Abstract
Since 1990, the frequency of Neisseria meningitidis serogroup C (NMSC) outbreaks in the United States has increased. Based on multilocus enzyme electrophoresis (MEE), the current molecular subtyping standard, most of the NMSC outbreaks have been caused by isolates of several closely related electrophoretic types (ETs) within the ET-37 complex. We chose 66 isolates from four well-described NMSC outbreaks that occurred in the United States from 1993 to 1995 to evaluate the potential of pulsed-field gel electrophoresis (PFGE) to identify outbreak-related isolates specific for each of the four outbreaks and to differentiate between them and 50 sporadic isolates collected during the outbreak investigations or through active laboratory-based surveillance from 1989 to 1996. We tested all isolates collected during the outbreak investigations by four other molecular subtyping methods: MEE, ribotyping (ClaI), random amplified polymorphic DNA assay (two primers), and serotyping and serosubtyping. Among the 116 isolates, we observed 11 clusters of 39 NheI PFGE patterns. Excellent correlation between the PFGE and the epidemiological data was observed, with an overall sensitivity of 85% and specificity of 71% at the 95% pattern relatedness breakpoint using either 1.5 or 1.0% tolerance. For all four analyzed outbreaks, PFGE would have given public health officials additional support in declaring an outbreak and making appropriate public health decisions.
Studies in the areas of population biology, pathogenesis, epidemiology, and molecular microbiology have demonstrated the clonal nature of relationships among Neisseria meningitidis isolates. While some clones have been isolated repeatedly, spanning long periods of time and from multiple locations throughout the world, others have been isolated rarely and then only from certain geographic areas. Meningococci associated with epidemics and outbreaks generally belong to uniform clonal groups, in contrast to meningococci causing sporadic disease, which are more variable (1). Multilocus enzyme electrophoresis (MEE) has been proven to be a suitable method to detect such clones and therefore has been the “gold standard” for molecular subtyping of N. meningitidis for over two decades (5, 19). Molecular subtyping of isolates may provide useful information in determining whether a group of cases actually represents an outbreak or simply represents a change in the incidence of sporadic cases. Even though the annual incidence of meningococcal disease caused by all serogroups in the United States has remained stable at 1/100,000 to 1.5/100,000 over the past 30 years, since 1990 the number of N. meningitidis serogroup C (NMSC) outbreaks has increased (11, 21; C. R. Woods, N. E. Rosenstein, and B. A. Perkins, Abstr. 38th Annu. Meet. Infect. Dis. Soc. Am., abstr. 125FR, 1998). In the United States a threshold attack rate is used to differentiate outbreaks from sporadic meningococcal disease cases and serves as the basis for initiation of mass vaccination campaigns (6). Most of the NMSC outbreaks in the United States have been caused by isolates of the closely related electrophoretic types (ETs) within the ET-37 complex, but the same ETs are also commonly found in endemic isolates. Also, MEE is labor-intensive, time-consuming, and difficult for interlaboratory comparison and therefore is limited to very few laboratories that currently maintain this capacity. Recently, pulsed-field gel electrophoresis (PFGE), being rapid and easy to perform, has been successfully applied to the molecular subtyping of numerous bacteria (24). It currently serves as the basis for the PulseNet, a network for nationwide monitoring of several food-borne disease pathogens, such as Escherichia coli O157:H7, Salmonella spp., Listeria monocytogenes, and Shigella sonnei. Reports on the use of PFGE to differentiate isolates of N. meningitidis of serogroups A and B (4, 13, 14, 25, 29) indicate that, especially for serogroup B, PFGE provides a substantial level of discrimination comparable to that of ribotyping and better than those of serosubtyping, MEE, and PCR-restriction fragment length polymorphism analysis. However, the epidemiology of meningococcal disease associated with serogroups A and B differs from that of disease associated with NMSC. N. meningitidis serogroup A has long been the main cause of meningococcal disease outbreaks worldwide, especially in the African “meningitis belt.” It has been associated with a particular clonal group, identified by MEE as subgroup III-1, and with only a few PFGE patterns identified over extended periods of time. In contrast, N. meningitidis serogroup B has demonstrated the most diversity of all serogroups, with over 1,000 ETs identified to date. Due to that diversity and to the fact that in the United States it is mainly associated with sporadic meningococcal disease, PFGE analysis would not be particularly useful for epidemiological purposes. PFGE has been used as a tool to molecularly characterize NMSC isolates from sporadic cases of meningococcal disease and in NMSC outbreak investigations (3, 7, 8, 18, 28; T. L. Bannerman, K. B. Sohner, S. Karam, S. Nowicki, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. C-396, p. 186, 1999; R. Danila, R. Rainbow, D. Boxrud, J. Besser, K. Moore, M. Osterholm, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. L-2094, p. 691, 1999). However, isolates from a single outbreak either have not been analyzed along with isolates from sporadic cases or have not been extensively compared with the epidemiological data.
We chose four well-described NMSC outbreaks occurring from 1993 to 1995 in Texas, New Mexico, Arizona, and California (15, 22, 26) to systematically evaluate the potential of PFGE to identify outbreak-associated isolates. We also included in the study 26 NMSC isolates, representing sporadic disease in the United States, collected through the active laboratory-based surveillance program from 1989 to 1996. Since an outbreak is defined based on the epidemiological criteria and supported by the current gold standard for subtyping, MEE, we assayed all isolates by MEE as well. To further assess the relative differentiation capabilities of PFGE, we tested all isolates collected during the outbreak investigations by three additional molecular subtyping methods: ribotyping, serotyping and serosubtyping, and random amplified polymorphic DNA (RAPD) assay.
MATERIALS AND METHODS
Serogroup C meningococcal disease (SCMD) outbreaks.
An outbreak is defined, by the Advisory Committee on Immunization Practices of the Centers for Disease Control and Prevention (CDC), as an occurrence of three or more confirmed or probable cases of meningococcal disease during a period of <3 months in persons who have a common affiliation but no close contact with one another (an organization-based outbreak) or in persons residing in the same area who are not close contacts and who do not share a common affiliation (a community-based outbreak), resulting in a primary disease attack rate of at least 10 cases per 100,000 persons (6). A total of 90 isolates were collected during the investigations of four well-defined NMSC outbreaks. Epidemiological information was solicited from state and local health authorities as well as from CDC investigators, and all available reports concerning these outbreaks were reviewed. Isolates epidemiologically defined as being a part of a particular outbreak will be referred to as outbreak-associated, isolates, while those that were identified as not being part of an outbreak will be referred to as sporadic isolates. For easy reference, the prefix OA or SP, respectively, is used as part of each isolate's designation.
(i) Texas.
Between 14 February 1993 and 2 September 1995, 41 cases of meningococcal disease were reported in Gregg County, Tex. (22). We analyzed isolates from 26 of these outbreak-associated cases and nine additional isolates from sporadic cases throughout Texas from 1995 to 1996.
(ii) New Mexico.
Between 2 and 16 March 1994, seven confirmed cases of meningococcal disease occurred in five rural New Mexico communities, yielding an attack rate of 200 per 100,000 persons (15). In addition to the seven isolates epidemiologically defined as outbreak associated, we included in this study five isolates from patients with sporadic meningococcal disease in New Mexico isolated in the same year.
