Abstract
Background:
Despite improvements in disease diagnosis, treatment, and prognosis, breast cancer is still a leading cause of cancer death for women. Compelling evidence suggests that targeting cancer stem cells (CSCs) have a crucial impact on overcoming the current shortcomings of chemotherapy and radiotherapy. In the present study, we aimed to study the effects of T cells and a critical anti-tumor cytokine, interferon-gamma (IFN-γ), on breast cancer stem cells.
Methods:
BALB/c mice and BALB/c nude mice were subcutaneously injected with 4T1 tumor cells. Tumor growth and pulmonary metastasis were assessed. ALDEFLOUR™ assays were performed to identify aldehyde dehydrogenasebright (ALDHbr) tumor cells. ALDHbr cells as well as T cells from tumor-bearing BALB/c mice were analyzed using flow cytometry. The effects of CD8+ T cells on ALDHbr tumor cells were assessed in vitro and in vivo. The expression profiles of ALDHbr and ALDHdim 4T1 tumor cells were determined. The levels of plasma IFN-γ were measured by enzyme-linked immunosorbent assay, and their associations with the percentages of ALDHbr tumor cells were evaluated. The effects of IFN-γ on ALDH expression and the malignancy of 4T1 tumor cells were analyzed in vitro.
Results:
There were fewer metastatic nodules in tumor-bearing BALB/c mice than those in tumor-bearing BALB/c nude mice (25.40 vs. 54.67, P < 0.050). CD8+ T cells decreased the percentages of ALDHbr 4T1 tumor cells in vitro (control vs. effector to target ratio of 1:1, 10.15% vs. 5.76%, P < 0.050) and in vivo (control vs. CD8+ T cell depletion, 10.15% vs. 21.75%, P < 0.001). The functions of upregulated genes in ALDHbr 4T1 tumor cells were enriched in the pathway of response to IFN-γ. The levels of plasma IFN-γ decreased gradually in tumor-bearing BALB/c mice, while the percentages of ALDHbr tumor cells in primary tumors increased. IFN-γ at a concentration of 26.68 ng/mL decreased the percentages of ALDHbr 4T1 tumor cells (22.88% vs. 9.88%, P < 0.050) and the protein levels of aldehyde dehydrogenase 1 family member A1 in 4T1 tumor cells (0.86 vs. 0.49, P < 0.050) and inhibited the abilities of sphere formation (sphere diameter <200 μm, 159.50 vs. 72.0; ≥200 μm, 127.0 vs. 59.0; both P < 0.050) and invasion (89.67 vs. 67.67, P < 0.001) of 4T1 tumor cells.
Conclusion:
CD8+ T cells and IFN-γ decreased CSC numbers in a 4T1 mouse model of breast cancer. The application of IFN-γ may be a potential strategy for reducing CSCs in breast cancer.
Keywords: Cancer stem cells, Aldehyde dehydrogenase, Interferon-gamma, CD8+ T cells
Introduction
Breast cancer is still the most common malignancy and the second leading cause of cancer-related death in women.[1] One reason that patients succumb to breast cancer is treatment resistance, which leads to metastasis and relapse.[2,3] Several studies have suggested that breast cancer stem cells (BCSCs) mediate metastasis, are resistant to radiation and chemotherapy, and contribute to relapse.[4–6] Cancer stem cells (CSCs) are a small population of cells in the tumor that have unique characteristics, such as self-renewal and the ability to generate heterogenic lineages of cancer cells. Furthermore, CSCs exhibit characteristics of epithelial-to-mesenchymal transition, a known mechanism of metastasis.[5,7,8] Therefore, BCSCs are considered to be the key population leading to resistance to radiotherapy and chemotherapy for breast cancer.[4,9]
Researchers have found several potential markers that are suggested for the identification of BCSCs in clinical practice, such as CD44/CD24 and aldehyde dehydrogenase (ALDH).[5,10–12] ALDH activity is a hallmark of BCSCs that is measurable by the ALDEFLOUR™ (Stem Cell Technologies, Vancouver, BC, Canada) assay.[13,14] In studies using murine breast cancer models, increased ALDH activity has been commonly used to define CSCs.[15] ALDH is a member of the nicotinamide adenine dinucleotide-dependent enzyme family that catalyzes the oxidation of aldehydes to acids.[16] Aldehyde dehydrogenase 1 family member A1 (ALDH1A1) is an ALDH that can catalyze the oxidation of retinaldehyde to retinoic acid, which has been associated with the stemness of CSCs as well as normal tissue stem cells.[13,17] ALDH can prevent elevated levels of reactive oxygen species in drug-tolerant cancer cells[18] and protect cells from cytotoxic drugs.[19] Suppressing ALDH1A1 through specific small interfering RNA (siRNA) sensitizes colon cancer cells to chemotherapy.[20] ALDH1A1-positive lung cancer cells displayed more resistance to gefitinib than ALDH1A1-negative lung cancer cells.[16] Inhibition of ALDH activity with the non-specific inhibitor diethylaminobenzaldehyde (DEAB) significantly suppressed the tumorsphere formation and lung metastasis abilities of breast cancer cells, including 4T1 cells. Silencing aldehyde dehydrogenase isoform 1 with a specific siRNA showed a similar inhibitory effect on tumorsphere formation.[21] Therefore, ALDH may be used as not only a marker for stem cells but also to regulate cellular functions related to self-renewal, expansion, differentiation, and resistance to drugs and radiation.[18] In our previous study, aldehyde dehydrogenasebright (ALDHbr) tumor cells were identified as CSCs for the 4T1 murine breast cancer cell line.[22] ALDHbr could be used as a CSC marker in the 4T1 mouse model of breast cancer, and its increased activity was a major mechanism that led to treatment resistance in BCSCs.[19,23–25]
The immunoediting theory consisting of immune elimination, equilibrium, and escape, is proposed to explain the dynamic interplay between the immune system and cancer cells. Immune cells, especially T lymphocytes, affect cancer cells, and even CSCs are associated with the prognosis of patients with breast cancer.[26] A major mechanism by which immune cells function is by releasing cytokines. Cytokines play an essential role in the development and propagation of a range of cell types, including cancer cells and CSCs.[6,27] Interferon-gamma (IFN-γ) is primarily secreted by activated T cells, natural killer cells, natural killer T cells, and gamma delta T (γδT) cells, which play a pivotal role in systemic and local immunity and are involved in host anti-tumor immunity.[28] IFN-γ has been found to promote the apoptosis of CSCs in colon cancer,[29] but it increases the percentages of CD133+ CSCs in hepatocellular carcinoma and does not promote the apoptosis of cell lines with high percentages of CD133.[30] It has also been reported that IFN-γ increases the proliferation and colony formation abilities of CD34+ stem/progenitor cells from patients with chronic myeloid leukemia in vitro.[31] The effect of IFN-γ on CSCs seems to be cancer- and context-dependent,[32] and the effects of IFN-γ on CSCs in breast cancer is still unclear. IFN-γ could inhibit the growth and metastasis of 4T1 tumor cells through phagocytic cells in vivo[33]; however, the specific mechanisms are unknown. Whether it functions by inhibiting CSCs in 4T1 tumor cells remains to be identified. In addition, the effect of IFN-γ on ALDH expression in tumor cells has not been reported. Here, we found that IFN-γ inhibited ALDH expression in 4T1 tumor cells and decreased the percentages of ALDHbr 4T1 tumor cells, thus revealing a potential beneficial effect of IFN-γ on reducing CSCs in breast cancer.
