Abstract
A restriction fragment length polymorphism (RFLP) assay was developed to identify and differentiate Old World, African-Eurasian orthopoxviruses (OPV): variola, vaccinia, cowpox, monkeypox, camelpox, ectromelia, and taterapox viruses. The test uses amplicons produced from virus genome DNA by PCR with a consensus primer pair designed from sequences determined for the cytokine response modifier B (crmB) gene of 43 different OPV strains of known taxonomic origin. The primer pair amplified a single specific product from each of the 115 OPV samples tested. Size-specific amplicons identified and differentiated ectromelia and vaccinia virus strains, which contain a truncated crmB gene, and enabled their differentiation from other OPV species. Restriction digests of amplified products allowed the identification and differentiation of variola, monkeypox, camelpox, vaccinia, and cowpox virus species and strains.
Orthopoxviruses (OPV) are large, morphologically identical, antigenically related vertebrate viruses that replicate in the cytoplasm of host cells. These viruses possess a linear, hairpin-terminated, double-stranded DNA genome of approximately 200 kbp containing different-sized inverted terminal repeats, with a G+C content of approximately 35% (7, 9, 12, 24). Eight Old World African-Eurasian OPVs are known, and four of them infect humans: variola virus (VAR), the causative agent of smallpox, now eradicated in nature; monkeypox virus (MPV), cowpox virus (CPV), and vaccinia virus (VAC) (12). Camelpox virus (CML) and ectromelia virus (ECT) are important animal pathogens (12). The two other OPVs, taterapox virus (TPV) and Uasin Gishu virus, are not well characterized, and DNA of the latter is not available.
OPV taxonomy is based on biological characteristics, such as host range in animals, the reproductive ceiling temperature in cell culture or on chicken embryo chorioallantoic membrane (CAM), and the morphology of viral pocks produced on the CAM, and on genomic DNA restriction maps (2, 7, 8, 19, 26, 27). Other diagnostic methods have included differentiation by viral protein-profile following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and by various serologic assays (1, 28). All of these methods required virus isolation and propagation, and clear-cut differentiation was often difficult. Therefore, a test that can directly detect OPV in clinical specimens is desirable. More recently, PCR methods have been developed which eliminate the need to propagate virus (17, 22, 23, 31), but they were not suitable for strain differentiation. OPV strain differentiation is important for several practical applications including epidemiological investigations. OPV PCR tests were developed based on the determination of complete DNA sequences for VAC (13, 16) and VAR (20) and the availability of partial genomic sequences from other OPVs. These sequence data have shown that a large central genomic region is highly conserved among OPV isolates, which explains the significant degree of cross-reactivity in various tests (9, 11). However, the data also revealed that genes within the terminal regions have both conserved and variable segments, and several of these genes have been associated with host range, tissue tropism, and/or virulence of different OPV species and strains (13, 20, 21). Thus, the species-specific genes make these terminal variable regions ideal targets for use in PCR-based diagnostics.
The OPV cytokine response modifier B (crmB) gene encodes a tumor necrosis factor-binding protein that probably plays a significant role in virus pathogenesis of certain OPVs (data not shown). The crmB gene, encoding a functional full-length protein of CPV and VAR, has been sequenced (14, 20); however, the sequences of VAC strains indicated that the VAC crmB gene cognate is truncated (13). We determined the crmB gene sequences of 43 different OPVs and used them as the basis for a PCR-restriction fragment length polymorphism (RFLP) test and have examined various clinical samples to evaluate the ability of this PCR-RFLP assay to identify Old World OPVs. The test represents a rapid and accurate method for the detection and differentiation of these OPVs and was used at the World Health Organization Collaborating Center for Smallpox and Other Poxvirus Infections (Atlanta, Ga.) as a routine component of laboratory-based epidemiologic investigations, including the human monkeypox outbreaks in the Democratic Republic of Congo (Congo) in 1996 to 1997 (25).
MATERIALS AND METHODS
Viruses, DNA preparation and sequencing.
The origins of the 115 viruses, representing seven OPV species, used in this study have been described previously (2, 4, 5, 7, 12, 15, 18–26, 29, 30). Isolation of OPV DNA from purified virions has also been described in detail elsewhere (6). DNA from human lesion material was prepared using NucleoSpin tissue kits (Clontech Laboratories Inc., Palo Alto, Calif.). The primary structures of approximately 2-kbp amplicons overlapping the crmB gene region for 43 OPV strains of known taxonomic origin were sequenced with an ABI Prism Dye Terminator cycle-sequencing kit (Applied Biosystems, Foster City, Calif.) as specified by the manufacturer. These data were used to design a universal primer pair for OPV crmB region amplification. To evaluate the specificity of PCR amplification, the nucleotide sequences of selected amplicons were determined and compared with the crmB gene region sequences of the 43 OPV reference strains used to design the consensus PCR primers. The Genetics Computes Group, Madison, Wis. (3), DNASIS 2.1 (Hitachi Software, San Bruno, Calif.), and OLIGO 5 (NBI, Plymouth, Minn.) software suites were used for computer analysis of nucleotide sequences.
