Abstract
Understanding the long-term maintenance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immunity is critical for predicting protection against reinfection. In an age- and gender-matched cohort of 24 participants, the association of disease severity and early immune responses on the maintenance of humoral immunity 12 months post-infection is examined. All severely affected participants maintain a stable subset of SARS-CoV-2 receptor-binding domain (RBD)-specific memory B cells (MBCs) and good neutralizing antibody breadth against the majority of the variants of concern, including the Delta variant. Modeling these immune responses against vaccine efficacy data indicate a 45%–76% protection against symptomatic infection (variant dependent). Overall, these findings indicate durable humoral responses in most participants after infection, reasonable protection against reinfection, and implicate baseline antigen-specific CD4+ T cell responses as a predictor of maintenance of antibody neutralization breadth and RBD-specific MBC levels at 12 months post-infection.
Keywords: SARS-CoV-2, 12 months after SARS-CoV-2 infection, RBD-specific memory B cells longevity, protection efficacy prediction, SARS-CoV-2 antibody duration, SARS-CoV-2 neutralization, variants of concern, Delta variant, SARS-CoV-2 OX40 CD4 T cell assay
Graphical abstract
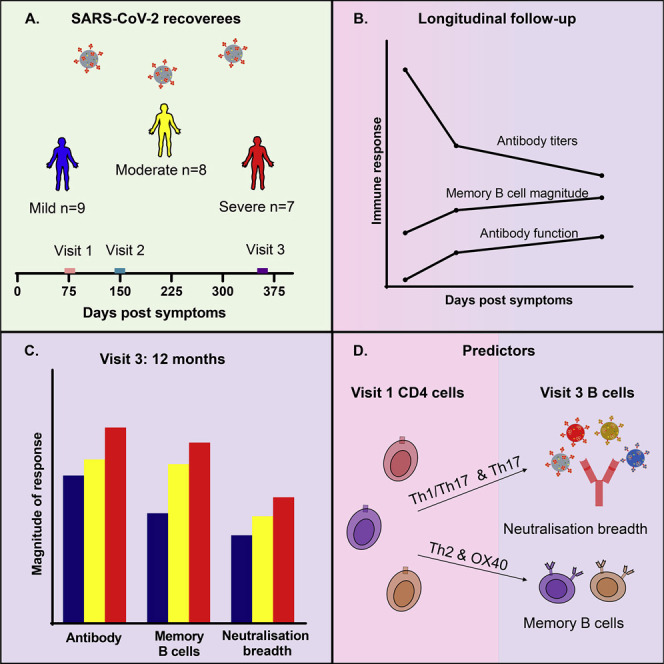
One year after SARS-CoV-2 infection, despite declining antibody titers, Balachandran et al. report a disease-severity-associated maintenance of antibody functions and memory B cell magnitudes. The convalescent serum could neutralize most VOCs, and the initial CD4 T cell levels predict the antibody neutralization breadth and memory B cell durability.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has continued to spread globally, with an estimated 259 million people naturally infected and an overlapping 7.5 billion now vaccinated 18 months after the infection first appeared (Center for Systems Science and Engineering, and JHU.edu, 2021). The cost and logistics of the vaccine rollouts and boosters, combined with the likely heterogeneity of immune responses in vulnerable groups and the emergence of new variants, suggests that some subpopulations will remain susceptible for some time to come, especially in low-middle-income countries. Immunity from natural infection may become an important component of achieving herd immunity, which is estimated to be at least 70% of the population (Anderson et al., 2020; McDermott, 2021). However, the susceptible population will be greatly increased if protective immunity after infection or immunization wanes rapidly.
The presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific neutralizing antibodies, even at low titers, has been associated with protection from severe disease (Dan et al., 2021; Dispinseri et al., 2021; Lau et al., 2021; Mumoli et al., 2020; Stephens and McElrath, 2020). These antibodies act by binding to the receptor-binding domain (RBD) on the virus Spike glycoprotein and by blocking access to the angiotensin-converting enzyme 2 (ACE2) receptor on host cells (Letko et al., 2020; Pandey et al., 2021). Beyond neutralization, the Fc portion of the antibody also mediates additional functions such as antibody-dependent cellular phagocytosis (ADCP), which has been reported to have a protective effect in SARS-CoV-2 infection (Adeniji et al., 2021; Atyeo et al., 2020; Bartsch et al., 2021). Following acute infection, neutralizing antibody titers decline significantly during the convalescent phase and continue to slowly decline 3–8 months post-infection (Abayasingam et al., 2021; Sakharkar et al., 2021; Seow et al., 2020). Encouragingly, a recent paper reported that the initial steep decline in antibody titers after natural infection is followed by a relatively stable maintenance phase lasting up to 12 months post-infection (Petersen et al., 2021). This plateau has been putatively attributed to the generation of long-lived plasma cells localized in the bone marrow (Turner et al., 2021).
Our group, and others, have shown that a robust RBD-specific memory B cell (MBC) repertoire is also maintained despite the decline in antibody titers in most individuals 6–8 months post-infection (Abayasingam et al., 2021; Dan et al., 2021; Sakharkar et al., 2021). One study reported an evolving population of RBD-specific MBCs at 1 year post-infection with selective retention of broad and potent antibody-encoding MBC clones that expanded dramatically post-vaccination (Wang et al., 2021). Maintenance of stable T cell immunity and RBD-specific MBCs has been reported 1 year after mild SARS-CoV-2 infection (Garcia Valtanen et al., 2021). Upon re-exposure, MBCs have the capacity to divide, undergo germinal center (GC) reactions, and differentiate into antibody-secreting cells (ASCs), leading to a subsequent wave of protective antibody production. It is well established in influenza that the type of help B cells receive from CD4+ T follicular helper (Tfh) cells during the acute phase of infection influences the quality of neutralizing antibodies and the magnitude of B cell responses that lead to differentiation into short- and long-lived plasma cells secreting high titers of neutralizing antibodies and MBCs (Grifoni et al., 2020; Juno et al., 2020; Koutsakos et al., 2019). Similar data in SARS-CoV-2 infection also suggest that CD4+ Tfh responses are key (Adeniji et al., 2021; Boppana et al., 2021; Seow et al., 2020; Zhang et al., 2019). However, the characteristics of these Tfh responses at, or soon after, infection have not been related to the MBC and antibody responses for SARS-CoV-2 1 year after infection.
Although B cells and CD4+ Tfh cells interact within the GC, a subset of CXCR5-expressing CD4+ Tfh cells are found in circulation (cTfh) and are often studied as a representative of the lymphoid-derived Tfh cells (Morita et al., 2011). Subpopulations of CXCR5+ Tfh cells are defined by the expression of CXCR3 and CCR6 as Th1 (CXCR3+ CCR6–), Th2 (CXCR3– CCR6–), Th17 (CXCR3– CCR6+), and Th1/Th17 (CXCR3+ CCR6+). cTfh Th2 and Th17 cells are known to help naive B cells (Morita et al., 2011), while cTfh Th1 provide help to the MBCs (Bentebibel et al., 2013; Koutsakos et al., 2019).
One limitation of the studies evaluating longitudinal responses to SARS-CoV-2 infection is the overrepresentation of participants with mild or moderate illness (Juno et al., 2020; Rodda et al., 2020). Given the very different levels of immune activation and immune subset profiling in acute severe infection, it is likely that the longevity of protective immunity will also vary with disease severity. In this study, we evaluated longitudinal humoral immune responses among 24 age- and gender-matched participants in the 12 months following mild, moderate, or severe SARS-CoV-2 infection in a setting with low rates of infection and reinfection in Australia. The data collected were used to model correlates of protection and identify predictors of immune longevity.
Results
Cohort
Antibody and cellular assays were performed on three samples collected longitudinally over 12 months in 24 adult participants (12 females, median age 61.5 [range: 35–87]) stratified for mild (n = 9), moderate (n = 8), or severe (n = 7) acute infection as designated by NIH guidelines (https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/) and with comparable age and gender matching (Table 1 ). The median numbers of days post-symptom onset (DPS) for visits 1 (baseline), 2, and 3 were 76.5 (range: 39–152), 149.5 (range: 106–253), and 358 (range: 312–424), respectively. The participants all had RT-PCR-confirmed acute infection when the SARS-CoV-2 614D variant was dominant in Sydney (March 6, 2020, and September 17, 2020). At the time of collection of the 12-month samples, Australia had had very limited community transmission since the first wave, when these participants were infected (NSW Government, 2021), and none of these participants had received COVID immunization.
Table 1.
Characteristics of COSIN participants (n = 24)
| Total study population | Infection severity |
|||
|---|---|---|---|---|
| Mild (n = 9) | Moderate (n = 8) | Severe (n = 7) | ||
| Age, median (range) | 61.5 (35–87) | 61 (46–82) | 62 (35–67) | 62 (36–87) |
| Days post-symptom onset, median (range) | ||||
| Visit 1 | 76.5 (39–152) | 73 (57–81) | 67 (42–86) | 93 (39–152) |
| Visit 2 | 149.5 (106–253) | 141 (106–247) | 134 (114–238) | 228 (156–253) |
| Visit 3 | 358 (312–424) | 353 (339–424) | 363 (312–415) | 357 (334–396) |
| Gender, n (%) | ||||
| Female | 12 (50) | 5 (55.6) | 4 (50) | 3 (42.9) |
| Male | 12 (50) | 4 (44.4) | 4 (50) | 4 (57.1) |
Maintenance of highly functional RBD-specific serum antibodies
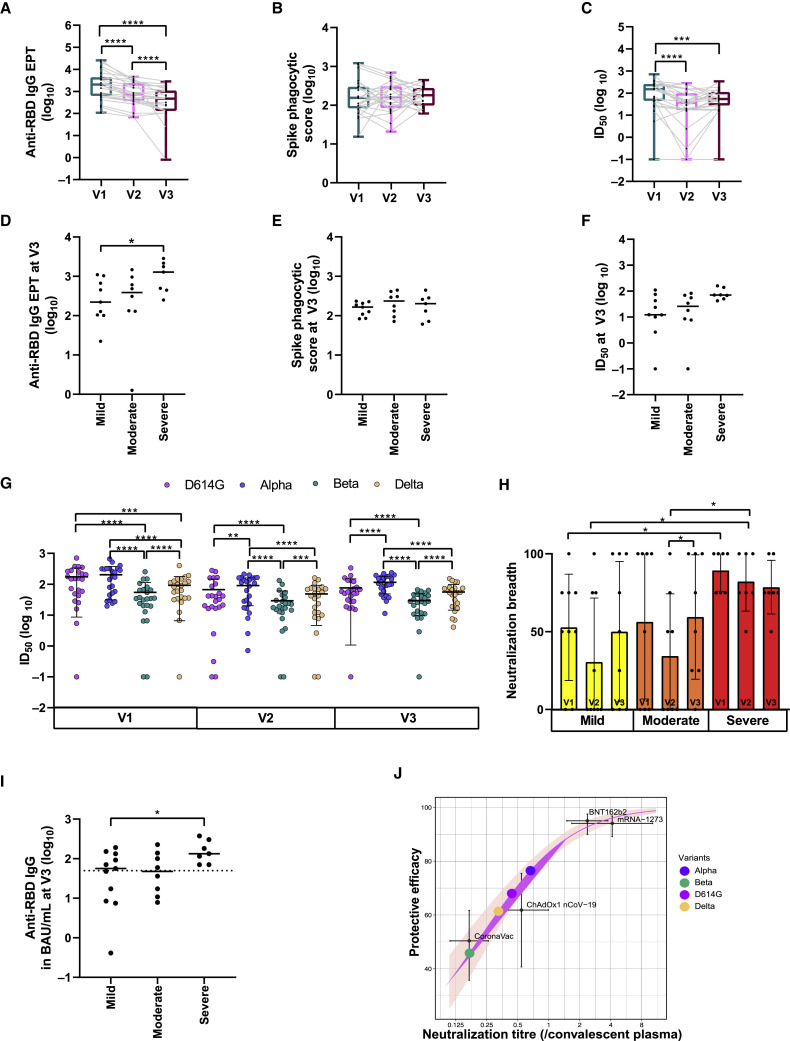
The anti-RBD immunoglobulin G (IgG) endpoint titers (EPTs) in the serum were determined by ELISA for all 24 participants across all three timepoints. All participants mounted and maintained detectable levels of anti-RBD IgG antibodies during the first two visits, with only one participant having undetectable anti-RBD IgG antibodies at visit 3 (Figure 1 A). The EPTs declined significantly between visits 1 and 2 (V1: 2,040 [range: 108–25,538], V2: 709 [range: 68–4,614], p < 0.0001) and between visits 2 to 3 (V3: 468 [range: 0–2,848], p < 0.0001). The participants with severe disease generated and maintained higher titers of anti-RBD IgG antibodies at visit 3 than did those with mild disease (severe: 1,281 [range: 251–2,848], mild: 222 [range: 22–1,104], p = 0.0311) (Figures 1D and S1B).
