Abstract
Acidity is an important factor influencing the organoleptic quality of apple fruits. In this study, an apple pyrophosphate-energized proton pump (PEPP) gene was isolated and designated MdMa12. On the basis of a phylogenetic analysis in Rosaceae species, PEPP genes were divided into three groups, with apple PEPP genes most closely related to pear PEPP genes. Gene expression analysis revealed that high malic acid content was generally accompanied by high MdMa12 expression levels. Moreover, MdMa12 was mainly expressed in the fruit. A subcellular localization analysis suggested that MdMa12 is a mitochondrial protein. The ectopic expression and overexpression of MdMa12 in “Micro-Tom” tomato and apple calli, respectively, increased the malic acid content. One (MDH12) of four malate dehydrogenase genes highly expressed in transgenic apple calli was confirmed to encode a protein localized in mitochondria. The overexpression of MDH12 increased the malate content in apple calli. Furthermore, MdMa12 overexpression increased MdDTC1, MdMa1, and MdMa10 expression levels, which were identified to transport malate. These findings imply that MdMa12 has important functions related to apple fruit acidity. Our study explored the regulatory effects of mitochondria on the complex mechanism underlying apple fruit acidity.
Introduction
Organic acids are important fruit components that affect nutritional quality and taste. Malate, the dominant organic acid in mature apple fruit, is mainly synthesized in the cytoplasm and mitochondria [1, 2]. More specifically, malate is synthesized from oxaloacetate via a reaction catalyzed by NAD-dependent cytoplasmic malate dehydrogenase (NAD-cytMDH) in the cytosol. The excessive production of NAD-cytMDH in apple leads to increased malic acid, fructose, and sucrose contents in apple calli [2]. Malate is also metabolized in mitochondria through the glyoxylate and GABA shunts. In mitochondria, malate is oxidized in two pathways. In a reversible reaction, it is converted to oxaloacetate by NAD-dependent mitochondrial malate dehydrogenase (NAD-mtMDH) [1]. Alternatively, it is converted to pyruvate by the NAD-dependent mitochondrial malic enzyme (NAD-mtME) [3], which ultimately leads to citrate synthesis and an altered malate:citrate ratio. Malate is also synthesized in mitochondria via mtMDH. Nevertheless, the role of mtMDH in regulating fruit malate content remains unclear.
Organic acids are eventually stored in vacuoles. Because they must cross a biological membrane to enter vacuoles, only the ionic forms of organic acids are transported [4]. Many organic acid transporters have been identified in various species, including the tonoplast malate transporter (tDT) in Arabidopsis thaliana [4] and the aluminum-activated malate transporter (ALMT) in grape [5] and apple [6]. The Ma1 gene, which encodes an ALMT localized in the vacuolar membrane, affects the organic acid content of apple fruit. Moreover, the ma1 mutation disrupts malate transport because of the resulting lack of a conserved C-terminal domain in the encoded protein, which leads to decreased apple fruit acidity [6, 7]. Additionally, in citrus fruit, CsCit1 reportedly transports citrate through the vacuolar membrane [8].
Proton pumps, which also influence fruit acidity, are mostly localized in the vacuolar and cytoplasmic membranes. The two main types of proton pumps are H+-ATPase and H+-pyrophosphatase (H+-PPase), which hydrolyze ATP and pyrophosphate, respectively, to facilitate ion transport. Differences in fruit acidity among species and varieties may be associated with the diversity in proton pumps [9, 10]. For example, PhPH5 encodes a petunia P-type proton pump in the vacuolar membrane and it is critical for the vacuolar acidification of petals [11]. The PH1 and PH5 proton pumps can form a complex that promotes the acidification of petunia vacuoles [12]. In apple, Ma10 is a proton pump crucial for fruit vacuolar acidification [13].
In addition to vacuolar transporters, mitochondrial organic acid transporters have been identified. The solute carriers in the inner mitochondrial membrane connect the internal metabolism with that of the surrounding cell [14]. For example, CjDTC is involved in mitochondrial dicarboxylic acid/tricarboxylic acid transport [15]. Regalado et al. [16] identified three genes for mitochondrial dicarboxylic acid/tricarboxylic acid carriers (VvDTC1, 2, and 3), which are likely to be involved in the mitochondrial transport of malate, citrate, and other di/tricarboxylates in grapes. Vacuolar proton pumps have been investigated much more extensively than mitochondrial proton pumps.
Pyrophosphatase (H+-PPase) hydrolyzes pyrophosphate and is important for regulating cell swelling, the H+ electrochemical gradient, and the secondary activities of inorganic ions, organic acids, and sugars. The A. thaliana AVP1 gene encodes a PPase proton pump, and its overexpression enhances the salt tolerance and drought resistance of A. thaliana and peanut [17, 18]. Moreover, H+-PPase plays a key role in the sweet potato response to Fe deficiency [19]. Furthermore, PPase helps regulate sucrose accumulation in citrus fruit [20]. A previous study proved that overexpressing a bacterial PPase increases vitamin C, sucrose, and glucose contents of mature fruit but has the opposite effect on the starch content [21]. More recently, the sweet potato H+-PPase IbVP1 was revealed to influence starch metabolism and yield by regulating carbon flux [22]. In apple, the overexpression of a vacuolar proton pump gene, MdVHP1, increases the malate content of calli via the regulation of the transporter gene MdtDT [23]. However, the functions of mitochondrial PPases are relatively poorly characterized.
An earlier investigation confirmed the differential expression of a pyrophosphate-energized proton pump (PEPP) gene in apple varieties that varied in terms of the mature fruit malate contents [13]. In this study, we analyzed the apple Ma12 gene, which encodes a PEPP localized in mitochondria. The overexpression and ectopic expression of MdMa12 in apple calli and “Micro-Tom” tomato, respectively, led to increased malic acid contents. The results of this study will strengthen the characterization of the complex molecular regulation of fruit acidity.