The OPV DNA sequences have been deposited in GenBank under accession no. U90225 to U90235 for CPV strains OPV89/1 (cat)-M5, OPV89/4 (cat)-M6, OPV89/5 (cat)-M7, OPV90/1 (cat)-M8, OPV90/5 (cat)-M9, Brighton clone Atlanta, M1, 58, OPV90/2 (human), and OPV89/2 (cat), respectively; U87837 to U87840 for CML strains Somalia-1978, Iran-M2 (CP-1), Saudi-M3, and Dubai CP5 1992, respectively; U88145 to U88152 for VAR major strains Harvey-1944, Bangladesh-1975, Congo-1970, and “Whitepox” ch9-2; African VAR minor strain Somalia-1977 and VAR minor alastrim strains Sierra Leone-1968, Butler-1952, and Garcia-1966, respectively; U87841 to U88147, U87994, U87995, U88142 to U88144, and U88543 for MPV strains Zaire-1996 (96-16), Liberia 1970(70-187), Sierra Leone 1970 (70-0266), Nigeria 1971 (71-0082), Zaire 1977 (77-0666), Benin 1978 (78-3945), Zaire 1979 (79-0005), Copenhagen 1958 clones CW-N1 and CV1, Zaire 1970 (CN8), Washington DC-1961 (WMP), Rotterdam 1965 (UTC), and Zaire-1996 (96-17), respectively; U86874 for TPV Dahomey-1971; U86380 and U86381 for ECT strains Moscow (MOS) and Mill Hill (MLH), respectively; and U86871 to U86873, U87232, U87233, U87584, and U87585 for VAC strains Columbia, Lister, Venezuela, Copenhagen (COP) wt, VAC subspecies Buffalopoxvirus (BUF) strains BUF 81 and BUF 3906, and rabbitpoxvirus (RPV) Utrecht, respectively.
PCR assay.
In standard PCRs 0.5 μg of each oligonucleotide of the primer pair VL2N (5′-ACATGCATGCCAGGAC) and VL33 (5′-ACCATTACAAACATTATCC), plus 50 ng of template DNA extracted from virion or cell culture were used. For clinical samples, PCRs employed the same amount of the primers plus a ∼1:50 portion of the DNA purified from a single crusted scab (∼5 mm in diameter). Reaction mixtures were passed through 25 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and polymerization at 72°C for 1 min, followed by a single extension step at 72°C for 5 min (model 9600 thermal cycler; Perkin-Elmer Cetus). Amplicons were resolved by electrophoresis in 1% agarose gels in TAE buffer (40 mM Tris-acetate [pH 8.0], 1 mM disodium EDTA). Restriction reactions were done at 37°C with 50μl of the PCR product adjusted to recommended endonuclease buffer and 10 U of NlaIII, AciI, NciI, or Sau3A (New England Biolabs, Inc., Beverly, Mass.). Endonuclease-cleaved DNA products were resolved by gel electrophoresis in TAE buffer using a mixture of 3% NuSieve-GTG agarose gels and 1% SeaKem-GTG agarose (FMC Corp., Marine Colloids Division, Rockland, Maine) in TAE buffer. The 1-kb and 100-bp DNA ladders (Gibco, Gaithersburg, Md.) were used as DNA size markers.
RESULTS AND DISCUSSION
crmB region PCR amplification of different OPV species.
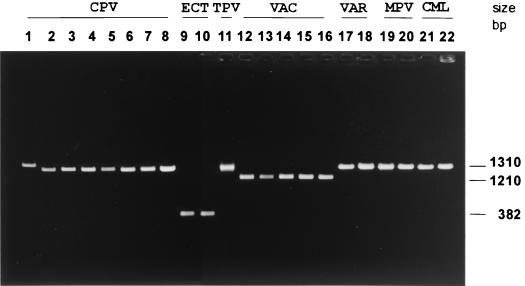
As shown in Fig. 1, the consensus primers VL2N and VL33 successfully amplified the crmB region from DNA of eight CPV, two ECT, one TPV, five VAC, two VAR, two MPV, and two CML strains, all established OPV isolates with known histories and taxonomic designations, and these were selected as reference strains for the present study. A single amplicon was produced for each strain of CPV (1,291 to 1,369 bp), ECT (380 bp), TPV (1,313 bp), VAC (1,201 to 1,212 bp), VAR (1,308 to 1,311 bp), MPV (1,317 to 1,323 bp), or CML (1,315 bp). The distinct size of the ECT and VAC amplicons differentiated these species without further manipulation; however, the amplicons produced from CPV, TPV, VAR, MPV, and CML have similar electrophoretic mobilities and cannot be used to identify these species.
FIG. 1.