Figure 1.
SARS-CoV-2 antibody analysis
(A–C) Longitudinal (A) anti-RBD IgG antibody titer calculated from technical duplicates, (B) ADCP activity, and (C) neutralization ID50 trend for D614G variant at visits 1, 2, and 3 calculated from technical duplicates.
(D–F) Impact of disease severity at visit 3 on (D) anti-RBD IgG antibody titer, (E) ADCP activity, and (F) neutralization ID50 for D614G variant.
(G) Neutralization ID50 for all variants of concern calculated from technical duplicates across three visits.
(H) Breadth of neutralization across three visits stratified by disease severity.
(I and J) Estimated protective efficacy of convalescent serum against symptomatic reinfection based on (I) anti-RBD IgG antibody levels at visit 3 (dotted line indicates the estimated 60% vaccine efficacy against symptomatic infection) and (J) neutralization titers at visit 3 as a proportion of early convalescent titers plotted on a logistic model developed for comparing vaccine efficacy studies (purple line indicates best fit for the logistics model, and pink shading indicates the 95% prediction interval) (Mean and SD indicated; ∗∗∗∗p < 0.0001; ∗∗∗p between 0.0001 and 0.001; ∗∗p between 0.001 and 0.01; ∗p between 0.01 and 0.05).
Also see Figures S1 and S2.
ADCP activity was measured by incubating participant plasma with fluorescent beads labeled with recombinant Spike before the measurement of phagocytosis by THP-1 cells. The phagocytosis (P) scores against Spike were consistent throughout the course of all three visits (Figure 1B). At visit 1, the severe group had significantly higher P scores than the mild group (severe: 287 [range: 94–1,212], mild: 98 [range: 51–367], p = 0.031). However, during subsequent visits, there were no significant differences between disease severity groups, and the level of ADCP activity remained stable over time (Figures 1E and S1C).
To determine the longevity and breadth of neutralizing activity in the serum, the infective dose (ID)50 was generated for each sample using a live virus assay against four SARS CoV-2 variants, D614G, Alpha, Beta, and Delta, in HEK-ACE2/TMPRSS cells. A slow decline in neutralizing activity against D614G was observed from visit 1 to visit 2 (V1: ID50 149.85 [range: 0–709.4], V2: ID50 36.21 [range: 0–281.5], p < 0.0001), followed by a plateauing from visit 2 to visit 3 (Figures 1C and S1E). The visit 3 neutralization activity was still significantly lower than visit 1 but not visit 2 (V3: ID50 53.64 [range: 0–333.7], p = 0.0007). The disease severity was not associated with the neutralizing activity against D614G at visit 3 (Figure 1F) despite being significantly different between mild and severe disease at visits 1 and 2 (V1: mild ID50 59.08 [range: 15.85–277.9] and severe ID50 202.5 [range: 140–370] p = 0.0311; V2: mild ID50 18.06 [range: 0–160] and severe ID50 102.6 [range: 21.06–281.5] p = 0.002).
Neutralizing activity against the other three variants was still present in 23 of 24 participants (96%) at 12 months (Figure 1G). As expected, the mean ID50 was higher in the Alpha variant (V1: ID50: 172.05 [range: 19.6–658.1], V3: ID50: 102.6 [range: 11.3–236.9]) and lower in the Delta variant (V1: ID50: 72.7 [range: 0–400.8], V3: ID50: 52.96 [range:4.1–151.9]) (Figures 1G, S1F, and S1G). The mean ID50 against the Beta variant was lowest among all the variants (V1: ID50: 34.36 [range: 0–253.7], V3: ID50: 27.4 [range: 0–66.2]) (Figures 1G and S1H). At visit 1, the severe group had significantly higher neutralizing activity than the mild group for Alpha (mild ID50 58.1 [range:19.61–404.1] and severe ID50 284.2 [range: 168.1–658.1] p = 0.031), Delta (mild ID50 38.7 [range: 0–148.3] and severe ID50 107.6 [range: 63.2–191.7] p = 0.016), and Beta (mild ID50 23.7 [range:0–66.57] and severe ID50 81.05 [range: 20.8–149.4] p = 0.0079). At visit 2, the severe group had significantly higher neutralizing activity than the moderate and the mild for Alpha (severe ID50 160 [range: 107.9–213.9], moderate ID50 58.99 [range: 7.881–166.2], and mild ID50 28.08 [range: 0.7–160] p = 0.04 and p = 0.002), Delta (severe ID50 70.69 [range: 42.4–160.1], moderate ID50 25.93 [range: 0–82.1], and mild ID50 12.15 [range: 0–110.1] p = 0.04 and p = 0.003), and Beta (severe ID50 37.3 [range: 19.34–132.3], moderate ID50 17.16 [range: 0–64.27], and mild ID50 12.89 [range: 0–44.8] p = 0.02 and p = 0.003). However, at visit 3, disease severity did not associate with the neutralizing activity for these variants, similar to the D614G variant.
Using a scoring system similar to that used for HIV (Mishra et al., 2020), neutralization at greater than 50% at 1/40 dilution was defined as significant. For each sample, observations of significant neutralization against each of the four variants were summed and expressed as a percentage score (i.e., a score of 25% or 1 out of 4 for each of the four variants neutralized at 50% or greater at a dilution of 1/40). A comparison of this score between timepoints and disease severity groups revealed that more severe disease was associated with broader neutralizing activity at visit 1, which was then followed by a non-significant decline by visit 3. For participants with mild and moderate disease, only half the variants could be neutralized at a 1/40 dilution, and this remained stable over time (Figures 1H and S1D). Overall, these results indicated that despite a decline in antibody titers, antibody functions were retained in most participants through 12 months post-infection.
Estimating the level of protection at 12 months
Two studies to date have provided numerical estimates for levels of protection from COVID vaccines (Feng et al., 2021; Khoury et al., 2021). We applied these estimates to our natural immunity data to estimate the level of protection that might be expected at 12 months post-infection. Both studies used values that can be universally applied. The first study associated the binding antibody titers (BAU/mL) with vaccine efficacy against symptomatic breakthrough infections following ChAdOx1 vaccination (Feng et al., 2021). In this analysis, 62.5% of participants had binding titers above the 60% vaccine efficacy level (50 BAU/mL anti-RBD IgG), indicating that a similar efficacy of protection against symptomatic disease from natural immunity may be expected in the participant group reported here. Since these titers were measured against the Wuhan variant RBD, the percentage of participants protected may be expected to be slightly lower for the other variants (Figure 1I).
Neutralization is another correlate of vaccine efficacy, and a recent study used the reported convalescent titers as an internal standard to enable a comparison between vaccine studies (Khoury et al., 2021). Following this approach, the 12-month titers for each variant were standardized against the neutralization titer determined for the D614G variant at visit 1. Based on this observation, the findings predict that 76.5% (95% confidence interval [CI]: 72.7–80.3), 68.0% (95% CI: 63.0–73.2), 61.4% (95% CI: 55.0–67.7), and 45.8% (95% CI: 36.9–54.7) of the participants in the present study would have a 50% protective efficacy against symptomatic infection with the Alpha, D614G, Delta, and Beta variants, respectively (Figure 1J).
Severe disease was associated with maintenance of RBD MBCs at 12 months
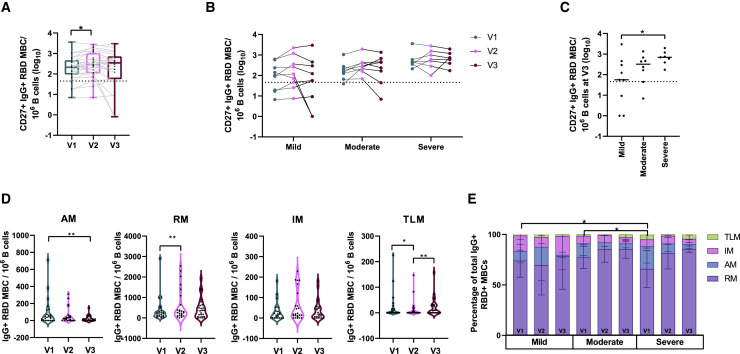
Longitudinal SARS-CoV-2-specific MBC responses (RBD-specific MBCs) were analyzed from stored peripheral blood mononuclear cells (PBMCs). B cells were stained for flow cytometry, and the CD19+ CD20+ CD10 population was analyzed for RBD binding and the expression of CD27, CD21, IgD, and IgG surface phenotypes. The threshold of detection of RBD-specific MBCs was set at mean +2 × standard deviation of the values obtained from 6 healthy donor PBMCs (representative figure in Figure S3A). Of the 24 participants, 20 (83%) had responses above the detection threshold for RBD-specific CD27+ IgG+ MBCs at visit 1. Furthermore, similar to reports in other studies (Rodda et al., 2020; Sakharkar et al., 2021), the number of RBD-specific CD27+ IgG+ MBCs increased significantly from visit 1 to visit 2 (V1: 211 [range: 7–3,562], V2: 289 [range: 7–2,761], p = 0.0214). No significant change from visit 2 to visit 3 (V3: 344 [range: 0–2,979]) was observed (Figure 2 A). Of the 20 participants who had detectable responses at visit 1, 16 (80%) still had detectable responses at visit 3. Interestingly, two participants who did not have responses above the threshold of detection at visit 1 did have just above detection threshold responses at visit 3, while a third participant who did not have detectable responses at visit 1 mounted a well-above-threshold response at visit 3. This participant was not a vaccine recipient, and since the study had been conducted in a low-prevalence setting, reinfection was highly unlikely. In four participants, despite being recorded at high frequencies at visit 1, the MBC responses dropped below the detection threshold at visit 3. Overall, class-switched classical memory (CD27+ IgG+) B cells specific to RBDs were identified in 19 of the 24 participants (79%) 12 months post-infection (Figure 2B).