Methods
Cell culture
4T1 tumor cells, a mammary gland tumor cell line with autonomous metastatic potency, were purchased from American type culture collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco's modified Eagle medium: nutrient mixture F-12 (DMEM/F12) media (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Zhejiang Tianhang Biotechnology, Huzhou, Zhejiang, China), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Gibco, Grand Island, NY, USA) in a humidified atmosphere of 5% CO2 at 37°C.
Establishment of a subcutaneous 4T1 tumor model
Female BALB/c and BALB/c nude mice (5–6 weeks) were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) and maintained under specific pathogen-free conditions in the animal facility of Jiangsu University and Cancer Institute, Chinese Academy of Medical Sciences (Approval no.: SYXK[SI]2013-0036). All animal experiments were approved by the institutional review board of Jiangsu University and the Animal Care and Use Committee of the Cancer Institute and Hospital, Chinese Academy of Medical Sciences.
A total of 5 × 104 4T1 tumor cells in 50 μL of phosphate-buffered saline (PBS) were injected subcutaneously into the right flank of a female BALB/c mouse or BALB/c nude mouse (five mice per group). Tumors were measured using digital calipers once every 3 to 4 days and volumes were calculated using the following formula: volume = 0.5 × length × width2. Three weeks later, the mice were sacrificed. Their lungs were dissected and fixed and dyed with Bouin fixation solution. Then, the lungs were weighed and the number of lung metastatic nodules was counted.
Flow cytometry assay of ALDHbr 4T1 tumor cells and tumor-infiltrating cells (TILs) in primary tumors
Single-cell suspensions of primary tumor tissues were obtained as previously described.[22] Briefly, four 4T1 tumor-bearing mice per group were sacrificed at the indicated time points, and the primary tumor masses were isolated, minced, and digested with type IV collagenase (1 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA) and DNase I (300 U/mL) (Sigma-Aldrich) for approximately 1.5 h in a humidified atmosphere of 5% CO2 at 37°C. Cell suspensions were filtered through a 200-mesh screen and lysed with ammonium chloride solution (Stem Cell Technologies, Vancouver, BC, Canada) to exclude red blood cells. The ALDEFLOUR™ assay for ALDH activity was performed according to the manufacturer's instructions (ALDEFLOUR™ Kit, Stem Cell Technologies) as previously described.[22] In brief, cells were incubated in ALDEFLOUR™ assay buffer containing activated ALDEFLOUR™ substrate at 37°C for 45 min. For negative controls, activated ALDEFLOUR™ substrates were added into cells and mixed, and DEAB was immediately added. Then, the cells were stained with allophycocyanin-conjugated anti-CD45 (APC-CD45) and its isotype antibody at 4°C in the dark for 30 min. For measuring TILs, a total of 106 cells in 100 μL staining buffer were incubated with CD16/CD32 (eBioscience, San Diego, CA, USA) blocking antibodies at 4°C for 10 min and then stained with APC-CD45, fluorescein isothiocyante-conjugated anti-CD3 (FITC-CD3), peridinin-chlorophyll-protein complex/cyanine5.5-conjugated anti-CD4 (PerCP/Cy5.5-CD4), and phycoerythrin-conjugated anti-CD8 (PE-CD8) antibodies at 4°C in the dark for 30 min according to the manufacturer's instructions. All antibodies were purchased from BioLegend (San Diego, CA, USA) unless otherwise mentioned [Supplementary Table 1]. Stained cells were detected with a flow cytometer (BD LSRII, BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed using FlowJo software (TreeStar, Ashland, OR, USA).
Characterization of the splenic immune cell by flow cytometry
Splenocytes from four 4T1 tumor-bearing BALB/c mice per group were lysed with ammonium chloride solution (Stem Cell Technologies) and filtered through a 70-μm cell strainer to obtain single-cell suspensions. A total of 106 splenocytes in 100 μL of staining buffer from each sample were incubated with CD16/CD32 blocking antibodies at 4°C for 10 min and then stained with fluorescence-conjugated antibodies at 4°C in the dark for 30 min. In brief, APC-CD45, FITC-CD3, PerCP/Cy5.5-CD4, and PE-CD8 were used to stain CD4 T cells and CD8 T cells. PerCP/Cy5.5-CD4, allophycocyanin-conjugated anti-programed death-1 (APC-PD-1), and phycoerythrin-conjugated anti-C-X-C motif chemokine (PE-CXCR5) were used to stain follicular helper T (Tfh) cells. FITC-CD3 and phycoerythrin-conjugated anti-gamma delta T-cell receptor (PE-TCRγδ) were used to stain γδT cells. For intracellular Foxp3 staining of regulatory T cells (Tregs), cells were fixed and permeabilized according to the manufacturer's protocol (FOXP3 Fix/Perm Buffer Set, BioLegend) and incubated with PE-Foxp3 antibodies after surface staining with PerCP/Cy5.5-CD4. Stained cells were detected with a flow cytometer (BD LSRII). All antibodies were purchased from BioLegend unless otherwise mentioned. Data from flow cytometry assays were analyzed using FlowJo software (TreeStar).
In vivo lymphocyte depletion assays
For lymphocyte depletion, anti-CD4 (800 μg/200 μL, homemade) monoclonal antibodies (mAbs) (clone GK1.5) or anti-CD8 (800 μg/200 μL, homemade) mAbs (clone 2.43) were injected via the tail vein on day −5 and day −1 before 4T1 tumor inoculation (four mice per group) and thereafter once every 4 or 5 days until the end of the experiment, according to a previous protocol[34] with some modifications. Control groups of mice received an equivalent volume of PBS. Two weeks after 4T1 tumor inoculation, splenocytes were obtained as described above and the depletion efficiencies of splenic CD4+ T cells and CD8+ T cells were determined using flow cytometry.
Percentages of ALDHbr 4T1 tumor cells after coculturing with T lymphocytes
Splenocytes from 4T1 tumor-bearing BALB/c mice were prepared as described above. Splenic CD8+ T cells were isolated using a CD8a+ T Cell Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. 4T1 tumor cells were incubated with isolated CD8+ T cells at the indicated ratios. After 24-h incubation in a humidified atmosphere with 5% CO2 at 37°C, culture supernatants were discarded and cells adhering to culture plates were washed twice with PBS, digested, and assessed using the ALDEFLOUR™ assay. ALDHbr 4T1 tumor cells were detected with a flow cytometer (BD LSRII, BD Biosciences).
Gene expression microarray analysis of ALDHbr and ALDHdim 4T1 tumor cells
The ALDEFLOUR™ assay was used to detect ALDH activity in 4T1 tumor cells as mentioned above. ALDHbr and ALDHdim cells with high and low/negative ALDH activity were sorted using a FACSDiVa flow cytometer (BD Biosciences). Total RNA from both kinds of sorted cells was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and expression profiles were analyzed using Affymetrix GeneChip mouse Transcriptome Array 2.0 (Shanghai Baygene Biotechnology Company Limited, Shanghai, China). Gene ontology (GO) annotation was used to determine the functions of differentially expressed genes between ALDHbr 4T1 tumor cells and ALDHdim 4T1 tumor cells. Prediction terms with a P value < 0.05 were selected and ranked by their P values. The −log10 (P value) yielding an enrichment score was used to rank the significant GO terms of differentially expressed genes.
