Abstract
The development of a rhinovirus (RV)-RNA-specific reverse transcription (RT)-PCR assay is complicated by the close homology between the RV and enterovirus (EV) genomes in the highly conserved 5′-noncoding region, which is chosen for primer design in most RT-PCR assays. We have developed a sensitive, rapid, and RV-specific nested RT-PCR assay and have used it to test nasopharyngeal aspirates from 556 patients presenting with acute respiratory tract infections. RV RNA was detected by nested RT-PCR not only in all of 52 samples that were RV positive by virus isolation methods but also in 124 of 367 samples that were negative by virus isolation methods and enzyme-linked immunosorbent assay (ELISA). In addition, in 23 of 137 samples that were positive for a different respiratory virus by virus isolation and/or ELISA, RV RNA was detected by RT-PCR. EVs, adenoviruses, respiratory syncytial viruses, coronaviruses, and influenza and parainfluenza viruses, including clinical isolates as well as stock viruses, were not amplified in our RV-specific RT-PCR assay, indicating that this assay was highly specific. The processing time was less than 2 days for the RT-PCR, as opposed to up to 2 weeks for virus isolation. These results indicate that nested RT-PCR is more sensitive than conventional methods for the detection of RV in patients experiencing acute respiratory tract infections and represents the only reliable tool for the early laboratory diagnosis of RV infections. This is especially important in light of new opportunities for therapy currently being developed.
Human rhinoviruses (RVs) cause an estimated one-third to one-half of all acute respiratory tract infections throughout the year (5, 17) and account for the majority of respiratory illnesses during spring and fall (6). Infections with RVs are usually limited to the upper respiratory tract. However, these viruses have also been shown to be involved in acute otitis media (2), sinusitis (22), and lower respiratory tract infections (5, 21).
Infants, the elderly, and people with conditions associated with preexisting airway inflammation such as asthma, cystic fibrosis, and tobacco smoking are at especially high risk for severe complications in the course of RV infections, including wheezing, exacerbations of asthma, small-airway obstruction, bronchitis, and pneumonia (6, 10). Due to the existence of more than 100 different RV serotypes, previous infection does not confer complete immunity, and individuals therefore frequently experience reinfection by an other serotype.
Enteroviruses (EVs), a genus within the family Picornaviridae, that is distinct from the RVs, may cause quite similar clinical symptoms of acute respiratory tract infections, particularly in young children (16). Both EVs and RVs are single-stranded RNA viruses with a highly conserved 5′-noncoding region (5′-NCR) common to all serotypes of EVs and RVs.
Previously developed reverse transcription (RT)-PCR assays for the detection of RV RNA in respiratory secretions took advantage of the 5′-NCR by using several short conserved stretches in this region as binding sites for oligonucleotide probes and primers (1, 4, 7, 9, 11, 23). Due to the close homology between EVs and RVs within this region, viruses of both genera are amplified by these RT-PCR assays. Therefore, additional steps for differentiation between these viruses are required, either by hybridization with RV-specific probes (1, 4, 9, 11, 22) or by restriction enzyme digestion (12). These steps are laborious and time-consuming and consequently reduce the usefulness of these assays for therapeutic decisions.
Virus isolation in tissue culture followed by an acid lability test represents the standard method in the laboratory for confirmation of clinically suspected RV infections (27), but the time required for results to become available is up to 2 weeks, also making this assay of limited value for therapeutic decisions.
Because new therapeutic medications will soon be available, cost-effective and sensitive methods for the rapid detection of RVs in clinical samples are urgently required. In addition, a sensitive and specific virus detection assay is a prerequisite for reliable epidemiological data and for clinical studies on the sequelae of RV infections. The aim of this study, therefore, was to develop a highly sensitive and specific PCR assay for the rapid and cost-effective diagnosis of RV infections.
MATERIALS AND METHODS
Patients and specimens.