Results
Identification of MdMa12 and analysis of phylogenetic relationships
The MdMa12 gene encoding a PEPP was identified as a candidate gene affecting fruit acidity [13]. A structural analysis revealed eight exons and seven introns in MdMa12 (Fig. S1a). The 2304-bp coding sequence of MdMa12 was predicted to encode a protein comprising 767 amino acids (Fig. S1a), with a molecular weight, total average hydrophilicity, theoretical isoelectric point, and instability index of 80448.66 kD, 0.622, 5.25, and 22.84, respectively. Additionally, MdMa12 was predicted to contain 13 transmembrane domains, six of which are located in the N-terminus, whereas the other seven are present in the C-terminus (Fig. S1c). A phylogenetic analysis conducted to explore the evolutionary relationships of PEPPs in four Rosaceae species indicated that apple and pear PEPPs are closely related (Fig. 1a). On the basis of their phylogenetic relationships, PEPP proteins can be divided into three subgroups, with MdMa12 belonging to subgroup I (Fig. 1a).
Figure 1.
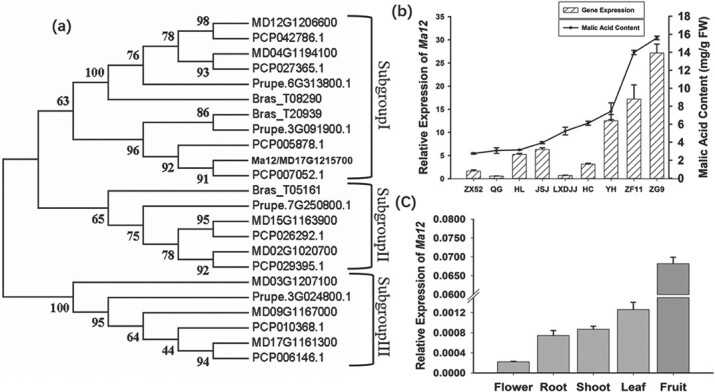
Phylogenetic analysis of genes encoding PEPP in Rosaceae species and a comparison of the MdMa12 expression profiles in apple. a. Phylogenetic tree of PEPP-encoding genes in Rosaceae species. Bootstrap values are provided near the branched lines. b. MdMa12 expression profiles and malic acid contents in fruit from different species. All fruit matured similarly at approximately 90 days after full bloom. FW, fresh weight; ZG9, Malus toringoides; ZX52, Malus spectabilis; ZF11 Malus mandshurica; QG, Qinguan; HL, Honglu; JSJ, Jinshiji; LXDJJ, Liangxiangdejijie; HC, Honeycrisp; YH, Yuhua. c. MdMa12 expression levels in the flower, root, shoot, leaf, and fruit.
Examination of MdMa12 expression profiles
Quantitative real-time (qRT)-PCR analysis was performed to investigate the MdMa12 expression profiles in the mature fruit of six apple cultivars [“Qinguan” (QG), “Honglu” (HL), “Jinshiji” (JSJ), “Liangxiangdejijie” (LXDJJ), “Honeycrisp” (HC), and “Yuhua” (YH)] and three wild relatives [Malus toringoides (ZG9), Malus spectabilis (ZX52), and Malus mandshurica (ZF11)] that vary significantly regarding their malic acid contents. The examined apple cultivars and their wild relatives have similar ripening periods, with fruit maturation occurring approximately 90 days after full bloom. The highest MdMa12 expression levels were detected in ZG9, ZF11, and YH, which is consistent with the malic acid contents of the mature fruits (Fig. 1b). However, MdMa12 was more highly expressed in HL and JSJ than in LXDJJ and HC, whereas the malic acid contents of the mature fruit were lower for HL and JSJ than for LXDJJ and HC (Fig. 1b). Furthermore, MdMa12 was similarly expressed in QG and LXDJJ, but the mature fruit malic acid contents differed significantly between these two cultivars. Similar expression patterns were detected in ZX52 and HC, which is in contrast to the significant differences in their malic acid contents.
An examination of the tissue specificity of MdMa12 expression indicated that the gene is the most highly expressed in fruit, followed by the leaf, shoot, root, and flower (Fig. 1c). The MdMa12 expression level in flowers was almost 300 times lower than that in fruit (Fig. 1c). Accordingly, it is likely that MdMa12 contributes to the regulation of fruit acidity.
Subcellular localization of MdMa12
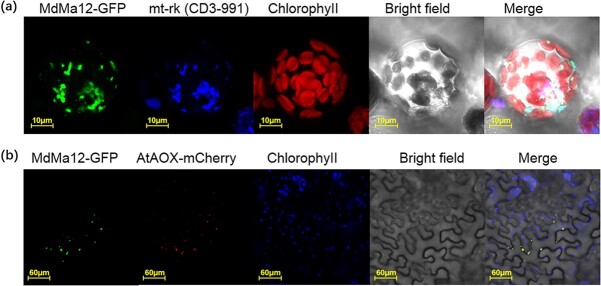
To investigate the subcellular localization of MdMa12, a MdMa12-GFP fusion construct under the control of the CaMV 35S promoter was inserted into A. thaliana mesophyll protoplasts. The MdMa12-GFP fusion protein was detected in the mitochondrial membrane (Fig. 2a). To confirm that MdMa12 is localized in the mitochondrial membrane, Nicotiana benthamiana leaves transiently transformed with the MdMa12-GFP fusion construct were examined. The fluorescence of MdMa12-GFP merged completely with that of AtAOX-mCherry, which is a mitochondrial membrane marker (Fig. 2b). These results indicate that MdMa12 is located in the mitochondrial membrane.
Figure 2.
Subcellular localization of MdMa12. a. Subcellular localization of MdMa12 in Arabidopsis thaliana protoplasts. b. Subcellular localization of MdMa12 in Nicotiana benthamiana leaves. MdMa12-GFP fluorescence was detected using a laser confocal scanning microscope, with mt-rk (CD3–991) and AOX-mCherry used as mitochondrial markers.
