Abstract
The prevalence of recently described mutation V176F, located in the beginning of the rpoB gene and associated with rifampin resistance and the wild-type cluster I sequence, was determined by analyzing the distribution of rpoB mutations among 80 rifampin (RIF)-resistant Mycobacterium tuberculosis strains isolated in Germany during 1997. The most frequent rpoB mutations were changes in codon 456 (52 isolates, 65%), followed by changes in codon 441 (13 isolates, 16%) and codon 451 (11 isolates, 14%). The V176F mutation was detected in one isolate of the study population and in 5 of 18 RIF-resistant strains with no cluster I mutation from six previously published studies. In three isolates, a mixture of resistant and susceptible subpopulations (heteroresistance) prohibited the detection of rpoB mutations in the initial analysis; however, in these isolates, cluster I mutations could be verified after a passage on RIF-containing medium. IS6110 DNA fingerprinting of 76 strains revealed eight clusters comprising 27 strains with identical restriction fragment length polymorphism patterns that mainly also show identical rpoB mutations and identical or similar drug resistance patterns. In conclusion, our results indicate that the V176F mutation should be included in molecular tests for prediction of RIF resistance in M. tuberculosis. We further demonstrated that heteroresistance caused by a mixture of mycobacterial subpopulations with different susceptibilities to RIF may influence the sensitivity of molecular tests for detection of resistance.
One of the most alarming trends concerning tuberculosis (TB) is the emergence of drug-resistant Mycobacterium tuberculosis strains, which has become a worldwide health care problem (13). Drug-resistant TB is increasing in many parts of the world, and high rates of drug-resistant and multidrug-resistant TB-causing (MDR-TB) isolates (resistant to at least isoniazid [INH] and rifampin [RIF]) have been reported in several countries (16). So far in Germany, the incidence of TB, and also of drug resistance, is low compared to international data (16), but this situation may change because of importation of drug-resistant strains from countries with high rates of drug resistance, e.g., the former Soviet Union (11).
Early detection of drug resistance in clinical M. tuberculosis isolates is crucial for appropriate treatment to prevent the development of further resistance and the spread of resistant strains. Compared to conventional methods using solid media, the introduction of manual and automated methods for susceptibility testing in liquid media has resulted in a reduction of turnaround times for susceptibility results from 4 to 6 weeks to 3 to 15 days (6, 17). More recently, the identification of resistance mutations, e.g., the genetic basis for RIF resistance, enables the development of molecular tests which may allow the detection of resistant strains within 1 day (18).
RIF is one of the most potent antituberculous drugs. More than 90% of RIF-resistant TB-causing isolates are also resistant to INH, and RIF resistance is therefore a valuable surrogate marker for MDR-TB (15), which is a tremendous obstacle for TB therapy. In contrast to INH resistance, resistance to RIF is associated mainly with single point mutations in a small 81-bp hot-spot region (codons 432 to 458, cluster I) of the RNA polymerase gene (rpoB) (18) that can easily be amplified by PCR. Thus, several rapid molecular assays for prediction of RIF resistance targeting these mutations have been established or are under investigation (12). However, the detection of RIF resistance by these methods fails to match with a resistant phenotype in about 5% of all cases (15). Recently, we have reported a mutation in the beginning of the rpoB gene conferring resistance to RIF on Helicobacter pylori (V149F) and M. tuberculosis (V176F) (5). The amino acid exchange is located amino terminal to cluster I in M. tuberculosis and may account for false-negative test results obtained by molecular methods that only detect mutations within the cluster I region. Furthermore, we were able to show that, as described for Escherichia coli, mutations in clusters II and III located carboxy terminal to cluster I can reduce the susceptibility of H. pylori to rifamycins (4).
To determine the frequency of the V176F mutation, we analyzed the prevalence of mutations inside and outside of the cluster I region of the rpoB gene in a collection of RIF-resistant clinical M. tuberculosis isolates during a 1 year (1997) survey at the German National Reference Center for Mycobacteria (NRC). Moreover, 18 samples of RIF-resistant M. tuberculosis strains, 16 DNA samples, and 2 culture samples described in six previous reports (2, 3, 7, 10, 21, 22) to have no cluster I mutation were obtained and analyzed.