A total of 556 nasopharyngeal aspirates (NPAs) obtained from patients with various forms of acute respiratory tract infection were tested for the presence of RV RNA by RT-PCR. The patients were 1 week through 88 years of age with a median age of 2.5 years (mean, 11.3; standard deviation, 23.3). The clinical diagnoses of these patients included rhinitis (6.4%), laryngitis (9.4%), bronchitis (46.4%), pneumonia (18.5%), asthma with coincident respiratory tract infection (1.3%), nonspecific febrile illness (15.0%), otitis media (0.9%), coughing (1.7%), and pharyngitis (0.4%).
The aspirates were diluted 1:3 in RPMI medium and were screened by tissue culture virus isolation and enzyme-linked immunosorbent assay (ELISA) for the presence of the following viruses: RVs, influenza viruses A and B, parainfluenza viruses, respiratory syncytial viruses (RSVs), adenoviruses, and EVs. Seventy-four samples collected from patients with acute respiratory tract infection in the period between January 1988 and August 1999 had been stored at −70°C and were retrospectively tested for the presence of RV RNA by RT-PCR. These samples were selected according to the following criteria: (i) positive for RV by virus isolation (n = 35), negative for RV by virus isolation but obtained from patients with acute respiratory tract infection (n = 16), and positive for a virus other than RV (EV, n = 5; RSV, n = 6; parainfluenza, n = 4; and/or adenovirus, n = 9) by virus isolation methods or ELISA.
From August 1999 to January 2000, clinical specimens from 482 patients with acute respiratory tract infection were prospectively tested by RT-PCR in addition to antigen-ELISA and virus isolation methods.
Testing of nasopharyngeal secretions for respiratory viruses. (i) Virus isolation in tissue culture.
The RV-sensitive “Ohio” strain of HeLa cells (25), kindly provided by D. A. I. Tyrrell (Clinical Research Centre, Common Cold Unit, Salisbury, Wiltshire, England), was used for virus isolation as described previously (13). The cells were inoculated before they were confluent, 24 h after seeding. Prior to inoculation, the NPAs were diluted 1:3 in RPMI medium containing 200 μg of neomycin (GIBCO, Life Technologies, Lofer, Austria) per ml and 200 U of streptomycin (GIBCO, Life Technologies, Lofer, Austria) per ml and incubated for 2 h at 4°C. Then 200 μl of the NPA-RPMI dilution was inoculated into each of two cell culture tubes and incubated on a roller drum at 33°C for 2 h. After the tubes had been washed with Hanks' balanced salt solution, 1.5 ml of the maintenance medium (minimal essential medium [MEM], 1% heat-inactivated fetal calf serum, 10 mg of neomycin per ml, 1% MEM nonessential amino acids, 0.03 M MgCl2, and 1% glutamine) was added. The tubes were then incubated on the roller drum for 10 days at 33°C, and all cultures were examined daily for cytopathic effects.
Classification of EVs and RVs was done by the acid lability test. The test for pH lability was considered RV positive if the virus was inactivated at pH 3 and simultaneously showed a characteristic cytopathic effect at pH 7. RSVs; adenoviruses; parainfluenza viruses 1, 2, and 3, and influenza viruses A and B were further identified by indirect fluorescent-antibody assay or by ELISA.
(ii) ELISA.
The ELISA for detection of antigens from RSVs; adenoviruses; parainfluenza viruses 1, 2, and 3; and influenza viruses A and B in NPAs was performed as previously described (14, 24).
Preparation of stock viruses.
HeLa cells were used for the propagation of RV and EV stocks. Supernatants of RV-infected and uninfected HeLa cells served as positive and negative controls, respectively. The different RV serotypes were obtained from the American Type Culture Collection (serotypes 1A, 1B, 2 through 15, 17 through 19, 21 through 24, and 26 through 89). Stock virus preparations of RV 14, RV 72, and RV 87 contained virus concentrations of 105 50% tissue culture-infective doses (TCID50) per ml.