(iii) Arizona.
We included a total of 23 isolates from cases of SCMD from Maricopa County, Ariz., collected in 1993 and 1994. Using a combination of epidemiological data and PFGE analysis (carried out with a different restriction enzyme) at the Arizona State Health Department, 19 of the isolates were identified as outbreak associated and four were identified as sporadic isolates from other geographic locations in Arizona.
(iv) California.
Between 1 January and 31 March 1993, 54 cases of meningococcal disease occurred in Los Angeles County, Calif.; 45 were among community residents and 9 were in inmates in the county's jail system (26). In this study we included 10 isolates from nine inmates (OA29, OA30, OA31, OA32, OA33, OA140, OA141, OA145, OA148, and OA149), 4 isolates from community residents with reported contact with inmates (OA34, OA147, OA151, and OA152), and 6 isolates from community residents with no contact with inmates (SP128, SP131, SP143, SP144, SP146, and SP153).
(v) Active surveillance.
Population-based surveillance for invasive disease caused by N. meningitidis is part of an ongoing multistate active surveillance project coordinated by the CDC. Between 1989 and 1996, CDC collaborated on active surveillance with investigators in state and local health departments or universities in as many as seven geographically dispersed areas of the United States with an aggregate population of 22 million. A surveillance case of meningococcal disease was defined as the isolation of N. meningitidis from a normally sterile site, such as blood or cerebrospinal fluid, in a resident of a surveillance area, and 807 cases of meningococcal disease were detected, for an average annual incidence of 1.1/100,000 persons during this period. Twenty-six isolates were selected randomly to represent a temporal and geographic distribution of all isolates, and these will be consistently referred to as surveillance isolates, with the prefix SU as part of the isolate designation.
Laboratory methods.
All isolates of N. meningitidis were characterized at CDC using standard microbiological procedures (16). Initial serogrouping was done at hospital microbiology laboratories or at state health departments, and the serogrouping was then repeated at CDC.
(i) PFGE.
The PFGE method used in this study is based upon procedures for testing E. coli O157:H7 as described by Barrett et al. (2) and by Gautom (9). Deviations from the published protocols are as follows. Overnight growth from a sheep blood agar plate was harvested with a 1-μl disposable loop. The loop was gently rubbed against the side wall of a test tube containing 2 ml of cell suspension buffer, followed by gentle mixing to result in a final suspension with a reading of 0.48 to 0.52 in a Dade Microscan turbidity meter. Plugs were prepared with 400 μl of cell suspension, 20 μl of a 2% proteinase K solution (Amresco, Solon, Ohio), and an equal volume of 1% SeaKem Gold–1% sodium dodecyl sulfate-agarose and dispensed into reusable plug molds. Incubation for 1.5 to 2 h in a shaker water bath at 50 to 54°C range allowed for lysing the cells of one plug in a 2-ml round-bottom tube containing 1.5 ml of cell lysis buffer. After washing of the plugs, microtubes containing endonuclease NheI diluted in restriction buffer to 50 U per plug slice were incubated in a 37°C water bath for 1.5 to 2 h. Restricted plug slices were placed at the bottom of each tooth of a 10-tooth comb and submerged in 95 ml of molten 1.0% SeaKem Gold agarose in 0.5× Tris-borate-EDTA buffer until the gel hardened. After comb removal, the wells were sealed with molten agarose and the gel was placed in the prepared electrophoresis chamber. Electrophoresis conditions were as follows: chamber, CHEF-DR III (Bio-Rad Laboratories, Hercules, Calif.) containing 2 liters of freshly prepared 0.5× Tris-borate-EDTA buffer cooled to 14°C, initial switch time, 2.2 s; final switch time, 35 s; duration of run, 18 h, angle, 120°; gradient, 6 V/cm with a linear ramping factor.
For data analysis, Tiff images of the gels were normalized using the Molecular Analyst Fingerprinting Plus software, version 1.11 (Bio-Rad Laboratories), by aligning the standard NMSC isolate (M413), located in lanes 1, 5, and 10 of each gel, with the global standard for the database (M413). Isolates were tested two times and bands that were faint, not consistent on repeat testing of the isolate, and larger than the largest band on the standard were not included in the analysis. Analysis of band patterns was performed with the Dice coefficient using 0.5, 1, and 1.5% tolerances for the band migration distance. Patterns were defined as indistinguishable using the Molecular Analyst software if the patterns had the same number of marked bands and if the band positions were within 2, 4, or 6 points of each other for a 0.5, 1.0, or 1.5% tolerance, respectively. All patterns were visually inspected after computer analysis. Patterns that were identified as indistinguishable by the computer and were indistinguishable after visual inspection were assigned a pattern designation. Four exceptions were seen where patterns were identified as indistinguishable by the computer but were visually distinguishable after inspection. These patterns were assigned a different designation. PFGE patterns were designated using the organism, enzyme, and pattern number scheme recommended by the CDC PFGE committee and used by PulseNet. Clustering of patterns was performed by unweighted pair group with arithmetic averaging (UPGMA). The percentage of similarity between patterns is used only to address the relatedness among the PFGE patterns and is not an indication of (quantitative) genetic relatedness among respective isolates, a measurement that can be appropriately determined by other techniques such as MEE.
(ii) MEE.
MEE, using 24 enzymes, was performed as described previously (23). Numbers were assigned to enzyme alleles on the basis of enzyme mobilities, and each unique set of alleles was defined as an ET. An index of genetic relatedness was determined by weighting the degree of diversity at each of the 24 enzyme loci, and similarities among the ETs were assessed by dendrogram analysis (12).
(iii) RAPD assay.
The RAPD assay was carried out as described previously (S. E. Schmink, M. W. Reeves, B. Plikaytis, and T. Popovic, submitted for publication).
(iv) Serotyping and serosubtyping.
The meningococcal isolates were tested with a set of 15 monoclonal antibodies (MAbs) against the variable regions of PorB outer membrane proteins (serotyping) and against 15 murine MAbs produced against the variable regions of PorA outer membrane proteins (serosubtyping) by dot blot analysis as previously described (27). Briefly, the whole-cell suspensions were dotted on nitrocellulose, and strips were blocked for 30 min using bovine serum albumin (3% in phosphate-buffered saline). MAbs were pipetted into the blocking buffer at dilutions ranging from 1 in 4,000 to 1 in 32,000. After overnight incubation, the strips were incubated for 2 h with goat anti-mouse immunoglobulin G conjugated to peroxidase (1 in 4,000) (Sigma, St. Louis, Mo.) and developed with the substrate 3-amino-9-ethylcarbazole (Sigma) and hydrogen peroxidase. MAbs against serotypes 2a (5D4-5), 2b (2H10-2), 2c (6-D9-5.6-F3), 4 (5DC4-C8-G8), 5 (7BG5-H2), 11 (9-1-P11), 15 (8-B5-5-B9), and 21 (6B11-C2-F1) and serosubtypes P1.2 (OD6-4), P1.3 (5G8-B2-F9), P1.16 (OF11-4), and P1.19 (7A2-11) were supplied by W. D. Zollinger, Walter Reed Army Medical Center, Washington, D.C. MAbs against serotypes 1 (MN3C6B) and 14 (MN5C8C) and serosubtypes P1.1 (MN14C2.3), P1.4 (MN20B9.34), P1.5 (MN22A9.19), P1.6 (MN19D6.13), P1.7 (MN14C11.6), P1.9 (MN5A10.7), P1.10 (MN20F4.17), P1.12 (MN20A7.10), P1.13 (MN25H10.75), P1.14 (MN21G3.17), and P1.15 (95-718) were supplied by I. M. Feavers, National Institute for Biological Standards and Control, Herts, United Kingdom. MAbs against serotypes 7 (F22-8B5/1D10), 9 (F24-11F5/3B4), 10 (F11-6D12/1C5), and 17 (F4-3C1/1A6) were provided by C. T. Sacchi, Institute Adolfo Lutz, San Paulo, Brazil. MAb against serotype 22 (IA5D90) was supplied by P. Kriz, National Institute of Public Health, Prague, Czech Republic.