Figure 2.
Class-switched RBD-specific MBC analysis
(A and B) Longitudinal (A) classical MBC levels (dotted line indicates the healthy control threshold) and (B) trends of classical MBC levels across three visits stratified by disease severity.
(C) Impact of disease severity on classical MBCs at visit 3.
(D) Number of class-switched RBD-specific MBCs per million B cells in each subset per visit.
(E) Percentages of MBC subsets within the class-switched RBD-specific MBC group across three visits stratified by disease severity (mean + SD of participants).
AM, activated memory; RM, resting memory; IM, intermediate memory; TLM, tissue-like memory (∗∗p between 0.001 and 0.01;∗p between 0.01 and 0.05).
Also see Figures S1–S3 and Table S1.
When stratified by disease severity, individual RBD-specific MBC trajectories were quite varied, particularly between visits 2 and 3 (Figures 2B and S1A). At visit 1, 100% (7/7) of participants in the severely ill group, 88% (7/8) participants in the moderately ill group, and 67% (6/9) participants in the mildly ill group mounted a class-switched RBD-specific classical MBC response. At visit 2, 100% of the severe (7/7) and the moderately ill (8/8) groups and 67% (6/9) of the mildly ill groups maintained the response. At visit 3, 100% (7/7) of the severe, 75% (6/8) of the moderate, and 67% (6/9) of the mildly ill groups had retained detectable MBC responses. In the mildly and moderately ill groups, two participants from each, despite maintaining an above-threshold response for the first two visits, lost this response at visit 3. Conversely, two participants in the mildly ill group who did not have detectable responses at visit 1 had low but detectable responses at visit 3.
The severely ill group had higher levels of RBD-specific CD27+ IgG+ MBCs at 12 months post-infection compared with the mildly ill group (severe: 736 [range: 173–1,947], mild: 56 [range: 0–2,979], p = 0.0223) (Figure 2C). Representative flow plots are shown in Figure S3A. Participant age and gender did not influence these levels (Figures S2A and S2B).
Disease severity is associated with expansion of non-classical MBC subsets
The RBD-specific IgG+ B cell subpopulations, including activated memory (AM) (CD21– CD27+), resting memory (RM) (CD21+ CD27+), intermediate memory (IM) (CD21+ CD27–), and tissue-like memory (TLM) (CD21– CD27–), were also examined over time (Moir et al., 2008). As expected, a significant decline in the AM subset was observed from visit 1 to visit 3, while the RM population showed a significant expansion from visit 1 to visit 2. The TLM subset decreased significantly from visit 1 to visit 2, followed by a significant increase from visit 2 to visit 3. There were no significant changes in the IM subset over time (Figure 2D). Examining the various subsets as a percentage of the total RBD-specific IgG+ MBCs at each visit across the different disease severity groups revealed a significant enrichment of the TLM subset in the severely ill group compared with in the mild and moderate disease groups at visit 1 (mild: 0% [range: 0%–8%], moderate: 0% [range: 0%–8%], severe: 3% [range: 0%–11%], p = 0.0253 and p = 0.0441) (Figure 2E).
Early levels of CD4+ T cell subsets, antibodies, and MBCs correlate with neutralizing breadth and MBC magnitude 12 months after infection
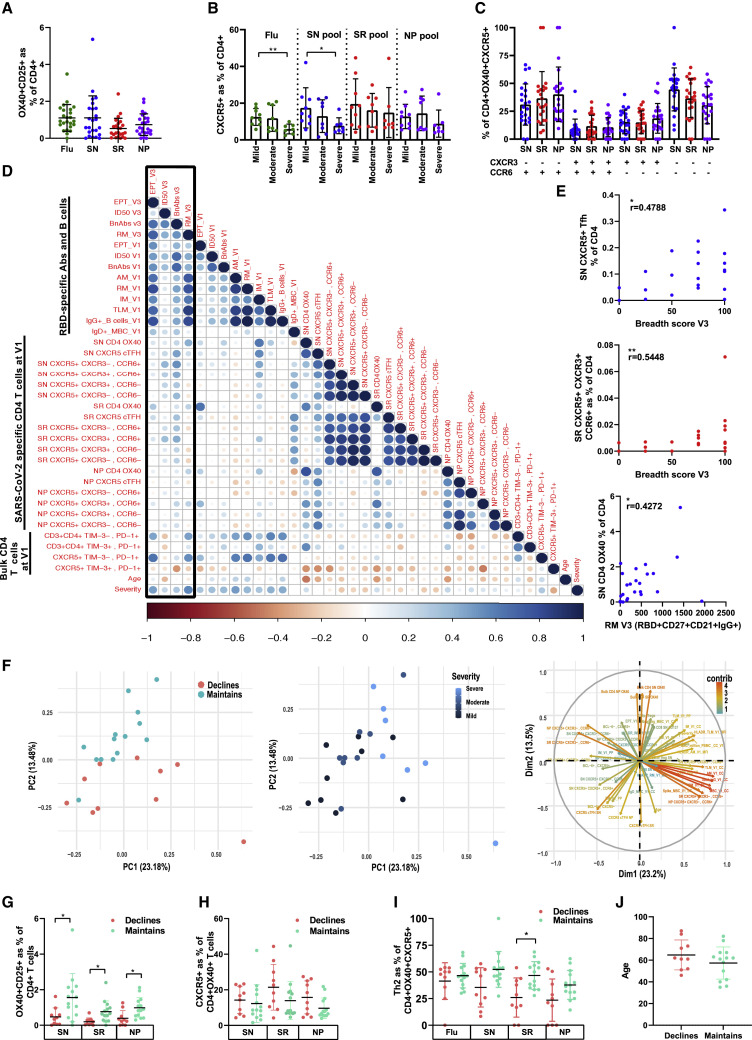
As maintenance of broadly neutralizing antibodies in the serum and RBD-specific MBCs are key features of long-lived protective immunity, the features of the early immune response after infection resolution (visit 1) were examined to determine if they were predictive of a sustained humoral immune response at 12 months (visit 3). As CD4+ T cells have a key role in the generation of antibody and B cell responses, SARS-CoV-2-specific CD4+ T cell responses were measured at visit 1 with an activation induced marker (AIM) assay using peptide pools against the SARS-CoV-2 nucleoprotein (NP), Spike-excluding RBD (SN), RBD only (SR), and influenza (Flu, as a control). Exhaustion markers Tim-3+ and PD-1+ were also determined ex vivo on total CD4+ T cell subsets. Antigen-specific OX40+ CD4+ T cells (CD134+ CD25+) cells were detected in 100% of participants (Figure 3 A), and the frequencies of cTfh cells (CXCR5+) identified were similar to other studies (Rydyznski Moderbacher et al., 2020). As previously reported (Shuwa et al., 2021), severe disease was associated with a general reduction in CD4+ OX40+ CXCR5+ cTfh across all four antigens tested but was only significantly different between mild and severe disease for the Spike (SN) (r = 0.0115) and Flu (r = 0.0036) antigens (Figure 3B). The cTfh cells were enriched for Th17 cells (CXCR3– CCR6+), as previously reported (Juno et al., 2020; Rydyznski Moderbacher et al., 2020), but also for the cTfh Th2 subset (CXCR3– CCR6–) (Figure 3C).
Figure 3.
T cell subset analysis
(A) Antigen-specific OX40+ CD4+ T cells in the 24 participants at visit 1.
(B) Percentage of OX40+ CXCR5+ cTfh cells of total CD4+ T cells examined by disease severity and antigen: Flu, Spike (SN), RBD (SR), and nucleocapsid protein (NP).
(C) Proportion of cTfh Th1 (CXCR3+ CCR6–), Th2(CXCR3– CCR6–), Th17 (CXCR3– CCR6+), and Th1/Th17 (CXCR3+ CCR6+) subsets by antigen.
(D) Spearman's correlation matrix comparing immune cell frequencies. Boxed area indicates comparison with the visit 3 immune measurements.
(E) Representative Spearman's correlation plots.
(F) PCA comparing all immune components and certain clinical or immunological characteristics.
(G–J) A loading plot indicates the impact of the different variables on the clustering. Column graph comparing the influence of (G) OX40 CD4+ T cells, (H) OX40+ CXCR5+ T cells, (I) cTfh Th2 CD4 T cells, and (J) age on maintenance versus decline of MBC levels at visit 3, significance was tested with a logistic regression (Mean and SD indicated; ∗∗p between 0.001 and 0.01; ∗p between 0.01 and 0.05).
Also see Figures S2 and S3 and Table S1.
In order to determine if the magnitude of the CD4+ T cell and humoral (antibody and B cells) responses at visit 1 predicted the magnitude of the EPT titer, neutralizing breadth, neutralizing ID50, and RBD-specific resting MBCs (RM) at 1 year (visit 3), the frequency of the bulk and antigen-specific CD3+ CD4+ PD-1± Tim3±, OX40+ (CD134+ CD25+), cTfh (CXCR5+), and cTfh subsets (Th1: CXCR3+ CCR6–; Th2: CXCR3– CCR6–; Th17: CXCR3– CCR6+; Th1/Th17: CXCR3+ CCR6+) were compared with the magnitude of the humoral response measurements at visits 1 and 3 (Figure 3D). The different humoral immune response measurements shared a strong positive correlation within visit 1 and between visits 1 and 3, as may be expected. In particular, the ID50 and neutralizing breadth at visit 1 positively correlated with the ID50 and neutralizing breadth at visit 3 (ID50 V1 and ID50 V3, r = 0.67, p = 0.0004; breadth V1 and breadth V3, r = 0.67, p = 0.0004). The magnitude of the different B cell memory subsets at baseline also correlated with the magnitude of RBD-specific IgG+ CD27+ MBCs at 12 months (visit 3) (AM: r = 0.71, p < 0.0001; RM: r = 0.70, p = 0.0001; IM: r = 0.52, p = 0.009; TLM: r = 0.73, p < 0.0001) (Figure 3D). Disease severity correlated strongly with the magnitude of the EPT titer (r = 0.45, p = 0.03) and RBD-specific resting MBCs (RM) (r = 0.59, p = 0.002) at 12 months (visit 3).