Determination of ALDH1A1 messenger RNA (mRNA) levels by real-time polymerase chain reaction (PCR)
A total of 500,000 4T1 tumor cells were seeded into a 10-cm culture dish and cultured with or without IFN-γ (26.68 ng/mL, PeproTech, Rocky Hill, NJ, USA). After 24-h incubation, total RNA was extracted using TRIzol reagent (Invitrogen). Reverse transcription was performed according to the manufacturer's instructions (PrimeScript™ RT reagent Kit with genomic DNA (gDNA) Eraser, Takara, Dalian, China). mRNA levels were determined using TB Green™ Fast qPCR Mix (Takara) with a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA). Primers for ALDH1A1 were purchased from Sangon Biotech Co. Ltd. (Shanghai, China) (F: TGTGAAGGCTGCAAGACAGG R: TGTAGCCAGCAGCAGACGAT).
Determination of plasma IFN-γ using enzyme-linked immunosorbent assay (ELISA)
Plasma was collected from 4T1 tumor-bearing mice at the indicated time point. The concentrations of plasma IFN-γ were determined using a Mouse IFN-γ ELISA Kit (Dakewe Biotech Co., Ltd., Shenzhen, China) according to the manufacturer's instructions.
Western blotting assay
A total of 500,000 4T1 tumor cells were seeded into a 10-cm culture dish and cultured with or without IFN-γ (26.68 ng/mL, PeproTech) for 24 h. The cells were harvested and lysed on ice with radioimmunoprecipitation assay lysis buffer (Solarbio, Beijing, China) containing a cocktail of proteinase inhibitors (Roche, Basel, Switzerland) for 30 min. Total proteins (30 μg/sample) were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene difluoride membrane. Rabbit-anti-mouse ALDH1A1 (Abcam, Cambridge, MA, UK) and glyceraldehyde-3-phosphate dehydrogenase (CST, Danvers, MA, USA) were used at a dilution of 1:1000. After washing, horseradish peroxidase-coupled goat-anti-rabbit immunoglobulin G (IgG) antibodies (Abcam) were used at a dilution of 1:2000. Protein bands were visualized using an enhanced chemiluminescence detection system (Millipore, Billerica, MA, USA) on a Tanon-5200 biomolecular imager (Tanon, Shanghai, China).
Immunocytochemistry (ICC)
A total of 60,000 4T1 tumor cells per well were seeded into a six-well plate in the presence or absence of IFN-γ (26.68 ng/mL, PeproTech) for 24 h. After washing with PBS, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 1% bovine serum albumin (Solarbio). Then, treated cells were incubated with rabbit-anti-mouse ALDH1A1 (Abcam) at a dilution of 1:100 at 4°C overnight. After washing with PBS with Tween-20, cells were incubated with Alexa Fluor 488-conjugated goat-anti-rabbit IgG antibodies (Beyotime, Shanghai, China) at a dilution of 1:500 at room temperature for 1 h. After washing, 4′,6-diamidino-2-phenylindole (Beyotime) was added for 5 min and washed and slides were mounted. Cells were observed under a fluorescence microscope (Leica, Wetzlar, Germany) at 400× magnification.
Sphere formation assay
A total of 15,000 4T1 tumor cells per well were seeded in an ultralow attachment six-well plate and cultured in plasma-free DMEM/F12 medium supplemented with 100 IU/mL penicillin, 100 μg/mL streptomycin (Gibco), epidermal growth factor (20 ng/mL, PeproTech), and basic fibroblast growth factor (20 ng/mL, PeproTech) with or without IFN-γ (26.68 ng/mL). One week later, the numbers of spheres were counted.
Migration and invasion assays
A total of 40,000 4T1 tumor cells per well were seeded into a 12-well Transwell insert with a pore size of 8 μm (CoStar, Corning, NY, USA) coated with or without 80 μL of Matrigel (Corning, Corning, NY, USA). DMEM/F12 supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco) with or without IFN-γ (26.68 ng/mL, PeproTech) were added into the lower chamber of the well. After 24-h incubation, migrated or invaded cells on membranes were fixed, stained, and counted under a microscope at 200× magnification. Five fields on a membrane were viewed, cell numbers in each field were counted, and an average number was calculated.
Proliferation assay
A total of 3000 4T1 tumor cells per well were seeded into a 96-well plate and cultured with or without IFN-γ (26.68 ng/mL, PeproTech). After 24-h incubation, cell counting kit-8 reagents (Dojindo, Kumamoto, Japan) were added according to the manufacturer's instructions and the optimal density value at 450 nm was measured with a microplate reader.
Statistical analyses
Statistical analyses were performed using SPSS version 10.0 (IBM SPSS Statistics, Armonk, NY, USA) or GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA). Data are presented as the means ± standard error. Independent Student's t test or one-way analysis of variance followed by Dunnett multiple comparison test or Tukey multiple comparison test was performed to analyze differences between two groups. A P value of < 0.050 was considered statistically significant.
Results
Fewer pulmonary metastases were observed in BALB/c mice than those in BALB/c nude mice
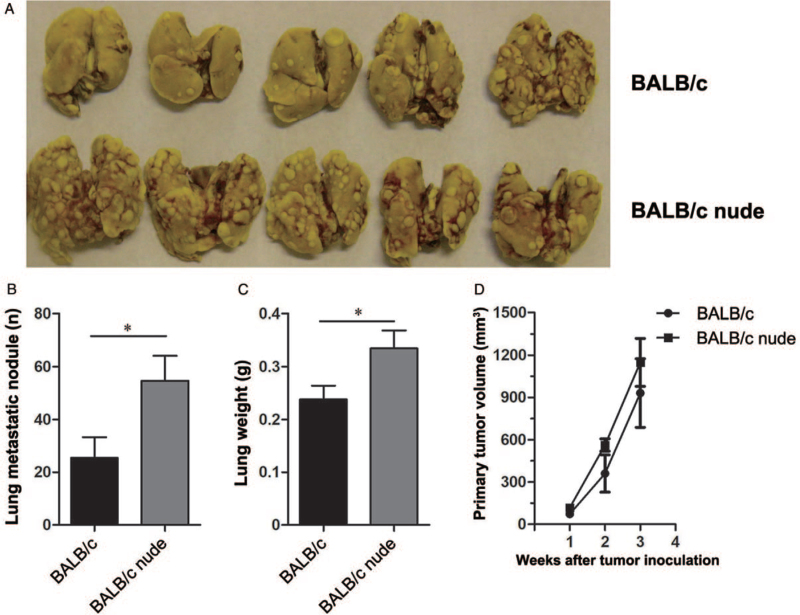
BALB/c mice with a complete immune system and BALB/c nude mice without a thymus, and therefore, lacking αβT cells were injected subcutaneously with 4T1 tumor cells. We observed significantly fewer pulmonary metastatic nodules (25.40 vs. 54.67, t = 2.32, P < 0.050) [Figure 1A and 1B] and lower lung weights (0.24 vs. 0.33, t = 2.25, P < 0.050) [Figure 1C] in the BALB/c group than those in the BALB/c nude group. Although the primary tumor grew slowly in the BALB/c group, there was no significant difference between the BALB/c group and the BALB/c nude group in primary tumor volumes [Figure 1D].
Figure 1.
