ABSTRACT
The opportunistic pathogen Pseudomonas aeruginosa often adapts to its host environment and causes recurrent nosocomial infections. The extracytoplasmic function (ECF) sigma factor enables bacteria to alter their gene expression in response to host environmental stimuli. Here, we report an ECF sigma factor, HxuI, which is rapidly induced once P. aeruginosa encounters the host. Host stresses such as iron limitation, oxidative stress, low oxygen, and nitric oxide induce the expression of hxuI. By combining RNA-seq and promoter-lacZ reporter fusion analysis, we reveal that HxuI can activate the expression of diverse metabolic and virulence pathways which are critical to P. aeruginosa infections, including iron acquisition, denitrification, pyocyanin synthesis, and bacteriocin production. Most importantly, overexpression of the hxuI in the laboratory strain PAO1 promotes its colonization in both murine lung and subcutaneous infections. Together, our findings show that HxuI, a key player in host stress-response, controls the in vivo adaptability and virulence of P. aeruginosa during infection.
IMPORTANCE P. aeruginosa has a strong ability to adapt to diverse environments, making it capable of causing recurrent and multisite infections in clinics. Understanding host adaptive mechanisms plays an important guiding role in the development of new anti-infective agents. Here, we demonstrate that an ECFσ factor of P. aeruginosa response to the host-inflicted stresses, which promotes the bacterial in vivo fitness and pathogenicity. Furthermore, our findings may help explain the emergence of highly transmissible strains of P. aeruginosa and the acute exacerbations during chronic infections.
KEYWORDS: Pseudomonas aeruginosa, ECF sigma factor, HxuI, host stress-response, virulence
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that causes various health care-associated infections, including pneumonia, burn wound infections, sepsis, urinary tract infections, and surgical site infections (1–3). To establish an effective infection, pathogens have to contend with host-inflicted stresses, such as iron deprivation (4), hypoxia (5), oxidative stress (1), and nitrosative stress (6, 7). The cell-surface signaling (CSS) system is a membrane-spanning signaling pathway that allows Gram-negative bacteria to transduce extracellular stimuli into coordinated transcriptional responses, and thus plays an important role in regulating bacterial adaptability and pathogenicity in response to diverse niches (8).
Typically, the CSS system is a tripartite molecular device that is composed of (i) an outer membrane TonB-dependent receptor, which senses the extracellular stimulus; (ii) a cytoplasmic membrane-spanning anti-σ factor involved in signal transduction from the periplasm to the cytoplasm; and (iii) an extracytoplasmic function (ECF) sigma factor that initiates transcription by directing core RNA polymerase (cRNAP) to the stimulus-responsive target gene(s) (8). The ECFσ family is highly diverse, and a comprehensive classification has been reported based on more than 2,700 ECFσ from hundreds of bacterial genomes (9). These ECFσ often act orthogonally with limited cross talk and allow the partitioning of the transcriptional space. The high stringency of ECFσ promoter recognition restricts the number of target genes to mount specific responses (10). In P. aeruginosa, the strains PAO1 and PA14 encode 19 and 21 ECFσ factors, respectively. They mediate the functions of cell envelope stress response, production of the exopolysaccharide alginate, iron uptake, and pathogenicity (8, 11).
The Hxu CSS pathway, which consists of three adjacent genes hxuIRA encoding ECFσ factor, anti-σ factor, and TonB-dependent outer membrane receptor, respectively (Fig. 1A), has been recently shown to be involved in heme signaling in P. aeruginosa, and mediates heme acquisition from host hemopexin (12, 13). However, the target genes of the ECFσ factor HxuI remain unknown. In the present study, we found that HxuI is highly conserved in different P. aeruginosa strains. In addition to heme, the host stresses of iron limitation, oxidative stress, hypoxia, and nitric oxide can all induce the expression of HxuI which, in turn, controls a variety of physiological functions associated with P. aeruginosa infection, including iron acquisition, anaerobic respiration, pyocyanin synthesis, and pyocin production. Most importantly, overexpression of hxuI in PAO1 promoted bacterial colonization and long-term infection in various murine infection models. Together, these studies suggest that HxuI is an important ECFσ factor contributing to the in vivo fitness and pathogenicity of P. aeruginosa.
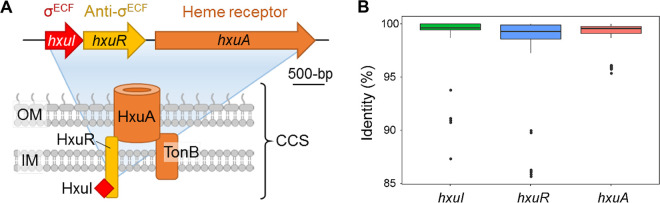
FIG 1.
Conservation of Hxu system in P. aeruginosa. (A) Schematic representation of Hxu system. (B) Conservation analysis of hxuAIR genes in 723 P. aeruginosa strains. Dots represent outliers from the respective groups.
RESULTS
HxuI is highly conserved in P. aeruginosa.
To analyze the conservation of the Hxu system, 723 P. aeruginosa clinical isolates with available genome sequences were analyzed by BLASTn (14). All strains possessed the huxIRA genes, and the hxuI gene is highly conserved among various P. aeruginosa strains (Fig. 1B), reflecting its important physiological functions.
Host stresses induce the expression of ECFσ factor HxuI.
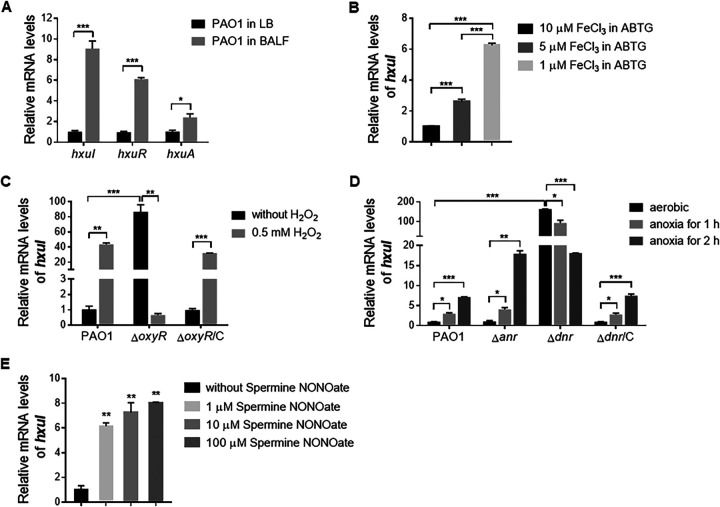
To test whether Hxu responds to the host environment during infection, we infected mice with a laboratory strain PAO1 intranasally and collected bacterial cells from the bronchoalveolar lavage fluid (BALF) 6 h postinfection (pi). Quantitative real-time PCR (qPCR) assays showed that the hxuIRA genes were upregulated 9-, 6.1-, and 2.4-fold, respectively (Fig. 2A), indicating that Hxu indeed responds to the host environment. To address the in vivo inducing signals, we tested a number of well-known host stress conditions to determine their effects on huxI gene expression. First, we tested hxuI expression under iron-deficient conditions. In the PAO1 strain, hxuI expression was increased with the decrease of Fe(III) in ABTG medium (Fig. 2B). During host infection, phagocytic cells generate reactive oxygen species (ROS) such as superoxides, which are involved in antibacterial activity (15). Next, we tested hxuI expression under oxidative stress. In the wild-type (wt) PAO1 strain, exposure to 0.5 mM H2O2 for 30 min induced a 43-fold increase in hxuI expression (Fig. 2C). OxyR is an H2O2-responsive regulator which activates the expression of defense genes against oxidative stress in P. aeruginosa (16). In an oxyR mutant, the expression of hxuI increased 86-fold even without the H2O2 treatment, indicating a repressive effect of OxyR on hxuI expression, which was restored by complementation with the oxyR gene (Fig. 2C). Possible explanations revolve around the significant regulatory cross-talk in the management of redox-stress and iron homeostasis through ferric uptake regulator (Fur) (17). Nonetheless, these data indicated that hydrogen peroxide induces the expression of HxuI in the presence of OxyR.