(Parts of the results presented here were presented as a poster at the 100th General Meeting of the American Society for Microbiology in Los Angeles, Calif. [section U-2]).
MATERIALS AND METHODS
Strains analyzed.
In total, 93 RIF-resistant M. tuberculosis isolates from 80 patients obtained during 1997 by the NRC were analyzed in this study. Moreover, 18 DNA samples, including 2 culture samples, of RIF-resistant strains with no known cluster I mutation provided by six other centers were investigated (Table 1).
TABLE 1.
rpoB codon exchanges in DNA samples from RIF-resistant clinical isolates without cluster I mutations described in recent studies
| Reference | Isolatea | MICc (μg/ml) | Mutation | Origin | Total no. of Rifr strains analyzed |
|---|---|---|---|---|---|
| 7 | 1Mya105 | >40 | Asia | 90 | |
| 2Ye16 | >40 | ||||
| 3Indn2 | >40 | ||||
| 21 | 69 | 1 | Japan | 95 | |
| 71 | 2 | ||||
| 72 | 4 | ||||
| 75 | 8 | V176F | |||
| 83 | 32 | V176F | |||
| 84 | 32 | V176F | |||
| 3 | 8872 | >2/>1 | V176F | Rwanda | 177 |
| 3662–84 | >2/>1 | New York | |||
| 3518–84 | >2/>1 | ||||
| 1875–65 | >2/>1 | Colorado | |||
| 2 | F364/90 | >2 (256)b | V176F | Mozambique | 41 |
| 362/91 | >2 (64)b | I497F (cluster II) | |||
| 10 | 3200 | >2 | Durban, South Africa | 113 | |
| 1630 | >8 | ||||
| 22 | R17 | >256 | Australia | 33 |
Strain designations are those in the original publications.
Available for MIC determination in this study.
RIF MICs determined or resistance breakpoint concentrations used in the respective study.
Primary isolation and culturing of mycobacterial isolates obtained from the NRC were performed as described elsewhere (9). All isolates were identified as M. tuberculosis using gene probes as described by the manufacturer (ACCUProbe; Gen-Probe, San Diego, Calif.) and standard biochemical procedures (9). Drug susceptibility was determined by the proportion method on Löwenstein-Jensen medium in accordance with the Deutsches Institut für Normung (DIN) guidelines and/or the modified proportion method in BACTEC 460TB in accordance with the manufacturer's instructions. MIC testing of selected isolates was performed with Löwenstein-Jensen slants with serial dilutions of RIF and rifabutin by following the DIN guidelines (DIN 58943-8).
PCR amplification conditions.
For analysis of the presence of the V176F mutation, a 365-bp fragment of the rpoB gene was amplified using primers TB-176-F (5′-CTTCTCCGGGTCGATGTCGTTG-3′) and TB-176-R (5′-CGCGCTTGTCGACGTCAAACTC-3′) as recently described (5). Five microliters of a 200-fold dilution (in double-distilled water) of total bacterial DNA (see DNA genotyping techniques) was used per PCR mixture.
Amplification of a 749-bp rpoB DNA fragment comprising the complete region from cluster I to cluster III was carried out with primers TBB-1 (5′ ATCACACCGCAGACGTTG-3′) and TBB-2 (5′TGCATCACAGTGATGTAGTCG-3′). The 50-μl reaction mixture contained 5 μl of 1:200-diluted total DNA, 2.5 μl of dimethyl sulfoxide, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 30 pmol of each primer, and one U of Ampli Taq DNA polymerase (Perkin-Elmer, Foster City, Calif.). PCR amplifications were performed using the following protocol: initial denaturation at 94°C for 5 min; 40 cycles of denaturation at 94°C for 45 s, annealing at 64°C for 1 min, and extension at 72°C for 2 min; and a final extension at 72°C for 7 min.
DNA sequencing analysis.