Identification and differentiation of OPVs by PCR amplification of the crmB gene region from genomic DNA. Amplicons of the crmB gene region were obtained with consensus sequence primer pair VL2N-VL33. The sizes of PCR products were determined by direct sequencing of amplified products (18) to be as follows: CPV Brighton, Atlanta (1,369 bp) (lane 1), CPV strain M1 (1,307 bp) (lane 2), CPV strain M5 (1,297 bp) (lane 3), CPV strain M6 (1,322 bp) (lane 4), CPV strain M7 (1,312 bp) (lane 5), CPV strain M8 (1,309 bp) (lane 6), CPV strain M9 (1,291 bp) (lane 7), CPV strain 58 (1,309 bp) (lane 8), ECT MOS (382 bp) (lane 9), ECT MLH (382 bp) (lane 10), TPV Dahomey-1971 (1,303 bp) (lane 11), VAC Lister (1,208 bp) (lane 12), VAC Columbia (1,208 bp) (lane 13), BUF India 81-1985 (1,212 bp) (lane 14), BUF India-3906 (1,208 bp) (lane 15), RPV Utrecht (1,208 bp) (lane 16), VAR major Bangladesh-1975 (1,308 bp) (lane 17), VAR minor alastrim Brazil-Garcia-1966 (1,311 bp) (lane 18), human MPV Congo-8 (1,317 bp) (lane 19), MPV Copenhagen (1,318 bp) (lane 20), CML Somalia-1978 (1,315 bp) (lane 21), and CML (CP-5) Dubai-M4 (1,315 bp) (lane 22).
We defined the limit of crmB DNA detection as the amount of sample template DNA that produced an amplicon clearly visible by standard ethidium bromide staining of the resolving gel. Serial dilutions of highly purified DNA from VAR strain Bangladesh, VAC strain Columbia, CML strain Somalia, CPV strain Brighton, and ECT strain Moscow indicated that ∼150 genomes of template DNA were needed to produce a visible amplicon (data not shown). In addition, we confirmed that the VL2N and VL33 primers are specific for the seven OPV species, since they failed to amplify any PCR product from genomic DNA of parapoxvirus, yatapoxvirus, suipoxvirus, leporipox, and avipoxvirus isolates or from cellular DNA isolated from uninfected human, monkey, rabbit, mouse, or rat tissue culture cells (data not shown). Moreover, template DNA from the New World OPVs raccoonpox, volepox, and skunkpox viruses did not produce amplicons using the VL2N and VL33 primers. In this regard, we reported a protocol based on amplification of the gene for the hemagglutinin protein with a primer pair specifically designed to detect the New World OPVs (31).
RFLP analysis.
Amplicons from CPV, TPV, VAR, MPV, and CML DNAs were further analyzed by RFLP. We used a number of restriction enzymes to digest the 43 determined sequences, searching for an endonuclease that would produce different digest profiles for each OPV species. Sequence analysis indicated that the restriction endonuclease NlaIII would be useful for the identification of all seven Old World OPV species and additionally useful for the differentiation of CML, VAC, and CPV strains. For MPV and VAR strain differentiation, Sau3A and AciI restriction enzymes, respectively, were identified. In the following paragraphs, the comparative results of RFLP assays for each OPV species tested are described. For this phase of the study, we used DNAs from 115 different Old World OPVs to establish the reproducibility and fidelity of the method.
(i) VAR.
Figure 2 (lanes 1 and 2) shows NlaIII RFLP profiles of the crmB gene region for two reference VAR strains (VAR major Bangladesh-1975 and VAR minor alastrim Garcia-1966) compared with RFLP profiles of other OPV species. For these VAR strains, three NlaIII RLFP digest fragments of 798, 336, and 85 bp or 821, 339, and 85 bp were resolved, respectively. These data reflect the patterns seen during examination of all 34 VAR samples from the Centers for Disease Control and Prevention Smallpox Virus Specimen and DNA Repository, collected worldwide from 1944 to 1977, including VAR major strains Harvey-1944, Bangladesh-1975 (isolates VAR-Jalaluddin, VAR-Hawa, VAR-Parvin, VAR-Mannan, VAR-Shahzaman, VAR-Chandra, and VAR-Solaiman), and Congo-1970; African VAR minor strain Somalia-1977; VAR minor alastrim strains Sierra Leone-1968, Butler-1952, Garcia-1966, and isolates VAR-Kudano and Nigeria-1961; VAR-Level, Liverpool-1958; VAR-Ethiopia no.17 (R14-1x-72); VAR-Ethiopia no.16 (R14-1x-72); VAR-Ilem Bula Tanzania-1965; Variolator no.4 Afghanistan-1971; Variolator no.2 Afghanistan-1972; Variolator no.3 Afghanistan-1972; VAR-Ratia-1969; VAR-696 Madras DJB; VAR-Nur Islam (28-I-75); VAR-Aba Hashem (28-I-75); VAR-Herrlich, Bombey-1958; and “Whitepox” viruses ch9, ch9-2, ch9-4, and ch9-18 (data not shown).
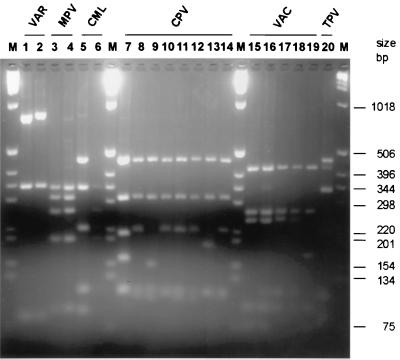
FIG. 2.