Metastases and primary tumor growth in BALB/c and BALB/c nude mice. 4T1 tumor cells (5 × 104) were injected subcutaneously into the right flank of female BALB/c mice or BALB/c nude mice. Tumor growth was monitored, and volumes were calculated. Three weeks after tumor inoculation, the mice were sacrificed and their lungs were isolated. (A) A representative image of pulmonary metastasis of 4T1 tumor-bearing BALB/c mice and BALB/c nude mice. (B) The numbers of pulmonary metastatic nodules and (C) lung weights of 4T1 tumor-bearing BALB/c mice and BALB/c nude mice. (D) Primary tumor growth of BALB/c mice and BALB/c nude mice. Five mice per group were used and experiments were replicated at least twice. Data are presented as the mean ± standard error (SE). ∗P < 0.050.
CD8+ T cells are associated with decreased proportions of ALDHbr 4T1 tumor cells
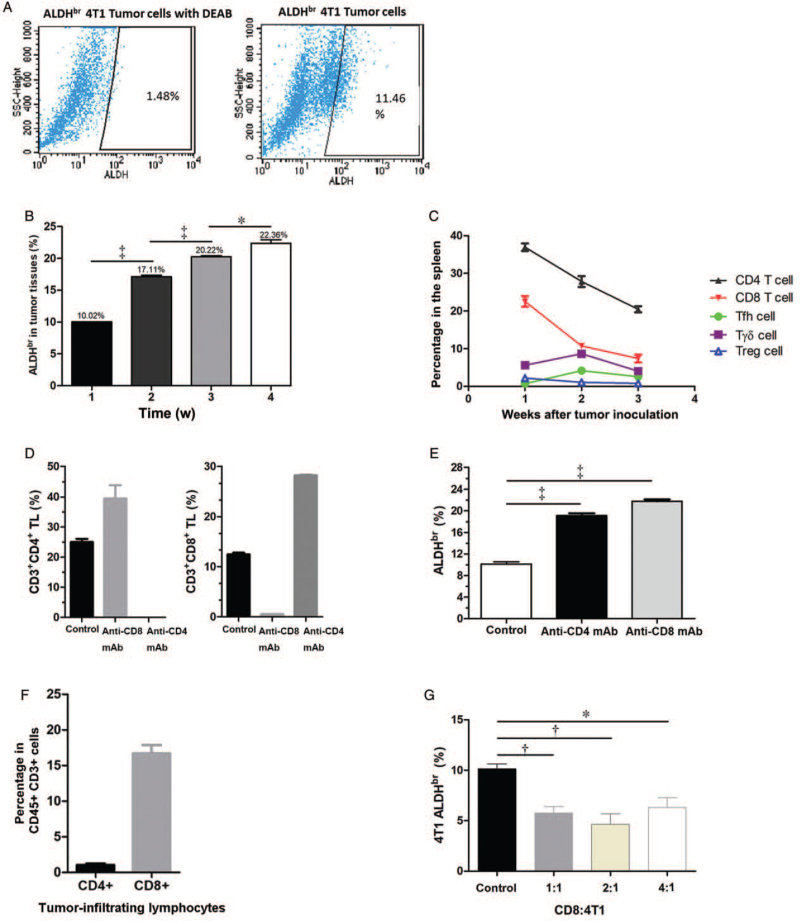
CSCs are considered responsible for metastasis.[4] In our previous study, ALDHbr 4T1 tumor cells were identified as CSCs for the 4T1 murine breast cancer cell line.[22] In the present study, we also performed an ALDEFLOUR™ assay to identify ALDHbr 4T1 tumor cells [Figure 2A] and found that the proportions of ALDHbr 4T1 tumor cells in tumors from BALB/c mice increased with tumor growth (week 1 vs. week 2, 10.02% vs. 17.11%; week 2 vs. week 3, 17.11% vs. 20.22%; week 3 vs. week 4, 20.22% vs. 22.36%; F = 297.20, all P < 0.050) [Figure 2B and Supplementary Figure 1A]. Moreover, in the spleens, the dominant CD4+ T cells and CD8+ T cells decreased gradually but the percentages of γδT cells and two kinds of specific CD4+ T cells, that is, Tfh cells and Treg cells, were low and did not vary obviously [Figure 2C].
Figure 2.
CD8+ T cells are associated with decreased proportions of ALDHbr 4T1 tumor cells. (A) Representative flow cytometry profiles of a negative control and a positive sample of ALDHbr 4T1 tumor cells. The proportion of ALDHbr 4T1 tumor cells was increased in the primary tumors of 4T1 tumor-bearing BALB/c mice. 4T1 tumor cells (5 × 104) were injected subcutaneously into BALB/c mice. (B) The primary tumors were isolated and digested at 1, 2, 3, and 4 weeks after tumor inoculation. At each week, four mice per group were determined. The ALDEFLOUR™ assay was performed and ALDHbr 4T1 tumor cells were determined by flow cytometry and gating of CD45− cells. (C) Dynamic changes in T lymphocyte subsets in the spleens of 4T1 tumor-bearing BALB/c mice. Splenic CD4+ T cells, CD8+ T cells, γδT cells, Tfh cells, and Tregs were analyzed by flow cytometry at 1, 2, and 3 weeks after tumor inoculation, gating of living cells. At each week, four mice per group were analyzed. (D) Depletion efficiencies for CD4+ T cells and CD8+ T cells in vivo. Anti-CD4 monoclonal antibodies or anti-CD8 monoclonal antibodies were injected via the tail vein on day −5 and day −1 before 4T1 tumor inoculation and thereafter once every 4 to 5 days. Two weeks after tumor inoculation, mice were sacrificed, and the percentages of splenic CD4+ T cells and CD8+ T cells were analyzed by flow cytometry, gating living cells. Four mice per group were detected. (E) The percentages of ALDHbr tumor cells increased in tumors of 4T1 tumor-bearing BALB/c mice after CD4+ T cell and CD8+ T cell depletion. The primary 4T1 tumors were isolated and digested. The ALDEFLOUR™ assay was performed, and ALDHbr 4T1 tumor cells in tumors were determined by flow cytometry and gating of CD45− cells. Four mice per group were assessed. (F) The percentage of CD4+ T cells and CD8+ T cells in tumors from 4T1 tumor-bearing BALB/c mice. The primary tumors were isolated and digested 2 weeks after tumor inoculation, and CD4+ TILs and CD8+ TILs were analyzed by flow cytometry, gating CD45+ and CD3+ cells. Four mice per group were analyzed. (G) The percentages of ALDHbr 4T1 tumor cells after coculturing of 4T1 tumor cells with splenic CD8+ T cells at effector-to-target ratios of 1:1, 2:1, and 4:1. Twenty-four hours later, the ALDEFLOUR™ assay was performed and ALDHbr 4T1 tumor cells were determined by flow cytometry and gating of CD45− cells. Data are presented as the mean ± SE. ∗P < 0.050; †P < 0.010; ‡P < 0.001. ALDHbr: Aldehyde dehydrogenasebright; TILs: Tumor-infiltrating cells.