Functional analysis of MdMa12 using transgenic tomato plants and apple calli
To verify the regulatory effects of MdMa12 on fruit acidity, “Micro-Tom” tomato was transformed with MdMa12. Three transgenic lines were generated, with no differences in fruit size or color between the wild-type and transgenic tomato plants (Fig. 3a). The overexpression of MdMa12 in the three transgenic lines and the lack of MdMa12 expression in the wild-type control were confirmed by RT-PCR (Fig. 3b). Malic and citric acid contents were significantly higher in the transgenic fruit than in the wild-type fruit (Fig. 3c, d). The total fruit acidity was also higher in the transgenic lines than in the wild-type control (Fig. 3e). These observations imply that MdMa12 plays an important role in increasing fruit acidity via the accumulation of organic acids.
Figure 3.
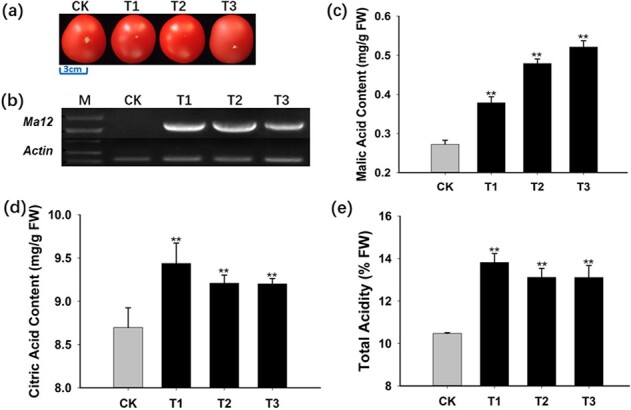
Functional analysis of MdMa12 through its overexpression in “Micro-Tom” tomato. a. Mature tomato fruit. b. Analysis of MdMa12 expression in tomato fruit by RT-PCR. c–e. Malic acid and citric acid contents and the total acidity of fresh mature tomato fruit. FW, fresh weight; CK, wild-type; T1, 2, and 3, transgenic lines 1, 2, and 3. **, significant difference from the wild-type control (P < 0.01; t-test).
To further evaluate the role of MdMa12 in fruit malic acid accumulation, MdMa12-overexpressing apple calli were obtained via Agrobacterium tumefaciens-mediated transformation (Fig. 4a). Three transgenic lines were generated and validated by PCR (Fig. 4b). qRT-PCR analysis indicated that MdMa12 was more highly expressed in the transgenic lines than in the wild-type control (Fig. 4c). The malic acid contents of the transgenic and wild-type apple calli were measured (Fig. 4d). The average malic acid content was 1.7 times higher in the transgenic calli [0.63 mg/g fresh weight (FW)] than in the wild-type control (0.23 mg/g FW). These results suggest that MdMa12 contributes to malic acid accumulation in apple fruit.
Figure 4.
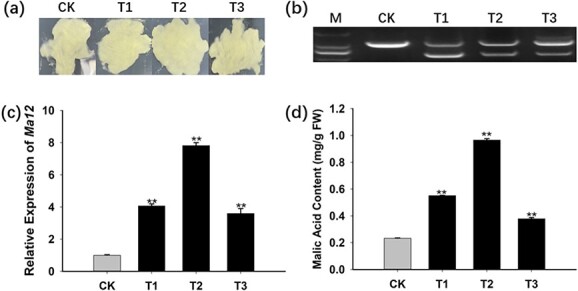
Functional analysis of MdMa12 through its overexpression in “Orin” apple calli. a. MdMa12-overexpressing apple calli and wild-type apple calli. b. Confirmation of transgene expression in transgenic apple calli by PCR amplification of a DNA template. A primer pair crossing an intron was designed to amplify the coding sequence and the genome sequence. Only the genome sequence was amplified for the wild-type control, whereas both the coding sequence and the genome sequence were amplified for the transgenic lines. c. Relative MdMa12 expression levels in apple calli. d. Malic acid contents in fresh apple calli. FW, fresh weight; CK, wild-type; T1, 2, and 3, transgenic lines 1, 2, and 3. **, significant difference from the wild-type control (P < 0.01; t-test).
MdMDH gene expression in MdMa12-overexpressing apple calli and predicted localization of the encoded protein
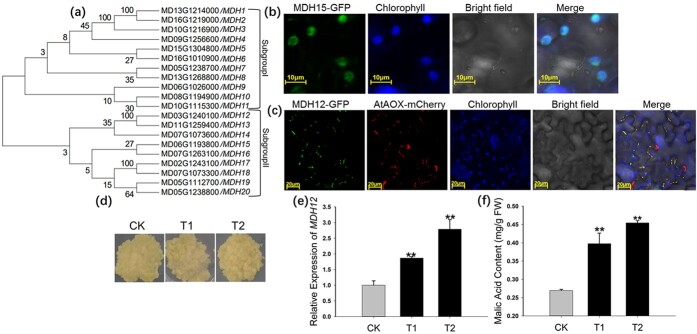
Because malate is primarily synthesized through reactions catalyzed by MDHs, the MdMDH expression profiles in wild-type and MdMa12 transgenic apple calli were analyzed by qRT-PCR. Evolutionary analysis was conducted of these 20 MdMDHs found in apple, and their names were defined according to their evolutionary relationships (Fig. 5a). Of the 20 MdMDH genes, 9 were expressed in MdMa12 transgenic and wild-type apple calli, and four genes (MDH8, MDH11, MDH12, and MDH15) had obviously increased expression levels in MdMa12 transgenic apple calli compared with those in the wild-type control (Table 1). The proteins encoded by two of these genes (MDH12 and MDH15) were predicted to be localized in the mitochondrial membrane (Fig. S2). To verify the localization of MDH12 and MDH15, MDH12-GFP and MDH15-GFP constructs were generated and inserted into A. tumefaciens cells. In subcellular localization experiments conducted using N. benthamiana leaves, MDH12 and AtAOX (i.e. mitochondrial markers) were detected at the same location (Fig. 5c), whereas MDH15 was localized in the chloroplast (Fig. 5b). We inferred that an increased expression level of mitochondrial MDH (MDH12) might have an important role in regulating malic acid content in MdMa12-overexpressing apple calli.