Ribotyping.
Ribotyping was performed as previously described (17) with the following modifications. DNA was extracted from the isolates using the Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.) according to the manufacturer's instructions. The extracted DNA was restricted using ClaI at 37°C for 4 h. Hybridization was performed using a mixture of five oligonucleotide probes as described by Renault et al. (20).
RESULTS
A total of 116 NMSC isolates were included in this study. Of 90 isolates collected during the investigations of four epidemiologically well-defined outbreaks of SCMD, 66 were epidemiologically defined as outbreak associated, while the remaining 24 were from sporadic cases of meningococcal disease. An additional 26 isolates were from the laboratory-based surveillance.
Texas.
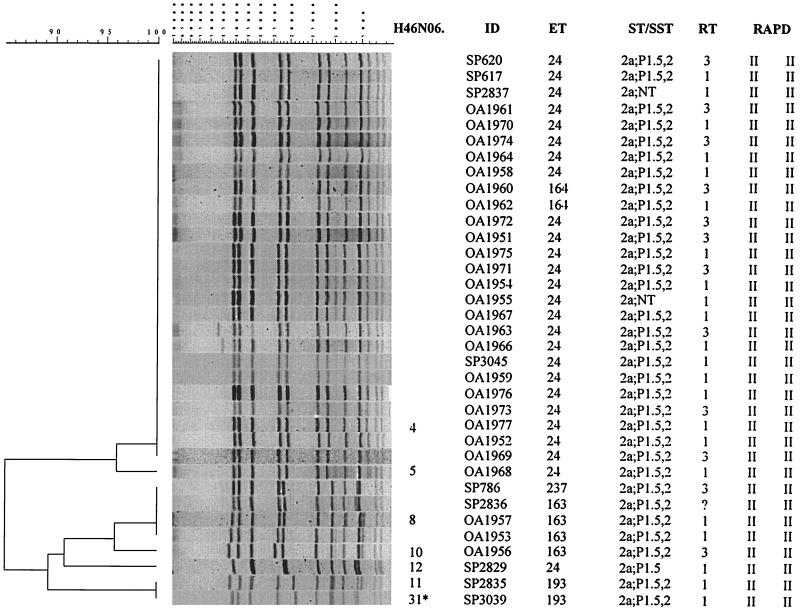
Among the 35 isolates from Texas (26 outbreak associated and nine sporadic), two distinct PFGE clusters composed of seven PFGE patterns were identified, with 85% overall relatedness (Fig. 1). The larger cluster contained 27 isolates and two very similar PFGE patterns (96% relatedness). Twenty-two of the 26 outbreak-associated isolates had indistinguishable PFGE patterns (H46N06.0004), which they shared with four sporadic isolates. In addition, these 22 outbreak-associated isolates were either of ET-24 (20 isolates) or of ET-164 (2 isolates). There is only a single enzyme difference between these two ETs. Pattern H46N06.0005, seen in isolate OA1968, differed from the predominant H46N06.0004 pattern by a single band. The smaller cluster contained eight isolates and five PFGE patterns. Three isolates (OA1953, OA1956, and OA1957) were epidemiologically identified as part of the outbreak, while the remaining five were isolated from sporadic cases of meningococcal disease in Texas. Two of these three outbreak-associated isolates (OA1953 and OA1957) had an H46N06.0008 pattern indistinguishable from that of two sporadic isolates (SP786 and SP2836). The H46N06.0008 pattern showed only 85% relatedness to the predominant H46N06.0004 pattern. ET-163, identified in isolates OA1953 and OA1957, differed from ET-24, seen in the outbreak-associated isolates, in a single enzyme (relatedness between these two ETs was >98%). Patterns of two sporadic isolates, SP2835 and SP3039, were identified as indistinguishable in the computer analysis, but visual inspection showed spacing differences between bands in these two isolates. The pattern of isolate SP2835 was designated H46N06.0011 and the pattern of isolate SP3039 was designated H46N06.0031. This is one of four exceptions made in the assigning of patterns.
FIG. 1.
Designations, molecular characterization, and PFGE analysis (with NheI) of chromosomal DNAs of 35 NMSC isolates collected during an investigation of a meningococcal disease outbreak in Texas in 1995. The PFGE patterns of isolates SP2835 and SP3039 were identified as indistinguishable using the 1.5% tolerance, but based on visual differences in spacing between the bands, isolate SP3039 was defined as distinct and was given a pattern designation of H46N06.0031 (asterisk). ID, isolate identification number; ST/SST, serotype and serosubtype; RT, ribotype; RAPD, RAPD type (with two primers).
New Mexico.
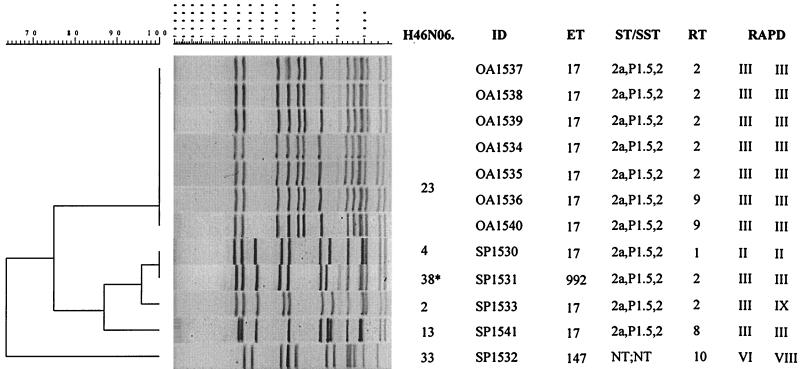
Analysis of the 12 New Mexico isolates resulted in three separate clusters of PFGE patterns at the 85% relatedness breakpoint (Fig. 2). One cluster contained only a single PFGE pattern (H46N06.0023), which was identified in all seven outbreak-associated isolates. These isolates also had identical ETs (ET-17), identical serotype and serosubtype, and identical RAPD types. The five sporadic isolates were grouped within the two remaining PFGE clusters. The first sporadic cluster had four PFGE patterns (H46N06.0004, H46N06.0038, H46N06.0002, and H46N06.0013). Isolates SP1530 and SP1531, with patterns H46N06.0004 and H46N06.0038 (identified as indistinguishable by the computer analysis but defined as different upon visual inspection, based on band spacing differences), were distinguishable from the New Mexico outbreak-associated isolates by MEE (SP1531) or ribotyping and RAPD analysis (SP1530). Another isolate within this cluster (SP1541) with an unique PFGE pattern (H46N06.0013) had an ET, serotype and serosubtype, and RAPD type identical to those of the outbreak-associated isolates. A fourth sporadic isolate (SP1533) in this cluster had a unique PFGE pattern (H46N06.0002), but its ET, serotype and serosubtype, and ribotype were indistinguishable from those of the outbreak isolates. The second sporadic cluster contained only a single isolate (SP1532), whose H46N06.0033 pattern showed less than 65% relatedness with all other PFGE patterns. Results from the other molecular subtyping methods also suggested that this isolate was unique.