In order to evaluate the specificity of our RT-PCR with regard to the coamplification of EV sequences, 10-fold serial dilutions of tissue culture-propagated EVs obtained from the American Type Culture Collection (polioviruses 1, 2, and 3; echoviruses 6, 18, 24, and 30; coxsackieviruses B1 through B6; and coxsackievirus A9) were tested by RT-PCR. In addition, tissue-culture-grown adenoviruses, RSVs, and coronaviruses were used as negative controls. All stock virus preparations of EVs and controls contained virus concentrations of ≥105 TCID50 per ml.
Detection of RV RNA sequences in stock virus preparations and in clinical specimens. (i) Primer selection and sequences.
The genomic sequence of RV 14 was used for the selection of primers, and this serotype was our reference strain in all of our RV-specific RT-PCR assays. The sequences of the primers chosen for RT and the first step of PCR, amplifying a fragment of 106 base pairs, were as follows: cDNA, 5′-CCC CTG AAT G(CT)G GCT AAC CT-3′; reverse, 5′-CGG ACA CCC AAA GTA GT(CT) GGT C-3′. For nested PCR we used primers amplifying a fragment of 93 base pairs: cDNA, 5′-GAA TG(CT) GGC TAA CCT TAA (AC)CC-3′; reverse, 5′-CAA AGT AGT (CT)GG TCC C (AG)T CC-3′.
The National Center for Biotechnology Information (Bethesda, Md.)'s sequence similarity search tool “BLAST” was used for the selection of RV and EV prototype strains to test the specificity and sensitivity of the RT-PCR. RVs and EVs were chosen according to their homologies to the amplified sequences of RV 14 (bases 460 to 580). In this analysis the following EVs exhibited the closest homology to RV 14 (ordered from highest to lowest homology): poliovirus 3, echoviruses 6 and 7, and coxsackieviruses B3 and B6. With regard to the homology between RV 14 and published sequences of other RV serotypes, RV 72 exhibited intermediate and RV 87 the lowest homology to RV 14.
(ii) Preparation of samples for RT-PCR.
Immediately after the addition of 1 μl of RNase inhibitor (Boehringer GmbH, Mannheim, Germany) to a final concentration of 0.01 U/μl, viral RNA was extracted from cell culture supernatants and NPAs by QIAamp viral RNA kits (Qiagen, Hilden, Germany).
(iii) RT.
For RT, an aliquot (10 μl) of the extracted RNA was added to the reaction mixture, yielding a total volume of 50 μl. The mixture consisted of 10 μl of EZ buffer (5× buffer [Gene Amp Kit; Perkin-Elmer/Cetus Corp., Norwalk, Conn.]), 4 μl of Mn(OAc)2 (25 mM solution), 8 μl of deoxynucleoside triphosphates (dNTPs: dATP, dCTP, dGTP, and dUTP), 25 pmol of each primer, 2 μl of rTth DNA polymerase (2.5 U/μl [Gene Amp kit; Perkin-Elmer/Cetus]), 0.25 μl of AmpErase UNG (1 U/μl; catalog no. N808-0068; Perkin-Elmer, Branchburg, New Jersey), and 15 μl of double-distilled H2O. The reaction mixtures were incubated at 60°C for 30 min.
(iv) Amplification.
Immediately after RT at 60°C, the reaction mixture (cDNA) was incubated in a DNA Thermal Cycler (Perkin-Elmer/Cetus) through 50 cycles of programmed amplification (denaturation at 94°C for 20 s, annealing at 60°C for 30 s, extension at 72°C for 9 s, and final incubation for 5 min at 72°C). The second amplification step was performed with 2 μl of the RT-PCR mixture, consisting of 5 μl of EZ buffer (10× buffer; Perkin-Elmer/Cetus), 4 μl of MgCl2 (25 mM solution), 8 μl of dNTPs (dATP, dCTP, dGTP, and dUTP), 25 pmol of each primer, 0.2 μl of Taq-Gold DNA polymerase (5 U/μl; 250 U of AmpliTaq Gold DNA polymerase; Perkin-Elmer/Cetus), and 29 μl of double-distilled H2O in a final volume of 50 μl. Ten minutes of denaturation was followed by 45 cycles of thermocycling (denaturation at 94°C for 15 s, annealing at 60°C for 30 s, extension at 72°C for 8 s, and final incubation for 5 min at 72°C). Each PCR experiment included at least one positive and several negative controls, interposed between the samples tested.