Figure 5.
Analyses of MdMDH expression levels and functions. a. Evolutionary analysis of MdMDHs. A phylogenetic tree of MDH genes in apple was constructed using MEGA (version 7). Bootstrap values are provided near branched lines. b, c. Subcellular localization of MDH15 and MDH12 in Nicotiana benthamiana leaves. The MDH15-GFP and MDH12-GFP signals were detected using a laser confocal scanning microscope, with AOX-mCherry serving as a mitochondrial marker. The chloroplasts in Nicotiana benthamiana leaf cells were identified on the basis of their chlorophyll autofluorescence. d. MDH12-overexpressing apple calli and wild-type apple calli. e, f. Analyses of MDH12 expression and malic acid contents in MDH12-overexpressing apple calli and wild-type calli. CK, wild-type; T1 and 2, MDH12 transgenic lines 1 and 2. **, significant difference from the wild-type control (P < 0.01; t-test).
Table 1.
Relative expression of 12 MdMDHs
| CK | T1 | T2 | T3 | |
|---|---|---|---|---|
| MDH1 | 1 ± 0.04 | 0.15 ± 0.00 | 1.5 ± 0.1 | 0.03 ± 0.00 |
| MDH5 | 1 ± 0.04 | 0.40 ± 0.00 | 1.95± 0.02 | 0.25 ± 0.06 |
| MDH8 | 1 ± 0.06 | 1.5 ± 0.06** | 1.6 ± 0.07** | 1.43 ± 0.02** |
| MDH9 | 1 ± 0.03 | 0.42 ± 0.01 | 1.9 ± 0.29 | 0.64 ± 0.04 |
| MDH11 | 1 ± 0.02 | 2 ± 0.05** | 1.7 ± 0.09** | 1.98 ± 0.01** |
| MDH12 | 1 ± 0.02 | 1.82 ± 0.03** | 3.54 ± 0.17** | 1.86 ± 0.15** |
| MDH13 | 1 ± 0.03 | 0.46 ± 0.03 | 0.18 ± 0.04 | 0.36 ± 0.01 |
| MDH15 | 1 ± 0.03 | 2.26 ± 0.01** | 4.39 ± 0.20** | 1.89 ± 0.01** |
| MDH17 | 1 ± 0.04 | 0.29 ± 0.01 | 0.98 ± 0.02 | 0.2 ± 0.01 |
Note: Values are presented as the mean ± s.d. of determinations on three biological replicates. CK, wild-type; T1, 2, and 3, MdMa12 transgenic lines 1, 2, and 3. **, significant difference from the wild-type control (P < 0.01; t-test).
Functional analysis of MdMDH12 using apple calli
To determine whether MdMDH12 overexpression can increase the malic acid content, apple calli overexpressing MdMDH12 were obtained. (Fig. 5d). A qRT-PCR assay indicated that the MdMDH12 expression level was significantly higher in transgenic lines than in wild-type calli (Fig. 5e). The average malic acid content was 1.4 times higher in the transgenic apple calli (0.62 mg/g FW) than in the wild-type calli (0.26 mg/g FW) (Fig. 5f). The overexpression of MdMDH15 did not significantly affect the malic acid content between wild-type and transgenic apple calli (Fig. S4). These results indicate that MdMDH12 probably affects the apple fruit malic acid content.
Expression levels of genes involved in malate transport
Malate is eventually stored in vacuoles [24]. Malate transport from mitochondria to vacuoles is mostly dependent on mitochondrial dicarboxylate/tricarboxylate transporters (DTCs) [16], proton pumps, and ALMT on the vacuolar membrane [6]. Two apple DTC genes were identified, but only MdDTC1 was more highly expressed in transgenic apple calli than in wild-type calli (Fig. 6). Previous research proved that MdMa1, which encodes an ALMT, modulates fruit malic acid accumulation [6]. In the current study, the MdMa1 (MD16G1045200) expression level was higher in transgenic lines than in wild-type apple calli (Fig. 6). Additionally, an analysis of the expression levels of six vacuolar proton pump genes related to fruit acidity [13, 23, 25, 26] revealed that only MdMa10 was more highly expressed in all three transgenic lines than in wild-type apple calli (Fig. 6). Hence, the overexpression of MdMa12 appears to induce MdDTC1, MdMa1, and MdMa10 expression to promote malate transport and uptake by vacuoles.
Figure 6.
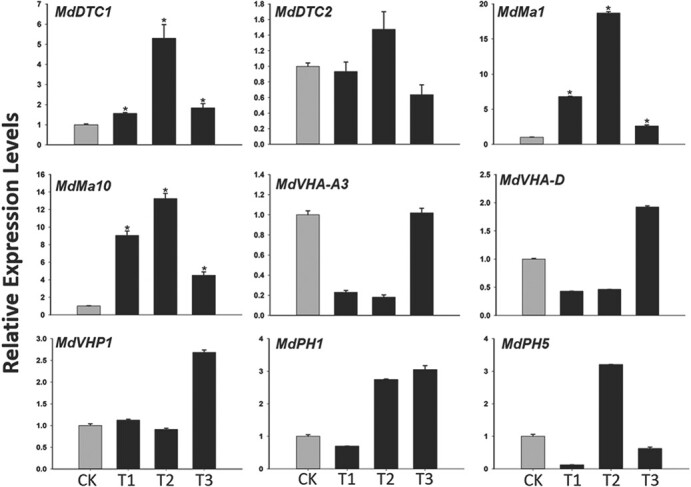
Expression levels of genes involved in malate and proton transport in MdMa12-overexpressing apple calli. CK, wild-type; T1, 2, and 3, transgenic lines 1, 2, and 3. Data are presented as the mean ± SE (n ≥ 3). *, significant difference (P ≤ 0.05).