The different CD4+ T cell response measurements at visit 1 correlated positively between most of the different CD4+ T cell subsets at visit 1. One notable exception was that the ex vivo CXCR5+ Tim-3+ PD-1+ exhausted CD4+ T cell subset inversely correlated with most antigen-specific CD4+ T cells detected (SN: r = −0.60, p = 0.0019; SR: r = −0.54, p = 0.067). Several Spike (SN)- and RBD (SR)-specific cTfh subsets measured at visit 1 positively correlated with some of the humoral immune responses measured at visit 3. In particular, the breadth of neutralization at visit 3 correlated positively with all Spike and RBD cTfh subsets, with several reaching statistical significance. These included SN CXCR5+ cTfh (r = 0.48, p = 0.02); SN cTfh Th17 (r = 0.57, p = 0.004); SN cTfh Th1/Th17 (r = 0.62, p = 0.001); SN cTfh Th2 (r = 0.49, p = 0.02); SR cTfh Th17 (r = 0.43, p = 0.04); and SR cTfh Th1/Th17 (r = 0.55, p = 0.006) (Figure 3E). Interestingly, the breadth of neutralization at visit 3 did not correlate with the NP cTfh antigen. ID50 at visit 3 had a comparable profile to neutralizing breadth and was significantly correlated with SN CXCR5+ cTfh (r = 0.45, p = 0.03); SN cTfh Th1/Th17 (r = 0.57, p = 0.004); SN cTfh Th17 (r = 0.53, p = 0.008); SN cTfh Th1 (r = 0.45, p = 0.03)’ SR cTfh Th1/Th17 (r = 0.48, p = 0.02); and SR cTfh Th17 (r = 0.43, p = 0.04). The classical resting MBC responses at visit 3, the RBD+ CD27+ CD21+ IgG+ RM B cells, showed no significant correlation with the SARS-CoV-2-specific cTfh subsets but instead correlated with the SN CD4+ OX40+ (r = 0.43, p = 0.04) (Figure 3E) and bulk CD4+ CXCR5+ Tim-3– PD-1+ (r = 0.43, p = 0.04) subsets. The EPT at visit 3 correlated with the CD4+ T cell subsets SR CXCR5+ Th1/Th17 (r = 0.47, p = 0.02) (Figure S2C), CD4+PD-1+Tim-3– (r = 0.41, p = 0.05), and CXCR5+ Tim-3– PD-1+ (r = 0.43, p = 0.03).
MBCs are a source of rapid production of high titer antibodies upon reinfection. As a dichotomy in the maintenance of the MBC responses was observed over time in the cohort, the participants were divided based on whether the magnitude of the MBC responses were maintained or declined over the 12 months (Figure S2D) (with decline defined as a decrease in the response that was 2-fold greater than the coefficient of variation of the assay). Based on this criterion, 10 out 24 participants (41.67%) were categorized into the “declines” category. In order to investigate whether immune parameters at visit 1 were associated with the maintenance versus decline of the RBD+ IgG+ MBC response at visit 3, a principal-component analysis (PCA) was performed on the visit 1 immunological variables and revealed clustering based on either maintained or declined MBC responses (Figure 3F). Interestingly, no clustering based on the disease severity was observed. A loading plot identified several variables that were positively associated with the maintenance of RBD+ IgG+ MBCs, which were then confirmed with logistic regression. These included CD4+ OX40+ T, cTfh, and Th2 cells. In general, the CD25+ OX40+ CD4+ T cell frequencies were higher against all three antigens in the group that maintained their MBC responses and were significant for the SN (p = 0.0305), SR (p = 0.0221), and NP (p = 0.0293) antigens (Figure 3G). Within the SARS-CoV-2 reactive subset (OX40+), the group that maintained their responses generally had a lower proportion of cTfh cells, with this trend being consistent across all three antigen groups though not reaching statistical significance (Figure 3H). However, within the cTfh subset, the group that maintained responses were enriched for the Th2 subset, this trend was observed for all SARS-CoV-2 antigens, but not the Flu antigen, and was significant for the RBD (SR) antigen (p = 0.0221) (Figure 3I).
In summary, these data suggest that the maintenance of high titer and broad neutralizing antibodies was associated with strong cTfh responses, in particular the Th1/Th17 subset (CXCR3+ CCR6+), whereas maintenance of MBCs was associated with a strong general Spike- and RBD-specific CD4+ T cell responses.
Discussion
It is becoming increasingly evident that achieving herd immunity against SARS-CoV-2 infection through vaccination alone is hard, but should a durable natural immune response to infection be generated, this could become a key contributor to this goal. Further, maintenance of a high-quality protective response is becoming increasingly important with ongoing development and transmission of new variants of concern (VOCs). In this study, we report several findings, some of which confirm key findings reported by other groups regarding the duration of the different components of the humoral immune response during convalescence after SARS-CoV-2 infection. At 12 months, despite declining anti-RBD IgG antibody titers, the majority of participants maintained ADCP and neutralizing activity. The retention of neutralizing breadth against most VOCs, including the currently circulating Delta variant, is especially encouraging. RBD-specific MBC responses were also maintained in just over half of the participants. In this study, we found that the frequency of the antigen-specific CD4+ T cell subsets at baseline predicted the maintenance of the MBC subsets at 12 months.
There was a sharp decline in the anti-RBD antibody titers during the initial convalescent phase followed by stabilization out to 12 months consistent with previous reports (Li et al., 2021; Turner et al., 2021). As reported by others in early infection (Chen et al., 2020), the antibody levels in our study remained significantly higher in participants with severe disease when compared with those with mild disease even out to 12 months post-infection in our age- and gender-matched subcohorts. This is an important finding, as in most studies, the severe disease group is biased toward an older population. This finding in our cohort therefore suggests that the antibody level is specifically associated with disease severity. During SARS-CoV-2 infection, the rapid differentiation of some B cells into short-lived plasma cells, under the influence of activated cTfh (CD4+ CXCR5+ PD1+) cells, leads to an initial influx of antibodies, which declines during convalescence. The stabilization of antibody levels is reported to be attributable to long-lived plasma cells in the bone marrow sustaining low-circulating levels of antibodies (Turner et al., 2021). In this study, the overall maintenance of the neutralizing activity and breadth, as well as ADCP functions, despite the decline in overall magnitude, suggest that the quality of the antibodies being generated is improving over time, consistent with repeated GC cycling and affinity maturation. This finding is concordant with a recent report of continued maturation in the circulating MBC population over 12 months post-SARS-CoV-2 infection (Wang et al., 2021).
We also observed that most of the convalescent sera had detectable neutralizing activity against the VOCs, with higher neutralizing activity against the D614G and Alpha variants than against the Delta followed by the Beta variants (Bates et al., 2021; Cele et al., 2021; Gallais et al., 2021; Xiang et al., 2021). The retention of neutralizing activity in the majority of the participants against the current Delta variant, albeit at a lower level, is promising and has been reported by other groups (Edara et al., 2021; Liu et al., 2021). Although the precise titer of neutralizing antibodies required to infer protection is unclear, even low titers of neutralizing antibodies in humans have been associated with protection against severe disease (Dispinseri et al., 2021; Lau et al., 2021).
At this stage, there is no study that has directly correlated antibody titers at the time of exposure and protection. Among people who received the ChAdOx1 vaccine, antibodies were measured at approximately 28 days after the second dose and then correlated with the risk of symptomatic infection in participants in the subsequent 4–6 months (Feng et al., 2021). As antibody titers are likely to decline after immunization, the titers at the time of infection were likely to have been lower than those present when measured at the peak of the response. In our study, antibody titers at 12 months were similar to those associated with a vaccine efficacy of 60% against mild disease; therefore, we believe our estimated correlate of protection is quite conservative, and our cohort may have a protective efficacy against symptomatic disease that is greater than 60%. Our analysis of the neutralizing titers against the different variants predicts that this level of protection will be impacted by the variant of the infecting strain, with a low level of protection predicted for the known immune escape variant, Beta. Encouragingly, the predicted level of protection against symptomatic disease from the currently circulating Delta virus was higher, at approximately 60%. In concordance with new modeling that was published recently, we speculate that boosting our cohort with one immunization dose will lead to increased protection (Cromer et al., 2021).
The neutralizing breadth of the antibodies at 12 months in our cohort generally correlated with baseline Spike- and RBD-specific cTfh subsets. Similar to other reports in early convalescence (Boppana et al., 2021), we confirm the presence of a robust cTfh subset even at late convalescence and that the CXCR3+ cells are responsible for enhanced neutralization breadth (Zhang et al., 2019). Importantly, in this study, the early baseline measurements predicted the later 12-month neutralization activity and MBCs. Specifically, the neutralization breadth was predicted by the levels of cTfh Th1/Th17 (CXCR3+ CCR6+) and cTfh Th17 (CXCR3– CCR6+) present during the early convalescent phase. Broad neutralizing activity has been associated with both CXCR3+ and CXCR3–cTfh subsets in people with HIV infection (Locci et al., 2013; Martin-Gayo et al., 2017) but only with CXCR3+ in people with hepatitis C virus (HCV) infection (Zhang et al., 2019) and following influenza (Bentebibel et al., 2013) and human papillomavirus (HPV) (Matsui et al., 2015) vaccinations. CCR6 expression on cTfh has not previously been associated with breadth of neutralization. In SARS-CoV-2 infection, the secretion of the Th17 cytokine, interleukin (IL)-17a, was not observed in the cTfh Th17 subset, hence the CCR6+ expression is inferred to be a lung-homing indicator (Rydyznski Moderbacher et al., 2020). A possible explanation for the CCR6 association in our study is that CCR6 expression on cTfh has been shown to increase during convalescence (Dan et al., 2021), whereas influenza-specific cTfh Th1 have been reported to peak quite early and then decline (Juno et al., 2020; Koutsakos et al., 2019). In this context, differences in sampling times may effect a direct comparison of cTfh cells; in the influenza study, which reported an association of cTfh Th1 (CXCR3+ CCR6–), the baseline data were collected at around 1 month post-infection (Juno et al., 2020), while the baseline data reported here were obtained around 2.5 months. However, since the GCs for SARS-CoV-2 persist to 6 months post-infection (Gaebler et al., 2021), understanding the evolving kinetics of the CD4+ T cell subsets longitudinally is important. One additional difference in our study in comparison to two other similar studies is that we had a larger representation of severe disease (29% versus 10%–12%) (Juno et al., 2020; Wang et al., 2021), and this group was age matched, thereby minimizing age-associated biases in the immunological analysis, allowing a more focused look at the long-term immunological impact of severe disease.
The rationale behind reporting the RBD-specific MBCs instead of the more commonly reported Spike-specific MBCs was that the latter tend to be more cross-reactive against other common coronaviruses. However, a strong correlation between the RBD-specific and the Spike-specific MBC numbers was observed at visit 3 (Figure S2E). The initial increase in MBC levels from visit 1 to visit 2 was followed by a relative stabilization at 12 months, in line with another analysis (Wang et al., 2021). As the second line of defense, the maintenance of the magnitude of the MBC repertoire is critical for mounting an anamnestic response in the event of reinfection.
The magnitude of the RBD-specific MBC responses at each visit correlated with the anti-RBD IgG antibody levels at that visit (Figure S2F). The initial anti-RBD IgG antibody levels also correlated with the RBD-specific MBC levels at 12 months (Figure S2G). The severity of the disease had a significant impact on the magnitude of the MBC subset, with those recovering from severe COVID having higher levels of RBD-specific MBCs than the mild or moderate disease groups, indicating that a small proportion of these two groups may benefit from COVID-19 vaccination at 12 months. The increased levels of the atypical or TLM (CD21– CD27–) RBD-specific MBCs observed in the severe disease group in the present study confirms previous findings (Oliviero et al., 2020; Pušnik et al., 2021). This subset then normalized over time, as observed by Wildner et al. (2021). The MBC analysis also identified a declining trend in the CD27+ IgD+ RBD-specific MBCs (Figure S2H), implying an ongoing recruitment of this repertoire into the IgG+ compartment. This persistent contraction of the unswitched CD27+ IgD+ MBC subset has been reported by others as well (de Campos-Mata et al., 2021).