To determine whether CD4+ T cells and CD8+ T cells affect ALDHbr 4T1 tumor cells in vivo, we depleted these two kinds of T cells in tumor-bearing BALB/c mice with antibodies. The depletion efficiencies for CD4+ T cells and CD8+ T cells in the spleen were >97% [Figure 2D and Supplementary Figure 1B]. With either CD4+ T cell or CD8+ T cell depletion, the percentages of ALDHbr 4T1 tumor cells in primary tumors were increased significantly compared with those in the control group without T cell depletion (control vs. CD4+ T cell depletion with anti-CD4 mAb, 10.15% vs. 19.13%; control vs. CD8+ T cell depletion with anti-CD8 mAb, 10.15% vs. 21.75%; F = 214.00, both P < 0.001) [Figure 2E]. Therefore, both CD4+ and CD8+ T cells could inhibit the proportions of ALDHbr 4T1 tumor cells in tumors in vivo.
Then, we detected CD4+ T cells and CD8+ T cells in TILs that may affect CSCs directly. We found that TILs contained both CD4+ T cells and CD8+ T cells, while CD8+ T cells accounted for a much higher proportion than CD4+ T cells, which may directly affect 4T1 tumor cells [Figure 2F and Supplementary Figure 2A]. Therefore, splenic CD8+ T cells from tumor-bearing BALB/c mice were isolated and cocultured with 4T1 tumor cells to investigate whether CD8+ T cells reduced the proportions of ALDHbr 4T1 tumor cells directly. We found that the percentages of ALDHbr 4T1 tumor cells decreased after coculturing 4T1 tumor cells with CD8+ T cells at different ratios (control vs. effector to target [E:T] ratio of 1:1, 10.15% vs. 5.76%; control vs. E:T ratio of 2:1, 10.15% vs. 4.64%; control vs. E:T ratio of 4:1, 10.15% vs. 6.32%; F = 10.46, all P < 0.050) [Figure 2G and Supplementary Figure 2B], which indicates that CD8+ T cells may play a crucial role in inhibiting ALDHbr 4T1 tumor cells.
ALDH expression in 4T1 tumor cells is associated with the response to IFN-γ
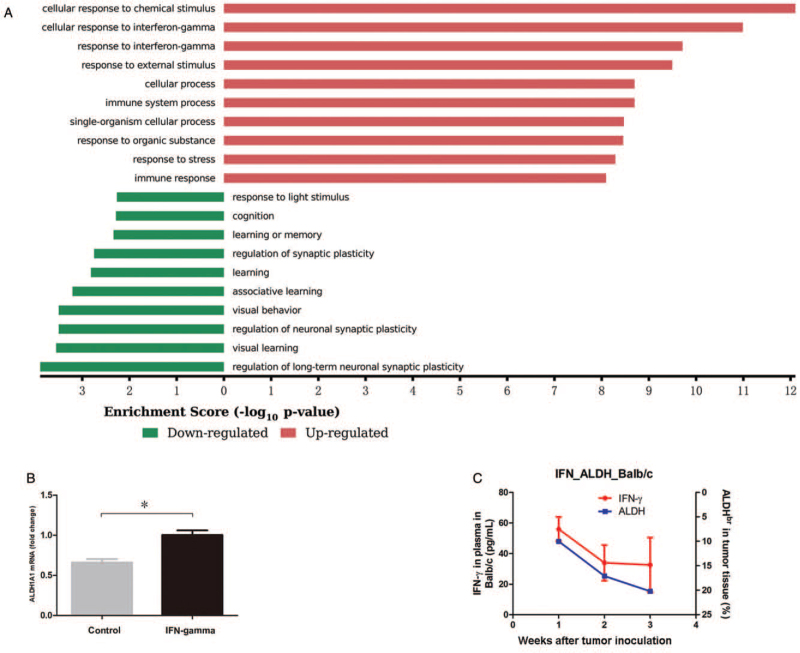
On the other hand, to identify differences in gene expression between ALDHbr 4T1 tumor cells and 4T1 tumor cells expressing low to negative ALDH (ALDHdim), we sorted ALDHbr and ALDHdim 4T1 tumor cells by flow cytometry and determined their expression profiles using a gene chip assay. Significantly dysregulated genes in ALDHbr 4T1 tumor cells were identified, and GO annotation was used to determine which functions these significantly dysregulated genes were enriched. The top ten dysregulated GO terms for functional categories ranked by enrichment scores are shown in Figure 3A. We found that two of the top three processes for upregulated genes in ALDHbr 4T1 tumor cells were referred to as the response to IFN-γ [Figure 3A]. This means that many genes associated with the function of response to IFN-γ were upregulated in ALDHbr 4T1 tumor cells, which indicates that the ALDHbr 4T1 tumor cells were more sensitive or resistant to IFN-γ than ALDHdim 4T1 tumor cells. Therefore, we detected the effect of IFN-γ on ALDH1A1 (a member of the ALDH family) expression in 4T1 tumor cells and found that IFN-γ treatment increased the mRNA levels of ALDH1A1 (1.00 vs. 0.50, t = 8.23, P < 0.010) [Figure 3B]. We also detected the plasma IFN-γ levels and determined whether they were associated with the percentages of ALDHbr 4T1 tumor cells in vivo. The data showed that the concentrations of plasma IFN-γ decreased gradually in the 4T1 tumor-bearing BALB/c mice, while the proportions of ALDHbr 4T1 tumor cells in tumor tissue increased [Figure 3C].
Figure 3.
ALDH expression in 4T1 tumor cells is associated with the response to IFN-γ. (A) The top ten GO terms for upregulated and downregulated genes in ALDHbr 4T1 tumor cells. ALDHbr and ALDHdim 4T1 tumor cells were sorted with flow cytometry and expression profiles were detected using an Affymetrix GeneChip mouse transcriptome array. GO annotations were used to determine the functions of differentially expressed genes in ALDHbr 4T1 tumor cells compared with ALDHdim 4T1 tumor cells. Prediction terms with a P value < 0.05 were selected and ranked by their P values. The −log10 (P value) yields an enrichment score representing the significance of GO term enrichment among differentially expressed genes. The top ten enriched GO terms for dysregulated genes in ALDHbr 4T1 tumor cells were ranked by enrichment score. (B) Relative expression of ALDH1A1 (a member of the ALDH family) in 4T1 tumor cells treated with or without IFN-γ (26.68 ng/mL) for 24 h. The mRNA levels of ALDH were determined by real-time PCR. (C) The levels of plasma IFN-γ are associated with the proportions of ALDHbr 4T1 tumor cells in BALB/c mice. The plasma IFN-γ levels of 4T1 tumor-bearing mice were determined by ELISA at 1, 2, and 3 weeks after tumor inoculation. Data are presented as the mean ± SE. ∗P < 0.010. ALDHbr: Aldehyde dehydrogenasebright; ELISA: Enzyme-linked immunosorbent assay; GO: Gene ontology; IFN-γ: Interferon-gamma.