FIG. 2.
Designations, molecular characterization, and PFGE analysis (with NheI) of chromosomal DNAs of 12 NMSC isolates collected during an investigation of a meningococcal disease outbreak in New Mexico in 1994. The PFGE patterns of isolates SP1530 and SP1531 were identified as indistinguishable using the 1.5% tolerance, but based on visual differences in spacing between the bands, isolate SP1531 was defined as distinct and given a pattern designation of H46N06.0038 (asterisk). ID, isolate identification number; ST/SST, serotype and serosubtype; RT, ribotype; RAPD, RAPD type (with two primers).
Arizona.
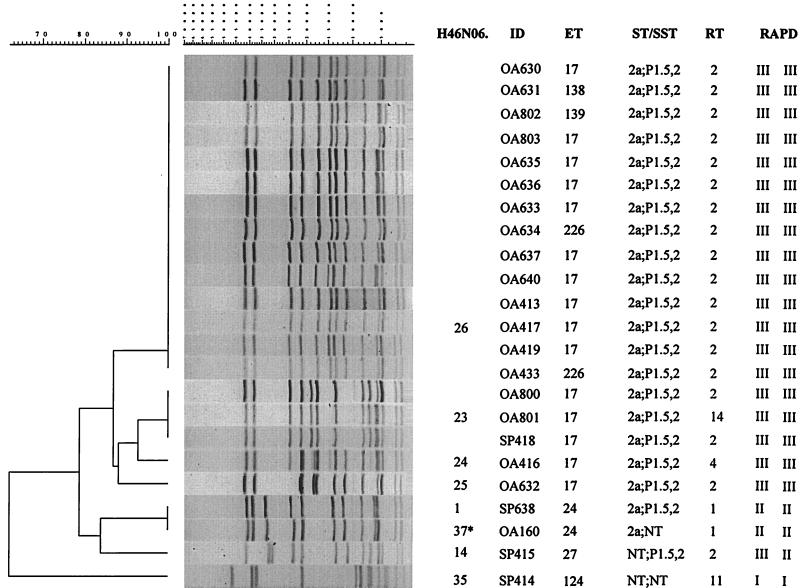
Among the 23 isolates from the Arizona outbreak investigation, four PFGE pattern clusters were identified at the 85% relatedness level (Fig. 3). The largest cluster contained 19 isolates and four PFGE patterns; 14 of the 19 outbreak-associated isolates had pattern H46N06.0026. Ten of these 14 isolates were ET-17. The other four isolates had three different ETs, which differed from ET-17 by three or four enzymes. Results from serotyping and serosubtyping, ribotyping, and RAPD analysis gave indistinguishable results for all 14 isolates. The remaining three PFGE patterns of this cluster were identified in five isolates: pattern H46N06.0023 was identified in OA800, OA801, and SP418; pattern H46N06.0024 was identified in OA416; and pattern H46N06.0025 was identified in OA632. The results from the molecular subtyping for OA800, OA632, and SP418 were identical to the results for isolates with the H46N06.0026 pattern, while OA801 and OA416 differed from the isolates with the H46N06.0026 pattern only in their ribotype. The final four isolates, SP638, OA160, SP415, and SP414, had four different PFGE patterns, which were related to the H46N06.0026 pattern by less than 78%. Isolates SP638 and OA160 were identified as indistinguishable by the computer analysis but after visual inspection were assigned patterns H46N06.0001 and H46N06.0037 due to spacing differences between bands. These four isolates had molecular markers that were diverse and in unique, individual combinations.
FIG. 3.
Designations, molecular characterization, and PFGE analysis (with NheI) of chromosomal DNAs of 23 NMSC isolates collected during an investigation of a meningococcal disease outbreak in Arizona in 1993 to 1994. The PFGE patterns of isolates SP638 and OA160 were identified as indistinguishable using the 1.5% tolerance, but based on visual differences in spacing between the bands, isolate OA160 was defined as distinct and given a pattern designation of H46N06.0037 (asterisk). ID, isolate identification number; ST/SST, serotype and serosubtype; RT, ribotype; RAPD, RAPD type (with two primers).
California.
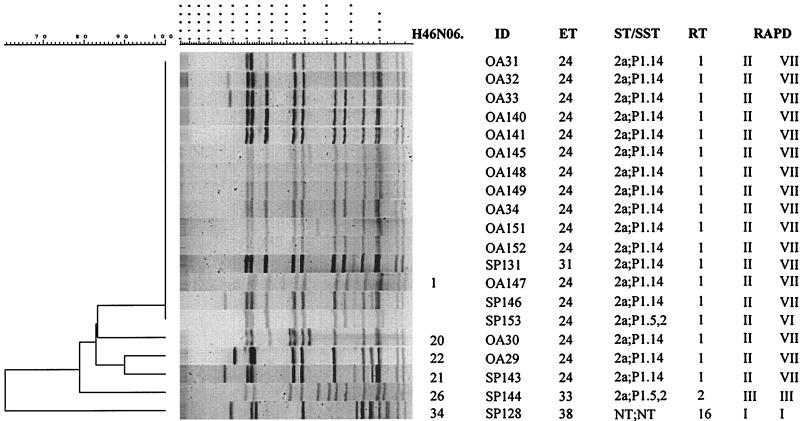
Among the 20 isolates tested, 15 isolates had pattern H46N06.0001 (Fig. 4). Eight of those isolates were from inmate cases, four were from community cases with reported contact with the inmates (OA34, OA147, OA151, and OA152), and three were from community cases with no reported contact with the inmates (SP131, SP146, and SP153). With two exceptions (SP153 and SP131), 13 of the isolates were also indistinguishable as defined by the four other molecular subtyping methods (MEE, serotyping and serosubtyping, ribotyping, and RAPD assay). The PFGE patterns of the remaining five isolates were 84% similar to the PFGE patterns of the outbreak-associated isolates. Two of these five isolates were substantially different by both PFGE and all other molecular subtyping methods. These two were isolated from community cases without contact with the inmates (SP128 and SP144). The PFGE patterns of the three additional isolates (OA29, OA30, and SP143) were closer to the H46N06.0001 pattern of the outbreak-associated isolates (but still less than 85% related) and had all other molecular markers identical to those of the outbreak-associates isolates.
FIG. 4.