Comparative NlaIII RLFP analysis of PCR-amplified crmB fragments from intact genomes of OPV isolates. NlaIII fragment sizes noted in parentheses were determined by direct sequence analysis of the crmB gene region (18) and agreed with sizes of fragments calculated by comparison to the 100-bp DNA ladder size marker (M). Lanes: 1, VAR major Bangladesh-1975; 2, VAR minor alastrim Brazil-Garcia-1966; 3, human MPV Congo-8; 4, human MPV 71-0082; 5, CML Somalia-1978; 6, CML (CP-5) Dubai-M4; 7, CPV Brighton; 8, CPV strain M1; 9, CPV strain M5; 10, CPV strain M6; 11, CPV strain M7; 12, CPV strain M8; 13, CPV strain M9; 14, CPV strain 58; 15, VAC Lister; 16, VAC Columbia; 17, BUF India-81-1985; 18, BUF India-3906; 19, RPV Utrecht; 20, TPV Dahomey-1971.
As shown in Fig. 2, the NlaIII RFLP assay clearly differentiated VAR from other OPV species. No significant differences were observed in NlaIII digest patterns of VAR major, minor, or alastrim strains, which suggested a high degree of conservation of crmB in VAR. However, nucleotide sequence of VAR crmB amplicons indicated that VAR major and alastrim strains can be differentiated by RFLP using AciI (Fig. 3). The crmB gene of most VAR amplicons showed four AciI sites, except for the alastrim isolates, which showed three AciI sites. Indeed AciI RFLP profiles differentiated alastrim from other VAR strains. Alastrim VAR minor strain crmB amplicons from Garcia-1966, Sierra Leone-1968, and Butler-1952 all produced digest fragments of 626, 374, 170, and 168 bp following AciI cleavage. VAR major strains Bangladesh-1975, Congo-1970, and Harvey-1944 and the low case-fatality VAR virus strain Somalia-1977 produced 377-, 347-, 249-, 170-, and 168-bp AciI fragments. These RFLP results agreed with the epidemiological and biological analysis of VAR alastrim strains (12). It is also noteworthy that 22 of the DNA samples used in this study were isolated from lesions of smallpox patients. These indicated that the current method is suitable for VAR detection and differentiation in clinical material. The other important finding was that VAR DNA can be isolated from scabs, even after 40 years of cold storage, as shown for the VAR-Herrlich (Bombey-1958) sample.
FIG. 3.
RLFP analysis after AciI digestion of PCR-amplified crmB fragments from genomes of VAR isolates. Lanes: 1, VAR major strain Bangladesh-1975; 2, VAR major strain Congo-1970; 3, VAR major Harvey-1944; 4, African VAR minor Somalia-1977; 5, “Whitepoxch9-2; 6, “Whitepox” ch9-4; 7, alastrim VAR minor strain Sierra Leone-1968; 8, alastrim VAR minor Garcia-1966.
We also were able to confirm that sample “Lenny,” taken from a set of suspected VAR scab materials, contained only VAC DNA.
(ii) MPV.
Figure 2 (lanes 3 and 4) shows NlaIII RFLP profiles of crmB amplicons for two reference strains of human MPV in comparison with other OPV species. NlaIII digestion of MPV amplicons produced fragments of 336, 304, 265, 200, and 93 bp. This set of fragments was observed for all 21 MPV isolates collected from 1958 to 1998, including MPV strains isolated from humans: Congo strains MPV 74-0226, MPV 77-0666, MPV 79-0005, Congo-8, MPV CnCR, MPV CnCwN1, MPV CnCwN2, MPV Zaire 96-16, MPV DR Congo 97, and MPV DR Congo 98; and West African strains MPV Liberia 70-0187, Sierra Leone 70-0266, MPV Nigeria 71-0082, and MPV Benin 78-3945; and strains isolated from monkeys (MPV PVM, MPV Copenhagen, MPV CpCR, MPV CpCWN1, MPV CpCWN2, MPV PCH, MPV WMP, MPV UTC, MPV HMX, and MPV CpMR). In addition, MPV DNA was detected by characteristic NlaIII RFLP profiles in 15 DNA template samples that were extracted from crusted scabs or vesicular pustule fluid from patients infected during 1997 (25) in the largest human MPV outbreak ever observed in the Democratic Republic of Congo. These results suggested a high level of MPV crmB sequence conservation and clear-cut differentiation of MPV from the other OPVs.
Further sequence analysis indicated that it should be possible to differentiate MPV strains by digesting crmB amplicons with Sau3A. This was verified by the detection of 404-, 365-, 356-, and 191-bp digest fragments using MPV crmB amplicons for all but the Congo isolates, which included a predicted additional Sau3A site for production of 299- and 66-bp fragments, instead of the 365-bp fragment (Fig. 4). This analysis confirmed the utility of DNA genome restriction mapping for differentiating Congo and non-Congo MPV isolates (7, 12, 19).
FIG. 4.
RLFP analysis after Sau3A digestion of PCR-amplified crmB fragments from genomes of MPV isolates. Lanes: 1, MPV Zaire 74-0226; 2, MPV Zaire 77-0666; 3, MPV Zaire 79-0005; 4, MPV Congo-8; 5, MPV Liberia 70-187; 6, MPV Zaire 96-16; 7, MPV Sierra Leone; 8, MPV Nigeria 71-0082; 9, MPV Benin-1978 78-3945; M, size marker (100-bp DNA ladder).