IFN-γ inhibited ALDH protein expression in 4T1 tumor cells
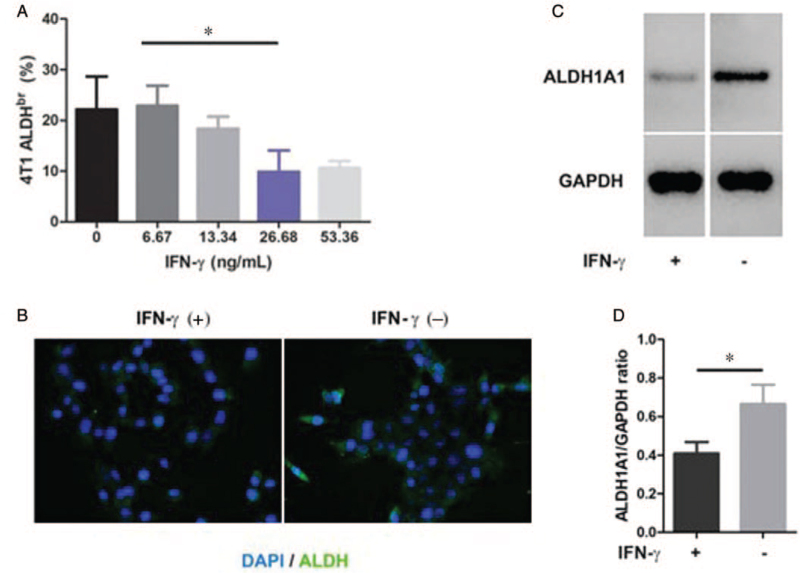
To further identify whether IFN-γ affected ALDHbr 4T1 tumor cells directly, we treated 4T1 tumor cells with IFN-γ at different concentrations in vitro. We found a dose-dependent inhibitory effect of IFN-γ on the percentages of ALDHbr 4T1 tumor cells [Figure 4A and Supplementary Figure 2C]. Compared with a concentration of 6.67, 26.68 ng/mL of IFN-γ significantly decreased the percentages of ALDHbr 4T1 tumor cells (22.88% vs. 9.88%, t = 2.25, P < 0.050). Then, we investigated ALDH expression in 4T1 tumor cells treated with IFN-γ (26.68 ng/mL). The results from ICC [Figure 4B] and Western blotting [Figure 4C] showed that the protein levels of ALDH1A1 in 4T1 tumor cells were significantly decreased after IFN-γ (26.68 ng/mL) treatment (0.86 vs. 0.49, t = 2.61, P < 0.050) [Figure 4D].
Figure 4.
IFN-γ decreases the protein expression of ALDH in 4T1 tumor cells. (A) Dose-dependent inhibitory effect of IFN-γ on the proportions of ALDHbr 4T1 tumor cells. 4T1 tumor cells were treated with IFN-γ at the indicated concentrations. After 24-h incubation, ALDHbr cells were assessed using the ALDEFLOUR™ assay and analyzed by flow cytometry. (B) Images of ICC for the expression of ALDH1A1 in 4T1 tumor cells treated with or without IFN-γ (26.68 ng/mL) for 24 h (400×). (C) Western blotting analysis of the protein levels of ALDH1A1 in 4T1 tumor cells treated with or without IFN-γ (26.68 ng/mL) and (D) the relative protein expression of ALDH1A1 in contrast to GAPDH after quantifying the protein bands. Data are presented as the mean ± SE. ∗P < 0.050. ALDHbr: Aldehyde dehydrogenasebright; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; ICC: Immunocytochemistry; IFN-γ: Interferon-gamma.
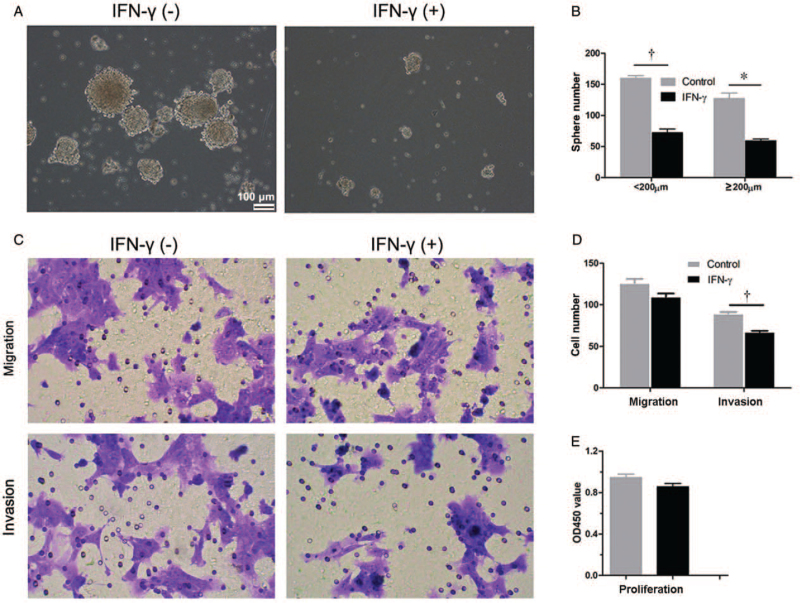
IFN-γ attenuated sphere formation and the invasion of 4T1 tumor cells
To investigate whether IFN-γ attenuated the malignancy of 4T1 tumor cells directly, we measured the sphere formation, migration, invasion, and proliferation of 4T1 tumor cells treated with or without IFN-γ. We found that the number of formed spheres was reduced significantly in 4T1 tumor cells cultured in the presence of IFN-γ (sphere diameter <200 μm, control vs. IFN-γ treatment, 159.50 vs. 72.0, t = 10.23; ≥200 μm, control vs. IFN-γ treatment, 127.0 vs. 59.0, t = 7.95; both P < 0.050) [Figure 5A and 5B]. The invasion of 4T1 tumor cells was also inhibited significantly after IFN-γ treatment (89.67 vs. 67.67, t = 10.31, P < 0.001) [Figure 5C and 5D]. IFN-γ also inhibited the migration of 4T1 tumor cells to some degree, although there was no significant difference (P = 0.063) [Figure 5C and 5D]. No significant difference in cellular proliferation was observed between 4T1 tumor cells treated with and without IFN-γ in vitro [Figure 5E], which may also suggest that decreased ALDH expression in 4T1 tumor cells [Figure 4B–D] is due to the inhibition of ALDH expression in individual cells instead of cytotoxicity of IFN-γ to ALDHbr 4T1 tumor cells. The decreased sphere formation and invasion after IFN-γ treatment indicates that IFN-γ attenuated the cancer stem-like character of 4T1 tumor cells, which may play a role in inhibiting tumor metastasis.
Figure 5.
IFN-γ attenuates sphere formation and the invasion of 4T1 tumor cells. (A) Phase-contrast images of sphere formation of 4T1 tumor cells treated with or without IFN-γ (100×). A total of 15,000 4T1 tumor cells per well were seeded in an ultralow attachment six-well plate with or without the addition of IFN-γ (26.68 ng/mL). One week later, numbers of spheres with a diameter <200 μm and no <200 μm were counted (B). Images (C) and quantification (D) of migrated and invaded 4T1 tumor cells. A total of 40,000 4T1 tumor cells per well were seeded into a 12-well Transwell insert with or without Matrigel. Medium supplemented with or without IFN-γ (26.68 ng/mL) was added to the lower chamber of the well. After 24-h incubation, cells on the underside of a membrane were fixed, stained, and counted under a microscope at 200× magnification. (E) The proliferation of 4T1 tumor cells treated with IFN-γ or not. A total of 3000 4T1 tumor cells per well were cultured with or without IFN-γ (26.68 ng/mL) for 24 h and then, cell viabilities were measured using CCK-8. Data are presented as the mean ± SE. ∗P < 0.050, †P < 0.010. CCK-8: Cell counting kit-8; IFN-γ: Interferon-gamma.