Designations, molecular characterization, and PFGE analysis (with NheI) of chromosomal DNAs of 20 NMSC isolates collected during an investigation of a meningococcal disease outbreak in California in 1993. ID, isolate identification number; ST/SST, serotype and serosubtype; RT, ribotype; RAPD, RAPD type (with two primers).
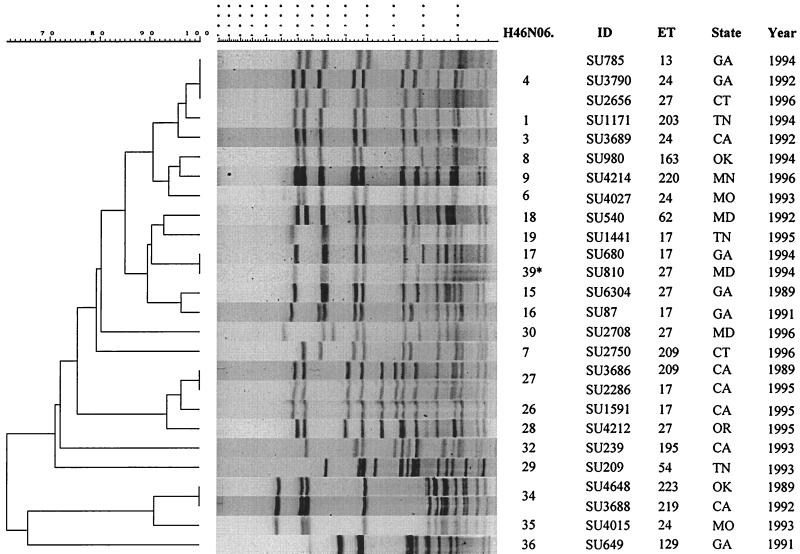
Surveillance isolates.
Among the 26 surveillance isolates, 22 different PFGE patterns in eight clusters (at 85% relatedness) were identified (Fig. 5). No specific geographic or temporal grouping was observed. Patterns of two surveillance isolates, SU680 and SU810, were defined as distinguishable upon visual inspection (based on the spacing between two mid-sized bands) and designated H46N06.0017 and H46N06.0039, respectively, even though they were defined as indistinguishable by computer analysis.
FIG. 5.
Designations, molecular characterization, and PFGE analysis (with NheI) of chromosomal DNAs of 26 NMSC isolates collected from sporadic cases of meningococcal disease through active laboratory-based surveillance, 1989 to 1996. The PFGE patterns of isolates SU680 and SU810 were identified as indistinguishable using the 1.5% tolerance, but based on visual differences in spacing between the bands, isolate SU810 was defined as distinct and given a pattern designation of H46N06.0039 (asterisk). ID, isolate identification number. GA, Georgia; CT, Connecticut; TN, Tennessee; CA, California; OK, Oklahoma; MN, Minnesota; MO, Missouri; MD, Maryland; OR, Oregon.
Overall analysis.
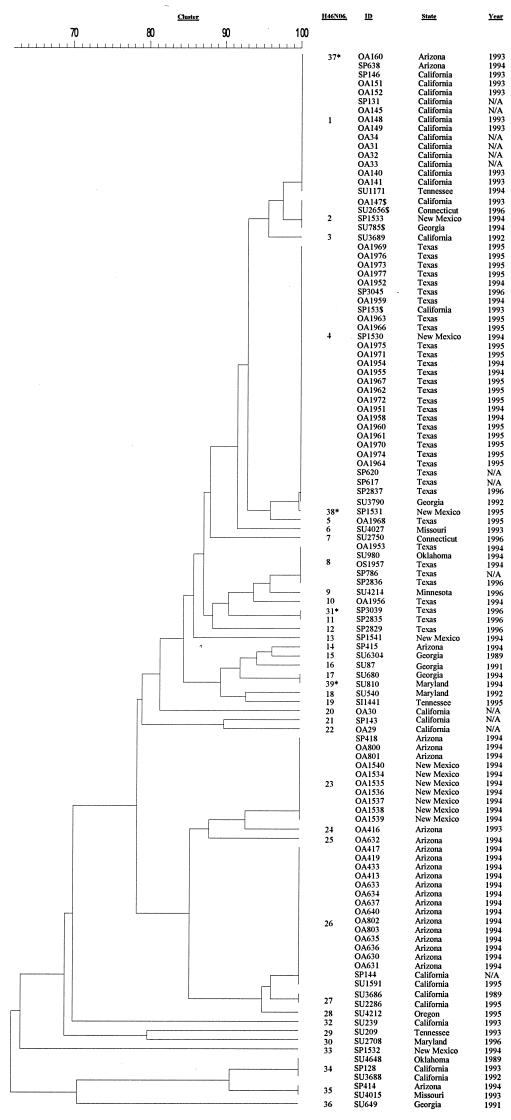
Among our 116 isolates, 39 PFGE patterns were identified, 35 by the computer analysis and 4 additional ones based on the visual inspection, due to the differences in spacing between particular bands. At the 85% relatedness breakpoint they formed 11 clusters. Six of the clusters contained only a single isolate, while the remaining 110 isolates, characterized by 33 PFGE patterns, formed five clusters, containing from 2 to 65 isolates per cluster (Fig. 6). Clear grouping of the isolates within these clusters was observed based on their designation as being either outbreak associated or not. With only two exceptions (OA29 and OA30), two clusters (clusters 1 and 5) contained all outbreak-associated isolates from the four analyzed outbreaks (64 isolates). They also contained 19 sporadic isolates and 13 surveillance isolates: 14 of these 32 isolates had a PFGE pattern indistinguishable from that of one of the four major outbreak-associated patterns. The presence of such indistinguishable PFGE patterns in some sporadic isolates is not unusual; the margins of an outbreak are somewhat arbitrary, and several chains of transmission may be occurring at once. Also, sporadic cases can continue to occur while an outbreak is taking place. In two additional clusters (clusters 2 and 10) that contained multiple isolates (seven and five isolates, respectively), no outbreak-associated isolates were found. Cluster 4 contained two isolates from California, one outbreak associated and one sporadic. Finally, within the six clusters that contained only a single isolate, only isolate OA30 was epidemiologically defined as outbreak associated. All others were isolated from patients with sporadic meningococcal disease, collected either as a part of the outbreak investigations (one isolate) or through the active laboratory-based surveillance (four isolates).
FIG. 6.
Dendrogram showing relatedness by PFGE analysis (with NheI) of 116 NMSC isolates collected during investigations of four meningococcal disease outbreaks in Texas, New Mexico, Arizona, and California and from sporadic cases of meningococcal disease collected through the active laboratory-based surveillance, 1989 to 1996. Pairs of isolates (SP2835 and SP3039, SP1530 and SP1531, SP638 and OA160, and SU680 and SU810) were identified as having indistinguishable PFGE patterns using the 1.5% tolerance. Based on visual inspection, differences in spacing between the bands were observed, and therefore the additional patterns H46N06.0031, H46N06.0037, H46N06.0038, and H46N06.0039, respectively, were established (asterisks). $, isolates on the smaller dendrograms identified as follows: OA147, pattern H46N06.0001; SP153, pattern H46N06.0001; SU785, pattern H46N06.0004; SU2656, pattern H46N06.0004. Their designations as given in Fig. 4 and 5 were used throughout the analysis. ID, isolate identification number.