Direct sequencing of the rpoB PCR fragments was performed by cycle sequencing using the BigDye Ready Reaction Terminator Cycle Sequencing Kit (Perkin-Elmer) and the ABI Prism 377 DNA sequencer (Perkin-Elmer) as instructed by the manufacturer. The PCR primers were also used as sequencing primers. The DNASIS program (version 2.1; Hitachi, San Bruno, Calif.) was used for DNA sequence comparisons. DNA sequences were compared with the most recent version of the GenBank NR data bank using the BLASTN algorithm (1).
The sequence of DNA sample 84 (Table 1), provided by Yang et al. (21) and showing mutation V176F, is in the GenBank database under accession number AF177294.
DNA genotyping techniques.
Extraction of genomic DNA from mycobacterial strains and DNA fingerprinting using IS6110 as a probe were performed in accordance with a standardized protocol described elsewhere (11, 19). The IS6110 fingerprint patterns of mycobacterial strains were analyzed using the Gelcompare software (Windows 95, version 4.2; Applied Maths, Kortrijk, Belgium) as described previously (11, 19).
RESULTS AND DISCUSSION
Patient demographics.
Of the 80 patients studied, data on age and sex were available for 76 patients, of whom 53 (70%) were males. The male-to-female ratio was 2.3:1 and comparable to the ratio described for the total number of patients from whom M. tuberculosis strains were isolated in Germany in 1997 (16). Most (75%) of the patients were between 20 and 50 years old. Indications of foreign birth were seen in up to 58% of the patients, strongly suggesting that MDR-TB, to a great extent, is a problem of certain risk groups in Germany so far.
First-line drug resistance patterns.
The susceptibilities to the first-line drugs INH, RIF, ethambutol (EMB), and pyrazinamide (PZA) of the clinical M. tuberculosis isolates obtained from the NRC were determined by conventional methods. Of the 80 strains tested (1 isolate per patient) only 7 showed resistance to only RIF, while 73 isolates (91%) were at least resistant to INH and RIF. Of these MDR-TB isolates, a majority of 60 strains (82%) showed further resistance to at least one other first-line drug and 21 (28%) were resistant to RIF, INH, PZA, and EMB. Since 91% of all RIF-resistant strains also were resistant to INH, our data confirm the use of RIF resistance as a surrogate marker for the rapid detection of MDR-TB in clinical M. tuberculosis isolates.
Distribution and frequency of rpoB mutations.
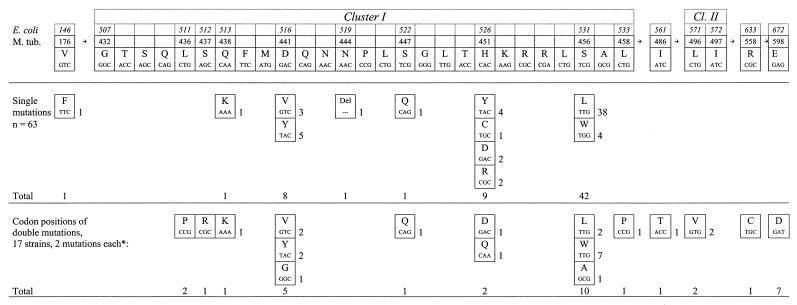
The rpoB genes of the 80 RIF-resistant M. tuberculosis strains were analyzed by direct sequencing of two rpoB DNA fragments either containing codon 176 or encompassing the whole region from cluster I to cluster III (4). The distribution and frequency of the rpoB mutations are shown in Fig. 1 using two classifications, (i) the codon numbering system used by Telenti et al. (18) deriving from homologous mutations in E. coli and (ii) the codon designations deriving from the M. tuberculosis genome. Only the latter is used in the text. In the initial analyses, a cluster I mutation could be detected in 76 of the 80 RIF-resistant strains. In accordance with previous data, mutation of codon 456 was most frequent (52 isolates, 65%) among the strains analyzed. However, in contrast to other studies, e.g., in the United States (8) and Australia (22), the rate of mutation in codon 451 was low (11 isolates, 14%) and nearly identical to the rate of mutation in codon 441 (13 isolates, 16%). Double mutations were detected in 17 isolates (22%), of which 11 isolates possessed a second mutation outside the cluster I region (Fig. 1). Although the combination of two single point mutations has been described previously for RIF-resistant M. tuberculosis strains (8, 20), the high percentage of double mutations found among the strains isolated in Germany differed clearly from the lower prevalence of double mutations in other studies. It might be of interest that several amino acid exchanges which occurred in combination with a second mutation in clusters I and II or between clusters II and III have been described to be very rare and/or to induce a lower level of resistance (15). Mutation S456A (S531A in the E. coli numbering system) might represent a newly described allele.