Discussions
In the present study, we observed fewer pulmonary metastatic nodules in tumor-bearing BALB/c mice than those in tumor-bearing BALB/c nude mice. CD8+ T cells decreased the percentages of ALDHbr 4T1 tumor cells in vitro and in vivo. Upregulated genes associated with the response to IFN-γ were enriched in ALDHbr 4T1 tumor cells. The plasma IFN-γ levels were decreased as the percentage of ALDHbr tumor cells increased in tumor-bearing BALB/c mice. IFN-γ treatment decreased the protein levels of ALDH1A1 in 4T1 tumor cells and inhibited the sphere formation and invasion abilities of 4T1 tumor cells.
The results of fewer pulmonary metastatic nodules in BALB/c mice than those in BALB/c nude mice confirmed [Figure 1A–C] that T cells play a vital role in inhibiting tumor invasion, which was consistent with the inhibitory effect of IFN-γ, a critical anti-tumor effector of T cells, on tumor invasion in vitro [Figure 5C and 5D]. The decreased percentages of ALDHbr 4T1 tumor cells after coculturing CD8+ T cells with 4T1 tumor cells [Figure 2G] were also consistent with the decreased percentages of ALDHbr 4T1 tumor cells and ALDH expression in 4T1 tumor cells treated with IFN-γ [Figure 4]. Pulaski et al[33] showed that IFN-γ may inhibit metastasis of 4T1 tumor cells in vivo via host-derived phagocytic cells. Therefore, both IFN-γ and CD8+ T cells could attenuate the stemness of 4T1 tumor cells to some degree. After depletion of either CD4+ T cell type, the proportions of ALDHbr cells also increased significantly. CD4+ T cells are critical for the priming of tumor-specific CD8+ T cells and the secondary expansion and memory of CD8+ T cells.[35,36] In TILs of the 4T1 tumor model, CD4+ T cells changed their dominant subsets from Th1 to Treg and Th17 during tumor growth.[37] In the 4T1 tumor model, depletion of Tregs with anti-CD25 antibodies induced tumor regression, and the number of infiltrated CD8+ T cells increased after Treg depletion. However, the further depletion of CD4+ T cells with anti-CD4 antibodies after Treg depletion did not increase the number of infiltrated CD8+ T cells, and the level of IFN-γ secreted by infiltrated CD8+ T cells was decreased, which ascribes a primary role of the CD4 Th cells in the process of tumor regression.[38] Without CD4+ effector T cells, CD8+ T cell activity and the production of IFN-γ are impaired, which may confer increased proportions of ALDHbr cells in vivo.
Regarding the growth of primary tumors in BALB/c mice and BALB/c nude mice, no significant difference was observed between the two groups [Figure 1D]. This result was similar to that of the proliferation assay in vitro, in which IFN-γ did not show significant inhibitory effects on cell proliferation [Figure 5E]. Different effects of IFN-γ on cancer cells may be explained by the varied levels of IFN-γ receptors. Kmieciak et al[39] showed that tumor cells that express high levels of IFN-γ Rα were eliminated by CD8+ T cells, whereas those with low levels did not die and remained dormant in the presence of IFN-γ-producing CD8+ T cells until losing the neu antigen, whereupon the tumor relapsed. 4T1 tumor cells express the IFN-γ receptor and the expression levels were not varied by IFN-γ treatment,[40] which may partially explain the lack of an inhibitory effect of IFN-γ on the proliferation of 4T1 tumor cells in vitro. The lack of a difference in tumor growth between BALB/c mice and BALB/c nude mice may also be due to the effects of various suppressive immune factors in vivo, such as myeloid-derived suppressive cells, which are IFN-γ-promoted, and suppress the anti-tumor effector T cell response,[41] which were not analyzed in the present study.
CSCs are relatively resistant to conventional chemotherapeutic regimens and radiation and are very likely to be the origin of cancer metastasis, relapse, and progression.[42–45] Specifically, the expression of the ALDH1A1 isoform correlates with a higher tumor grade and tumor metastasis.[19,46] In this study, we found that CD8+ T cells and IFN-γ decreased the percentages of CSCs in breast cancer, that is, ALDHbr 4T1 tumor cells. In addition, IFN-γ directly inhibited ALDH1A1 expression in 4T1 tumor cells, which may contribute to attenuated sphere-formation and invasion of 4T1 tumor cells. Thus, IFN-γ can be combined with chemotherapy and radiotherapy, which may enrich CSCs in cancer treatment. In addition, blocking immunosuppressive factors may be necessary if IFN-γ is used in vivo.
The limitations of the present study should be noted. The percentage changes in ALDHbr 4T1 tumor cells in mice after IFN-γ depletion could be assessed to confirm the effects of IFN-γ on ALDHbr 4T1 tumor cells in vivo. IFN-γ blockade in the coculture of CD8+ T cells with 4T1 tumor cells will suggest whether CD8+ T cells decrease the percentages of ALDHbr 4T1 tumor cells through IFN-γ. In the future, whether IFN-γ is secreted by CD8+ T cells and subsequently favors the downregulation of ALDH expression in 4T1 tumor cells needs to be determined. The mechanisms behind the effects of IFN-γ on 4T1 stem cells need further study.
To conclude, we found that IFN-γ may play a crucial role in inhibiting BCSCs. Combining IFN-γ therapy with chemotherapy or radiotherapy and strategies for blocking immunosuppressive factors may improve the prognosis of breast cancer patients.
Acknowledgements
The authors are grateful to Mr. David A Snouffer for his helpful and critical comments during the preparation of this manuscript and sincerely thank Prof. Shuren Zhang for his suggestions on the organization and discussions of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81402442 and No. 81903637) and the Fundamental Research Funds for the Central Universities (WK9110000027).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Zhuang X, Shi G, Hu X, Wang H, Sun W, Wu Y. Interferon-gamma inhibits aldehyde dehydrogenasebright cancer stem cells in the 4T1 mouse model of breast cancer. Chin Med J 2022;135:194–204. doi: 10.1097/CM9.0000000000001558
Xiufen Zhuang and Guilan Shi contributed equally to this work.