In addition to the magnitude, the maintenance of the responses is critical. Interestingly, in this study, participants that maintained their responses were enriched for total antigen-specific CD4+ T cell subsets (CD25+ OX40+) and had a lower proportion of SARS-CoV-2-specific cTfh cells recognizing epitopes. Examination of cTfh subsets revealed that the Th2 subset was positively associated with maintenance across all SARS-CoV-2 antigens, but not influenza, and was significant for the SR antigen. The observation that cTfh Th2 cells during early convalescence associate with the maintenance of MBCs may be attributed to the B cell proliferation and growth induced by the IL-4 secretion by the Th2 cells (Vazquez et al., 2015). Overall, the data from this study support the proposed model that B cells that receive limited help from Tfh cells will differentiate into MBCs, whereas those with a higher level of help will re-enter the GC reaction for further maturation and potential differentiation into antibody-secreting cells.
In conclusion, the quality of the virus-specific humoral responses and the magnitude of the cellular responses are well maintained through to 12 months post-SARS-CoV-2 infection. In concordance with other reports, the antibody titers declined, but the convalescent sera maintained ADCP responses and had detectable neutralizing responses against the VOCs tested. A vast majority of our cohort (96%) still had detectable neutralization activity against the Delta variant at 12 months, and based on our modeling, 61.4% of our cohort would have a 50% protective efficacy against symptomatic infection with the Delta variant. The maintenance of serum neutralization breadth suggests that most individuals will be protected from severe disease in the event of reinfection 1 year after primary infection. This ongoing enhancement of antibody functions, even in the mildly infected participants, can be attributed to ongoing somatic hypermutation. Further, the high magnitude of RBD-specific MBCs, especially in the severe group, should provide this population with an additional level of protection against reinfection. Among people with prior mild or moderate acute SARS-CoV-2 infection, the variability of these correlates suggests that a benefit may be derived from vaccination. The association between the maintenance of the B cell responses with the early CD4+ T cell subsets reported by us may prove to be useful markers for predicting vaccine longevity and for our general understanding of the relationship of CD4+ T cell subsets with different components of the humoral immune response.
Limitations of the study
The relatively small sample size limits the scope of the findings reported in this study. However, since our aim was to understand the kinetics of the durability of SARS-CoV-2-specific immune responses and identify the predictors of durable responses, the study focused on an age- and gender-matched cohort stratified by disease severity, purposively selected from a larger cohort. This design ensured that we were comparing disease severity groups rather than age, which is known to impact immune responses and is proportionately enriched in those suffering severe disease. This overcomes some of the limitations of other studies that have a limited representation of severe disease in general and younger people with severe disease specifically. Additionally, for visit 1, three participants in the severe group were sampled between 145 and 152 DPS, which overlapped with the time window of visit 2 (106–253 DPS). Since our investigation was focused on analyzing the trajectory of MBCs in individual participants, we stratified the three timepoints as visits instead of reporting on DPS. Another limitation is the dynamic nature of the maturing immune subsets over time, in particular the CD4+ Tfh subsets, and we suggest that follow-up studies exploring the findings from this study should ensure that comparisons be closely time matched to ensure an accurate comparison of the predictive variables. Finally, in the protective efficacy analysis, the immunization studies often compared the neutralization titers measured at baseline with the probability of being infected weeks to months later, whereas we were inferring the level of protection at the titers measured at 12 months, and hence, we may have underestimated the level of protection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Streptavidin-PE | ThermoFisher Scientific | Cat#S21388 |
| Streptavidin-APC | BD Biosciences | Cat#554067; RRID:AB_10050396 |
| Fixable Viability Stain 700 | BD Biosciences | Cat#564997; RRID:AB_2869637 |
| Human Fc block | BD Biosciences | Cat#564220; RRID:AB_2869554 |
| Stain brilliant buffer | BD Biosciences | Cat#566349; RRID:AB_2869750 |
| B-ly4 (BV421) [Anti-CD21] | BD Biosciences | Cat#562966; RRID:AB_2737921 |
| FA6-2 (BV510) [Anti-IgD] | BD Biosciences | Cat#563034; RRID:AB_2737966 |
| HI10A (BV605) [Anti-CD10] | BD Biosciences | Cat#562978; RRID:AB_2737929 |
| SJ25C1 (BV711) [Anti-CD19] | BD Biosciences | Cat#563036; RRID:AB_2737968 |
| 2H7 (APC-H7) [Anti-CD20] | BD Biosciences | Cat#560734; RRID:AB_1727449 |
| G18-145 (BV786) [Anti-IgG] | BD Biosciences | Cat#564230; RRID:AB_2738684 |
| N-T271 (PE-CF594) [Anti-CD27] | BD Biosciences | Cat#562297; RRID:AB_11154596 |
| HIT2 (PE-Cy7) [Anti-CD38] | BD Biosciences | Cat#560677; RRID:AB_1727473 |
| G46-6 (BB515) [Anti-HLA-DR] | BD Biosciences | Cat#564516; RRID:AB_2732846 |
| SK7 (BB700) [Anti-CD3] | BD Biosciences | Cat#566575; RRID:AB_2860004 |
| AffiniPure Goat Anti-Human IgG, F(ab')2 Fragment Specific | Jackson ImmunoResearch | Cat#109-005-097; RRID:AB_2337540 |
| 2A3 (APC) [Anti-CD25] | BD Biosciences | Cat#340939; RRID:AB_400551 |
| L106 (PE) [Anti-CD134] | BD Biosciences | Cat#340420; RRID:AB_400027 |
| RPA-T4 (Alexa 700) [Anti-CD4] | Biolegend BD Biosciences | Cat#557922; RRID:AB_396943 |
| RPA-T8 (BV650) [Anti-CD8a] | Biolegend | Cat#301041; RRID:AB_11125174 |
| 4B4-1 (BV421) [Anti-CD137 or 4-1BB] | Biolegend | Cat#309819; RRID:AB_10895902 |
| FN50 (PE-Cy7) [Anti-CD69] | Biolegend | Cat#310911; RRID:AB_314846 |
| A1 (FITC) [Anti-CD39] | Biolegend | Cat#328205; RRID:AB_940423 |
| G034 × 103 (PerCP Cy5.5) [Anti-CD196/CCR6] | Biolegend | Cat#353406; RRID:AB_10918437 |
| G025H7 (Alexa Fluor 647) [Anti-CXCR3] | Biolegend | Cat#353712; RRID:AB_10962948 |
| MU5UBEE (SB436) [Anti-CD185/CXCR5] | Thermo Fisher | Cat#62-9185-4 2; RRID: AB_2724064 |
| UCHT1 (AF532) [Anti-CD3] | Thermo Fisher | Cat#58-0038-4 2; RRID: AB_11218675 |
| 10.1 (FITC) [Anti-CD64/Fcγ receptor RI] | Invitrogen | Cat# 11-0649-42; RRID:AB_10544395 |
| B73.1 (PE) [Anti-CD16/FcγRIII] | BD Biosciences | Cat#561313; RRID: AB_10643606 |
| MφP9 (PerCP) [Anti-CD14] | BD Biosciences | Cat#340585; RRID:AB_400065 |
| FLI8.26 (ACP) [Anti-CD32/FcγRII] | BD Biosciences | Cat#559769; RRID: AB_398665 |
| Bacterial and virus strains | ||
| Staphylococcal Enterotoxin B from Staphylococcus aureus | Sigma-Aldrich Pty Ltd | S4881-1MG |
| SARS-Cov-2 Virus variant B.1.319 (D614G) | This paper | GISAID-ID: hCoV-19/Australia/NSW4713/2021 |
| SARS-Cov-2 Virus variant B.1.17 (Alpha) | This paper | GISAID-ID: hCoV-19/Australia/NSW-R0066/2021 |
| SARS-Cov-2 Virus variant B.1.351 (Beta) | This paper | GISAID-ID: hCoV-19/Australia/NSW4463/2021 |
| SARS-Cov-2 Virus variant B.1.617.2 (Delta) | This paper | GISAID-ID: hCoV-19/Australia/NSW4605/2021 |
| Biological samples | ||
| Convalescent donor blood samples | The Kirby Institute, UNSW | https://kirby.unsw.edu.au/project/natural-history-cohort-following-sars-cov-2-infection |
| Donor blood samples | Australian Red Cross Lifeblood | https://www.donateblood.com.au/ |
| Chemicals, peptides, and recombinant proteins | ||
| TMB Chromogen Solution | ThermoFisher Scientific | Cat#002023 |
| Imidazole | Sigma-Aldrich | Cat#I202 |
| ExpiFectinamine | ThermoFisher Scientific | Cat#A14524 |
| OptiMEM-1 | ThermoFisher Scientific | Cat#31985070 |
| Phosphate buffer saline | Gibco | Cat#10010023 |
| HEPES buffer | Gibco | Cat#15630080 |
| Sodium azide | Sigma | Cat# S2002 |
| Critical commercial assays | ||
| Biotin Protein Ligase | Genecopeia | Cat#BI001 |
| Biotinylation Kit NHS-Linker | ThermoFisher Scientific | Cat#20217 |
| Experimental models: Cell lines | ||
| Expi293-Freestyle cells | ThermoFisher Scientific | Cat#A14527 |
| ACE2-TMPRSS2-Hek293T cells | A/Prof Stuart Turville | N/A |
| VeroE6 | CellBank Australia | CODE: 85,020,020206 |
| THP-1 cells | ATCC | Cat#202 TIB |
| Recombinant DNA | ||
| pCAGGS-SARS-CoV-2-RBD | Dr Markus Hoffmann | N/A |
| pCAGGS-SARS-CoV-2-Spike | Dr Markus Hoffmann | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | N/A |
| FlowJo version 10.7.1 | Tree Star, Inc | N/A |
| SPSS 25 | IBM | N/A |
| R 4.0.2 | The R Foundation | N/A |
| Incarta Image analysis software | Cytiva | N/A |
| Other | ||
| Streptavidin coated Alexa-488 microbeads | Spherotech | Cat# VFP-0552-5 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, A/Professor Rowena Bull (r.bull@unsw.edu.au).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
Convalescent COVID-19 donors
Commencing enrolment in February 2020, COSIN (Collection of COVID-19 Outbreak Samples in NSW) is an ongoing prospective cohort study evaluating the natural history of SARS-CoV-2 infection among children and adults in Sydney, Australia. Participants were enrolled through seven hospitals in- and outpatient departments and referring microbiology laboratories. Visits post-acute infection were scheduled at one, four, eight, and 12-months following symptom onset or date of NAT-confirmed diagnosis (whichever came first). Standardized clinical data and blood samples (serum, PBMCs) were collected at each visit.
COVID disease severity was classified according to NIH criteria (www.covid19treatmentguidelines.nih.gov).
-
•
Asymptomatic: Individuals who test positive for SARS-CoV-2 using a virologic test (i.e., a nucleic acid amplification test or an antigen test) but who have no symptoms that are consistent with COVID-19.
-
•
Mild Illness: Individuals who have any of the various signs and symptoms of COVID-19 (e.g., fever, cough, sore throat, malaise, headache, muscle pain, nausea, vomiting, diarrhea, loss of taste and smell) but who do not have shortness of breath, dyspnea, or abnormal chest imaging.