To prevent carryover of nonspecific products produced prior to amplification, AmpErase UNG was added to the reaction mixture of the first-step PCR (1°PCR) and the mixture was incubated prior to RT for 2 min at 50°C. AmpErase UNG was gradually inactivated by exposure to 94°C for 20 s in every cycle of the 1°PCR and by a final incubation at 95°C for 2 min after the last cycle of the PCR. Evaluation of the influence of AmpErase UNG on the sensitivity of the RT-PCR assay was done by testing 10-fold dilutions of RV 14 stock virus preparations with and without this enzyme. The addition of AmpErase UNG did not influence the sensitivity of our RT-PCR.
(v) Evaluation of the effect of extension time on RT-PCR results.
Stock virus preparations of RV 14 (105 TCID50/ml) and poliovirus 3 (106 TCID50/ml) were tested by RT-PCR using extension times of 7, 8, 9, 10, 15, 20, 25, and 30 s. The prolongation of the extension time from 9 to 15 s resulted in the amplification of poliovirus 3 RNA (extracted from stock virus preparations containing ≥105 TCID50/ml) to detectable amounts, indicating a loss of specificity. Unfortunately, these PCR products were indistinguishable from RV RNA by gel electrophoresis. Reduction of the extension time from 15 to 10 s reduced amplification of EV RNA below detectable levels. An extension time of 9 s for the first step and 8 s for the second step was found to be optimal in terms of sensitivity and specificity and was therefore used for testing stock virus preparations and NPAs. Reduction of extension time beyond this threshold value reduced the sensitivity for the detection of RV RNA without increasing specificity.
(vi) Visualization of PCR amplicons.
The PCR amplicons, in 10-μl volumes, were analyzed by gel electrophoresis and ethidium bromide staining on 3% NuSieve agarose gel (FMC Bioproducts, Rockland, Maine) with 0.5 μg of ethidium bromide per ml in the gel.
Statistical analysis.
Statistical comparison for significant differences between groups of patients was carried out with the chi-square test and the Fisher exact test. The latter was used for two-by-two tables. A relationship was considered to be statistically significant if the P value was <0.05.
RESULTS
Sensitivity and specificity of the RT-PCR.
In order to determine the ability of the RT-PCR assay to detect a wide range of RV serotypes, 87 different tissue-culture-grown serotypes were tested, and the genomic sequences of all serotypes tested were amplified to detectable levels by the RT-PCR assay, as summarized in Table 1. The sensitivity of this assay for the different RV serotypes differed from that of RV 14, our prototype strain, in proportion to their genomic differences within the 5′-NCR, as determined by testing 10-fold dilutions of RV 14, RV 72, and RV 87 stock virus preparations. RV 14 could be detected up to a serial dilution of 1:109, whereas RV 72 could be detected up to a dilution of 1:107 and RV 87 up to a dilution of 1:105.
TABLE 1.
RV RT-PCR results obtained by testing stock virus preparations of reference virus strains
| RT-PCR result for strain(s)
| |
|---|---|
| Positive | Negative |
| RV 1a | Coxsackieviruses B1–B6 |
| RV 1b | Echoviruses 18, 24, 30 |
| RV 2–9 | Polioviruses 1–3 |
| RV 11–15 | Influenza virus A(H3N2) |
| RV 17–19 | Influenza virus B/Harbin |
| RV 21–24 | Adenovirus 5 |
| RV 26–89 | RSV (long strain of subgroup A) |
Likewise, specificity was determined (i) by testing 10-fold dilutions of EV stocks (coxsackieviruses B1 through B6; echoviruses 18, 24, and 30; and polioviruses 1 through 3), (ii) by testing tissue-culture-grown poliovirus 3 at a concentration of 106 TCID50/ml in 10-fold dilutions, and (iii) by testing stock virus preparations of other respiratory viruses (adeno-, influenza-, and parainfluenzaviruses, coronavirus OC43; and RSVs). Poliovirus 3 was selected because it exhibited the closest homology to the primer binding sites of RV 14 as revealed by BLAST analysis. Genomic sequences of EVs and the other respiratory viruses were never amplified in the RV-specific RT-PCR, and stock virus preparations of these viruses were used as negative controls for routinely testing NPAs and were never amplified in the RT-PCR assay (Table 1). The time required for testing by RT-PCR (including RNA extraction, amplification, and gel electrophoresis) was less than 2 days.