FIG 2.
Host stresses-response of ECFσ HxuI. (A) Mice were infected with 1 × 107 CFU of PAO1 intranasally. BALF was harvested from 16 mice at 6 h postinfection and pooled for bacterial cell isolation and subsequent RNA purification. Relative mRNA levels of hxuIRA genes of PAO1 in mouse BALF and LB medium were measured by qPCR. (B to E) qPCR determination of hxuI expression levels in wild-type PAO1, mutants, and the complemented strains (ΔoxyR/C and Δdnr/C) under conditions of Fe(III) limitation (panel B), hydrogen peroxide exposure (panel C), hypoxia (panel D), and NO donor Spermine NONOate treatment (panel E). Housekeeping gene ppiD was used as the internal reference. Error bars represent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
P. aeruginosa is able to grow in the absence of oxygen through anaerobic metabolism, which influences infectivity as well as biofilm formation (18). To investigate whether HxuI responds to hypoxia, we determined hxuI expression by qPCR after a short incubation under anaerobic conditions. As shown in Fig. 2D, hxuI expression increased along with the anaerobic culture time in the PAO1 strain. There are two well-known anaerobic sensors in P. aeruginosa: ANR and DNR (5). The expression of hxuI was increased under anaerobic growth conditions in an anr mutant, but not in a dnr mutant background (Fig. 2D). However, under aerobic conditions, hxuI expression increased by 164-fold in the dnr mutant but did not change in the anr mutant (Fig. 2D), indicating a negative regulation of hxuI by DNR. Complementation with a dnr gene restored hxuI expression levels in the Δdnr mutant (Fig. 2D). Since DNR is known to sense nitric oxide (NO), and NO-dependent DNR activity requires heme (18), we further tested whether NO directly induces the expression of HxuI. When the PAO1 wt strain was treated with 1 to 100 μM NO donor Spermine NONOate (19) for 30 min, the expression of hxuI was increased significantly in a dose-dependent manner (Fig. 2E). The above data suggested that oxygen limitation, likely via NO, induces the expression of HxuI.
Identification of the HxuI regulon genes.
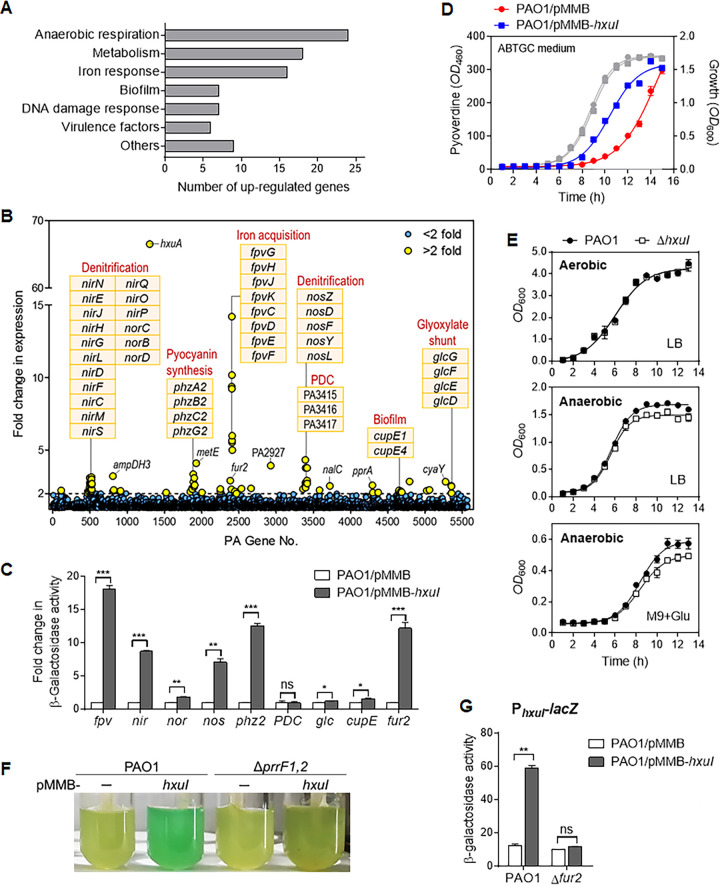
To gain insights into the HxuI regulons, a transcriptomic study was performed on P. aeruginosa PAO1 overexpressing the hxuI gene on an inducible expression vector pMMB. Most ECFσ are subject to positive auto-regulation and directly induce the expression of corresponding TonB receptor, thereby enhancing their signaling effect for as long as the inducing conditions prevail (11). The expression level of TonB receptor hxuA was monitored by qPCR at various isopropyl β-d-thiogalactopyranoside (IPTG) induction times, and it was found that hxuA expression peaked at 2 h postinduction (Fig. S1). Accordingly, total RNA samples of PAO1/pMMB-hxuI and PAO1/pMMB strains were collected 2 h after induction by 1 mM IPTG, and these were then subjected to RNA-seq analysis. The overexpression of hxuI resulted in the upregulation of 87 genes and the downregulation of 22 genes at rates of more than 2-fold (P value ≤ 0.05). Of the 87 genes significantly upregulated by HxuI, 24 genes are involved in anaerobic respiration and denitrifying redox chain, 18 are involved in metabolism, 16 in iron acquisition, 7 in biofilm formation, 7 in DNA damage response, and 6 in virulence (Fig. 3A and Table 1).
FIG 3.
The ECFσ HxuI regulon in P. aeruginosa. (A) Functional classification of upregulated genes in RNA-seq of HxuI-overexpressing strain. (B) Gene clusters that were remarkably upregulated in HxuI overexpressor. PDC, pyruvate dehydrogenase complex. (C) Analysis of the promoter-lacZ receptor expression. P. aeruginosa PAO1 containing the indicated lacZ transcriptional fusions, the plasmid pMMB (empty plasmid), or the plasmid pMMB-hxuI were grown in LB with 1 mM IPTG until late exponential growth phase and analyzed for β-galactosidase activity. Fold changes compared to PAO1/pMMB are shown. (D) Pyoverdine production (blue and red curves) and growth curves (gray) of indicated strains. (E) Growth curves of wt PAO1 and hxuI mutant under aerobic or anaerobic conditions. Glu, glucose. (F) PAO1 containing empty vector pMMB or pMMB-hxuI were grown in iron-deficient medium with 1 mM IPTG until late exponential growth phase; the presence of the green pigment indicates pyocyanin production. (G) Expression of hxuI promoter-lacZ receptor fusion in PAO1 and fur2 mutant backgrounds. Error bars represent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
TABLE 1.
