Abstract
Background
One of the key barriers preventing rapid diagnosis of leptospirosis is the lack of available sensitive point-of-care testing. This study aimed to develop and validate a clustered regularly-interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 12a (CRISPR/Cas12a) platform combined with isothermal amplification to detect leptospires from extracted patient DNA samples.
Methodology/Principal findings
A Recombinase Polymerase Amplification (RPA)-CRISPR/Cas12a-fluorescence assay was designed to detect the lipL32 gene of pathogenic Leptospira spp. The assays demonstrated a limit of detection (LOD) of 100 cells/mL, with no cross-reactivity against several other acute febrile illnesses. The clinical performance of the assay was validated with DNA extracted from 110 clinical specimens and then compared to results from qPCR detection of Leptospira spp. The RPA-CRISPR/Cas12a assay showed 85.2% sensitivity, 100% specificity, and 92.7% accuracy. The sensitivity increased on days 4–6 after the fever onset and decreased after day 7. The specificity was consistent for several days after the onset of fever. The overall performance of the RPA-CRISPR/Cas12a platform was better than the commercial rapid diagnostic test (RDT). We also developed a lateral flow detection assay (LFDA) combined with RPA-CRISPR/Cas12a to make the test more accessible and easier to interpret. The combined LFDA showed a similar LOD of 100 cells/mL and could correctly distinguish between known positive and negative clinical samples in a pilot study.
Conclusions/Significance
The RPA-CRISPR/Cas12 targeting the lipL32 gene demonstrated acceptable sensitivity and excellent specificity for detection of leptospires. This assay might be an appropriate test for acute leptospirosis screening in limited-resource settings.
Author summary
Clinical signs and symptoms of leptospirosis are similar to those of other infectious diseases such as dengue, sepsis, and malaria, making it difficult to diagnose. In this study, we developed an RPA-CRISPR/Cas12a -based detection platform to identify the lipL32 gene of pathogenic Leptospira spp. The results showed that the limit of detection (LOD) was approximately 102 cells/mL without cross-reactivity against other infectious diseases. The platform was validated using 110 patients from 15 hospitals in Sisaket province, Thailand. The sensitivity, specificity, and accuracy was found to be 85.2%, 100% and 92.7%, respectively, for the diagnosis of leptospirosis. Assay sensitivity increased at 4–6 d post-onset of fever, with a consistent specificity every day after the onset of fever. We also developed a lateral flow detection assay combined with RPA-CRISPR/Cas12a, which also had a LOD of 102 cells/mL and could correctly distinguish known positive and negative clinical samples in a pilot study. Findings from this study demonstrate the potential effectiveness of the RPA-CRISPR/Cas12a platform in improving speed and accuracy of leptospirosis diagnosis especially in limited-resource settings.
Introduction
Leptospirosis is a zoonotic disease that affects global health with over one million cases and 58,900 deaths annually [1]. The disease is caused by the pathogenic spirochete Leptospira spp. which can adapt to a broad spectrum of mammalian hosts and environments [2,3]. The clinical signs and symptoms of leptospirosis share similarities with other infectious diseases such as dengue, sepsis, and malaria, making it difficult to diagnose [4–6].
One of the key barriers in reducing the impact of leptospirosis is the lack of affordable sensitive diagnostic tools. There are currently standard methods recommended by the WHO [4]. The first is the microscopic agglutination test (MAT), a serological-based diagnosis method. Although the MAT is accurate, it requires a skilled technician, well-equipped laboratory, and is time-consuming. The second is dark field microscope diagnosis from sample cultures collected from the patient’s blood at the early stage of Leptospira spp. infection. However, because Leptospira spp. is a slow-growing bacterium, results can take several weeks. The third technique is the use of polymerase chain reaction (PCR), a nucleic acid detection method that is faster and more accurate. The quantitative polymerase chain reaction (qPCR) is an improvement of the PCR that is widely used as the primary diagnostic procedure [5]. The drawback of qPCR is the high-cost of equipment and the lack of availability in every hospital, especially in rural areas [5,6] where most leptospirosis cases occur. The WHO also recommends the enzyme-linked immunosorbent assay (ELISA) for antibody detection. However, antibody levels are often low during the first week after infection [5]. The development of a novel diagnostic tool that can detect pathogenic Leptospira spp. early, providing rapid and accurate results and is not cost-prohibitive to resource-limited hospitals and clinics is essential for public health [7].
Testing at the point of care is ideal for resource-constrained settings. Numerous studies have been conducted to investigate point-of-care tests for leptospirosis [8–10]. Loop-Mediated Isothermal Amplification for Diagnosis of both pathogenic Leptospira and intermediate Leptospira in humans by targeting rrs gene has been developed. However, this technique is limited due to an imprecise specificity and does not support routine clinical usage of LAMP [10]. Anti-leptospira IgM rapid diagnostic tests (RDTs) are commercially available. RDTs have a limited sensitivity and may be inefficient for screening acute leptospirosis [9].