Discussion
Pyrophosphate-energized proton pumps have various functions in many species. For example, the expression of A. thaliana AVP1 in transgenic A. thaliana and peanut is positively correlated with salt and drought resistance [17, 18]. A recent study on sweet potato proved that IbVP1 influences starch metabolism and yield by regulating carbon flux [22]. Although PEPPs have various functions, their effects on fruit acidity are relatively unknown. In apple, MdMa1 is the main gene controlling fruit acidity because the encoded protein is a malate transporter. A single nucleotide polymorphism (A/G) in the last exon of MdMa1 introduces an early stop codon (i.e. a loss-of-function mutation) [27, 28].
In a previous study, a PEPP gene was revealed to potentially control apple fruit acidity in ma1 homozygotes [13]. In the current study, we designated the PEPP gene MdMa12 and investigated its relationship with fruit acidity. We determined the malate contents and MdMa12 expression levels in nine apple species. The species with high malate contents usually had high MdMa12 expression levels (Fig. 1b). However, the MdMa12 expression levels were similar in QG and LXDJJ, whereas their malic acid contents were significantly different (Fig. 1b). This inconsistency suggests that genes other than MdMa12 are also involved in the regulation of malic acid accumulation in apple fruit. High organic acid levels are reasonable because many vacuolar proton pumps can increase organic acid contents through H+ flux into vacuoles [11–13]. An “acid trap” hypothesis has been proposed to explain how proton pumps increase organic acid contents [29]. This hypothesis assumes that proton pumps are localized in vacuolar membranes. Previous research confirmed that most proton pumps are found in vacuolar membranes, including PH1 and PH5 in petunia [12] and Ma10 and VHP1 in apple [13, 23]. However, in the current study, MdMa12 was localized in the mitochondrial membrane (Fig. 2b, c). Additionally, MdMa12-overexpressing transgenic apple calli and tomato contained more malic acid than the wild-type controls (Figs. 3, 4). Malate is primarily synthesized in mitochondria, but it is stored in vacuoles. Thus, the “acid trap” hypothesis does not fully explain how MdMa12 controls fruit acidity. Taken together, these results imply that the mechanism underlying the regulation of apple fruit acidity by mitochondrially localized MdMa12 is complex. Nevertheless, the data generated in our study confirm that MdMa12 is localized in mitochondria and affects fruit acidity.
H+-PPases are ubiquitous among Rosaceae species. A phylogenetic analysis revealed that MdMa12 and 22 PEPP genes from Rosaceae species can be classified into three groups (Fig. 1a). The apple and pear PEPP genes clustered together rather than with the peach and black raspberry genes (Fig. 1a). Similar results were reported for ALMT genes and MDH genes in Rosaceae species [30, 31]. The constructed phylogenetic tree included eight, eight, four, and three PEPP genes from apple, pear, peach, and black raspberry, respectively (Fig. 1a). Additionally, apple and pear have 17 haploid chromosomes, whereas peach and black raspberry contain eight and seven haploid chromosomes, respectively. Thus, it is likely that the apple and pear genomes underwent whole-genome duplication events [32, 33], with the duplicated genes retained under positive selection pressure during evolution and speciation. A similar genomic duplication probably did not occur during the evolution of peach and black raspberry.
The fruit acidity of cultivated apple species is associated with the malic acid content in mature fruit [34]. Proton pumps are important in malic acid accumulation in many fruit species [9–11, 13]. In this study, we isolated an apple PPase proton pump gene (MdMa12) encoding a protein that likely affects apple fruit acidity [13]. The mechanisms of MdMa12 are unclear because the encoded protein is localized in mitochondria [13]. Malate is synthesized in the cytoplasm and mitochondria in reactions catalyzed by cytMDH and mtMDH, respectively [35], but it is primarily synthesized in mitochondria [36]. Although cytMDH has been functionally characterized [2], the contribution of mtMDH to malate contents has not been studied. Four MDH genes were highly expressed in MdMa12-overexpressing transgenic apple calli (Table 1). The predicted subcellular localization indicated that only two of these genes encode proteins localized in mitochondria (Fig. S2). Of these two MDH genes, MDH12 was confirmed to encode a protein localized in mitochondria (Fig. 5c). In apple calli, the overexpression of MdMDH12 increased the malic acid content (Fig. 5d–f).
Therefore, we propose that MdMa12 controls fruit acidity by inducing malate synthesis in mitochondria. Malate produced in mitochondria is transported into the cytosol and is eventually stored in vacuoles. The mitochondrial carrier family (MCF) is critical for linking the metabolites of the mitochondria with the rest of the cell [14]. The DTCs of the MCF mediate the transport of unprotonated malate between mitochondria and the cytosol [37]. Although the apple genome includes two DTC genes, only the expression of MdDTC1 increased in all MdMa12-overexpressing transgenic apple calli (Fig. 6). Thus, malate synthesized in mitochondria should be transported out by MdDTC1. If cytosolic malate is to be stored in vacuoles, malate transporters and proton pumps in the vacuolar membrane must function cooperatively because only malate dianions can be transported into vacuoles, wherein they are immediately protonated [38].
Thus, we analyzed the expression of MdMa1, which encodes a protein that transports malate into vacuoles, and six proton pump genes related to fruit acidity in MdMa12-overexpressing transgenic apple calli [13, 23, 25, 26] (Fig. 6). Only MdMa1 and MdMa10 were highly expressed in all of the transgenic lines (Fig. 6). In contrast, the expression levels of the other five proton pump genes varied considerably among the examined transgenic lines (Fig. 6). This observation may be explained by the high malate content in the cytosol resulting from the transport of malate from the mitochondria and the activities of multiple proton pumps in the tonoplast.
Therefore, we developed a model of the regulatory effects of MdMa12 on apple fruit acidity (Fig. 7). The overexpression of MdMa12 accelerates the conversion of oxaloacetate to malate. The excess malate in mitochondria is transported into the cytosol through MdDTC1. Malate in the cytosol is transported into vacuoles via MdMa1 and MdMa10. In this study, we analyzed a mitochondrially localized proton pump and confirmed that it is involved in the regulation of apple fruit acidity.
Figure 7.