Upregulated genes of P. aeruginosa PAO1 overexpressing the ECFσ factor HxuI
| Locus tag | Gene | Description | Promoter region motif | Fold changea | P value |
|---|---|---|---|---|---|
| Anaerobic respiration | |||||
| PA0112 | Hypothetical protein | 2.2 | 6.13E-06 | ||
| PA0113 | Cytochrome C oxidase assembly factor | 2.21 | 6.07E-10 | ||
| PA0509 | nirN | Cytochrome C | Anr box | 2.61 | 4.52E-38 |
| PA0510 | nirE | Uroporphyrin-III C-methyltransferase | Anr box | 2.67 | 3.9E-30 |
| PA0511 | nirJ | Heme d1 biosynthesis protein | Anr box | 2.32 | 1.18E-28 |
| PA0512 | nirH | Heme d1 biosynthesis protein | Anr box | 2.25 | 2.72E-31 |
| PA0513 | nirG | Heme d1 biosynthesis protein | Anr box | 2.05 | 3.07E-19 |
| PA0515 | nirD | Heme d1 biosynthesis protein | Anr box | 2.05 | 1.85E-19 |
| PA0516 | nirF | Heme d1 biosynthesis protein | Anr box | 2.13 | 2.79E-20 |
| PA0518 | nirM | Cytochrome C-551 | Anr box | 2.96 | NA |
| PA0519 | nirS | Nitrite reductase | Anr box | 2.97 | 2.59E-20 |
| PA0520 | nirQ | Denitrification regulatory protein | Anr box | 3.13 | 2.71E-08 |
| PA0521 | nirO | Cytochrome C oxidase subunit | Anr box | 2.98 | 2.93E-07 |
| PA0522 | nirP | Hypothetical protein | Anr box | 2.18 | 0.000407 |
| PA0523 | norC | Nitric oxide reductase subunit C | Dnr binding site | 2.29 | 0.000129 |
| PA0524 | norB | Nitric oxide reductase subunit B | Dnr binding site | 2.61 | 1.04E-05 |
| PA0525 | norD | Denitrification protein | Dnr binding site | 2.68 | 8.35E-06 |
| PA1847 | nfuA | Fe/S biogenesis protein | 2.12 | 1.74E-10 | |
| PA3392 | nosZ | Nitrous-oxide reductase | Dnr box | 4.32 | 2.49E-39 |
| PA3393 | nosD | Copper-binding periplasmic protein | Dnr box | 2.74 | 1.7E-31 |
| PA3394 | nosF | Copper ABC transporter ATP-binding protein | Dnr box | 2.57 | 5.34E-27 |
| PA3395 | nosY | Membrane protein | Dnr box | 2.41 | 8.01E-16 |
| PA3396 | nosL | Accessory protein | Dnr box | 3.77 | 4.13E-14 |
| PA5275 | cyaY | Frataxin-like protein; iron-sulfur cluster assembly protein | 2.82 | 1.26E-25 | |
| Metabolism | |||||
| PA0494 | Probable acyl-CoA carboxylase (ACCase) subunit | 2.44 | 0.0000198 | ||
| PA0495 | Allophanate hydrolase | 2.72 | 1.72E-08 | ||
| PA0496 | Allophanate hydrolase | 3.08 | 8E-24 | ||
| PA1522 | xdhC | Xanthine dehydrogenase accessory protein | 2.07 | 3.09E-15 | |
| PA2003 | bdhA | 3-Hydroxybutyrate dehydrogenase | 2.07 | 7.56E-15 | |
| PA2249 | bkdB | Branched-chain alpha-keto acid dehydrogenase complex component | 2.17 | 4.46E-14 | |
| PA2250 | lpdV | Branched-chain alpha-keto acid dehydrogenase complex component | 2.31 | 8.19E-16 | |
| PA2446 | gcvH2 | Glycine cleavage system protein H | 2.14 | 3.3E-10 | |
| PA3415 | Probable dihydrolipoamide acetyltransferase | 3.76 | 3.12E-36 | ||
| PA3416 | Pyruvate dehydrogenase E1 component subunit beta | 2.9 | 5.1E-10 | ||
| PA3417 | Pyruvate dehydrogenase E1 component subunit alpha | 2.69 | 5.91E-22 | ||
| PA3582 | glpK | Glycerol kinase | GlpR binding site | 2.23 | 2.07E-06 |
| PA4792 | Putative glycerolphosphodiesterase | 2.83 | 1.42E-41 | ||
| PA5058 | phaC2 | Poly (3-hydroxyalkanoic acid) synthase; storage polymer polyhydroxyalkanoate (PHA) biosynthesis | 2.25 | 6.21E-16 | |
| PA5352 | glcG | Hypothetical protein | 2.02 | 0.000309 | |
| PA5353 | glcF | Glycolate oxidase iron-sulfur subunit | 2.53 | 2.42E-15 | |
| PA5354 | glcE | Glycolate oxidase FAD-binding subunit | 2.49 | 3.83E-14 | |
| PA5355 | glcD | Glycolate oxidase subunit | 2.02 | 0.0000669 | |
| Iron response | |||||
| PA0471 | fiuR | Anti-sigma factor | Fur box | 2.13 | 0.000138 |
| PA0472 | fiuI | ECF sigma factor; ferric uptake | Fur box | 2.01 | 6.87E-05 |
| PA1302 | hxuA | TonB-dependent receptor; heme receptor | 66.49 | 0 | |
| PA2384 | fur2 | Fur homologue | 2.89 | 2E-07 | |
| PA2398 | fpvA | TonB-dependent receptor; ferripyoverdine receptor | PvdS binding site | 2.3 | 0.000112 |
| PA2403 | fpvG | Iron dissociation from pyoverdine | PvdS binding site | 9.37 | 2.4E-29 |
| PA2404 | fpvH | Iron dissociation from pyoverdine | PvdS binding site | 14.2 | 8.67E-71 |
| PA2405 | fpvJ | Iron dissociation from pyoverdine | PvdS binding site | 9.23 | 1.32E-29 |
| PA2406 | fpvK | Iron dissociation from pyoverdine | PvdS binding site | 10.16 | 4.65E-38 |
| PA2407 | fpvC | Periplasmic binding protein | 5.52 | 7.38E-15 | |
| PA2408 | fpvD | ABC transporter ATPase | 5.65 | 2.14E-15 | |
| PA2409 | fpvE | ABC transporter permease | 5.98 | 5.47E-17 | |
| PA2410 | fpvF | Periplasmic binding protein | 4.99 | 4.89E-18 | |
| PA2467 | foxR | Anti-sigma factor FoxR | Fur box | 2 | 2.52E-05 |
| PA3530 | bfd | Bacterioferritin-associated ferredoxin | Fur box | 2.26 | 3.94E-05 |
| PA4688 | hitB | Iron (III)-transporter permease | 2.09 | 4.86E-12 | |
| Biofilm | |||||
| PA1875 | opmL | Type I toxin efflux outer membrane protein | AmrZ binding site | 2.3 | 4.39E-05 |
| PA2662 | Membrane protein | 2.36 | 1.99E-06 | ||
| PA4293 | pprA | Two-component sensor; regulation of membrane permeability and cupE | 2.57 | 2.18E-17 | |
| PA4298 | Assembly of type IVb pili | AmrZ/LasR binding site | 2.06 | 0.000718 | |
| PA4648 | cupE1 | Fimbriae assembly | 2.16 | 0.000132 | |
| PA4651 | cupE4 | Fimbriae assembly | 2.22 | 4.65E-05 | |
| PA4675 | chtA | TonB-dependent receptor; biofilm extracellular matrix | 2.15 | 2.5E-13 | |
| DNA damage response (pyocin- and cell lysis-related genes) | |||||
| PA0646 | F-type pyocin tail fiber protein | 2 | 2.42E-17 | ||
| PA0807 | ampDh3 | Peptidoglycan hydrolase, cell wall-targeting H2-T6SS effector, AlpA regulon | AmrZ binding site | 3.2 | 1.49E-45 |
| PA0808 | Auto-immunity protein for AmpDh3, AlpA regulon | 2.17 | 5.16E-12 | ||
| PA0819 | Hypothetical protein, AlpA regulon | PvdS binding site | 2.25 | 5.69E-05 | |
| PA0910 | alpD | Self-lysis, AlpA regulon | 2.09 | 2.38E-12 | |