Detection of rhinoviruses in NPAs by RT-PCR.
The overall detection rates for respiratory viruses in NPAs increased to 56.3% (313 of 556) when the RT-PCR assay was added, as opposed to 34% (189 of 556) when only ELISA and virus isolation were used. Of 556 clinical samples, 199 (35.8%) were positive for RV RNA by RT-PCR (Table 2), and in 137 NPAs a different respiratory virus could be detected. No virus could be detected in 243 of 556 patients. All of the NPAs that were RV positive by virus isolation (52 of 556) yielded positive results in our RT-PCR. In addition, RV-RNA was detected in 124 samples obtained from patients with acute respiratory tract infections that were negative by virus isolation and ELISA. RVs and a second respiratory virus could be detected in NPAs collected from 23 patients (RVs and RSVs, n = 13; RVs and influenza A viruses, n = 2; RVs and adenoviruses, n = 5; and RVs and parainfluenzaviruses, n = 3).
TABLE 2.
Results of the RV-specific RT-PCR compared with results of virus isolation and ELISA
| RV RT-PCR result | No. of cases by virus isolation and/or ELISA with result
|
||
|---|---|---|---|
| Negative | RV positive | OVRa | |
| Negative | 243 | 0 | 114 |
| Positive | 124 | 52 | 23 |
OVR, other viruses than RV (RSV, n = 36; influenza A virus, n = 69; adenoviruses, n = 12; parainfluenza viruses, n = 9; EVs, n = 11).
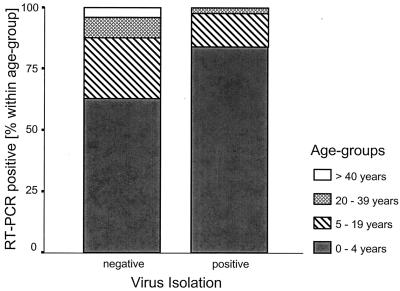
The highest detection rates of RVs in the 482 prospectively tested samples were observed in November 1999, within the peak incidence of RV infections (6, 18). In our study the failure of virus isolation to confirm RT-PCR-diagnosed RV infections increased significantly with increasing subject age (P = 0.029 [chi-square test]) (Fig. 1). In children younger than 5 years, the infecting RV could be detected by virus isolation in 37.8% (37 of 98) of RT-PCR-positive samples; in those between 5 and 19 years of age, in 20% (6 of 30); and in those older than 19 years, 7.7% (1 of 13). Failure rates of virus isolation were not significantly influenced by the affected part of the respiratory tract (lower versus upper) and the detection of a second respiratory virus within the same sample by ELISA or virus isolation (P = 0.74 and P = 0.66 [Fisher's exact test]).
FIG. 1.
Results of RT-PCR and virus isolation methods in relation to the age of the patients.
No significant association could be observed between a double infection (RV and a second respiratory virus) and the affected part of the respiratory tract (P = 0.46 [Fisher's exact test]). The frequency of double infections increased significantly with decreasing age, and the age group most commonly affected was those younger than 0.5 years (P < 0.01 [Fisher's exact test]). In this age group, 24.1% (13 of 54) of all patients experienced a double infection, in contrast to 6.5% (9 of 129) of patients older than 0.5 years.