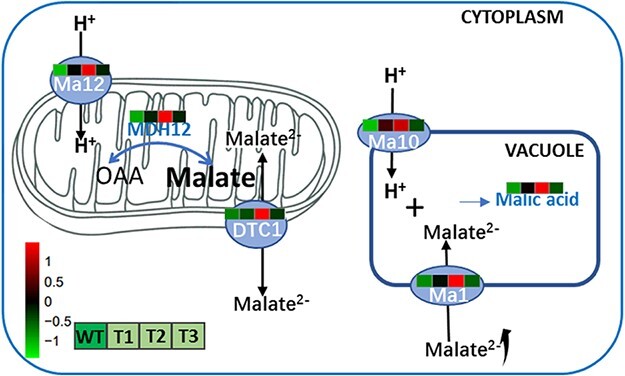
Proposed model for the regulatory effects of MdMa12 on apple fruit acidity. The overexpression of MdMa12 promotes the influx of H+ into mitochondria and induces the expression of MDH12-encoding genes. The conversion of oxaloacetate to malate is enhanced (i.e. malate accumulation). Malate is transported from mitochondria to the cytosol through MdDTC1, after which the vacuolar proteins MdMa1 and MdMa10 facilitate the transport of malate to vacuoles for storage.
Materials and methods
Plant materials
Nine apple accessions, including six apple cultivars (QG, HL, JSJ, LXDJJ, HC, and YH) and three wild relatives (ZG9, ZX52, and ZF11), were maintained at the Horticultural Experimental Station of Northwest A&F University, Yangling, Shaanxi Province, China. Five apple tissues (i.e. fully blooming flowers, roots, mature leaves, ripening fruit, and shoots) were collected from the plants for a qRT-PCR assay. The “Orin” apple callus was cultured on solid Murashige and Skoog culture medium supplemented with 1.5 mg/L 2,4-dichlorophenoxyacetic acid and 0.4 mg/L 6-benzylaminopurine at 25°C in darkness [25]. “Micro-Tom” tomato, N. benthamiana, and A. thaliana plants were grown at 25°C with a 16-h light/8-h dark cycle. The collected samples were immediately frozen in liquid nitrogen and stored at −80°C until analysis.
Identification of MdMa12 and analysis of phylogenetic relationships
The apple genome (GDDH13 V1.1) was used as the source for the MdMa12 sequence [39]. The MdMa12 amino acid sequence downloaded from the GDR database (https://www.rosaceae.org/) was used for predicting the physical and chemical properties as well as the transmembrane domains using online tools (https://web.expasy.org/protparam/ and http://www.cbs.dtu.dk/services/TMHMM, respectively). Additionally, PEPP amino acid sequences [AtAVP1(AT1G15690), AtVHP2(AT1G16780) and AtAVP2(AT1G78920)] were downloaded from the TAIR website (https://www.arabidopsis.org/) and then used to identify homologous genes in the reference genome sequences of apple [39], peach [40], pear [41], and black raspberry [42] with basic local alignment search tools. A phylogenetic tree was constructed according to the neighbor-joining method using previously described parameters of the MEGA (version 7) program (https://www.megasoftware.net/) [31].
Measurement of organic acid contents and total acidity
Organic acid contents were measured using a high-performance liquid chromatography (HPLC) system as previously described [43]. Briefly, approximately 0.1 g frozen powder was dissolved in 1 mL H2O, which was produced by the Milli-Q Element water purification system (Millipore, Bedford, MA, USA). The mixture was placed in an ultrasonic bath for 30 min at room temperature. The mixture was then centrifuged at 5000 g for 15 min at 4°C. The supernatant was collected and filtered using a 0.22 μm Sep-Pak water filter (Anpel, Shanghai, China). The total acidity of the filtrate was measured using an acidity meter (ATAGO, Guangzhou, China). The organic acid content of the filtrate was determined using a 1260 Infinity HPLC system (Agilent, Milford, MA, USA). Chromatographic separation was performed using an Athena C18 column (100 Å, 4.6 nm × 250 nm, 5 μm). The column temperature was maintained at 40°C. The mobile phase was a 0.02 M KH2PO4 solution (pH 2.4), and the flow rate was 0.8 mL/min. The analysis was completed using three biological replicates. The organic acid standards were purchased from Sigma (St. Louis, MO, USA) and dissolved in deionized water.
RNA isolation and qRT-PCR analysis
Total RNA was extracted using the Total RNA Extraction Kit (Wolact, Hong Kong, China). The RNA served as the template for synthesizing cDNA using TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China). The qRT-PCR assay was performed using a 20-μL SYBR Green-based reaction mixture consisting of 10 μL SYBR Green Master Mix (Takara, Dalian, China), 0.2 μM each primer, and 100 ng template cDNA. A LightCycler® 96 real-time PCR detection system (Roche, Basel, Switzerland) was used to complete the qRT-PCR analysis. A melting curve analysis was performed after 40 cycles to determine the specificity of the amplified products (Fig. S3). Apple and tomato actin genes described in a previous study [13] were used as internal reference controls. The qRT-PCR analysis involved three biological replicates, with relative expression levels calculated according to the 2−∆∆Ct method.
Subcellular localization analysis
The MdMa12, MdMDH12, and MdMDH15 coding sequences were amplified from cDNA templates prepared from HC fruit, after which they were inserted into the pMD19-T cloning vector. DNAMAN (version 8.0) was used for alignment. Additionally, the MdMa12 coding sequence was also inserted into the pHBT-GFP-NOS vector for subsequent expression under the control of the CaMV 35S promoter. A vector containing the mt-rk mitochondrial marker sequence (CD3–991) and the mCherry reporter gene was used for cotransformation experiments with MdMa12-GFP. A. thaliana protoplasts were obtained as previously described [44]. Chloroplasts in A. thaliana protoplasts were identified on the basis of chlorophyll autofluorescence (red signal).