| PA0911 | alpE | Self-lysis, AlpA regulon | 2.18 | 1.02E-11 | |
| PA0985 | pyoS5 | S-type pyocin | 2 | 8.87E-13 | |
| Virulence factors | |||||
| PA1871 | lasA | Protease LasA, staphylolysin | 2.37 | 2.92E-15 | |
| PA1899 | phzA2 | Phenazine biosynthesis protein PhzA | AmrZ binding site | 3.07 | 2.79E-31 |
| PA1900 | phzB2 | Phenazine biosynthesis protein PhzB | AmrZ binding site | 2.47 | 9.82E-17 |
| PA1905 | phzG2 | Pyridoxamine 5′-phosphate oxidase | AmrZ binding site | 2.58 | 0.00000117 |
| PA1927 | metE | 5-Methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | 4.09 | 4.19E-13 | |
| PA3361 | lecB | Fucose-binding lectin PA-IIL | Lux box | 2.31 | 8.99E-14 |
| Others | |||||
| PA0492 | Hypothetical protein | 2.06 | 0.000583 | ||
| PA1887 | Hypothetical protein | 3.34 | 6.76E-24 | ||
| PA1888 | Hypothetical protein | 2.79 | 7.61E-23 | ||
| PA2534 | Transcriptional regulator | 2.33 | 4.56E-19 | ||
| PA2927 | Hypothetical protein | 3.92 | 1.28E-54 | ||
| PA3721 | nalC | Repressor of MexAB-OprM efflux | 2.52 | 1.77E-24 | |
| PA4371 | Hypothetical protein | 2.08 | 0.000157 | ||
| PA5023 | Hypothetical protein | 2.2 | 0.000102 | ||
| PA5446 | Hypothetical protein | 2 | 9.77E-13 | ||
PAO1/pMMB-hxuI versus PAO1/pMMB with 1 mM IPTG. RNA-seq data were generated by three biological replicates.
As expected, the expression of TonB-dependent transducer hxuA was considerably increased (66.5-fold) in the hxuI overexpressor (Fig. 3B and Table 1). Beyond that, seven clusters of genes were remarkably upregulated in the hxuI-overexpressing strain (Fig. 3B). The upregulated genes, listed here in order from high to low, included the following: (i) the fpv gene cluster (PA2403-PA2410), which is involved in iron uptake via siderophore pyoverdine; (ii) the nir (PA0509-PA0522), nor (PA0523-PA0525), and (iii) nos (PA3391-PA3396) gene clusters, which are involved in denitrification of anaerobic respiration; (iv) the PA3415-PA3417 operon encoding putative pyruvate dehydrogenase complex (PDC) which converts pyruvate into acetyl-CoA; (v) the pyocyanin biosynthesis operon phz2 (PA1899-PA1905); (vi) the glc operon (PA5352-PA5355) associated with glycolate utilization and glyoxylate shunt; and (vii) two genes belonging to the cupE gene cluster (PA4648-PA4653) which encode fimbriae assembly that promotes biofilm formation (Fig. 3B). To further confirm the transcriptional activation effects of HxuI on the above genes, the promoter regions upstream of nirS, norC, phzA2, fpvG, nosR, PA3417 (PDC gene), cupE1, and glcD were fused to a lacZ reporter gene and introduced into a PAO1 strain harboring the hxuI overexpression plasmid pMMB-hxuI. Induction of hxuI expression by IPTG resulted in marked increases (7- to 18-fold) in β-galactosidase activity in Pfpv-lacZ, Pnir-lacZ, Pnos-lacZ, and Pphz2-lacZ fusions, and modest but significant increases in Pnor-lacZ, Pglc-lacZ, and PcupE-lacZ fusions (Fig. 3C).
Consistent with the above results, we further observed that (i) pyoverdine production in the hxuI-overexpressing strain was significantly higher than that of the wt strain during late exponential phases (Fig. 3D); (ii) under anaerobic condition, the growth rate of the hxuI deletion mutant was slower than that of the parent strain PAO1, while no growth defect was observed under aerobic conditions (Fig. 3E); and (iii) booming pyocyanin production was observed in PAO1 which overexpressed hxuI (Fig. 3F). In P. aeruginosa, a pair of tandem small RNAs, PrrF1 and PrrF2, promote the production of Pseudomonas quinolone signal (PQS), which activates pyocyanin production (20). In a prrF1,2 double mutant strain background, the activation of pyocyanin production by HxuI disappeared (Fig. 3F), indicating that HxuI-mediated activation of pyocyanin production requires the PrrF small RNAs.
PA2384 (Fur2) plays a major role in the regulation of hxuI regulon.
HxuI was classified as an iron-responsive ECFσ in previous studies, as its promoter region carries a Fur box (21). The ferric uptake regulator (Fur) plays a central role in iron response and is an essential gene in P. aeruginosa (22). The Fur protein employs Fe(II) as a cofactor and binds to a so-called “Fur box” in the promoters of iron-regulated genes, resulting in repression of the target genes; under low-iron conditions, the Fur protein is released from the operator sites and transcription takes place (21). Interestingly, RNA-seq data analysis showed that PA2384 encoding a Fur homologue (designated Fur2) was upregulated 2.89-fold in the PAO1 overexpressing HxuI (Table 1). A HxuI-mediated transcriptional activation was observed in Pfur2-lacZ reporter with a 12-fold increase in β-galactosidase activity (Fig. 3C). Fur2 shares 35% amino acid identity with the N-terminal DNA-binding domain of Fur (PA4764), but does not bear the C-terminal domain of Fur which is responsible for iron binding and dimerization (23). To determine whether Fur2 is involved in the regulation of hxuI regulon, we examined the transcriptional activation effects of HxuI on fpv, nir, nos, and phz2 promoters in a Δfur2 mutant. Overexpression of HxuI in the PAO1 strain led to significant increases in β-galactosidase activity in Pfpv-lacZ, Pnir-lacZ, Pnos-lacZ, and Pphz2-lacZ fusions in the wild-type strain (Fig. 3C); however, these HxuI-mediated activations were diminished in the Δfur2 mutant background (Fig. S2), suggesting that HxuI-mediated activation of the fpv, nir, nos, and phz2 genes requires the presence of Fur2. Similarly, overexpression of hxuI resulted in 5-fold increases in β-galactosidase activity in PAO1 harboring PhxuI-lacZ fusion reporter, but not in the fur2 mutant background (Fig. 3G), indicating that Fur2 is also required for HxuI self-regulation.
HxuI activates pyocin and bacterial cell lysis-related genes.