DISCUSSION
All previously described RV-specific RT-PCR assays either have required an additional step for differentiating between RVs and EVs (1, 4, 7, 9, 11, 23), or samples were differentiated by different lengths of RV and EV amplification products (21). All had in common a low number of RV serotypes tested. Our results clearly demonstrated that the establishment of a highly sensitive RV-specific RT-PCR for the early detection of most if not all RV serotypes in NPAs is possible, since all 87 different RV serotypes tested were detected by the RT-PCR. This result was achieved by careful primer selection and by optimizing cycling conditions. To avoid coamplification of EVs and to enable the sensitive simultaneous detection of all RV serotypes, the following primer selection strategy was pursued: (i) the 5′-NCR of the picornaviral genome was chosen as the primary target because of its high degree of conservation among all RV serotypes; (ii) multiple alignment was performed on the genomic sequences of 12 RV and 13 EV serotypes obtained from GenBank (Bethesda, Md.) using the commercial software MegAlign (DNASTAR Inc., Madison, Wis.). By this procedure, three highly conserved regions within the 5′-NCR were identified (regions A, B, and C [see Fig. 2]); (iii) of these three regions of the picornavirus genome, B and C were further selected as primer binding sites because they exhibited the closest homologies among all aligned RVs and contained scattered mismatches with the genomes of EVs. Region A exhibited less homology between aligned RVs, and potential primers for this region were predicted to form loops at relatively high annealing temperatures. Primer binding sites selected within regions B and C exhibited a maximum of mismatches to the EV genome located complementary to the 3′ terminus of each primer.
FIG. 2.
Diagrammatic representation of conserved regions A, B, and C within the 5′-noncoding region of the picornaviruses. The primer binding sites for the primers used are indicated in the expanded section in relation to the genome of RV 14. The positions of the two PCR products are represented by horizontal bars and product-lengths are indicated to the right in base pairs. 1°PCR, first-step PCR; 2°PCR, second-step PCR.
The effect of the mismatches within the primer binding sites was increased by a long annealing time and an extremely short extension time. Extension time was of utmost importance since a prolonged extension time negatively influenced the specificity of the RT-PCR, whereas a reduced extension time decreased sensitivity for the detection of RV RNA. Therefore, the additional step of differentiation between EVs and RVs following amplification was not necessary, and this significantly shortened the interval between the onset of symptoms and laboratory diagnosis. This time savings increased the usefulness of this assay for the treating physicians, who increasingly used the results for therapeutic decisions during the period of prospective investigation (personal communication).
In a considerable proportion of the clinical samples, this assay was the only reliable tool for diagnosing an RV infection, as two-thirds of all RV infections could not be confirmed by virus isolation methods. Although these positive results could not be confirmed by the far less sensitive conventional method of virus isolation in tissue culture, the high specificity of the RT-PCR when various stock virus preparations were tested and the consistently negative cross-contamination controls rendered nonspecific positive results very unlikely. Furthermore, none of the other viruses frequently implicated in acute respiratory tract infections could be detected in the majority of the NPAs that were positive for RVs by RT-PCR (except in patients with double infections), and the samples were predominantly obtained in the autumn, a seasonal peak of RV infection.
Our RT-PCR represents the method of choice especially for adult patients, since, in this patient group, virus isolation is able to diagnose only 7.7% of all RV infections. Consequently, application of RT-PCR provides the only reliable means to investigate the impact of RV infections on all patient groups.
Although RV infection rates decline with increasing age, the higher detection rate of RVs achieved by RT-PCR seems all the more valuable clinically for older age groups, since RVs tend to infect the lower respiratory tract more frequently with increasing age (18).
Double infections with RV and another respiratory virus were observed more frequently in our investigation (23 of 199 cases) than in other studies (14). Since cross-contamination controls were consistently negative and all positive RT-PCR results could be confirmed in a second run, false-positive results can be ruled out. The increased detection rate of double infections may be explained by the high sensitivity of the RT-PCR, which increases detection rates in general and therefore makes it possible to detect viral pathogens more frequently. This applies especially to very young patients, who usually experience multiple consecutive respiratory tract infections with a variety of respiratory viruses in a relatively short period of time (3, 19).