Additional analysis was possible when all isolates were incorporated into the overall database. Pattern H46N06.0004, characteristic for the Texas outbreak, was also identified in three isolates from Georgia (SU3790), New Mexico (SP1530), and California (SP153). Of the three outbreak-associated isolates (OA1953, OA1956, and OA1957) with patterns H46N06.0008 and H46N06.0010, the H46N06.0008 pattern was also seen in a surveillance isolate from Oklahoma (SU980). No other isolates with the H46N06.0010 pattern were identified, but this pattern was closely related to the H46N06.0009 pattern, which was seen in a single surveillance isolate from Minnesota (SU4214). Pattern H46N06.0023, seen in the New Mexico outbreak-associated isolates, was also identified in three isolates from Arizona (SP418, OA800, and OA801). Of the five New Mexico sporadic isolates, two were randomly distributed throughout the dendrogram (SP1541 and SP1532), and their unique patterns (H46N06.0013 and H46N06.0033) showed less than 88% relatedness to any other pattern in the database. The remaining two sporadic isolates (SP1530 and SP1531) had patterns H46N06.0004 and H46N06.0038, which were seen in 22 Texas outbreak-associated isolates as well as in a sporadic isolate from California (SP153) and a surveillance isolate from Georgia (SU3790). The last sporadic isolate (SP1533) from New Mexico, with pattern H46N06.0002, shared that pattern with three other isolates from California (OA147), Connecticut (SU2656), and Georgia (SU785). This pattern was closely related to the H46N06.0001 pattern, which is typical for the majority of California outbreak isolates. Pattern H46N06.0026, seen in 14 Arizona outbreak-associated isolates, was shared with a single surveillance isolate from California (SU1591) and another sporadic isolate from the California jail outbreak investigation (SP144). Additionally, two surveillance isolates from California (SU3686 and SU2286) and one from Oregon (SU4212) had patterns H46N06.0027 and H46N06.0028, which are very similar to H46N06.0026 found in the 14 Arizona outbreak-associated isolates (95% relatedness). Of the five isolates that were related to the 14 outbreak-associated isolates at the 87% relatedness level when only Arizona isolates were analyzed, three (SP418, OA800, and OA801) had pattern H46N06.0023, which is typical for the New Mexico outbreak-associated isolates. Of the 26 surveillance isolates, 22 isolates with 19 PFGE patterns were located in four multi-isolate clusters. The remaining four isolates made up clusters that contained a single isolate. Only three surveillance isolates had PFGE patterns indistinguishable from one of the outbreak-associated patterns. An isolate from Tennessee (SU1171) had a PFGE pattern indistinguishable from the H46N06.0001 pattern of the outbreak-associated isolates from California. An isolate from Georgia (SU3790) had a pattern (H46N06.0004) that was identified in the outbreak-associated isolates from Texas. A single surveillance isolate from California (SU1591) had an H46N06.0026 pattern indistinguishable from that of the outbreak-associated isolate from Arizona.
Effects of the tolerance window (0.5, 1.0, or 1.5%) on the clustering of the PFGE patterns.
Using the 1.5% tolerance window, 39 PFGE patterns were identified among the 116 isolates and were grouped in 11 clusters at the 85% relatedness level. Changing the window tolerances only slightly affected the clustering of the PFGE patterns of isolates from the investigated outbreaks and subsequent specificity and sensitivity (Table 1). The only significant change was seen in the grouping of the PFGE pattern from the California jail outbreak investigation. Overall, among the 90 isolates collected during the four outbreak investigations, 66 were epidemiologically defined as outbreak related; of those, 56 had PFGE patterns that were either indistinguishable (54 isolates) or related to each other at >95% (2 isolates). In 7 of 66 isolates PFGE patterns were related to the predominant outbreak-associated PFGE pattern at the 85 to 95% relatedness level. Using the epidemiological definition for an isolate to be outbreak associated or sporadic, this translates to a sensitivity of 85% (percentage of true outbreak-associated isolates correctly identified as such) and a specificity of 71% (percentage of sporadic isolates correctly identified as such) using the 95% relatedness breakpoint and either the 1.5 or 1.0% tolerance (Table 1).
TABLE 1.
Sensitivity and specificity of PFGE in differentiating outbreak-associated isolates from sporadic isolates of NMSCa
| Tolerance (%)b | PFGE profile relatedness (%) | Sensitivity (%)c | Specificity (%)c |
|---|---|---|---|
| 1.5 | 85 | 95 (87–99) | 46 (25–67) |
| 95 | 85 (74–93) | 71 (50–87) | |
| 1.0 | 85 | 88 (78–95) | 71 (49–87) |
| 95 | 85 (74–93) | 71 (49–87) | |
| 0.5 | 85 | 77 (65–87) | 71 (49–87) |
| 95 | 61 (48–72) | 79 (58–93) |
Based on analysis of 66 outbreak-related isolates and 24 sporadic isolates.
Tolerance for the band migration distance.
Numbers in parentheses are 95% confidence intervals.
DISCUSSION
The management of NMSC outbreaks differs from that of sporadic meningococcal disease and requires substantial public health action and resources. Vaccination with the currently available quadrivalent polysaccharide vaccine is not routinely indicated for prevention of sporadic disease, but it is useful for control of NMSC outbreaks (6). Looking for a rapid aid in identifying an outbreak and its extent, we focused on assessing PFGE to parallel the ability of MEE to reliably and consistently identify outbreak-associated isolates and then to differentiate outbreak-associated isolates from sporadic isolates. We chose isolates from four well-described NMSC outbreaks for analysis with PFGE, and for comparative purposes we analyzed all isolates by MEE, RAPD assay, ribotyping, and serotyping and serosubtyping as well. In addition, we randomly selected isolates representing sporadic disease in the areas where these four outbreaks have occurred, as well as isolates from the ongoing, national, active laboratory-based surveillance program.
Molecular subtyping of NMSC by MEE showed that most of the outbreaks in the United States have been caused by isolates of the ET-37 complex. Isolates of this complex are also the most common cause of sporadic disease in the United States. Indeed, all 66 outbreak-associated isolates from the four outbreaks in this study belonged to this complex: 36 were of ET-24, 21 were of the closely related ET-17, and 9 were of five other closely related ETs. However, among the 24 sporadic isolates collected during the investigations of these four outbreaks, 20 were also characterized by an ET of the ET-37 complex. Furthermore, among the 26 randomly chosen surveillance isolates, only 5 were not members of the ET-37 complex. Interestingly, only 3 of these 21 isolates had a PFGE pattern indistinguishable from one of the outbreak-associated PFGE patterns from the four analyzed outbreaks. This suggests that perhaps PFGE might distinguish outbreak-associated isolates from sporadic isolates, even when they are members of the same ET complex.