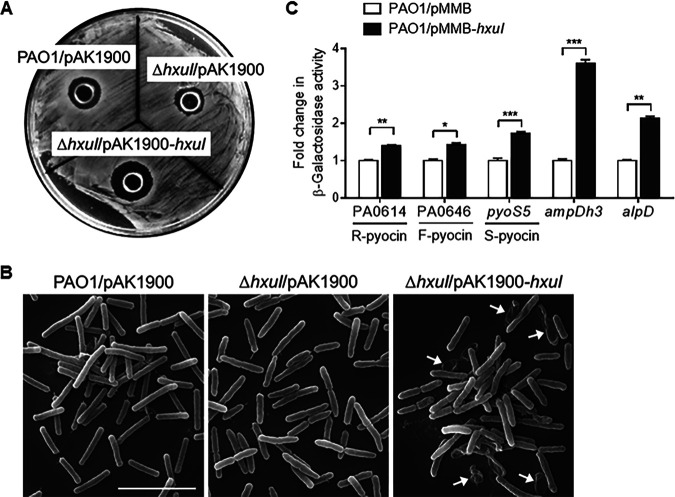
To establish infection, bacteria must establish a strong foothold for colony development and also outcompete resident microbes. One strategy that potentially addresses both needs is the use of phage tail-like bacteriocins, which are broadly called pyocins in P. aeruginosa (24). Pyocins are released into the environment through explosive cell lysis which kills the producer and nearby competitor bacteria (25). This event also releases extracellular DNA which structurally supports biofilm formation (26). Looking at the RNA-seq data, we noticed that the whole gene sets encoding all three types of pyocins in P. aeruginosa were upregulated in the HxuI-overexpressing strain (Table S1), including the soluble S-type pyocin S2, S4, S5 (PA0985 in Table 1), the contractile R-type pyocin, and the noncontractile F-type pyocin (PA0646 in Table 1). To test whether HxuI is involved in pyocin production, neat supernatants from wt PAO1, ΔhxuI, and the complemented strain ΔhxuI/pAK1900-hxuI were spotted onto an L agar overlay containing the indicator P. aeruginosa strain PAK. As shown in Fig. 4A, the growth inhibition zone of the ΔhxuI/pAK1900-hxuI strain was larger than that of the wt and the ΔhxuI mutant, indicating higher intraspecies competitiveness that might be mediated by pyocin production. In addition, two sets of cell lysis genes, PA0807 (ampDh3)-PA0808 (immunity of AmpDh3) and alpDE (27), were upregulated at average rates of ∼2.7-fold and ∼2.1-fold, respectively, in the HxuI overexpressor (Table 1). AmpDh3, a cell wall amidase, is thought to be delivered by the type VI secretion system locus II (H2-T6SS) to bacterial competitors and degrade the cell wall peptidoglycan of prey, thereby providing a growth advantage for P. aeruginosa (28). AlpDE belongs to the AlpBCDE self-lysis cassette which responds to DNA damage inflicted by the host immune system and enhances the virulence of P. aeruginosa (29). Under scanning electron microscopy (SEM), more bacterial cell lysis was observable in the ΔhxuI/pAK1900-hxuI culture than in the wt strain culture (Fig. 4B); cells that overexpressed hxuI were inclined to gather together on the coverslips and form colony-like architectures, while ΔhxuI cells were scattered evenly (Fig. 4B). To investigate the transcriptional activation effects of HxuI on the above genes, the promoter regions upstream of PA0614 (R-pyocin), PA0646 (F-pyocin), pyoS5, ampDh3, and alpD were fused to the lacZ reporter and introduced into a PAO1 strain harboring the plasmid pMMB-hxuI. Significant increases in β-galactosidase activity were observed in all five fusions when HxuI expression was induced (Fig. 4C).
FIG 4.
HxuI activates pyocin- and cell lysis-related genes. (A) Zones of clearance in P. aeruginosa PAK strain after exposure to the supernatant of wt PAO1/pAK1900 (empty vector), ΔhxuI/pAK1900, or ΔhxuI/pAK1900-hxuI (overexpress hxuI). (B) Scanning electron microscopy (SEM) of PAO1 and either ΔhxuI containing vector pAK1900 or ΔhxuI containing pAK1900-hxuI. The scale bar is 5 μm. (C) Promoter-lacZ fusions assay. P. aeruginosa PAO1 cells containing the lacZ reporter fusions in pDN19 and either plasmid pMMB (empty plasmid) or pMMB-hxuI were grown in LB with 1 mM IPTG until late exponential growth phase and analyzed for β-galactosidase activity. Error bars represent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DNA recognition sites of HxuI.
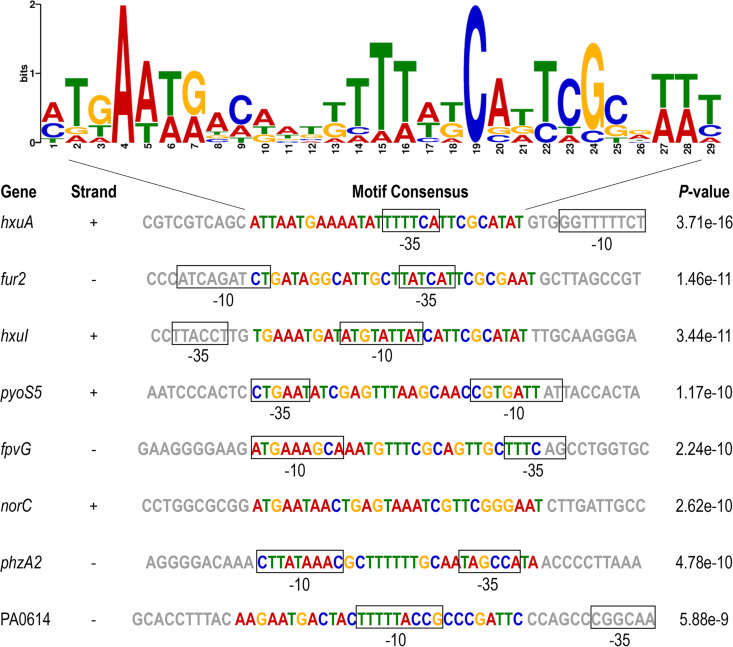
To accurately redirect gene expression, ECFσ select promoters with high stringency by combining sequence-specific interactions with the -10 and -35 promoter elements (10). To identify the specific DNA sequences that are recognized by HxuI, we analyzed the HxuI binding motif by using the MEME online tool (http://memesuite.org/tools/meme) (30) on the promoter regions of fpvG, nirS, norC, nosR, phzA2, glcD, cupE1, fur2, PA0614, PA0646, pyoS5, ampDh3, alpD, hxuA, and hxuI. The MEME analysis revealed a consensus motif of 5′-MTGAAWRACDWKKTTTWKCADTCGCRWWT-3′ as the potential HxuI binding site (Fig. 5). The genes hxuA, fur2, pyoS5, fpvG, phzA2, and PA0614 carry this motif in their promoter regions (Fig. 5), hinting these genes may be the direct targets of HxuI.
FIG 5.
HxuI recognition motif predicted by MEME. The positions of the -35 and -10 boxes in promoter DNAs are predicted by the BPROM online service (46). The potential promoter region of norC is not included in the indicated sequence.
HxuI promotes P. aeruginosa infection in mice.