FIG. 1.
Frequencies of rpoB, single and double mutations associated with RIF resistance in a collection of 80 clinical M. tuberculosis isolates from Germany. ∗, Patterns and numbers of double mutations: L436P plus D441G, one; and L436P plus H451Q, one; S437R plus D441V, one; Q438K plus H451D, one; D441Y plus L496V, two; D441V plus L458P, one; S447Q plus S456A, one; S456L plus I486T, one; S456L plus R558C, one; S456W plus E598D, seven.
Additional (serial) isolates were obtained from 13 patients, and 12 had the same mutation as the first isolate. In one case, a strain initially carrying a single cluster I mutation (S447Q) apparently had acquired a second mutation within cluster I (S456A). In this case, the second isolate was included in the evaluation (Fig. 1) rather than the first.
Prevalence of mutation V176F.
In one of four isolates showing no mutation in the cluster I to cluster III region in the initial analysis, the V176F mutation could be found in the beginning of the rpoB gene. This mutation was also found in 5 out of the 18 samples with no mutation in cluster I, II, or III, respectively, obtained from six other studies comprising M. tuberculosis cultures, of strains from Asia, Africa, the United States, and Australia (Table 1). For two M. tuberculosis cultures, of which one had the V176F mutation and the other had a cluster II (I497F) mutation (kindly provided by D. Caugant; Table 1), MIC testing was performed. Mutation V176F in the first strain was associated with a RIF MIC of 256 μg/ml and a rifabutin MIC of 16 μg/ml. The second strain showed a cluster II mutation (I497V) so far observed only in one isolate from Australia (22); the RIF and rifabutin MICs were 64 and 2 μg/ml, respectively. These data confirm our previous observation that the V176F mutation in the rpoB gene is associated with high-level resistance to RIF in clinical M. tuberculosis isolates and may be responsible for more than 1% of resistant isolates.
Heteroresistance.
No rpoB mutation was detected by PCR and sequencing in 3 of the 80 isolates. However, re-examination using bacterial cultures grown on RIF-containing medium led to the identification of cluster I mutations in all three cases. Hence, a mixture of wild-type and resistant subpopulations in the initial culture (heteroresistance) presumably prohibited the detection of resistance mutations by means of PCR in the initial analyses of bacterial cultures grown on medium without RIF. In five other isolates, mixed bacterial populations showing the wild-type sequence and a typical point mutation could be detected in the initial sequence analysis. These results indicate that heteroresistance seems to occur in a considerable number of clinical M. tuberculosis isolates (8 out of 80 in our study) and may influence the sensitivity of molecular tests for the prediction of drug resistance if the percentage of the resistant subpopulation is comparably low. Methods which are able to detect small resistant subpopulations in a clinical sample, e.g., by specific hybridization, might be more sensitive in these cases.
IS6110 restriction fragment length polymorphism analysis.
To analyze the genetic relationship of the clinical M. tuberculosis strains obtained from the NRC, DNA fingerprinting using IS6110 as a probe was performed for 76 isolates (11). Forty-nine strains showed unique IS6110 patterns, confirming independent acquisition of resistance. However, 27 strains (35%) clustered in eight groups comprising two to seven isolates with identical restriction fragment length polymorphism patterns (Table 2). Transmission of the same RIF-resistant M. tuberculosis strain among the patients of one cluster could further be confirmed by the fact that in most clusters all of the isolates showed the same rpoB mutations and identical or very similar drug resistance patterns (Table 2). In only two clusters did isolates display different rpoB mutations, indicating independent acquisition of RIF resistance. Thus, recent transmission of resistant M. tuberculosis strains may have occurred in Germany. The results presented here indicate that at least the rpoB mutations found in the clustered strains seem not to affect the virulence of M. tuberculosis isolates in a way that transmission is not possible. Frequent transmission of resistant M. tuberculosis strains might also explain the disequilibrium in the distribution of rpoB mutations observed in RIF-resistant M. tuberculosis isolates from different countries (8, 14, 22).