(iii) CML.
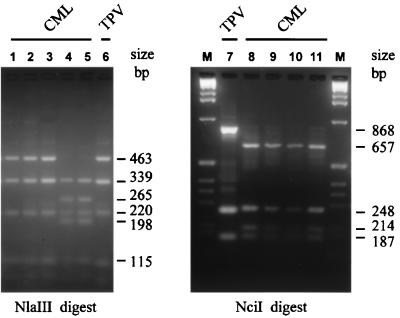
Figure 2 (lanes 5 and 6) shows the NlaIII profiles for CML Somalia and Dubai, and Fig. 5 shows the CML Somalia NlaIII profile compared with the profile of crmB amplicons from the DNA of three other CML isolates and TPV. Figure 5 also shows CML Somalia NciI profiles compared with the NciI profiles of three other CML isolates and TPV. The origin of the CML isolates has been discussed previously (29, 30). The NlaIII digest of crmB of the African CML strains Somalia, Mauretania, and Niger produced fragments of 463, 339, 220, 115, and 91 bp. The presence of an additional NlaIII site in the amplicons of Asian CML strains Iran, Saudi Arabia, and Dubai results 265- and 198-bp fragments, replacing the 463-bp fragment found in African CML strains. Sequence analysis of CML crmB indicated that the DNA homology between CML strains was about 99%, equivalent to the percent crmB identity between VAR and MPV isolates.
FIG. 5.
RLFP analysis of PCR-amplified crmB fragments from genomes of CML and TPV isolates. NlaIII digest fragments of the PCR products of CML Mauretania (lane 1), CML Somalia-1978 (lane 2), CML Niger (lane 3), CML (CP-1) Iran-M2 (lane 4), CML-Saudi-M3 (lane 5), and TPV Dahomey-1971 (lane 6) are shown in the left panel. NciI digest fragments of the PCR products of TPV Dahomey-1971 (lane 7), CML Somalia-1978 (lane 8), CML Niger (lane 9), CML Mauretania (lane 10), and CML Iran-M2 (lane 11) are shown in the right panel. Size markers are provided by a 100-bp DNA ladder (lane M).
(iv) TPV.
Interestingly, NlaIII digestion of the TPV crmB amplicon produced a restriction pattern that differed from the NlaIII digest patterns of most of the OPVs tested in this study, except for those of the African strains of CML (Fig. 2 and 5). DNA sequence analysis indicated that NciI digestion would enable the separation of TPV and African CML crmB, and, as shown in Fig. 5 (lanes 7 to 11), the NciI digest patterns indeed differentiated these viruses.
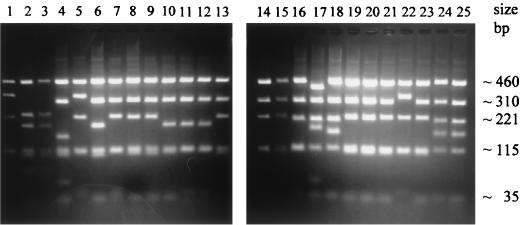
(v) CPV.
Figure 2 (lanes 7 to 14) shows NlaIII digest profiles of crmB amplicons for CPV Brighton and seven other CPV isolates in comparison with other OPV species, and Fig. 6 shows the NlaIII digest profiles of reference strain CPV Brighton compared with 23 other CPV isolates and CPV Brighton strain that has been maintained in Germany. A variation in size of the CPV crmB amplicons was reflected in results of the RFLP assay. Several different RFLP patterns were detected among 24 CPV isolates from humans and seven animal species. However, NlaIII digestion of the crmB amplicon from most CPV isolates produced fragments of 460, 310, 221, 115, and 108 bp (Fig. 2, lanes 7 to 14). However, as Fig. 6 shows, digestion of the crmB amplicons of 24 CPV isolates revealed seven different RFLP patterns that were grouped as A to G: A (lanes 1, 5, and 22) produced fragments of 460, 345, 223, and 115 bp; B (lanes 2 and 3) produced fragments of 460, 223, 190, 115, 109, and 75 bp; C (lane 4) produced fragments of 463, 308, 153, 115, and 108 bp; D (lanes 6 and 10 to 12) produced fragments of 463, 310, 190, 115, and 108 bp; E (lanes 7 to 9, 13 to 16, 19 to 21, and 23) produced fragments of 460, 310, 223, 115, and 108 bp; F (lane 17) produced fragments of 400, 310, 220, 190, and 115 bp; and G (lanes 18, 24, and 25) produced fragments of 472, 310, 212, 168, and 115 bp. Thus, RFLP of crmB amplicons appears useful for strain differentiation of CPV isolates.
FIG. 6.