Clarifying the etiology of every acute respiratory tract infection and providing adequate treatment based on laboratory diagnosis is of particular importance to prevent overprescription of antibiotics (20). In addition, antiviral compounds for the treatment of picornavirus infections may soon be available (8, 15, 26). Considered in this light, rapid and reliable detection of the infecting agents in acute respiratory tract infections is not only of academic interest but will provide the prerequisite for specific treatment of RV infections.
ACKNOWLEDGMENTS
We thank Barbara Dalmatiner and Sylvia Malik for their excellent technical assistance and Steven L. Allison for critical reading of the manuscript.
REFERENCES
- 1.Andeweg A C, Bestebroer T M, Huybreghs M, Kimman T G, de Jong J C. Improved detection of rhinoviruses in clinical samples by using a newly developed nested reverse transcription-PCR assay. J Clin Microbiol. 1999;37:524–530. doi: 10.1128/jcm.37.3.524-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arola M, Ziegler T, Ruuskanen O, Mertsola J, Näntö-Salonen K, Halonen P. Rhinovirus in acute otitis media. J Pediatr. 1988;113:693–695. doi: 10.1016/S0022-3476(88)80380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman S, McIntosh K. Selective primary health care: strategies for control of disease in the developing world. XXI. Acute respiratory infections. Rev Infect Dis. 1985;7:674–691. doi: 10.1093/clinids/7.5.674. [DOI] [PubMed] [Google Scholar]
- 4.Blomqvist S, Skyttä A, Roivainen M, Hovi T. Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. J Clin Microbiol. 1999;37:2813–2816. doi: 10.1128/jcm.37.9.2813-2816.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couch R B. Rhinoviruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T P, Meinick J L, Roizman B, editors. Fields' virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 713–734. [Google Scholar]
- 6.Gern J E, Busse W W. Association of rhinovirus infections with asthma. Clin Microbiol Rev. 1999;12:9–18. doi: 10.1128/cmr.12.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halonen P, Rocha E, Hierholzer J, Holloway B, Hyypiä T, Hurskainen P, Pallansch M. Detection of enteroviruses and rhinoviruses in clinical specimens by PCR and liquid-phase hybridization. J Clin Microbiol. 1995;33:648–653. doi: 10.1128/jcm.33.3.648-653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hueston W J, Mainous A G, Ornstein S, Pan Q, Jenkins R. Antibiotics for upper respiratory tract infections: follow-up utilization and antibiotic use. Arch Fam Med. 1999;8:426–430. doi: 10.1001/archfami.8.5.426. [DOI] [PubMed] [Google Scholar]
- 9.Ireland D C, Kent J, Nicholson K G. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. J Med Virol. 1993;40:96–101. doi: 10.1002/jmv.1890400204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston S L, Pattemore P K, Sanderson G, Smith S, Campbell M J, Josephs L K, Cunningham A, Robinson B S, Myint S H, Ward M E, Tyrrell D A, Holgate S T. The relationship between upper respiratory infections and hospital admissions for asthma: a time-trend analysis. Am J Respir Crit Care Med. 1996;154:654–660. doi: 10.1164/ajrccm.154.3.8810601. [DOI] [PubMed] [Google Scholar]
- 11.Johnston S L, Sanderson G, Pattemore P K, Smith S, Bardin P G, Bruce C B, Lambden P R, Tyrrell D A, Holgate S T. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J Clin Microbiol. 1993;31:111–117. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kämmerer U, Kunkel B, Korn K. Nested PCR for specific detection and rapid identification of human picornaviruses. J Clin Microbiol. 1994;32:285–291. doi: 10.1128/jcm.32.2.285-291.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellner G, Popow-Kraupp T, Kundi M, Binder C, Wallner H, Kunz C. Contribution of rhinoviruses to respiratory viral infections in childhood: a prospective study in a mainly hospitalized infant population. J Med Virol. 1988;25:455–469. doi: 10.1002/jmv.1890250409. [DOI] [PubMed] [Google Scholar]