TABLE 2.
IS6110 fingerprint clusters among the 76 RIF-resistant strains analyzed
| Cluster | Isolate | Drug resistance patterna | rpoB mutation(s) | No. of IS6110 bands |
|---|---|---|---|---|
| A | 1 | INH, RIF, EMB, PZA | D441Y, L496V | 12 |
| 2 | INH, RIF, EMB, PZA | D441Y, L496V | 12 | |
| B | 3 | INH, RIF, PZA | S456L | 11 |
| 4 | INH, RIF | S456L, R558C | 11 | |
| C | 5 | INH, RIF, EMB | H451D | 15 |
| 6 | INH, RIF, EMB, | H451D | 15 | |
| 7 | INH, RIF, EMB, PZA | S447Q | 15 | |
| 8 | INH, RIF, EMB, PZA | S447Q | 15 | |
| 9 | INH, RIF | S456L | 15 | |
| D | 10 | INH, RIF | S456L | 16 |
| 11 | INH, RIF | S456L | 16 | |
| 12 | INH, RIF, EMB, PZA | S456L | 16 | |
| E | 13 | INH, RIF | S456L | 16 |
| 14 | INH, RIF | S456L | 16 | |
| 15 | INH, RIF, EMB, PZA | S456L | 16 | |
| 16 | INH, RIF, EMB, PZA | D441V, S437R | 16 | |
| F | 17 | INH, RIF | S456W, E598D | 10 |
| 18 | INH, RIF | S456W, E598D | 10 | |
| 19 | INH, RIF | S456W, E598D | 10 | |
| 20 | INH, RIF, EMB | S456W, E598D | 10 | |
| 21 | INH, RIF, PZA | S456W, E598D | 10 | |
| 22 | INH, RIF, EMB, PZA | S456W, E598D | 10 | |
| 23 | INH, RIF, EMB, PZA | S456W, E598D | 10 | |
| G | 24 | INH, RIF, EMB | D441Y | 8 |
| 25 | INH, RIF, EMB | D441Y | 8 | |
| F | 26 | INH, RIF, EMB | S456L | 2 |
| 27 | INH, RIF, EMB, PZA | S456L | 2 |
Only first-line drugs.
Conclusion.
In the M. tuberculosis isolates evaluated in this study, RIF resistance was associated with the V176F mutation of the rpoB gene when cluster I to III mutations and heteroresistance were excluded. In the remaining 12 DNA samples from the literature with unexplained resistance, additional independent resistance mechanisms, heteroresistance, or even rpoB mutations outside the regions investigated cannot be ruled out. Mutation V176F seems to confer high-level resistance in clinical M. tuberculosis isolates and may account for more than 1% of all RIF-resistant strains. Appropriate molecular tests should be able to detect such mutations for early and reliable prediction of RIF susceptibility in clinical M. tuberculosis samples or isolates. The presence of a mixture of susceptible and resistant subpopulations in mycobacterial cultures isolated from clinical specimens, so-called heteroresistance, seems to represent an important obstacle to molecular drug resistance testing and also to successful therapy.