The MdMa12, MdMDH12, and MdMDH15 coding sequences were amplified by PCR to add EcoRI and KpnI sites at the 5′ and 3′ ends, respectively. Following double digestion, the sequences were inserted into separate pART27 vectors [45] carrying the GFP gene. Then, recombinant plasmids were generated and inserted into A. tumefaciens cells. Nicotiana benthamiana leaves were transiently transformed with the MdMa12-GFP, MdMDH12-GFP, and MdMDH15-GFP constructs via agroinfiltration. For the subsequent determination of subcellular localization, AtAOX-mCherry was used as the fluorescent mitochondrial marker [46]. Fluorescence was observed using a laser confocal scanning microscope (Leica TCS-SP8 SR) at 24–48 h after agroinfiltration. More specifically, GFP and mCherry fluorescence and chlorophyll autofluorescence were detected at wavelengths of 488, 610, and 750 nm, respectively. Chloroplasts in N. benthamiana leaf cells were identified on the basis of chlorophyll autofluorescence (blue signal).
Functional analysis of MdMa12, MdMDH12 and MdMDH15
The MdMa12, MdMDH12 and MdMDH15 coding sequences were amplified by PCR to add PacI and AscI sites at the 5′ and 3′ ends, respectively. After double digestion, the sequences were inserted into separate pMDC83 [13] vectors for subsequent expression under the control of the CaMV 35S promoter. The 35S-MdMa12, 35S-MdMDH12 and 35S-MdMDH15 constructs were inserted into A. tumefaciens GV3101 cells, which were then used to transform tomato plants and apple calli as described by Sun et al. [47] and Li et al. [48], respectively. Using a method developed by Masoodi et al. [49], DNA was extracted using cetyltrimethylammonium bromide. Tomato transformants were selected on the basis of hygromycin resistance. The transgenic seedlings were grown in soil in a greenhouse. Transgenic T1 plants were identified by RT-PCR. All primers used in this study are listed in Table S1.
Prediction of the subcellular localization of MdMDHs
The MdMDH mRNA and coding sequences were downloaded from the GDR database (https://www.rosaceae.org/). The coding sequences were translated using EditSeq (https://www.dnastar.com/software/lasergene/). Additionally, WoLF PSORT (https://wolfpsort.hgc.jp/) was used to predict the subcellular localization of all MdMDHs.
Statistical analyses
All statistical analyses were performed using SPSS Statistics 17.0 (SPSS Inc., Chicago, IL, USA). A two-tailed test was used to evaluate the significance of any differences (P ≤ 0.01). The heatmap was constructed using R.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Numbers 32072527 and 31701875) and the Program for the National Key Research and Development Program (2018YFD1000200). We thank the Horticulture Science Research Center at the College of Horticulture, NWAFU, for their technical support of this work.
Author contributions
B. M., M.L. and F.M. conceived and designed the experiments. M.G. and H.Z. performed the experiments and wrote the paper. L.Z., Y.P., W.M., R.T. and L.Z. analyzed the data. Y.Y. provided assistance with the GFP assay.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflict of interest
The authors declare no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research Journal online.
References
- 1. Sweetman C, Deluc LG, Cramer GRet al. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009;70:1329–44. [DOI] [PubMed] [Google Scholar]
- 2. Yao YX, Li M, Zhai Het al. Isolation and characterization of an apple cytosolic malate dehydrogenase gene reveal its function in malate synthesis. J Plant Physiol. 2011;168:474–80. [DOI] [PubMed] [Google Scholar]
- 3. Macrae AR, Moorhouse R. The oxidation of malate by mitochondria isolated from cauliflower buds. FEBS J. 2010;16:96–102. [DOI] [PubMed] [Google Scholar]
- 4. Rentsch D, Martinoia E. Citrate transport into barley mesophyll vacuoles—comparison with malate-uptake activity. Planta. 1991;184:532–7. [DOI] [PubMed] [Google Scholar]
- 5. Angeli A, Baetz U, Francisco Ret al. The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta. 2013;238:283–91. [DOI] [PubMed] [Google Scholar]
- 6. Ma BQ, Liao L, Zheng Het al. Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome. 2015;8:16. [DOI] [PubMed] [Google Scholar]
- 7. Li CL, Dougherty L, Coluccio AEet al. Apple ALMT9 requires a conserved C-terminal domain for malate transport underlying fruit acidity. Plant Physiol. 2020;182:992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shimada T, Nakano R, Shulaev Vet al. Vacuolar citrate/H+ symporter of citrus juice cells. Planta. 2006;224:472–80. [DOI] [PubMed] [Google Scholar]
- 9. Lu XP, Liu YZ, Zhou GFet al. Identification of organic acid-related genes and their expression profiles in two pear (Pyrus Pyrifolia) cultivars with difference in predominant acid type at fruit ripening stage. Sci Hortic. 2011;129:680–7. [Google Scholar]
- 10. Yang LT, Xie CY, Jiang HXet al. Expression of six malate-related genes in pulp during the fruit development of two loquat (Eriobotrya Japonica) cultivars differing in fruit acidity. Afr J Biotechnol. 2011;10:2414–22. [Google Scholar]
- 11. Verweij W, Spelt C, Vermeer Jet al. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower color. Nat Cell Biol. 2008;10:1456–62. [DOI] [PubMed] [Google Scholar]
- 12. Faraco M, Spelt C, Bliek Met al. Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep. 2014;6:32–43. [DOI] [PubMed] [Google Scholar]
- 13. Ma BQ, Liao L, Fang Tet al. A Ma10 gene encoding P-type ATPase is involved in fruit organic acid accumulation in apple. Plant Biotechnol J. 2019;17:674–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haferkamp I, Schmitz ES. The plant mitochondrial carrier family: functional and evolutionary aspects. Front Plant Sci. 2012;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng W, Luo KM, Li ZGet al. Molecular cloning and characterization of a mitochondrial dicarboxylate/tricarboxylate transporter gene in citrus junos response to aluminum stress. Mitochondrial DNA. 2008;19:376–84. [PubMed] [Google Scholar]
- 16. Regalado A, Leonardo Pierri C, Bitetto Met al. Characterization of mitochondrial dicarboxylate/tricarboxylate transporters from grape berries. Planta. 2013;237:693–703. [DOI] [PubMed] [Google Scholar]