-
•
Moderate Illness: Individuals who show evidence of lower respiratory disease during clinical assessment or imaging and who have saturation of oxygen (SpO2) ≥94% on room air at sea level.
-
•
Severe Illness: Individuals who have SpO2 <94% on room air at sea level, a ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) <300 mm Hg, respiratory frequency >30 breaths/min, or lung infiltrates >50%.
-
•
Critical Illness: Individuals who have respiratory failure, septic shock, and/or multiple organ dysfunction.
Healthy unexposed donors
Healthy controls for antibody studies had a median age of 45 (range 24-73). Blood from 11 of the healthy controls were collected prior to 2007 and the remaining 9 were collected between March and April 2020 in Sydney, Australia, where local transmission was very low at the time. None of these 9 healthy controls had a history of COVID-19, were not close contacts of cases of COVID-19 and were not health care workers. For the memory B cell assays stored PBMCs from two of these healthy controls and stored PBMCs from four Australian Red Cross Lifeblood donors collected prior to 2020 were used (median age 38, range 25–48).
Ethics statement
The protocol was approved by the Human Research Ethics Committees of the Northern Sydney Local Health District and the University of New South Wales, NSW Australia (ETH00520) and was conducted according to the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice (ICH/GCP) guidelines and local regulatory requirements. Written informed consent was obtained from all participants before study procedures.
Method details
RBD and spike protein production
SARS-CoV-2 Spike RBD (residues 319–541), with an N-terminal human Ig kappa leader sequence and C-terminal Avi- and His-tags, were cloned into pCEP4 (Invitrogen). Expi293-Freestyle cells (ThermoFisher Scientific) were cultured in Expi293 Expression Medium (ThermoFisher Scientific) at 37°C and 8% CO2. The plasmid was transiently transfected into Expi293-Freestyle cells as follows: 1.5 × 108 total cells (50mL transfection) were mixed with 50μg of plasmid, 160μL of Expifectamin and 6mL of OptiMEM-I and left overnight at 37°C in a shaking incubator. After 24 h, 300μL of ExpiFectamine Enhancer 1 and 3mL of ExpiFectamine Enhancer 2 was added to the cells and left in culture for a further 48 h. After a total of 72 h in culture, the cell culture is collected and centrifuged for 20 min at 4000xg, 4°C. The supernatant was clarified by passing through a 0.22μM filter twice. A HisTrap HP Column (GE Healthcare) was used to affinity purify the His-tagged protein from the cell supernatant and eluted with imidazole. The purified protein was buffer exchanged and concentrated in sterile DPBS by centrifuging at 4000xg for 30 min at 4°C in a 10,000 MWCO Vivaspin centrifugal concentrator (Sartorius) and stored at – 80°C. The recombinant RBD was biotinylated using the Avitag as described by the manufacturer (Genecopeia).
SARS-CoV-2 Spike, with a C-terminal His-tag, were cloned into pCAGGS (Krammer Research Group). Expi293-Freestyle cells cell transfection was performed same as SARS-CoV-2-RBD, with a single modification that the transfection was performed at 32°C. After a total of 72 h in culture, the cell culture is collected and centrifuged for 20 min at 4000xg, 4°C. Cellular debris was clarified by passing the supernatant twice through a 0.22μM filter. The His-tagged protein was then affinity purified from the cell supernatant using a HisTrap HP Column (GE Healthcare) and eluted with imidazole. The purified protein was then buffer exchanged and concentrated in sterile DPBS by centrifuging at 4000xg for 30 min at 4°C in a 10,000 MWCO Vivaspin centrifugal concentrator (Sartorius). The protein was then biotinylated with EZ-Link™ Sulfo-NHS-LC-Biotin (Thermo-Fischer) for 2 h at 4°C, then buffer exchanged and concentrated as above. The biotinylated recombinant SARS-CoV-2-Spike protein was stored at – 80°C.
RBD binding ELISA and limit of detection
Nunc-Immuno MicroWell plates, 96 well (ThermoFisher Scientific) were coated with recombinant SARS-2 RBD protein in DPBS at a concentration of 5μg/mL and incubated overnight at 4°C. The plates were washed three times with PBS containing 0.05% Tween 20 (PBS-T) and blocked with 5% non-fat milk in PBS-T for an hour at room temperature (RT). Patient serum was heat inactivated (56°C for 30 min), serially diluted (1/20 to 1/393,660) in 5% non-fat milk in PBS-T, added to the wells in duplicates and incubated for two hours at RT. Anti-human IgG-HRP (Jackson Immunoresearch) was added at a 1:3000 dilution to the plates for one hour at room temperature. The plates were developed with TMB Chromogen Solution (ThermoFisher Scientific) for 15 min at RT, followed by 1 M HCl to stop the reaction. CLARIOstar microplate reader (BMG Labtech) was used to measure the optical density (OD) at 450 nm. The limit of detection for this RBD binding ELISA was determined by adding 2 SD to the mean OD450 of the highest dilution (1/20) of sera from 19 healthy unexposed individuals.
We also used the commercially available Abbot Architect based SARS-CoV-2 IgG II Quant assay to detect the spike RBD-specific IgG antibody levels in sera at visit 3. The results obtained in RLU (Relative Light Units) were transformed into Abbot AU/mL (Arbitrary Units/mL). This was converted into the WHO standard of BAU/mL (Binding Antibody Units/mL) using the mathematical equation BAU/mL = 0.142 × AU/mL (Abbott Laboratories, 2021).
SARS-CoV-2 specific antibody dependent cellular phagocytosis assay
ADCP assays were performed as previously described (Adhikari et al., 2021) with the following modifications. Streptavidin coated Alexa 488 microbeads at a concentration of 1.5×109 beads/mL were mixed with recombinant biotinylated SARS-CoV-2 Spike protein (at 50μg/mL) in 1.5mL Eppendorf tubes and were incubated for 16 h at 4°C on a rotating chamber. Excess protein was removed by washing the beads with 1mL lipopolysaccharide minimized cold phosphate buffer saline (PBS) at pH 7.4 (Gibco, USA) with gentle centrifugation at 2292xg for 5 min (Beckman coulter microfuge 20R). Aliquots (50μL with 1.5 × 106 beads/μL) of the SARS-CoV-2 Spike coated beads were then incubated with 10μL of native plasma from SARS-CoV-2 patients for 2 h at 37°C or 10μL of plasma from healthy donors to form immune complexes. THP-1 cells in PBS (1×105 cells in 50μL) were added onto 60μL of immune-complex-coated microbeads in a 1.5mL Eppendorf tube and incubated for 2 h at 37°C, 5% CO2 in a final volume of 600μL assay buffer (RPMI 1640 including 0.1% Human serum and 100mM HEPES Thermofisher, USA). After the 2-h incubation, the cell-bead cocktails were washed once using 1mL of cold wash buffer (PBS including 0.5% Fetal bovine serum (FBS) and 0.005% of sodium azide) and subjected to centrifugation at 335xg (Beckman coulter X-15R), for 5 min at 4°C, fixed in 400μL of 1% paraformaldehyde (Sigma, USA) and transferred to 4°C in the dark. Cells incubated with SARS-CoV-2 Spike protein coated beads opsonized with healthy donor plasma were used as controls. A total of 2×104 events were acquired for THP-1 cells using BD FACSCaliburTM and the proportions of cells that phagocytosed the beads were analyzed using BD FlowJo version 10.5.0 software. Phagocytic scores (p-score) were then calculated as a percentage of bead positive cells multiplied by mean fluorescence intensity (MFI) divided by 1000 (Darrah et al., 2007). We utilized p-score to compare the fold change differences of percentage positive THP-1 cells with positive and negative controls in our assay.
To measure the THP-1 surface Fc-receptor expression, 5×105 THP-1 cells were incubated with 3μL each of Fcγ receptor RI (CD64)-FITC, FcγRIII (CD16)-PE, CD14-PerCP, FcγRII (CD32)-ACP and their isotype and fluorochrome matched negative control for 20min at room temperature. The cells were then washed twice with 1mL of cold wash buffer and centrifuged at 335xg (Beckman coulter X-15R) for 5 min, at 4°C and then fixed in 400μL of 1% paraformaldehyde until measured in BD FACSCaliburTM. A total of 2×104 events were acquired for THP-1 cells.
SARS-CoV-2 live virus neutralization assay
HEK293T cells stably expressing human ACE2 and TMPRSS2 were generated by transducing cells with lentiviral particles. Briefly, the ORFs for hACE2 (Addgene#1786) and hTMPRSS2a (Addgene#53887, synthetic gene fragment; IDT) were cloned into lentiviral expression vectors pRRLsinPPT.CMV.GFP.WPRE (Follenzi et al., 2002) and pLVX-IRES-ZsGreen (Clontech) respectively. Lentiviral particles expressing the above proteins were produced by co-transfecting expression plasmids individually with a second generation lentiviral packaging construct psPAX2 (courtesy of Dr Didier Trono through NIH AIDS repository) and VSVG plasmid pMD2.G (Addgene#2259) in HEK293T cells (Life Technologies) by using polyethyleneimine as previously described (Aggarwal et al., 2012). Two successive rounds of lentiviral transductions were then performed to generate the HEK293/ACE2/TMPRSS2a cells; clonal selection led to the identification of a highly permissive clone, HekAT24, which was then used to carry out SARS-CoV-2 neutralization assay as previously described (Tea et al., 2021).
On the day of the assay, HekAT24 cells were trypsinized, stained with Hoechst-33342 dye (5% v/v) (NucBlue, Invitrogen) in suspension and then seeded at 16,000 cells per well in a volume of 40μL of DMEM-5% FCS in a 384-well plate (Corning #CLS3985). Two-fold dilutions of patient plasma samples were mixed with an equal volume of SARS-CoV-2 virus solution (4 × 103 TCID50/ml) and incubated at 37°C for 1 h before transferring 40μL in duplicate to the cells (final MOI = 0.05). Viral variants used included the variants of concern; Alpha (B.1.1.7), Beta (B.1.351) and Delta (B.1.617.2), as well as control virus from the same clade with matching ‘D614G’ background (B1.319). Plates were incubated for a further 24 h following which entire wells were imaged by high-content fluorescence microscopy and cell counts obtained with automated image analysis software. Percentage of virus neutralization was calculated with the formula: %N = (D-(1-Q)) × 100/D, where Q = nuclei count normalized to mock controls and D = 1-Q for average of infection controls as previously described (Tea et al., 2021). An average %N > 50% was defined as having neutralizing activity. The 50% inhibitory concentration (ID50 for serum) titer was calculated using non-linear regression model of normalized data (GraphPad Prism 8). The ID50 cutoff defined by the 19 healthy controls for the live virus was 0.6 (0.06 + 2 × 0.27), as previously published (Abayasingam et al., 2021).
Detection of RBD and spike specific memory B cells
A tetramer bait based flow cytometry technique was used to detect the SARS-CoV-2 specific memory B cells (MBCs) (Abayasingam et al., 2021). The tetramerization method was adapted from a previously published method used for hepatitis C virus tetramers (Wu et al., 2019). Biotinylated RBD and Spike were incubated with Streptavidin-PE (SA-PE; Molecular probes; ThermoFisher Scientific) and Streptavidin-APC (SA-APC; BD Pharmingen) in a molar ratio of 4:1 and 2:1 respectively. The streptavidin dyes were added in 1/10th volume increments to the biotinylated protein, for a total of 10 times with a 10 min incubation at 4°C, in a rotating bioreactor, protected from light.