NlaIII RLFP analysis of PCR-amplified crmB fragments from DNA of CPV isolates from human and animals. NlaIII digest fragments after PCR amplification of OPV85 (human) (lane 1), OPV88/L (cat) (lane 2), OPV88/H (cat) (lane 3), OPV89/1 (cat)-M5 (lane 4), OPV89/2 (cat) (lane 5), OPV89/3 (cat) (lane 6), OPV89/4 (cat)-M6 (lane 7), OPV89/5 (cat)-M7 (lane 8), OPV90/1 (cat)-M8 (lane 9), OPV90/2 (human) (lane 10), OPV90/4 (dog) (lane 11), OPV90/5 (cat)-M9 (lane 12), OPV91/1 (cat) (lane 13), OPV91/2 (human) (lane 14), OPV91/3 (cow) (lane 15), CATPOX3 (lane 16), CATPOX5 (lane 17), RAT Moscow (rat) (lane 18), EP-1 (elephant) (lane 19), EP-2 (elephant)-M1 (lane 20), EP-3 (elephant) (lane 21), EP-4 (elephant) (lane 22), EP-5 (elephant) (lane 23), CPV BRT-Atlanta (lane 24), and CPV BRT-Munich (lane 25) are shown.
In contrast to data obtained for VAR, MPV, and CML strains described above, there was no correlation between the restriction patterns and the temporal or geographic origin of the CPV isolates. Moreover, the different animal origins of the CPV isolates did not correlate with the pattern differences. These results and extensive sequence data analysis suggested that the crmB gene may not be necessary for CPV virus propagation (at least for some strains) and is markedly divergent. In this respect, the mutation level for CPV crmB regions, which encode full-length crmB proteins, is comparable to the mutation level for the crmB gene regions of VAC strains (see below), which express truncated CrmB proteins due to several deletions and frameshifts in the crmB open reading frames. However, all of the CPV strains we examined contain full-length crmB open reading frames. It is possible that the divergence of CPV crmB reflects the adaptation to different hosts. We also cannot rule out the possibility that some of these CPV isolates represent new species of OPV. The phylogenetic relationships among these and other CPV strains available is under investigation to determine the extent of subspecies divergence among CPVs circulating in Europe.
(vi) VAC.
NlaIII digestion of VAC crmB amplicons (∼1,202 to 1,212 bp) from 18 different VAC strains, including 2 isolates each of the VAC subspecies BUF and RPV, produced four digest fragments (Fig. 2, lanes 15 to 19, and Fig. 7), which differentiate VAC strains. Most of the variation among strains was apparent in the different mobility associated with the third migrating NlaIII DNA fragment, which ranged in size from 243 to 97 bp. For example, VAC Lister (lane 3) showed a NlaIII fragment of 243 bp, VAC-VNV (lane 4) showed a 236-bp correlate, and RPV (lanes 4, 10, and 15) showed a 171-bp correlate. In addition, differences in NlaIII digest patterns for VAC COP amplicon (lane 13) and VAC Wyeth (lane 6) produced 426-, 267-, 171-, 115-, 96-, and 72-bp or 420-, 267-, 220-, 115-, and 97-bp fragment sets, respectively (lanes 6 and 13). Moreover, NlaIII digestion of crmB amplicons of BUF 81 and BUF 3906 (lanes 8 and 9) produced a third fragment of 115 bp, and the 77-bp fragment seen in most other strains was absent. In general, RFLP analysis of the crmB gene region from VAC, RPV, and BUF isolates suggests that deletions and truncations have occurred that cause significant variations in restriction patterns for the different isolates, allowing strain and subspecies differentiation. Sequence comparisons indicate that these alterations impair functional protein production due to several kinds of frameshifts (13, 16).
FIG. 7.
NlaIII RLFP analysis of PCR-amplified crmB fragments from DNA of VAC variants. NlaIII digest fragments of the PCR products of VAC COL (lane 1), VAC V (lane 2), VAC LIST (lane 3), VAC VNV (lane 4), VAC VCX (lane 5), VAC Wyeth (lane 6), VAC CV1-78 (lane 7), BUF 81 (lane 8), BUF 3906 (lane 9), RPV UTR (lane 10), Levaditi (lane 11), VAC M1 (lane 12), VAC COP wt (lane 13), VAC COP hr (lane 14), RPV Utrecht (lane 15), VAC Hagen (lane 16), VAC CVA (lane 17), VAC WR (lane 18), VAC Elstree (lane 19), and BUF BP-1 (lane 20) are shown.
As mentioned above, in the discussion of RFLP analysis of VAR samples, we determined that the suspected variola case Lenny, is a severely malnourished Nigerian woman who died after a smallpox-like illness with fever and generalized rash (10), was actually a VAC infection. The origin of this strain is obscure. However, as found (data not shown), the NlaIII digestion pattern of the Lenny crmB amplicon resembles the distinctive NlaIII electrophoretic profile of the crmB amplicon from the VAC Wyeth strain (Fig. 7, lane 6), which was being used as smallpox vaccine in Nigeria at that time. The initial investigation of this case showed differences between the Wyeth vaccine strain and the Lenny isolate in some of their biological properties. Presumably Lenny was a naturally occurring variant of the Wyeth vaccine strain or a contaminant selected during isolation of the virus from the infected patient (10). In general, the major conclusion of RFLP analysis for VAC samples is that the crmB gene is truncated and does not produce a functional protein; as a direct consequence, significant variations in restriction patterns are observed for different isolates of VAC, and these can be used for VAC strains and subspecies differentiation.