ACKNOWLEDGMENTS
We thank I. Radzio, F. Schaefer, B. Schlüter, and A. Zyzik, Borstel, Germany, and R. Birngruber, Regensburg, Germany, for excellent technical assistance and C. Abe, S. Kohno, D. Williams, D. Caugant, P. Kiepiela, and L. Yuen for providing DNA samples from recent surveys.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caugant D A, Sandven P, Eng J, Jeque J T, Tonjum T. Detection of rifampin resistance among isolates of Mycobacterium tuberculosis from Mozambique. Microb Drug Resist. 1995;1:321–326. doi: 10.1089/mdr.1995.1.321. [DOI] [PubMed] [Google Scholar]
- 3.Cockerill F R, Williams D E, Eisenach K D, Kline B C, Miller L K, Stockman L, Voyles J, Caron G M, Bundy S K, Roberts G D, Wilson W R. Prospective evaluation of the utility of molecular techniques for diagnosing nosocomial transmission of multidrug-resistant tuberculosis. Mayo Clin Proc. 1996;71:221–229. doi: 10.4065/71.3.221. [DOI] [PubMed] [Google Scholar]
- 4.Heep M, Odenbreit S, Beck D, Decker J, Prohaska E, Rieger U, Lehn N. Mutations at four distinct regions of the rpoB gene can reduce the susceptibility of Helicobacter pylori to rifamycins. Antimicrob Agents Chemother. 2000;44:1713–1715. doi: 10.1128/aac.44.6.1713-1715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heep M, Rieger U, Beck D, Lehn N. Mutations in the beginning of the rpoB gene can induce resistance to rifamycins in both Helicobacter pylori and Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2000;44:1075–1077. doi: 10.1128/aac.44.4.1075-1077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heifets L. Qualitative and quantitative drug-susceptibility tests in mycobacteriology. Am Rev Respir Dis. 1988;137:1217–1222. doi: 10.1164/ajrccm/137.5.1217. [DOI] [PubMed] [Google Scholar]
- 7.Hirano K, Abe C, Takahashi M. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis strains isolated mostly in Asian countries and their rapid detection by line probe assay. J Clin Microbiol. 1999;37:2663–2666. doi: 10.1128/jcm.37.8.2663-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapur V, Li L L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent P T, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 10.Kiepiela P, Bishop K, Kormuth E, Roux L, York D F. Comparison of PCR-heteroduplex characterization by automated DNA sequencing and line probe assay for the detection of rifampicin resistance in Mycobacterium tuberculosis isolates from KwaZulu-Natal, South Africa. Microb Drug Resist. 1998;4:263–269. doi: 10.1089/mdr.1998.4.263. [DOI] [PubMed] [Google Scholar]
- 11.Niemann S, Rüsch-Gerdes S, Richter E. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J Clin Microbiol. 1997;35:3015–3020. doi: 10.1128/jcm.35.12.3015-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother. 1996;40:1053–1056. doi: 10.1128/aac.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S, Kim S J, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994–1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 14.Pozzi G, Meloni M, Iona E, Orru G, Thoresen O F, Ricci M L, Oggioni M R, Fattorini L, Orefici G. rpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J Clin Microbiol. 1999;37:1197–1199. doi: 10.1128/jcm.37.4.1197-1199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 16.Rieder H L, Watson J M, Raviglione M C, Forssbohm M, Migliori G B, Schwoebel V, Leitch A G, Zellweger J P. Surveillance of tuberculosis in Europe. Working group of the World Health Organization (WHO) and the European region of the International Union against Tuberculosis and Lung Disease (IUATLD) for uniform reporting on tuberculosis cases. Eur Respir J. 1996;9:1097–1104. doi: 10.1183/09031936.96.09051097. [DOI] [PubMed] [Google Scholar]
- 17.Rüsch-Gerdes S, Domehl C, Nardi G, Gismondo M R, Welscher H M, Pfyffer G E. Multicenter evaluation of the mycobacteria growth indicator tube for testing susceptibility of Mycobacterium tuberculosis to first-line drugs. J Clin Microbiol. 1999;37:45–48. doi: 10.1128/jcm.37.1.45-48.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 19.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V, Gillis T P. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang B, Koga H, Ohno H, Ogawa K, Fukuda M, Hirakata Y, Maesaki S, Tomono K, Tashiro T, Kohno S. Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J Antimicrob Chemother. 1998;42:621–628. doi: 10.1093/jac/42.5.621. [DOI] [PubMed] [Google Scholar]
- 22.Yuen L K, Leslie D, Coloe P J. Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J Clin Microbiol. 1999;37:3844–3850. doi: 10.1128/jcm.37.12.3844-3850.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]