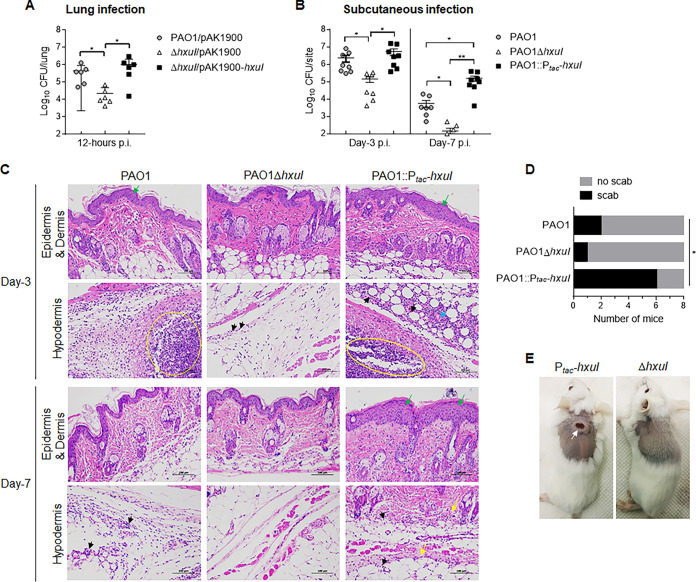
A mouse lung infection model was used to determine the role of HxuI in acute infection. Mice were intranasally infected with the same amount of wt PAO1, ΔhxuI mutant, and hxuI complementary strain, respectively. At 12 h postinfection, the hxuI deletion mutant exhibited a significantly lower bacterial load in lungs compared to that of wt PAO1, and complementation with hxuI restored bacterial colonization capacity to wt levels (Fig. 6A). These data indicated that HxuI is critical for colonization in P. aeruginosa. A murine cutaneous abscess model was further employed as a chronic infection model (31) to determine the role of HxuI in long-term infection. To avoid the loss of HxuI expression vector, hxuI driven by tac promoter was inserted into the PAO1 chromosome via a mini-Tn7 vector (PAO1::Ptac-hxuI), resulting in a constitutive expression of the hxuI gene (32, 33). Mice were subcutaneously inoculated with 5 × 106 CFU of wt PAO1, ΔhxuI, or PAO1::Ptac-hxuI. On day 3 postinfection, the ΔhxuI mutant-infected group exhibited a lower bacterial burden in lesions than those infected by wt PAO1 or PAO1::Ptac-hxuI (Fig. 6B). Histological examinations of skin abscesses indicated intense inflammatory infiltration, local tissue necrosis, and thickening of the epidermis in both PAO1 and PAO1::Ptac-hxuI infection groups, while infection by ΔhxuI resulted in very mild inflammations (Fig. 6C). On day 7, a large abscess with overlying crust/scab was formed on the dorsum skin of 75% (6/8) mice infected by PAO1::Ptac-hxuI, but on only 25% (2/8) and 12.5% (1/8) of those infected by PAO1 and ΔhxuI, respectively (Fig. 6D and E). Histological sections of the PAO1::Ptac-hxuI-infected group showed thickened epidermis, collagen fiber necrosis, lysis of subcutaneous muscle fibers, and inflammation (Fig. 6C). In comparison, the PAO1 and ΔhxuI infection groups exhibited much lower bacterial loads inside abscesses and fewer scattered inflammatory cells (Fig. 6B and C). These results indicated that forced expression of the HxuI enables P. aeruginosa to better adapt to the host environment, promoting the establishment of long-term infection.
FIG 6.
HxuI promotes P. aeruginosa infection in murine models. (A) In the acute pneumonia model, mice (n = 6/group) were intranasally inoculated with 1 × 107 CFU of the indicated bacterial cells. Bacterial loads in lungs were counted by plating at 12 h postinfection (pi). (B) In the cutaneous abscess model, mice (n = 8/group) were subcutaneously inoculated with 5 × 106 CFU of indicated bacterial cells. Bacterial loads in abscesses were counted on days 3 and 7 pi. Error bars represent SD. *, P < 0.05; **, P < 0.01. (C) Histological sections of cutaneous abscess. Yellow circles indicate inflammation and tissue injury, green arrows indicate thickening of the epidermis, black arrows indicate neutrophil infiltration, blue arrow indicates extravasated blood in capillaries, yellow arrows indicate fiber necrosis. (D) Scab formation on day 7 pi. P value was calculated using one-way ANOVA; *, P < 0.05. (E) Skin appearance of scab (white arrow) on day 7.
DISCUSSION
In this study, we found that ECFσ factor HxuI is highly conserved in different P. aeruginosa strains and can be induced by several host-inflicted stresses, including iron deprivation, oxidative stress, and hypoxia, as well as NO. Physiological adaptation to varied environmental stresses, such as changes in oxygen levels encountered within diverse niches, is an important capability for pathogenic bacterial species (34). The viability of P. aeruginosa within robust anaerobic biofilms requires NO reductase to modulate or prevent the accumulation of toxic NO, a byproduct of anaerobic respiration (35). Our data indicate that the NO sensor DNR negatively regulates HxuI, which further activates denitrification to reduce NO into nitrogen gas (36), revealing a novel ECFσ-mediated nitrosative stress-response pathway in P. aeruginosa.
Overexpression of HxuI remarkably activated the transcription of genes associated with pyoverdine-dependent iron acquisition, denitrification, pyocyanin biosynthesis, and the production of pyocins involved in intraspecies competition. Fur2 is positively regulated by HxuI and plays a critical role in HxuI-mediated transcriptional regulation, and even in the auto-activation of HxuI. Most notably, forced expression of the hxuI gene promotes the establishment of long-term P. aeruginosa infection in vivo; therefore, HxuI functions as an important regulator that senses host stresses and enables P. aeruginosa to tune metabolic strategies for adaptation to the host environment and express virulence factors which promote persistent infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table S2. Gene deletion and complementation were performed as previously described (37, 38). Bacterial cells were grown at 37°C in LB (Luria-Bertani) broth or in M9 medium with 0.1% (wt/vol) glucose (39). The following concentrations of antibiotics were used: for P. aeruginosa, gentamicin at 30 μg/mL in LB, tetracycline at 50 μg/mL in LB, and carbenicillin at 150 μg/mL in LB; for Escherichia coli, tetracycline at 10 μg/mL, gentamicin at 10 μg/mL, kanamycin at 50 μg/mL, and ampicillin at 100 μg/mL. For iron-limitation condition, strains were grown in ABTG medium [15.1 mM (NH4)2SO4, 33.7 mM Na2HPO4, 22.0 mM KH2PO4, 0.05 mM NaCl, 1 mM MgCl2, 100 μM CaCl2, 0.5% (wt/vol) glucose, and 1 to 10 μM FeCl3] (40). Anaerobic conditions were established by an anaerobic workstation (Don Whitley Scientific) with an oxygen content of 0.07%, and bacteria were statically cultured in various media supplemented with 50 mM NaNO3. All experiments were done in the biosafety level 2 laboratory at Nankai University.
Gene conservation analysis.
The population structure of P. aeruginosa can be divided into five groups (41). The complete genomes of 723 P. aeruginosa strains that covered all five groups were analyzed in this study. The nucleotide sequences of the huxIRA of PAO1 were used as reference. We aligned each genome sequence of the 723 strains against the reference using BLASTn (14) with the criteria set as E value < 1e-5 and length coverage of the gene > 85% to find the homologous sequences. Finally, the identities between each strain and reference were illustrated using the R package (http://www.r-project.org/).
Ethics statement.
All animal studies complied with National and Nankai University guidelines regarding the use of animals in research. All animal experiment protocols were approved by the Institutional Animal Care and Use Committee of the College of Life Sciences of Nankai University with the permit number NK-04-2012.
Murine lung infection.
The infection of mice was performed as previously described (42). Briefly, overnight bacterial culture was diluted 1:100 in fresh LB and grown at 37°C until the OD600 reached 1.0. Bacterial cells were collected by centrifugation and washed once with phosphate-buffered saline (PBS). The bacterial cell concentration was adjusted to 5 × 108 CFU/mL in PBS. Each female BALB/c mouse (Vital River, Beijing, China), at the age of 6 to 8 weeks, was anesthetized with an intraperitoneal injection of 7.5% chloral hydrate and inoculated with 20 μL of the bacterial suspension, resulting in 1 × 107 CFU per mouse. Bronchi alveolar lavage fluid (BALF) was collected as previously described (43). At 6 h postinfection, mice were euthanized via CO2 inhalation. One mL PBS containing 0.05 mM EDTA was injected into the lungs via the trachea by a vein detained needle (BD, Angiocath). After 1 min of detaining, BALF was collected.