- 14.Kellner G, Popow-Kraupp T, Popow C, Kundi M, Kunz C. Surveillance of viral respiratory tract infections over a one year period in mainly hospitalized Austrian infants and children by a rapid enzyme-linked immunosorbent assay diagnosis. Wien Klin Wochenschr. 1990;102:100–106. [PubMed] [Google Scholar]
- 15.Mainous A G, Hueston W J, Love M M. Antibiotics for colds in children: who are the high prescribers? Arch Pediatr Adolesc Med. 1998;152:349–352. [PubMed] [Google Scholar]
- 16.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Monath T P, Meinick J L, Roizman B, editors. Fields' virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 655–712. [Google Scholar]
- 17.Monto A S. Studies of the community and family: acute respiratory illness and infection. Epidemiol Rev. 1994;16:351–373. doi: 10.1093/oxfordjournals.epirev.a036158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monto A S, Bryan E R, Ohmit S. Rhinovirus infections in Tecumseh, Michigan: frequency of illness and number of serotypes. J Infect Dis. 1987;156:43–49. doi: 10.1093/infdis/156.1.43. [DOI] [PubMed] [Google Scholar]
- 19.Monto A S, Bryan E R, Rhodes L M. The Tecumseh study of respiratory illness. VII. Further observations on the occurrence of respiratory syncytial virus and Mycoplasma pneumoniae infections. Am J Epidemiol. 1974;100:458–468. doi: 10.1093/oxfordjournals.aje.a112058. [DOI] [PubMed] [Google Scholar]
- 20.Monto A S, Cavallaro J J. The Tecumseh study of respiratory illness. IV. Prevalence of rhinovirus serotypes, 1966–1969. Am J Epidemiol. 1972;96:352–360. doi: 10.1093/oxfordjournals.aje.a121466. [DOI] [PubMed] [Google Scholar]
- 21.Olive D M, Al-Mufti S, Al-Mulla W, Khan M A, Pasca A, Stanway G, Al-Nakib W. Detection and differentiation of picornaviruses in clinical samples following genomic amplification. J Gen Virol. 1990;71:2141–2147. doi: 10.1099/0022-1317-71-9-2141. [DOI] [PubMed] [Google Scholar]
- 22.Pitkäranta A, Arruda E, Malmberg H, Hayden F G. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. J Clin Microbiol. 1997;35:1791–1793. doi: 10.1128/jcm.35.7.1791-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santti J, Hyypiä T, Halonen P. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J Virol Methods. 1997;66:139–147. doi: 10.1016/s0166-0934(97)00049-9. [DOI] [PubMed] [Google Scholar]
- 24.Sarkkinen H K, Halonen P E, Arstila P P, Salmi A A. Detection of respiratory syncytial, parainfluenza type 2, and adenovirus antigens by radioimmunoassay and enzyme immunoassay on nasopharyngeal specimens from children with acute respiratory disease. J Clin Microbiol. 1981;13:258–265. doi: 10.1128/jcm.13.2.258-265.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stott E J, Tyrrell D A. Some improved techniques for the study of rhinoviruses using HeLa cells. Arch Gesamte Virusforsch. 1968;23:236–244. doi: 10.1007/BF01241896. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q M. Protease inhibitors as potential antiviral agents for the treatment of picornaviral infections. Prog Drug Res. 1999;52:197–219. doi: 10.1007/978-3-0348-8730-4_5. [DOI] [PubMed] [Google Scholar]
- 27.Yun B Y, Kim M R, Park J Y, Choi E H, Lee H J, Yun C K. Viral etiology and epidemiology of acute lower respiratory tract infections in Korean children. Pediatr Infect Dis J. 1995;14:1054–1059. doi: 10.1097/00006454-199512000-00005. [DOI] [PubMed] [Google Scholar]