Cryopreserved PBMCs were thawed rapidly in a 37°C waterbath and washed with pre-warmed RPMI media supplemented with 2mM L-glutamine, 50IU/mL penicillin, 50μg/mL streptomycin and 10% heat inactivated fetal calf serum (Sigma). The cells were resuspended in DPBS and counted using the trypan blue dye exclusion technique. A maximum of 1 × 107 cells per sample were stained with Fixable Viability Stain 700 (FVS700) (BD Bioscience in a 1:1000 dilution) and incubated at 4°C for 20 min, to exclude the dead cells. Samples were washed twice with FACS wash buffer (DPBS +0.1% BSA), followed by incubation with 5μL Human Fc block (BD Bioscience) per 2 × 106 cells at RT for 10 min, to block non-specific antibody binding. Following a wash, all consecutive steps were done either at 4°C or on ice. SARS-CoV-2-specific B cells were identified by simultaneously staining with a cocktail of RBD and Spike tetramers at a concentration of 1μg/mL each, at 4°C for 30 min followed by two washes. A staining mix of surface marker antibodies were used to identify the MBC population. This staining mixture contained 50μL stain brilliant buffer and a titrated combination of antibodies (all from BD Bioscience): 5μL each of CD21 BV421, IgD BV510, CD10 BV605, CD19 BV711 and CD20 APC-H7, 8μL of IgG BV786, 2μL each of CD27 PE-CF594 and CD38 PE-Cy7, 2.5μL HLA-DR BB515 and 0.5μL CD3 BB700. The cells are incubated with the staining cocktail at 4°C for 30 min. Following two wash steps the samples were resuspended in FACS wash buffer. A BD FACSAria™ III sorter was used to phenotype the samples. A minimum of 300,00 events were acquired for each sample. The data analysis was performed using FlowJo version 10.7.1 (TreeStar). The assay cut off was designated as mean +2 SD calculated from 6 healthy controls.
Ex vivo phenotyping and T cell activation assay (AIM assay)
Cryopreserved PBMCs were thawed using RPMI (+L-glut) medium (ThermoFisher Scientific, USA) supplemented with Penicillin/Streptomycin (Sigma-Aldrich, USW), and subsequently stained with antibodies binding to extracellular markers. Extracellular panel included: Live/Dead dye Near InfraRed, CXCR5 (MU5UBEE), CD38 (HIT2) (ThermoFisher Scientific, USA); CD3 (UCHT1), CD8 (HIL-72021), PD-1 (EH12.1), TIM-3 (TD3), CD27 (L128), CD45RA (HI100), IgD (IA6-2), CD25 (2A3), and CD19 (HIB19) (Biolegend, USA); CD4 (OKT4), CD127 (A019D5), HLA-DR (L234), GRP56 (191B8), CCR7 (G043H7) and CD57 (QA17A04) (BD Biosciences, USA). Perm Buffer II (BD Pharmingen) was used for intracellular staining of granzyme B (GB11, BD Biosciences). FACS staining of 48hr activated PBMCs were performed as described previously (Zaunders et al., 2009), but with the addition of CD137 (4B4-1) to the cultures at 24hrs. Final concentration of 10μg/mL of SARS-CoV-2 peptide pools (Genscript) were used, 1μg/mL of Fluarix tetra (Influenza vaccine-GSK) was used as a control antigen and SEB was used as a positive control (Thermo Fisher Scientific). In vitro activation panel included: CD3 (UCHT1), CD4 (RPA-T4), CD8 (RPA-T8), CD39 (A1), CD69 (FN50), CXCR3 (G025H7)- all Biolegend, CD25 (2A3), CD134 (L106)- BD Biosciences, and CXCR5 (MU5UBEE), and CCR6 (R6H1)- Thermo Fisher Scientific. Samples were acquired on an Cytek Aurora (Biolegend, USA) using the Spectroflo software. Prior to each run, all samples were fixed in 0.5% paraformaldehyde. Data analysis was performed using FlowJo version 10.7.1 (TreeStar).
Quantification and statistical analysis
Descriptive statistics of data arrays were summarized with measures of central tendency (mean or median) or dispersion (standard deviation or interquartile range) depending on normality of distributions. Absolute cell counts were used in the analysis unless mentioned otherwise. Statistically significant associations were explored with appropriate parametric or non-parametric tests (e.g., Wilcoxon matched pairs signed rank test to compare between visits, Mann Whitney U test to compare between severities, repeated measures ANOVA to compare the MBC subsets across visits and between severities), while adjusting for within (e.g., repeated measurements) and between host factors (e.g., disease severity, age, gender). Correlations between continuous variables were explored with spearman's rank correlation coefficient. For some analyses (e.g., factors associated with maintenance of RBD + MBC), continuous dependent variables were converted into categorical dichotomous outcomes (e.g., maintains vs declines) based on a cut-off value, and the relationship with independent variables (e.g., CD4+ T cell subsets) were explored using a logistic regression model with R package rms. Statistical analyses were done with GraphPad Prism (v8, GraphPad Software, USA) and SPSS (v25, IBM, USA). The Spearmanʼs correlation matrix was calculated using the corrplot function and the PCA was generated using the R package factoextra with the prcomp() function by first scaling the variables to have a unit variance, the data were visualized with the R package ggplot2 and its extension ggfortify, in Rstudio (v4.1.0 Integrated Development Environment for R, USA). Level of statistical significance was set at p < 0.05.
Acknowledgments
The authors would like to thank the study participants for their contribution to the research, as well as current and past researchers and staff. They would like to acknowledge members of the study groups: Protocol Steering Committee – Rowena Bull (Co-chair, The Kirby Institute, UNSW Sydney, Sydney, Australia), M.M. (Co-chair, The Kirby Institute, UNSW Sydney, Sydney, Australia), Andrew Lloyd (The Kirby Institute, UNSW Sydney, Sydney, Australia), John Kaldor (The Kirby Institute, UNSW Sydney, Sydney, Australia), Greg Dore (The Kirby Institute, UNSW Sydney, Sydney, Australia), Tania Sorrell (Marie Bashir Institute, University of Sydney, Sydney, Australia), W.R. (NSWHP), Jeffrey Post (POWH), B.H. (RNSH), Dominic Dwyer (NSWHP), A.B. (SCH), Sarah Sasson (UNSW), Nick Di Girolamo (UNSW), and Daniel Lemberg (SCHN); Coordinating Center - The Kirby Institute, UNSW Sydney, Sydney, Australia – Rowena Bull (coordinating Principal Investigator), M.M. (coordinating Principal Investigator), Marianne Byrne (Clinical Trials Manager), Mohammed Hammoud (Post-Doctoral Fellow and Data Manager), Andrew Lloyd (Investigator), and Roshana Sultan (Study coordinator); Site Principal Investigators – Jeffrey Post (Prince of Wales Hospital, Sydney, Australia), Michael Mina (Northern Beaches Hospital, Sydney, Australia), B.H. (Royal North Shore Hospital, Sydney, Australia), N.G. (Westmead Hospital, Sydney, Australia), W.R. (New South Wales Health Pathology, NSW, Australia), P.K. (St George Hospital, Sydney, Australia), G.A. and M.M. (Blacktown Mt Druitt Hospital, Sydney, Australia), A.B. (Sydney Children's Hospital, Sydney, Australia), and Gail Matthews (St Vincent's Hospital, Sydney, Australia); Site coordinators – Dmitrii Shek (Blacktown Mt Druitt Hospital), Katerina Mitsakos (Royal North Shore Hospital, Sydney, Australia), Renier Lagunday (Prince of Wales Hospital, Sydney, Australia), Sharon Robinson (St George Hospital, Sydney, Australia), Lenae Terrill (Northern Beaches Hospital, Sydney, Australia), Neela Joshi, (Lucy) Ying Li and Satinder Gill (Westmead Hospital, Sydney, Australia), and Rebecca Hickey and Alison Sevehon (St Vincent's Hospital, Sydney, Australia). Some of the data presented in this work were acquired by instruments at the Mark Wainwright Analytical Center (MWAC) of UNSW Sydney. The Kirby Institute is funded by the Australian Government Department of Health and Ageing. The views expressed in this publication do not necessarily represent the position of the Australian government. Research reported in this publication was supported by the Snow Medical Foundation as an investigator-initiated study. The content is solely the responsibility of the authors. R.A.B., M.M., C.R., and A.R.L. are fellows funded by National Health and Medical Research Council (NHMRC). MWAC is in part funded by the Research Infrastructure Program of UNSW.
Author contributions
Conceptualization, R.A.B., M.M., A.R.L., D.C., and N.T.; methodology, D.A., E.K., H.B., A. Adhikari, C.P., B.G.-B., S.T., and A. Aggarwal; investigation, H.B., C.P., D.A., A. Adhikari, A. Aggarwal, A.O.S., V.K., and C.R.; writing – original draft, R.A.B., H.B., C.K., V.K., C.P., B.H., L.B.S., M.G., and D.A.; writing – review & editing, A.B., M.M., A.R.L., D.C., S.C.S., J.J.P., A.K., and D.D.; funding acquisition, R.A.B., M.M., T.S., and A.R.L.; resources, M.H., W.R., M.M.M., M.M., J.J.P., B.H., P.K, G.A.,B.H., and N.G.; supervision, R.A.B., A.R.L., and M.M.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: February 08, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110345.