(vii) ECT.
Although ECT could be clearly differentiated from all other viruses tested by the distinctive size of the crmB amplicons (Fig. 1, lanes 9 and 10), the differentiation could be further confirmed using NlaIII RFLP analysis. On electrophoregrams (data not shown), cleavage of the ECT MOS and ECT MLH crmB amplicon with NlaIII produced fragments of 176 and 110 bp. Moreover, identical results were obtained with six other isolates of ECT tested : ECT MP-1 through ECT MP-5 (mouse) and ECT SF (farm silver fox). We noted that the relatively conservative primary structure of ECT crmB amplicons could be explained by complete elimination of crmB amino acid coding sequences. ECT amplicons in general contain only the regulatory signals for genes upstream and downstream of the crmB gene, which are important for ECT replication in infected hosts and are therefore conserved in these isolates (24).
This report describes a rapid and accurate PCR method based on the crmB gene for identifying OPVs and for differentiating species and strains in this group of viruses. The method uses RFLP analysis of PCR products produced from a consensus primer pair designed to amplify the crmB gene open reading frame. Depending on the OPV subgroup, amplification produces a PCR product that indicates the presence of OPV DNA and additionally distinguishes both ECT and VAC strains from other OPV species on the basis of amplicon size. Subsequent digestion with restriction endonucleases enables further differentiation, if required, of VAR, MPV, CML, VAC, and CPV species. In some instances, the resulting digestion fragments permit further classification of subspecies and strains.
Finally, our data confirm that PCR-RFLP analysis of the crmB gene region is a fast and accurate method for the identification and differentiation of Old World OPV isolates. The reliability and sensitivity of the new method were demonstrated by its successful use with VAR, VAC, and MPV clinical specimens. The RFLP patterns and sequencing data of the crmB gene region for numerous representative specimens of OPV isolates constitute valuable reference data for further genotyping of OPV isolates. It is important to note that where ambiguities prevent a clear-cut differentiation by RFLP, the amplicon should be sequenced and compared with reference OPV crmB sequences to confirm the identity of the strain. This and previously described PCR methods for OPV diagnostics (22, 31) provide a suitable assortment of tests for the detection and confirmation of OPV infections, thus increasing the specificity and confidence of new molecular assays as tools in the worldwide surveillance for emerging OPV infections.
ACKNOWLEDGMENTS
We are grateful to Janice Knight, Keith Dumbell, Deiter Burger, Mark Buller, Samuel Dales, and Yasuo Ichihashi for viruses and DNA samples used in the research. We thank the technical staff of the CDC Biotechnology Core Facility for oligonucleotide preparations.
REFERENCES
- 1.Arita M, Tagaya I. Structural polypeptides of several strains of orthopoxvirus. Microbiol Immunol. 1977;21:343–346. doi: 10.1111/j.1348-0421.1977.tb00298.x. [DOI] [PubMed] [Google Scholar]
- 2.Baxby D. Identification and interrelationships of the variola/vaccinia subgroup of poxviruses. Prog Med Virol. 1975;19:215–246. [PubMed] [Google Scholar]
- 3.Devereux J. The GCG sequence analysis software package. Madison, Wis: Genetics Computer Group; 1993. [Google Scholar]
- 4.Dumbell K R, Huq F. The virology of variola minor: correlation of laboratory tests with the geographic distribution and human virulence of variola isolates. J Epidemiol. 1986;123:403–415. doi: 10.1093/oxfordjournals.aje.a114255. [DOI] [PubMed] [Google Scholar]
- 5.Dumbell K R, Richardson M. Virological investigations of specimens from buffaloes affected by buffalopox in Maharashtra State, India, between 1985 and 1987. Arch Virol. 1993;128:257–267. doi: 10.1007/BF01309438. [DOI] [PubMed] [Google Scholar]
- 6.Esposito J J, Condit R, Obijeski J F. The preparation of orthopoxvirus DNA. J Virol Methods. 1981;2:175–179. doi: 10.1016/0166-0934(81)90036-7. [DOI] [PubMed] [Google Scholar]
- 7.Esposito J J, Knight J C. Orthopoxvirus DNA: a comparison of restriction profiles and maps. Virology. 1985;143:230–251. doi: 10.1016/0042-6822(85)90111-4. [DOI] [PubMed] [Google Scholar]
- 8.Esposito J J, Massung R F. Poxvirus infections in humans. In: Murray P R, Tenover F, Baron E J, Pfaller M A, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 1131–1138. [Google Scholar]
- 9.Fenner F. Poxviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press, Inc.; 1996. pp. 2673–2702. [Google Scholar]