We observed excellent correlation between the epidemiological data and PFGE results, and the overall analysis strongly suggests that PFGE patterns related to each other at >95% are typical for the isolates most likely to be a part of the same outbreak. Among the 90 isolates collected during the four outbreak investigations, 66 were epidemiologically defined as outbreak related. Fifty-six of these 66 isolates had PFGE patterns that were either indistinguishable (54 isolates) or related to each other at >95% (2 isolates). This high degree of similarity allows for defining an isolate as outbreak associated. However, in 7 of 66 isolates (11%), PFGE patterns were related to the predominant outbreak-associated PFGE pattern at the 85 to 95% relatedness level; similarities between PFGE patterns and clusters in that range do not always correlate with the epidemiological assessment. In such cases, analysis of PFGE patterns in the context of a larger database provides more information on their potential relatedness to patterns of isolates circulating in that particular geographic setting or beyond. However, the clustering of two PFGE patterns in an overall dendrogram, and consequently the calculated degree of their relatedness, may vary slightly from the actual degree of relatedness expressed when the two patterns are directly compared to each other. This is only true when the windows of tolerance are defined as a proportion of the size of a particular band, rather than a fixed value identical for all bands regardless of their size. Consequently, in some cases, patterns are identified as indistinguishable when compared to each other. However, when incorporated in the larger database they become parts of pattern clusters that are then compared one cluster to another, rather than each pattern against every other one, and these clusters are then presented as related to each other at less than 100% relatedness. In this study this was the case for OA147 and SP153, which were identified as H46N06.0001 when only California isolates were analyzed but were grouped with isolates having H46N06.0002 and H46N06.0004, respectively, in the overall dendrogram. This was also seen with SU785 and SU2656, which were identified as H46N06.0004 when only surveillance isolates were analyzed but were grouped with H46N06.0002 when incorporated in the overall dendrogram. All of these patterns were more than 90% similar. Sending Tiff images from gels run by this protocol, obtained in individual outbreak investigations, to CDC's N. meningitidis PFGE database is encouraged to allow for a better glimpse into the diversity of the circulating isolates and their PFGE patterns and to form a national database of N. meningitidis PFGE patterns so that each of the newly identified PFGE patterns could be compared to this database of PFGE patterns. In this way, local health authorities may be more rapid and effective in assessing whether or not they are dealing with an outbreak.
In this study we used the 1.5% tolerance window for analysis of PFGE pattern relatedness. The PFGE patterns in the Molecular Analyst program are compared using the band-matching option of the program. This option uses a band tolerance value in determining the relationship of the patterns. The tolerance value is expressed as a percentage of the total normalized gel length. Two bands on different PFGE patterns are considered to be the same if the difference in the position of the bands is less than the tolerance percentage. If the difference in the positions of the bands is greater than the tolerance percentage, then the bands are considered to be two different bands. A low tolerance (0.5%) allows for a minimal amount of shifting of the bands when comparing bands of different PFGE patterns. A tolerance of 1.5% allows for more shifting of the bands when comparing different PFGE patterns. In our analysis there were four exceptions due to the consistent differences in the spacing between two bands observed by visual inspection, i.e., four patterns were defined as distinct even though the computer analysis identified them as indistinguishable from four other PFGE patterns. In order to allow for differences observed in the band migration when identical isolates were tested on different gels, and especially for the interlaboratory comparison purposes, the 1.5% tolerance is more appropriate. An overall sensitivity of 85% and specificity of 71% were observed using the 1.5% tolerance for the band migration distance at the 95% relatedness level. Using the same tolerance, the sensitivity increased to 95%, while the specificity decreased to 46% at the 85% relatedness level among the clusters of PFGE profiles. Similar values, i.e., 88% sensitivity and 71% specificity at both the 85 and 95% relatedness levels, were observed using the 1.0% tolerance, and only a slight decrease in sensitivity (to 61%) was detected at the 95% relatedness level using the 0.5% tolerance (Table 1).
The range of genomic diversity differs in different species, and therefore, in very homogeneous species small differences may be even more significant and vice versa. However, a number of genetic events known as genetic drift (point mutations, insertions, deletions, and rearrangements, etc.) can significantly affect the applicability of a particular molecular approach to subtype the organism of interest (10). Horizontal exchange of genetic material in N. meningitidis is common, and it is reasonable to expect that such events would have greater effects on PFGE than on MEE or ribotyping, which are methods that deal with specific well-conserved genes, unlike PFGE. All of these factors need to be carefully considered in setting the criteria for using molecular markers as an aid in outbreak definition, and they may vary from one species to another. Therefore, we addressed the PFGE differentiation capability in relation to the epidemiological definition of an outbreak, as well as the comparison of its differentiation potential with those of other molecular subtyping methods, primarily MEE. We demonstrated that PFGE consistently provided a greater level of diversity than MEE. We recently experienced a situation where two cases of meningococcal disease were identified in contacts at a school in New Hampshire within a 1-week period. Isolates from these two cases were identified as identical by ET, but their PFGE patterns differed from each other by three bands, relating to each other at an 84.5% relatedness level, which is right at the level of 85% which was established as the borderline for definition of outbreak association in this study. Analysis of these isolates by other molecular subtyping methods showed that they also have an identical serotype and serosubtype (2a:P1.23,14) and RAPD type (II, II). These data suggest that the isolates were similar, consistent with an outbreak, and emphasize the need to analyze the PFGE patterns not only by band-to-band comparison but also according to the overall PFGE pattern relatedness.
The importance of the reproducibility of the method cannot be overstated. It is well known that factors such as different sources of reagents, laboratorians performing the method, and levels of experience, and even minimal deviations from the agreed-upon procedure, can have substantial effects on the interlaboratory comparison. Data from our small pilot study on 22 isolates from the Texas outbreak were quite encouraging. These isolates were analyzed by PFGE in four different laboratories (Florida Department of Health, Minnesota Department of Health, New York State Department of Health, and CDC) using the same restriction enzyme but with PFGE protocols that were developed or modified from already existing protocols in the individual laboratories. Regardless of the method used and the technical demands imposed by the software used for the data analysis, all isolates were grouped in the same manner. In developing this PFGE protocol, we followed the protocol used by the PulseNet (2). Laboratorians familiar with the PulseNet PFGE procedures should be able to easily analyze NMSC isolates by making only small adjustments to achieve the optimal technical results. At the recent annual PulseNet meeting (Minneapolis, Minn., May 2000), PFGE analysis of N. meningitidis was accepted as an integral part of the PulseNet activities. Data presented in the present study demonstrated that PFGE data correlated very well with epidemiological data for all four analyzed outbreaks. Subsequent inclusion of N. meningitidis into PulseNet will provide laboratorians, epidemiologists, and other public health officials throughout the country additional support in declaring an outbreak and thereby in making informed appropriate public health decisions.
ACKNOWLEDGMENTS
We are grateful to colleagues at the State Health Departments in Arizona, California, New Mexico, and Texas for providing us with the outbreak-associated isolates and relevant clinical and epidemiological information, as well as to the members of the Active Bacterial Core Surveillance Team for their efforts in providing us with the isolates collected from patients with sporadic meningococcal disease. We thank Paul Fiorella (Florida Department of Health, Jacksonville) and Dianna Schoonmaker-Bopp (New York State Department of Health, Albany) for their participation in the PFGE pilot interlaboratory comparison study. We also thank Chris Jambois and Gwen Barnett for their assistance in the design and production of the figures.