Supplemental information
Data and code availability
-
•
The published article includes all data generated or analyzed during this study, summarized in the accompanying tables, figures and supplemental information and are freely available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abayasingam A., Balachandran H., Agapiou D., Hammoud M., Rodrigo C., Keoshkerian E., Li H., Brasher N.A., Christ D., Rouet R., et al. Long-term persistence of RBD-positive memory B cells encoding neutralizing antibodies in SARS-CoV-2 infection. Cell Rep. Med. 2021;2:100228. doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott Laboratories . Abbott Laboratories; 2021. Abbott Architect SARS-CoV-2 IgG II Quant Instructions for Use; pp. 1–11. [Google Scholar]
- Adeniji O.S., Giron L.B., Purwar M., Zilberstein N.F., Kulkarni A.J., Shaikh M.W., Balk R.A., Moy J.N., Forsyth C.B., Liu Q., et al. Covid-19 severity is associated with differential antibody fc-mediated innate immune functions. mBio. 2021;12:e00281–e00321. doi: 10.1128/mBio.00281-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A., Eltahla A., Lloyd A.R., Rodrigo C., Agapiou D., Bull R.A., Tedla N. Optimisation and validation of a new method for antibody dependent cellular phagocytosis in hepatitis C virus infection. J. Immunol. Methods. 2021;495:113087. doi: 10.1016/j.jim.2021.113087. [DOI] [PubMed] [Google Scholar]
- Aggarwal A., Iemma T., Shih I., Newsome T., McAllery S., Cunningham A., Turville S. Mobilization of HIV spread by diaphanous 2 dependent filopodia in infected dendritic cells. PLoS Pathog. 2012;8:e1002762. doi: 10.1371/journal.ppat.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., Vegvari C., Truscott J., Collyer B.S. Challenges in creating herd immunity to SARS-CoV-2 infection by mass vaccination. Lancet. 2020;396:1614–1616. doi: 10.1016/S0140-6736(20)32318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch Y.C., Wang C., Zohar T., Fischinger S., Atyeo C., Burke J.S., Kang J., Edlow A.G., Fasano A., Baden L.R., et al. Humoral signatures of protective and pathological SARS-CoV-2 infection in children. Nat. Med. 2021;27:454–462. doi: 10.1038/s41591-021-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T.A., Leier H.C., Lyski Z.L., McBride S.K., Coulter F.J., Weinstein J.B., Goodman J.R., Lu Z., Siegel S.A.R., Sullivan P., et al. Neutralization of SARS-CoV-2 variants by convalescent and BNT162b2 vaccinated serum. Nat. Commun. 2021;121:1–7. doi: 10.1038/s41467-021-25479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentebibel S.-E., Lopez S., Obermoser G., Schmitt N., Mueller C., Harrod C., Flano E., Mejias A., Albrecht R.A., Blankenship D., et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 2013;5:176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppana S., Qin K., Files J.K., Russell R.M., Stoltz R., Bibollet-Ruche F., Bansal A., Erdmann N., Hahn B.H., Goepfert P.A. SARS-CoV-2-specific circulating T follicular helper cells correlate with neutralizing antibodies and increase during early convalescence. PLoS Pathog. 2021;17:e1009761. doi: 10.1371/journal.ppat.1009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Campos-Mata L., Tejedor Vaquero S., Tachó-Piñot R., Piñero J., Grasset E.K., Arrieta Aldea I., Rodrigo Melero N., Carolis C., Horcajada J.P., Cerutti A., et al. SARS-CoV-2 sculpts the immune system to induce sustained virus-specific naïve-like and memory B-cell responses. Clin. Transl. Immunol. 2021;10:e1339. doi: 10.1002/cti2.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Systems Science and Engineering, and JHU.edu . Johns Hopkins Coronavirus Resource Center; 2021. COVID-19 Map. [Google Scholar]
- Chen X., Pan Z., Yue S., Yu F., Zhang J., Yang Y., Li R., Liu B., Yang X., Gao L., et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal. Transduct. Target. Ther. 2020;51:1–6. doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer D., Steain M., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Kent S.J., Triccas J.A., Khoury D.S., Davenport M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet Microbe. 2021;3:e52–e61. doi: 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah P., Patel D., De Luca P., Lindsay R., Davey D., Flynn B., Hoff S., Andersen P., Reed S., Morris S., et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against leishmania major. Nat. Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- Dispinseri S., Secchi M., Pirillo M.F., Tolazzi M., Borghi M., Brigatti C., De Angelis M.L., Baratella M., Bazzigaluppi E., Venturi G., et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021;12:6–17. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.-V., Pinsky B.A., Suthar M.S., Lai L., Davis-Gardner M.E., Floyd K., Flowers M.W., Wrammert J., Hussaini L., Ciric C.R., et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.06.21.21258528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi A., Sabatino G., Lombardo A., Boccaccio C., Naldini L. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum. Gene Ther. 2002;13:243–260. doi: 10.1089/10430340252769770. [DOI] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais F., Gantner P., Bruel T., Velay A., Planas D., Wendling M.J., Bayer S., Solis M., Laugel E., Reix N., et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. 2021;71:103561. doi: 10.1016/j.ebiom.2021.103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Valtanen P., Hope C.M., Masavuli M.G., Yeow A., Balachandran H., Mekonnen Z.A., Al-Delfi Z., Abayasingam A., Agapiou D., Gummow J., et al. “One year later” - SARS-CoV-2-specific immunity in mild cases of COVID-19. SSRN Electron. J. 2021 doi: 10.2139/ssrn.3859298. [DOI] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Koutsakos M., Nguyen T.H.O., Kedzierska K. With a little help from T follicular helper friends: humoral immunity to influenza vaccination. J. Immunol. 2019;202:360–367. doi: 10.4049/jimmunol.1800986. [DOI] [PubMed] [Google Scholar]
- Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W., Chiu S.S., Ko R.L.W., Chan K.H., Cheng S.M.S., Perera R.A.P.M., et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021;121:1–7. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yu D., Wu X., Liang H., Zhou Z., Xie Y., Li T., Wu J., Lu F., Feng L., et al. Twelve-month specific IgG response to SARS-CoV-2 receptor-binding domain among COVID-19 convalescent plasma donors in Wuhan. Nat. Commun. 2021;121:1–9. doi: 10.1038/s41467-021-24230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M., Havenar-Daughton C., Landais E., Wu J., Kroenke M.A., Arlehamn C.L., Su L.F., Cubas R., Davis M.M., Sette A., et al. Human circulating PD-1+CXCR3−CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gayo E., Cronin J., Hickman T., Ouyang Z., Lindqvist M., Kolb K.E., Schulze zur Wiesch J., Cubas R., Porichis F., Shalek A.K., et al. Circulating CXCR5+CXCR3+PD-1lo Tfh-like cells in HIV-1 controllers with neutralizing antibody breadth. JCI Insight. 2017;2:e89574. doi: 10.1172/jci.insight.89574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Adelsberger J.W., Kemp T.J., Baseler M.W., Ledgerwood J.E., Pinto L.A. Circulating CXCR5+CD4+ T follicular-like helper cell and memory B cell responses to human papillomavirus vaccines. PLoS One. 2015;10:e0137195. doi: 10.1371/journal.pone.0137195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott A. Herd immunity is an important—and often misunderstood—public health phenomenon. Proc. Natl. Acad. Sci. U S A. 2021;118:1–4. doi: 10.1073/pnas.2107692118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Sharma S., Dobhal A., Kumar S., Chawla H., Singh R., Makhdoomi M.A., Das B.K., Lodha R., Kabra S.K., et al. Broadly neutralizing plasma antibodies effective against autologous circulating viruses in infants with multivariant HIV-1 infection. Nat. Commun. 2020;111:1–11. doi: 10.1038/s41467-020-18225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S., Ho J., Malaspina A., Wang W., DiPoto A., O’Shea M., Roby G., Kottilil S., Arthos J., Proschan M., et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R., Schmitt N., Bentebibel S.-E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N., et al. Human blood CXCR5+CD4+ T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumoli N., Vitale J., Mazzone A. Clinical immunity in discharged medical patients with COVID-19. Int. J. Infect. Dis. 2020;99:229. doi: 10.1016/j.ijid.2020.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSW Government NSW COVID-19 cases by location and likely source of infection, datasets. 2021. https://data.nsw.gov.au
- Oliviero B., Varchetta S., Mele D., Mantovani S., Cerino A., Perotti C.G., Ludovisi S., Mondelli M.U. Expansion of atypical memory B cells is a prominent feature of COVID-19. Cell Mol. Immunol. 2020;17:1101–1103. doi: 10.1038/s41423-020-00542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M., Ozberk V., Eskandari S., Shalash A.O., Joyce M.A., Saffran H.A., Day C.J., Lepletier A., Spillings B.L., Mills J., et al. Antibodies to neutralizing epitopes synergistically block the interaction of the receptor-binding domain of SARS-CoV-2 to ACE 2. Clin. Transl. Immunol. 2021;10:e1260. doi: 10.1002/cti2.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M.S., Hansen C.B., Kristiansen M.F., Fjallsbak J.P., Larsen S., Hansen J.L., Jarlhelt I., Pérez-Alós L., Steig B.Á., Christiansen D.H., et al. SARS-CoV-2 natural antibody response persists for at least 12 months in a nationwide study from the Faroe Islands. Open Forum Infect. Dis. 2021;8:ofab378. doi: 10.1093/ofid/ofab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pušnik J., Richter E., Schulte B., Dolscheid-Pommerich R., Bode C., Putensen C., Hartmann G., Alter G., Streeck H. Memory B cells targeting SARS-CoV-2 spike protein and their dependence on CD4+ T cell help. Cell Rep. 2021;35:109320. doi: 10.1016/j.celrep.2021.109320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2020;184:1–15. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C.C.C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar M., Rappazzo C.G., Wieland-Alter W.F., Hsieh C.-L., Wrapp D., Esterman E.S., Kaku C.I., Wec A.Z., Geoghegan J.C., McLellan J.S., et al. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci. Immunol. 2021;6:6916. doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuwa H.A., Shaw T.N., Knight S.B., Wemyss K., McClure F.A., Pearmain L., Prise I., Jagger C., Morgan D.J., Khan S., et al. Alterations in T and B cell function persist in convalescent COVID-19 patients. Med (N Y) 2021;2:720–735.e4. doi: 10.1016/j.medj.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens D.S., McElrath M.J. COVID-19 and the path to immunity. JAMA. 2020;324:1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tea F., Ospina Stella A., Aggarwal A., Ross Darley D., Pilli D., Vitale D., Merheb V., Lee F., Cunningham P., Walker G., et al. SARS-CoV-2 neutralizing antibodies: longevity, breadth, and evasion by emerging viral variants. PLoS Med. 2021;18:e1003656. doi: 10.1371/journal.pmed.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., Kim W., Kalaidina E., Goss C.W., Rauseo A.M., Schmitz A.J., Hansen L., Haile A., Klebert M.K., Pusic I., et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- Vazquez Monica I, Catalan-Dibene Jovani, Zlotnik Albert, et al. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine. 2015;74:318–326. doi: 10.1016/j.cyto.2015.02.007. PMC4475485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:1–10. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildner N.H., Ahmadi P., Schulte S., Brauneck F., Kohsar M., Lütgehetmann M., Beisel C., Addo M.M., Haag F., Schulze zur Wiesch J., et al. B cell analysis in SARS-CoV-2 versus malaria: increased frequencies of plasmablasts and atypical memory B cells in COVID-19. J. Leukoc. Biol. 2021;109:77–90. doi: 10.1002/JLB.5COVA0620-370RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B.R., Eltahla A.A., Keoshkerian E., Walker M.R., Underwood A., Brasher N.A., Agapiou D., Lloyd A.R., Bull R.A. A method for detecting hepatitis C envelope specific memory B cells from multiple genotypes using cocktail E2 tetramers. J. Immunol. Methods. 2019;472:65–74. doi: 10.1016/j.jim.2019.06.016. [DOI] [PubMed] [Google Scholar]
- Xiang T., Liang B., Fang Y., Lu S., Li S., Wang H., Li H., Yang X., Shen S., Zhu B., et al. Declining levels of neutralizing antibodies against SARS-CoV-2 in convalescent COVID-19 patients one year post symptom onset. Front. Immunol. 2021;12:2327. doi: 10.3389/fimmu.2021.708523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaunders J.J., Munier M.L., Seddiki N., Pett S., Ip S., Bailey M., Xu Y., Brown K., Dyer W.B., Kim M., et al. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40) J. Immunol. 2009;183:2827–2836. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu W., Wen B., Xie T., Tang P., Hu Y., Huang L., Jin K., Zhang P., Liu Z., et al. Circulating CXCR3 + Tfh cells positively correlate with neutralizing antibody responses in HCV-infected patients. Sci. Rep. 2019;91:1–10. doi: 10.1038/s41598-019-46533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The published article includes all data generated or analyzed during this study, summarized in the accompanying tables, figures and supplemental information and are freely available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.