Total RNA isolation and quantitative real-time PCR.
Total bacterial RNA was isolated using an RNAprep Pure Cell/Bacteria Kit (Tiangen Biotec, Beijing, China). cDNAs were synthesized with reverse transcriptase and random primers (Takara Bio, Dalian, China). Real-time (RT) PCR was performed using SYBR II Green Supermix (Bio-Rad, Beijing, China). Specific Primers (Table S3) were used for quantitative RT-PCR. The peptidyl-prolyl cis-trans isomerase D gene ppiD was used as an internal control.
Transcriptome sequencing and data analysis.
Both PAO1/pMMB and PAO1/pMMB-hxuI cultures (OD600=0.6) were grown in LB with 1 mM IPTG for 2 h. Total RNA was isolated using an RNAprep Pure Cell/Bacteria Kit (Tiangen Biotec, Beijing, China). Three replicates were prepared for each strain. Sequencing and analysis were performed as previously described (44).
Promoter-lacZ reporter assay.
The promoter region (500 bp upstream from the start codon) of each gene was cloned into pDN19lacΩ to construct the promoter-lacZ reporter construct. The reporter constructs, as well as the pMMB-hxuI or the empty plasmid pMMB, were introduced into PAO1 by electroporation, and the transformants were selected on an L agar plate containing Tc and Cb. After inducing the expression of HxuI with 1 mM IPTG for 2 h, bacterial cells were collected by centrifugation and resuspended in 500 μL of Z-buffer (16 g/L Na2HPO4·7 H2O, 4.8 g/L NaH2PO4, 0.746 g/L KCl, 0.246 g/L MgSO4·7 H2O, 3.5 mL/L β-mercaptoethanol [pH = 7]). To permeabilize the cells, 10 μL of 0.1% SDS and 10 μL of chloroform were added and vortex for 10 s. After this, 100 μL of 4 mg/mL ONPG (o-nitrophenyl-β-d-galactopyranoside) was added to the cells. The samples were incubated at 37°C until the yellow color became apparent, and 500 μL of Na2CO3 (0.5 M) was added to stop the reaction. Sample absorbance was read at 420 nm, and β-galactosidase activity was calculated as Miller units = 2,000 × OD420/OD600/incubation time (min). Each assay was repeated three times.
Measurement of pyoverdine production.
A microplate pyoverdine measurement was carried out in ABTGC medium [15.1 mM (NH4)2SO4, 33.7 mM Na2HPO4, 22.0 mM KH2PO4, 0.05 mM NaCl, 1 mM MgCl2, 100 μM CaCl2, 10 μM FeCl3, 0.2% (wt/vol) glucose and 0.2% (wt/vol) casamino acid] as previously described (45). The overnight P. aeruginosa cultures were adjusted to an OD600 of 0.01 in ABTGC medium. The cells were then incubated in 96-well plates at 37°C. Pyoverdine fluorescence (excitation maximum 400 nm, emission maximum 460 nm) and OD600 were recorded by the microplate reader (Tecan Group Ltd., Switzerland) every hour. Experiments were performed in triplicate, and results are shown as the mean ± SD (standard deviation).
Pyocin toxicity assays.
Zones of clearance were observed for the P. aeruginosa PAK strain using the supernatants of wt PAO1, hxuI mutant and hxuI-overexpressing strains. A 0.05 μg/mL volume of ciprofloxacin was used to induce the production of pyocins in the PAO1-derived strains. PAK was used as an indicator strain, diluted to OD600 = 0.6, and plated on LB agar. Finally, 200 μL of supernatants of the test strains were added to sterile Oxford cups placed on the PAK plate and cultured overnight at 37°C.
Scanning electron microscopy (SEM).
Bacterial cultures (OD600 = 1.0) were co-incubated with 0.1% gelatin-coated glass slides at 37°C for 4 h. The unattached bacterial cells were discarded. The glass slides with sessile bacteria were washed once with PBS and fixed with 4% paraformaldehyde. The bacterial cells were dehydrated with a gradient (30%, 50%, 70%, 90%, 100%) of alcohol, air dried, and imaged under an electron microscope.
Mouse cutaneous abscess model.
The infection of mice was performed as previously described (31). Briefly, mice were clipped in the dorsal area by a shaver and depilatory cream. Fifty μL of either 5 × 106 CFU bacterial suspension or saline were subcutaneously injected into the dorsum of each mouse. At 3 and 7 days postinfection, mice were euthanized with carbon dioxide, and then the skin abscesses were excised, homogenized in saline, and subjected to plating for CFU counting.
Statistical analysis.
Statistical evaluations were performed using GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA). P values were calculated using one-way analysis of variance (ANOVA), a two-tailed unpaired Student’s t test. Data were considered significant when P values were below 0.05, as indicated.
Data availability.
The transcriptome (RNA-Seq) data have been deposited in NCBI BioProject with the accession code PRJNA717102.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31870130, 31970680, 31970179, 32170199, and 82061148018), the National Key Research and Development Project of China (2017YFE0125600, 2021YFE0201300 and 2021YFE0101700), the National Research Foundation of Korea (NRF-2020K2A9A2A11102267), and the Guangdong Natural Science Foundation for Distinguished Young Scholars of China (2020B1515020003).
Footnotes
Supplemental material is available online only.
Contributor Information
Fang Bai, Email: baifang1122@nankai.edu.cn.