REFERENCES
- 1.Achtman M. Epidemic spread and antigenic variability of Neisseria meningitidis. Trends Microbiol. 1995;3:186–192. doi: 10.1016/s0966-842x(00)88918-0. [DOI] [PubMed] [Google Scholar]
- 2.Barrett T J, Lior H, Green J H, Khakhria R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondeau J M, Isaacson M, Yaschuck Y, Anderson C, Ducasse G. Neisseria meningitidis with decreased susceptibility to penicillin in Saskatchewan, Canada. J Clin Microbiol. 1995;33:1784–1786. doi: 10.1128/jcm.33.7.1784-1786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bygraves J A. Analysis of the clonal relationships between strains of Neisseria meningitidis by pulsed field gel electrophoresis. J Gen Microbiol. 1992;138:523–531. doi: 10.1099/00221287-138-3-523. [DOI] [PubMed] [Google Scholar]
- 5.Caugant D A. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS. 1998;106:505–525. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Control and prevention of meningococcal disease and control and prevention of serogroup C meningococcal disease: evaluation and management of suspected outbreaks. Morb Mortal Wkly Rep. 1997;46(RR-5):1–21. [Google Scholar]
- 7.Cookson S T, Loter J O, Regeira M, Binsztein N, Reeves M W, Ajello G, Jarvis W R. Disco fever: epidemic meningococcal disease in northeastern Argentina associated with disco patronage. J Infect Dis. 1998;178:266–269. doi: 10.1086/517450. [DOI] [PubMed] [Google Scholar]
- 8.Edmond M B, Houston A K, Wenzel R P. Molecular epidemiology of an outbreak of meningococcal disease in a university community. J Clin Microbiol. 1995;33:2209–2211. doi: 10.1128/jcm.33.8.2209-2211.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautom, R. K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977–2980. [DOI] [PMC free article] [PubMed]
- 10.Hobbs M M, Prasad P, Morelli G, Kusecek B, Heckels J E, Cannon J G, Achtman M. Recombinational reassortment among OPA genes from ET-37 complex of Neisseria meningitidis isolates of diverse geographical origins. Microbiology. 1998;144:157–166. doi: 10.1099/00221287-144-1-157. [DOI] [PubMed] [Google Scholar]
- 11.Jackson L A, Reeves M W, Wenger J D. Serogroup C meningococcal outbreaks in the United States. JAMA. 1995;273:383–389. [PubMed] [Google Scholar]
- 12.Jacobs D. Proceedings of the 15th annual SAS Users Group International Conference. Cary, N.C: SAS Institute; 1990. SAS/GRAPH software and numerical taxonomy; pp. 1413–1418. [Google Scholar]
- 13.Louie M, Rachlis A, Louie L. Nosocomial Neisseria meningitidis: molecular analysis of a clinical problem. Infect Control Hosp Epidemiol. 1997;18:203–204. doi: 10.1086/647589. [DOI] [PubMed] [Google Scholar]
- 14.Nicolas P, Martet G. Pulsed field gel electrophoresis analysis of clonal relationships among Neisseria meningitidis A strains from different outbreaks. Eur J Clin Microbiol Infect Dis. 1997;16:541–544. doi: 10.1007/BF01708241. [DOI] [PubMed] [Google Scholar]
- 15.Olson R K. State of New Mexico Department of Health Epidemiology Report. Santa Fe, N. Mex: State Department of Health; 1995. Outbreak of group C meningococcal meningitis in rural New Mexico; pp. 1–2. [Google Scholar]
- 16.Popovic T, Ajello G, Facklam D. Manual for the laboratory diagnosis of meningitis caused by Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae. Geneva, Switzerland: World Health Organization; 1999. [Google Scholar]
- 17.Popovic T, Kombarova S Y, Reeves M W, Nakao H, Mazurova I K, Wharton M, Wachsmuth I K, Wenger J D. Molecular epidemiology of diphtheria in Russia, 1985–1994. J Infect Dis. 1996;174:1064–1072. doi: 10.1093/infdis/174.5.1064. [DOI] [PubMed] [Google Scholar]
- 18.Raymond N J, Ajello G, Baughman W, Gheesling L L, Carlone G M, Wenger J D, Stephens D S. Molecular epidemiology of sporadic (endemic) serogroup C meningococcal disease. J Infect Dis. 1997;176:1277–1284. doi: 10.1086/514123. [DOI] [PubMed] [Google Scholar]
- 19.Reeves M W, Perkins B A, Diermayer M, Wenger J D. Epidemic-associated Neisseria meningitidis detected by multilocus enzyme electrophoresis. Emerg Infect Dis. 1995;1:53. doi: 10.3201/eid0102.950203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regnault B, Grimont R, Grimont P A D. Universal ribotyping method using a chemically-labeled oligonucleotide probe mixture. Res Microbiol. 1997;146:649–659. doi: 10.1016/S0923-2508(99)80064-3. [DOI] [PubMed] [Google Scholar]
- 21.Rosenstein N E, Stephens D, Lefkowitz L, Cartter M L, Danila R, Cieslak P, Shutt K A, Popovic T, Schuchat A, Harrison L H, Reingold A L the Active Bacterial Core Surveillance Team. The changing epidemiology of meningococcal disease in the United States, 1992–1996. J Infect Dis. 1999;180:1894–1901. doi: 10.1086/315158. [DOI] [PubMed] [Google Scholar]
- 22.Rosenstein N E, Levine O, Taylor J P, Evans D, Plikaytis B D, Wenger J D, Perkins B A. Efficacy of meningococcal vaccine and barriers to vaccination. JAMA. 1998;279:435–439. doi: 10.1001/jama.279.6.435. [DOI] [PubMed] [Google Scholar]
- 23.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan B, Matar G M. Molecular typing methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 26–50. [Google Scholar]
- 25.Swaminathan B, Reeves M W, Graves L M, Ajello G, Bibb W F, Helsel L O, Morales M, Dronavalli H, El-Swify M, DeWitt W, Hunter S. Molecular subtyping of Neisseria meningitidis serogroup B: comparison of five methods. J Clin Microbiol. 1996;34:1468–1473. doi: 10.1128/jcm.34.6.1468-1473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tappero J W, Reporter R, Wenger J D, Ward B A, Reeves M W, Missback T S, Plikaytis B D, Mascola L, Schuchat A. Meningococcal disease in Los Angeles County, California, and among men in the county jails. N Engl J Med. 1996;335:833–840. doi: 10.1056/NEJM199609193351201. [DOI] [PubMed] [Google Scholar]
- 27.Tondella M L C, Popovic T, Rosenstein N E, Lake D B, Carlone G M, Mayer L W, Perkins B A the Active Bacterial Core Surveillance Team. Distribution of Neisseria meningitidis serogroup B serosubtypes and serotypes circulating in the United States. J Clin Microbiol. 2000;38:3323–3328. doi: 10.1128/jcm.38.9.3323-3328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakubu D E, Pennington T H. Molecular epidemiology of recent United Kingdom isolates of Neisseria meningitidis serogroup C. Epidemiol Infect. 1994;113:53–65. doi: 10.1017/s0950268800051463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakubu D E. Epidemiological evaluation of Neisseria meningitidis serogroup B by pulsed-field gel electrophoresis. FEMS Immunol Med Microbiol. 1995;10:185–190. doi: 10.1111/j.1574-695X.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]