- 17. Gaxiola RA, Li S, Unduragga Set al. Drought and salt tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci U S A. 2001;98:11444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qin H, Gu Q, Sun Tet al. Expression of the Arabidopsis vacuolar H+-pyrophosphatase gene AVP1 in peanut to improve drought and salt tolerance. Plant Biotechnol Rep. 2013;7:345–55. [DOI] [PubMed] [Google Scholar]
- 19. Fan W, Wang H, Wu Yet al. H+-pyrophosphatase IbVP1 promotes efficient iron use in sweet potato [Ipomoea batatas (L.) lam.]. Plant Biotechnol J. 2017;15:698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bilal HS, Shi CY, Guo LXet al. Type I H+-pyrophosphatase regulates the vacuolar storage of sucrose in citrus fruit. J Exp Bot. 2020;71:5935. [DOI] [PubMed] [Google Scholar]
- 21. Osorio S, Nunes-Nesi A, Stratmann Met al. Pyrophosphate levels strongly influence ascorbate and starch content in tomato fruit. Front Plant Sci. 2013;4:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan W, Zhang Y, Wu Yet al. The H+-pyrophosphatase IbVP1 regulates carbon flux to influence the starch metabolism and yield of sweet potato. Horticulture research. 2021;8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yao YX, Dong QL, You CXet al. Expression analysis and functional characterization of apple MdVHP1 gene reveals its involvement in Na+, malate and soluble sugar accumulation. Plant Physiol Biochem. 2011;49:1201–8. [DOI] [PubMed] [Google Scholar]
- 24. Yamaki S. Isolation of vacuoles from immature apple fruit flesh and compartmentation of sugars, organic acids, phenolic compounds and amino acids. J Allergy Clin Immunol. 1984;25:151–66. [Google Scholar]
- 25. Hu DG, Li YY, Zhang QYet al. The R2R3-MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J. 2017;91:443–54. [DOI] [PubMed] [Google Scholar]
- 26. Jia DJ, Wu P, Shen Fet al. Genetic variation in the promoter of an R2R3-MYB transcription factor determines fruit malate content in apple (Malus domestica Borkh.). Plant Physiol. 2021;95:427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai Y, Dougherty L, Li Met al. A natural mutation-led truncation in one of the two aluminum-activated malate transporter-like genes at the Ma locus is associated with low fruit acidity in apple. Mol Gen Genomics. 2012;287:663–78. [DOI] [PubMed] [Google Scholar]
- 28. Khan SA, Beekwilder J, Schaart JGet al. Differences in acidity of apples are probably mainly caused by a malic acid transporter gene on LG16. Tree Genet Genomes. 2013;9:475–87. [Google Scholar]
- 29. Enrico M, Masayoshi M, Ekkehard NH. Vacuolar transporters and their essential role in plant metabolism. J Exp Bot. 2007;58:83–102. [DOI] [PubMed] [Google Scholar]
- 30. Ma BQ, Yuan Y, Gao Met al. Genome-wide identification, molecular evolution, and expression divergence of aluminum-activated malate transporters in apples. Int J Mol Sci. 2018;19:2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma BQ, Yuan Y, Gao Met al. Genome-wide identification, classification, molecular evolution and expression analysis of malate dehydrogenases in apple. Int J Mol Sci. 2018;19:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Velasco R, Zharkikh A, Affourtit Jet al. The genome of the domesticated apple (Malus domestica Borkh.). Nat Genet. 2010;10:833–9. [DOI] [PubMed] [Google Scholar]
- 33. Wu J, Wang Z, Shi Zet al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013;23:396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma BQ, Yuan Y, Gao Met al. Determination of predominant organic acid components in Malus species: correlation with apple domestication. Meta. 2018;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Etienne A, Genard M, Lobit Pet al. What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J Exp Bot. 2013;64:1451–69. [DOI] [PubMed] [Google Scholar]
- 36. Shangguan LF, Sun X, Zhang Cet al. Genome identification and analysis of genes encoding the key enzymes involved in organic acid biosynthesis pathway in apple, grape, and sweet orange. Sci Hortic. 2015;185:22–8. [Google Scholar]
- 37. Picault N, Palmieri L, Pisano Iet al. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria - bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2002;277:24204–11. [DOI] [PubMed] [Google Scholar]
- 38. Lüttge U, Ba LE. Electrochemical investigation of active malic acid transport at the tonoplast into the vacuoles of the CAM plant Kalanchoë daigremontiana. J Membr Sci. 1979;47:401–22. [Google Scholar]
- 39. Daccord N, Celton JM, Linsmith Get al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49:1099. [DOI] [PubMed] [Google Scholar]
- 40. Verde I, Jenkins J, Dondini Let al. The peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics. 2017;18:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chagne D, Crowhurst RN, Pindo Met al. The draft genome sequence of european pear (Pyrus communis L). PLoS One. 2014;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vanburen R, Bryant D, Bushakra JMet al. The genome of black raspberry (Rubus occidentalis). Plant J. 2016;87:535–47. [DOI] [PubMed] [Google Scholar]
- 43. Pilar F, Pilar H, José F. Determination of organic acids in fruits and vegetables by liquid chromatography with tandem-mass spectrometry. Food Chem. 2012;132:1049–54. [Google Scholar]
- 44. Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72. [DOI] [PubMed] [Google Scholar]
- 45. Gleave AP. A versatile binary vector system with a T-DNA organizational-structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–7. [DOI] [PubMed] [Google Scholar]
- 46. Narsai R, Law SR, Carrie Cet al. N-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 2011;157:1342–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun HJ, Sayaka U, Shin Wet al. A highly efficient transformation protocol for micro-tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006;47:426–31. [DOI] [PubMed] [Google Scholar]
- 48. Li D, Shi W, Deng X. Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep. 2002;21:153–6. [Google Scholar]
- 49. Masoodi KZ, Lone SM, Rasool RS. Genomic DNA extraction from the plant leaves using the CTAB method. Advanced Methods in Molecular Biology and Biotechnology. 2021;7:37–44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.