Joanna B. Goldberg, Emory University School of Medicine
REFERENCES
- 1.Mohamed FA, Shaker GH, Askoura MM. 2020. Oxidative stress influences Pseudomonas aeruginosa susceptibility to antibiotics and reduces its pathogenesis in host. Curr Microbiol 77:479–490. doi: 10.1007/s00284-019-01858-7. [DOI] [PubMed] [Google Scholar]
- 2.Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa: a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Barat L, Ferrer M, De Rosa F, Gabarrús A, Esperatti M, Terraneo S, Rinaudo M, Li Bassi G, Torres A. 2017. Intensive care unit-acquired pneumonia due to Pseudomonas aeruginosa with and without multidrug resistance. J Infect 74:142–152. doi: 10.1016/j.jinf.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Nelson CE, Huang W, Brewer LK, Nguyen AT, Kane MA, Wilks A, Oglesby-Sherrouse AG. 2019. Proteomic analysis of the Pseudomonas aeruginosa iron starvation response reveals PrrF small regulatory RNA-dependent iron regulation of twitching motility, amino acid metabolism, and zinc homeostasis proteins. J Bacteriol 201:e00754-18. doi: 10.1128/JB.00754-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trunk K, Benkert B, Quäck N, Münch R, Scheer M, Garbe J, Jänsch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the ANR and DNR regulons. Environ Microbiol 12:1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 6.Waite RD, Stewart JE, Stephen AS, Allaker RP. 2018. Activity of a nitric oxide-generating wound treatment system against wound pathogen biofilms. Int J Antimicrob Agents 52:338–343. doi: 10.1016/j.ijantimicag.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Buglino JA, Sankhe GD, Lazar N, Bean JM, Glickman MS. 2021. Integrated sensing of host stresses by inhibition of a cytoplasmic two-component system controls M. tuberculosis acute lung infection. Elife 10:e65351. doi: 10.7554/eLife.65351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llamas MA, Imperi F, Visca P, Lamont IL. 2014. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev 38:569–597. doi: 10.1111/1574-6976.12078. [DOI] [PubMed] [Google Scholar]
- 9.Staroń A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol 74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 10.Campagne S, Allain FHT, Vorholt JA. 2015. Extra Cytoplasmic Function sigma factors, recent structural insights into promoter recognition and regulation. Curr Opin Struct Biol 30:71–78. doi: 10.1016/j.sbi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Chevalier S, Bouffartigues E, Bazire A, Tahrioui A, Duchesne R, Tortuel D, Maillot O, Clamens T, Orange N, Feuilloley MGJ, Lesouhaitier O, Dufour A, Cornelis P. 2019. Extracytoplasmic function sigma factors in Pseudomonas aeruginosa. Biochim Biophys Acta Gene Regul Mech 1862:706–721. doi: 10.1016/j.bbagrm.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Zambolin S, Clantin B, Chami M, Hoos S, Haouz A, Villeret V, Delepelaire P. 2016. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat Commun 7:11590. doi: 10.1038/ncomms11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otero-Asman JR, García-García AI, Civantos C, Quesada JM, Llamas MA. 2019. Pseudomonas aeruginosa possesses three distinct systems for sensing and using the host molecule haem. Environ Microbiol 21:4629–4647. doi: 10.1111/1462-2920.14773. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Q, Minh PN, Dötsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, Häussler S, Cornelis P. 2012. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 18.Castiglione N, Rinaldo S, Giardina G, Cutruzzola F. 2009. The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology (Reading) 155:2838–2844. doi: 10.1099/mic.0.028027-0. [DOI] [PubMed] [Google Scholar]
- 19.Cai YM, Webb JS. 2020. Optimization of nitric oxide donors for investigating biofilm dispersal response in Pseudomonas aeruginosa clinical isolates. Appl Microbiol Biotechnol 104:8859–8869. doi: 10.1007/s00253-020-10859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhart AA, Nguyen AT, Brewer LK, Bevere J, Jones JW, Kane MA, Damron FH, Barbier M, Oglesby-Sherrouse AG. 2017. The Pseudomonas aeruginosa PrrF small RNAs regulate iron homeostasis during acute murine lung infection. Infect Immun 85:e00764-16. doi: 10.1128/IAI.00764-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochsner UA, Vasil AI, Vasil ML. 1995. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol 177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumas Z, Ross-Gillespie A, Kümmerli R. 2013. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc Biol Sci 280:20131055. doi: 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng P, Sun J, Geffers R, Zeng AP. 2007. Functional characterization of the gene PA2384 in large-scale gene regulation in response to iron starvation in Pseudomonas aeruginosa. J Biotechnol 132:342–352. doi: 10.1016/j.jbiotec.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Ghequire MGK, De Mot R. 2015. The tailocin tale: peeling off phage tails. Trends Microbiol 23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Vacheron J, Heiman CM, Keel C. 2021. Live cell dynamics of production, explosive release and killing activity of phage tail-like weapons for Pseudomonas kin exclusion. Commun Biol 4:87. doi: 10.1038/s42003-020-01581-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peña JM, Prezioso SM, McFarland KA, Kambara TK, Ramsey KM, Deighan P, Dove SL. 2021. Control of a programmed cell death pathway in Pseudomonas aeruginosa by an antiterminator. Nat Commun 12:1702. doi: 10.1038/s41467-021-21941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Hu Z, Du X, Shi Y, Dang J, Lee M, Hesek D, Mobashery S, Wu M, Liang H. 2020. A type VI secretion system delivers a cell wall amidase to target bacterial competitors. Mol Microbiol 114:308–321. doi: 10.1111/mmi.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFarland KA, Dolben EL, LeRoux M, Kambara TK, Ramsey KM, Kirkpatrick RL, Mougous JD, Hogan DA, Dove SL. 2015. A self-lysis pathway that enhances the virulence of a pathogenic bacterium. Proc Natl Acad Sci USA 112:8433–8438. doi: 10.1073/pnas.1506299112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 31.Pletzer D, Mansour SC, Wuerth K, Rahanjam N, Hancock RE. 2017. New mouse model for chronic infections by Gram-negative bacteria enabling the study of anti-infective efficacy and host-microbe interactions. mBio 8:e00140-17. doi: 10.1128/mBio.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terpe K. 2006. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 33.Choi K-H, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 34.Xiong L, Yang Y, Ye YN, Teng JL, Chan E, Watt RM, Guo FB, Lau SK, Woo PC. 2017. Laribacter hongkongensis anaerobic adaptation mediated by arginine metabolism is controlled by the cooperation of FNR and ArgR. Environ Microbiol 19:1266–1280. doi: 10.1111/1462-2920.13657. [DOI] [PubMed] [Google Scholar]
- 35.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock REW, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3:593–603. doi: 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 36.Arat S, Bullerjahn GS, Laubenbacher R. 2015. A network biology approach to denitrification in Pseudomonas aeruginosa. PLoS One 10:e0118235. doi: 10.1371/journal.pone.0118235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Cai Z, Fu W, Liu Y, Tian C, Wang H, Fu T, Wu Z, Wu D, Jin Y, Cheng Z, Terada N, Liu L, Wu W, Jin S, Bai F. 2020. High-efficiency protein delivery into transfection-recalcitrant cell types. Biotechnol Bioeng 117:816–831. doi: 10.1002/bit.27245. [DOI] [PubMed] [Google Scholar]
- 38.Shao X, Zhang X, Zhang Y, Zhu M, Yang P, Yuan J, Xie Y, Zhou T, Wang W, Chen S, Liang H, Deng X. 2018. RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J Bacteriol 200:e00205-18. doi: 10.1128/JB.00205-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X, Fan Z, Chen L, Liu C, Bai F, Wei Y, Tian Z, Dong Y, Shi J, Chen H, Jin Y, Cheng Z, Jin S, Lin J, Wu W. 2020. PvrA is a novel regulator that contributes to Pseudomonas aeruginosa pathogenesis by controlling bacterial utilization of long chain fatty acids. Nucleic Acids Res 48:5967–5985. doi: 10.1093/nar/gkaa377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J Molecular Biology 23:99–112. doi: 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- 41.Freschi L, Vincent AT, Jeukens J, Emond-Rheault JG, Kukavica-Ibrulj I, Dupont MJ, Charette SJ, Boyle B, Levesque RC. 2019. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol 11:109–120. doi: 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, Shi J, Liu C, Jin Y, Li K, Chen R, Jin S, Wu W. 2014. PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect Immun 82:1638–1647. doi: 10.1128/IAI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu W, Huang J, Duan B, Traficante DC, Hong H, Risech M, Lory S, Priebe GP. 2012. Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 186:420–427. doi: 10.1164/rccm.201202-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan X, Dong Y, Fan Z, Liu C, Xia B, Shi J, Bai F, Jin Y, Cheng Z, Jin S, Wu W. 2017. In vivo host environment alters Pseudomonas aeruginosa susceptibility to aminoglycoside antibiotics. Front Cell Infect Microbiol 7:83. doi: 10.3389/fcimb.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chua SL, Tan SY, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. 2013. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:2066–2075. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solovyev V. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p.61–78. In RW Li (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppage, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01620-21_Supp_1_seq9.pdf, PDF file, 0.3 MB (348.8KB, pdf)
Data Availability Statement
The transcriptome (RNA-Seq) data have been deposited in NCBI BioProject with the accession code PRJNA717102.