- 10.Fenner F, Henderson D A, Arita I, Jezek A, Ladnyi I D. Smallpox and its eradication. Geneva, Switzerland: World Health Organization; 1988. [Google Scholar]
- 11.Fenner F, Nakano J H. Poxviridae: the poxviruses. In: Lennette E H, Halonen P, Murphy F A, editors. Laboratory diagnosis of infectious diseases, vol. 2. Viral, rickettsial and chlamydial diseases. New York, N.Y: Springer-Verlag; 1988. pp. 177–207. [Google Scholar]
- 12.Fenner F, Wittek R, Dumbell K R. The orthopoxviruses. New York, N.Y: Academic Press, Inc.; 1989. [Google Scholar]
- 13.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 14.Hu F-Q, Smith C A, Pickup D J. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 15.Ichihashi Y, Dales S. Biogenesis of poxviruses: Interrelationship between hemagglutinin production and polykaryocytosis. Virology. 1971;46:533–543. doi: 10.1016/0042-6822(71)90057-2. [DOI] [PubMed] [Google Scholar]
- 16.Johnson G P, Goebel S J, Paoletti E. An update on the vaccinia virus genome. Virology. 1993;196:381–401. doi: 10.1006/viro.1993.1494. [DOI] [PubMed] [Google Scholar]
- 17.Knight J C, Massung R F, Esposito J J. Polymerase chain reaction identification of smallpox virus. In: Becker Y, Darai G, editors. Diagnosis of human viruses by polymerase chain reaction technology. 2nd ed. Berlin, Germany: Springer-Verlag KG; 1995. pp. 297–302. [Google Scholar]
- 18.Lourie B, Nakano J H, Kemp G E, Setzer H W. Isolation of poxvirus from an African rodent. J Infect Dis. 1975;132:677–681. doi: 10.1093/infdis/132.6.677. [DOI] [PubMed] [Google Scholar]
- 19.Mackett M, Archard L C. Conservation and variation in orthopoxvirus genome structure. J Gen Virol. 1979;45:683–701. doi: 10.1099/0022-1317-45-3-683. [DOI] [PubMed] [Google Scholar]
- 20.Massung R F, Esposito J J, Liu L, Jin Q, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, Selivanov N A, Cavallaro K F, Kerlavage A R, Mahy B W J, Venter J C. Potential virulence determinants in terminal regions of variola smallpox virus. Nature. 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 21.Massung R F, Loparev V N, Knight J C, Totmenin A V, Chizhikov V E, Parsons J M, Safronov P F, Gutorov V V, Shchelkunov S N, Esposito J J. Terminal region sequence variations in variola virus DNA. Virology. 1996;221:291–300. doi: 10.1006/viro.1996.0378. [DOI] [PubMed] [Google Scholar]
- 22.Meyer H, Pfeffer M, Rziha H. Sequence alterations within and downstream of the A-type inclusion protein genes allow differentiation of orthopoxvirus species by polymerase chain reaction. J Gen Virol. 1994;75:1975–1981. doi: 10.1099/0022-1317-75-8-1975. [DOI] [PubMed] [Google Scholar]
- 23.Meyer H, Ropp S L, Esposito J J. Gene for A-type inclusion body protein is useful for a polymerase chain reaction assay to differentiate orthopoxviruses. J Virol Methods. 1997;64:217–221. doi: 10.1016/S0166-0934(96)02155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields' virology. 3rd ed. New York, N.Y: Raven Press, Inc.; 1996. pp. 2637–2671. [Google Scholar]
- 25.Mukinda V B K, Mwema G, Kilundu M, Heymann D L, Khan A S, Esposito J J, Koen H, Delfi M, Muyembetamfum J J, Kweteminga T F, Moudi A, Mangindula L, Loparev V N, Parsons J M, Jue D L, Crews T W, Knight J C. Re-emergence of human monkeypox in Zaire in 1996. Lancet. 1997;349:1449–1450. doi: 10.1016/S0140-6736(05)63725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano J H. Poxviruses. In: Lennette E H, Schmidt N J, editors. Diagnostic procedures for viral, rickettsial, and chlamydial infections. 5th ed. Washington, D.C.: American Public Health Association, Inc.; 1979. pp. 257–308. [Google Scholar]
- 27.Nakano J H. Human poxvirus diseases and laboratory diagnosis. In: de la Maza L M, Peterson E M, editors. Medical virology. New York, N.Y: Elsevier; 1982. pp. 125–147. [Google Scholar]
- 28.Obijeski J F, Palmer E L, Gafford L G, Randall C C. Polyacrylamide gel electrophoresis of fowlpox and vaccinia virus proteins. Virology. 1973;51:512–516. doi: 10.1016/0042-6822(73)90454-6. [DOI] [PubMed] [Google Scholar]
- 29.Pfeffer M, Meyer H, Wernery U, Kaaden O-R. Comparison of camelpox viruses isolated in Dubai. Vet Microbiol. 1996;49:135–146. doi: 10.1016/0378-1135(95)00181-6. [DOI] [PubMed] [Google Scholar]
- 30.Renner-Muller I C E, Meyer H, Munz E. Characterization of camelpoxvirus isolates from Africa and Asia. Vet Microbiol. 1995;45:371–381. doi: 10.1016/0378-1135(94)00143-k. [DOI] [PubMed] [Google Scholar]
- 31.Ropp S L, Jin Q, Knight J C, Massung R F, Esposito J J. Polymerase chain reaction strategy for identification and differentiation of smallpox and other orthopoxviruses. J Clin Microbiol. 1995;33:2069–2076. doi: 10.1128/jcm.33.8.2069-2076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]