Lauzardo M, Fan ZH, et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int J Infect Dis 2020; 100:476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 27. Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 2020; 323:1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shlomai A, Ben-Zvi H, Glusman Bendersky A, Shafran N, Goldberg E, Sklan EH.. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID-19 patients. Crit Care 2020; 24:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zacharioudakis IM, Prasad PJ, Zervou FN, et al. Association of SARS-CoV-2 genomic load with COVID-19 patient outcomes. Ann Am Thorac Soc 2020; 18:900–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown KA, Joh E, Buchan S, et al. Inflection in prevalence of SARS-CoV-2 infections missing the N501Y mutation as a marker of rapid Delta (B.1.617.2) lineage expansion in Ontario, Canada [preprint]. medRvix 2021. https://www.medrxiv.org/content/10.1101/2021.06.22.21259349v1.article-info. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.