Supplemental digital content is available for this article.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, et al. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int 2018; 2018:5416923.doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bian L, Xu FR, Jiang ZF. Endocrine therapy combined with targeted therapy in hormone receptor-positive metastatic breast cancer. Chin Med J 2020; 133:2338–2345. doi: 10.1097/CM9.0000000000000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Angelis ML, Francescangeli F, Zeuner A. Breast cancer stem cells as drivers of tumor chemoresistance, dormancy and relapse: new challenges and therapeutic opportunities. Cancers (Basel) 2019; 11:1569.doi: 10.3390/cancers11101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep 2014; 2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest 2011; 121:3804–3809. doi: 10.1172/jci57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, et al. EMT and tumor metastasis. Clin Transl Med 2015; 4:6.doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell 2019; 49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunes T, Hamdan D, Leboeuf C, El Bouchtaoui M, Gapihan G, Nguyen TT, et al. Targeting cancer stem cells to overcome chemoresistance. Int J Mol Sci 2018; 19:4036.doi: 10.3390/ijms19124036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shima H, Yamada A, Ishikawa T, Endo I. Are breast cancer stem cells the key to resolving clinical issues in breast cancer therapy? Gland Surg 2017; 6:82–88. doi: 10.21037/gs.2016.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horimoto Y, Arakawa A, Sasahara N, Tanabe M, Sai S, Himuro T, et al. Combination of cancer stem cell markers CD44 and CD24 is superior to ALDH1 as a prognostic indicator in breast cancer patients with distant metastases. PLoS One 2016; 11:e0165253.doi: 10.1371/journal.pone.0165253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riaz N, Idress R, Habib S, Azam I, Lalani EM. Expression of androgen receptor and cancer stem cell markers (CD44(+)/CD24(−) and ALDH1(+)): prognostic implications in invasive breast cancer. Transl Oncol 2018; 11:920–929. doi: 10.1016/j.tranon.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcato P, Dean CA, Giacomantonio CA, Lee PWK. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011; 10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 14.Vassalli G. Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells. Stem Cells Int 2019; 2019:3904645.doi: 10.1155/2019/3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sultan M, Vidovic D, Paine AS, Huynh TT, Coyle KM, Thomas ML, et al. Epigenetic silencing of TAP1 in Aldefluor(+) breast cancer stem cells contributes to their enhanced immune evasion. Stem Cells 2018; 36:641–654. doi: 10.1002/stem.2780. [DOI] [PubMed] [Google Scholar]
- 16.Huang CP, Tsai MF, Chang TH, Tang WC, Chen SY, Lai HH, et al. ALDH-positive lung cancer stem cells confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Lett 2013; 328:144–151. doi: 10.1016/j.canlet.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 2011; 29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 18.Raha D, Wilson TR, Peng J, Peterson D, Yue P, Evangelista M, et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res 2014; 74:3579–3590. doi: 10.1158/0008-5472.CAN-13-3456. [DOI] [PubMed] [Google Scholar]
- 19.Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016; 7:11018–11032. doi: 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozovska Z, Patsalias A, Bajzik V, Durinikova E, Demkova L, Jargasova S, et al. ALDH1A inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer 2018; 18:656.doi: 10.1186/s12885-018-4572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim RJ, Park JR, Roh KJ, Choi AR, Kim SR, Kim PH, et al. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2alpha. Cancer Lett 2013; 333:18–31. doi: 10.1016/j.canlet.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang X, Zhang W, Chen Y, Han X, Li J, Zhang Y, et al. Doxorubicin-enriched, ALDH(br) mouse breast cancer stem cells are treatable to oncolytic herpes simplex virus type 1. BMC Cancer 2012; 12:549.doi: 10.1186/1471-2407-12-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res 2009; 15:4234–4241. doi: 10.1158/1078-0432.ccr-08-1479. [DOI] [PubMed] [Google Scholar]
- 24.Kida K, Ishikawa T, Yamada A, Shimada K, Narui K, Sugae S, et al. Effect of ALDH1 on prognosis and chemoresistance by breast cancer subtype. Breast Cancer Res Treat 2016; 156:261–269. doi: 10.1007/s10549-016-3738-7. [DOI] [PubMed] [Google Scholar]
- 25.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med 2009; 13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai YG, Gao GX, Zhang H, Zhang S, Liu YH, Duan XN, et al. Prognostic value of tumor-infiltrating lymphocyte subtypes in residual tumors of patients with triple-negative breast cancer after neoadjuvant chemotherapy. Chin Med J 2020; 133:552–560. doi: 10.1097/CM9.0000000000000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bharti R, Dey G, Mandal M. Cancer development, chemoresistance, epithelial to mesenchymal transition and stem cells: a snapshot of IL-6 mediated involvement. Cancer Lett 2016; 375:51–61. doi: 10.1016/j.canlet.2016.02.048. [DOI] [PubMed] [Google Scholar]
- 28.Mandai M, Hamanishi J, Abiko K, Matsumura N, Baba T, Konishi I. Dual faces of IFNgamma in cancer progression: a role of PD-L1 induction in the determination of pro- and antitumor immunity. Clin Cancer Res 2016; 22:2329–2334. doi: 10.1158/1078-0432.ccr-16-0224. [DOI] [PubMed] [Google Scholar]
- 29.Ni C, Wu P, Zhu X, Ye J, Zhang Z, Chen Z, et al. IFN-gamma selectively exerts pro-apoptotic effects on tumor-initiating label-retaining colon cancer cells. Cancer Lett 2013; 336:174–184. doi: 10.1016/j.canlet.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Chen JN, Zeng TT, He F, Chen SP, Ma S, et al. CD133+ liver cancer stem cells resist interferon-gamma-induced autophagy. BMC Cancer 2016; 16:15.doi: 10.1186/s12885-016-2050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schurch C, Riether C, Amrein MA, Ochsenbein AF. Cytotoxic T cells induce proliferation of chronic myeloid leukemia stem cells by secreting interferon-gamma. J Exp Med 2013; 210:605–621. doi: 10.1084/jem.20121229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mojic M, Takeda K, Hayakawa Y. The dark side of IFN-gamma: its role in promoting cancer immunoevasion. Int J Mol Sci 2017; 19:89.doi: 10.3390/ijms19010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulaski BA, Smyth MJ, Ostrand-Rosenberg S. Interferon-gamma-dependent phagocytic cells are a critical component of innate immunity against metastatic mammary carcinoma. Cancer Res 2002; 62:4406–4412. [PubMed] [Google Scholar]
- 34.Laky K, Kruisbeek AM. In vivo depletion of T lymphocytes. Curr Protoc Immunol 2016; 113:411–419. doi: 10.1002/0471142735.im0401s113. [DOI] [PubMed] [Google Scholar]
- 35.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 2003; 421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 36.Pardoll DM, Topalian SL. The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol 1998; 10:588–594. doi: 10.1016/s0952-7915(98)80228-8. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, Ma C, Zhang Q, Ye J, Wang F, Zhang Y, et al. CD4+ and CD8+ T cells have opposing roles in breast cancer progression and outcome. Oncotarget 2015; 6:17462–17478. doi: 10.18632/oncotarget.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaput N, Darrasse-Jeze G, Bergot AS, Cordier C, Ngo-Abdalla S, Klatzmann D, et al. Regulatory T cells prevent CD8 T cell maturation by inhibiting CD4 Th cells at tumor sites. J Immunol 2007; 179:4969–4978. doi: 10.4049/jimmunol.179.8.4969. [DOI] [PubMed] [Google Scholar]
- 39.Kmieciak M, Payne KK, Wang XY, Manjili MH. IFN-gamma Ralpha is a key determinant of CD8+ T cell-mediated tumor elimination or tumor escape and relapse in FVB mouse. PLoS One 2013; 8:e82544.doi: 10.1371/journal.pone.0082544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.duPre SA, Redelman D, Hunter KW, Jr. Microenvironment of the murine mammary carcinoma 4T1: endogenous IFN-gamma affects tumor phenotype, growth, and metastasis. Exp Mol Pathol 2008; 85:174–188. doi: 10.1016/j.yexmp.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Guo Q, Lv Z, Fu Q, Jiang C, Liu Y, Lai L, et al. IFN-gamma producing T cells contribute to the increase of myeloid derived suppressor cells in tumor-bearing mice after cyclophosphamide treatment. Int Immunopharmacol 2012; 12:425–432. doi: 10.1016/j.intimp.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 42.Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci 2018; 25:20.doi: 10.1186/s12929-018-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, et al. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res 2008; 68:3243–3250. doi: 10.1158/0008-5472.can-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med 2017; 23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 45.Nandi S, Ulasov IV, Tyler MA, Sugihara AQ, Molinero L, Han Y, et al. Low-dose radiation enhances survivin-mediated virotherapy against malignant glioma stem cells. Cancer Res 2008; 68:5778–5784. doi: 10.1158/0008-5472.can-07-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007; 1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.