Recombinase polymerase amplification (RPA) is an isothermal nucleic acid amplification technology that can be operated in the field due to its low resource requirements. The RPA system utilizes three enzymes: recombinase, single-stranded DNA-binding protein (SSB), and strand-displacing polymerase [11]. The recombinase can pair oligonucleotide primers with homologous sequences in the target DNA. The SSB then binds to the replaced strand of DNA and protects the dissociation of primers. The strand-displacing polymerase initiates DNA synthesis. Amplification of the target DNA sequence by RPA can be accomplished at a constant temperature in less than 20 min. Moreover, RPA can work with the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas12a system that has shown promising results in nucleic acid detection [12–14]. The CRISPR/Cas12a system relies on a crispr RNA (crRNA) which acts as a targeting system for the effector function of the Cas12a enzyme to recognize and cleave specific DNA targets. After CRISPR/Cas12a detects its target and cleaves it, the collateral cleavage activity is activated resulting in the fluorescent reporter being cleaved from the quencher and creating a detectable fluorescent signal [14]. For this reason, RPA preamplification combined with the CRISPR/Cas12a detection system can be used for diagnostic screening in a limited-resource setting without the need for specialized instruments. The purpose of this study was to develop a new early rapid leptospirosis diagnostic tool using RPA and CRISPR/Cas12a targeting lipL32, which is considered to be a pathogenic species-specific gene that plays a critical role in inflammatory responses via binding to Toll-like receptor 2 [15,16]. Furthermore, the lipL32 gene is highly conserved across pathogenic Leptospira species [17].
Materials and methods
Ethics statement
The study protocol was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No.655/63). The study was performed under the international guidelines for human research protection of the Declaration of Helsinki, The Belmont Report, CIOMS Guideline, and International Conference on Harmonization in Good Clinical Practice. Written informed consent was obtained from each participant to participate in this study.
Culturing Leptospira interrogans
For the direct culture of Leptospira interrogans that was used as a positive control and limit of detection for the study, 1 mL of fresh whole blood was added into 4 mL of Ellinghausen, McCullough, Johnson, and Harris (EMJH) medium and incubated at 30°C for two weeks. The culture was examined using dark field microscopy to confirm the existence of Leptospira (18, 19). After confirmation, Leptospira cells were counted in a Petroff-Hauser chamber as previously described in [18,19]. DNA were then extracted at 108 cells/mL using the High Pure PCR Template Preparation Kit (Roche, USA).
Patients and study design
In this study, we tested the performance of the RPA-CRISPR/Cas12a system targeting lipL32 gene using blood samples from participants with a known leptospirosis status (infected or non-infected) from previous studies conducted in 15 hospitals in Sisaket province, Thailand (THAI-LEPTO study) [20]. The samples were collected between December 2015 to November 2016. The sample size was calculated when the sensitivity or specificity value of the standard method (qPCR) was known which is 86% and 100%, respectively [21]. A minimum of 110 samples were determined to be needed for this study according to the calculations.
Inclusion criteria for subjects were as follows: (i) older than 18 years old and admitted to a participating hospital; (ii) presenting with clinical suspicion of leptospirosis, high fever (body temperature higher than 38°C), severe myalgia; and (iii) history of exposure to reservoir animals [9,22]. The exclusion criteria were patients who suffered from other known infectious diseases. Following blood collection, total DNA was extracted from 200μL of whole blood samples for clinical sample validation using the High Pure PCR Template Preparation Kit (Roche, USA) according to the manufacturer’s instructions. A NanoDrop 2000 was used to determine the concentration and quality of the extracted DNA (Thermo Scientific, USA). Separation of serum from whole blood was accomplished by centrifuging at 3000 rpm for 10 minutes and storing at -80o C until the rapid diagnostic test (RDT) was performed. The samples from the first day of enrollment were selected and used as a blind test.
Detection by qPCR assay
Each positive sample based on the qPCR assay was defined as a leptospirosis confirmed case. The qPCR targeting the lipL32 gene was performed as previously described with minor modification (24). Briefly, 242 base pair products were amplified and detected using the forward primer (5′- AAG CAT TAC CGC TTG TGG TG -3′), reverse primer (5′- GAA CTC CCA TTT CAG CGA TT -3′) and Taqman probe (5′-FAM- AA AGC CAG GAC AAG CGC CG-BHQ1-3′). The qPCR mixture consisted of 5 μL of extracted DNA, 10 μL of SsoAdvanced Universal Probe Supermix (Bio-Rad Laboratories, USA), 1 μL of each primer (10 μM), 0.4 μL of Taqman probe (10 μM), and 2.6 μL of nuclease-free water in a final volume of 20 μL. The qPCR reactions were performed in duplicate. A no template control (NTC) with all the above reagents was used as the negative control. Extracted DNA of Leptospira interrogans from EMJH culture was used as positive control. Amplification and fluorescence detection were conducted in the StepOnePlus Real-Time PCR System (Applied Biosystems, USA). The amplification protocol consisted of 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. A negative result was considered with the threshold cycle (Ct) value higher than 40 cycles.
The RPA
The lipL32 gene amplification was performed using the TwistAmp Basic Kit (TwistDx, United Kingdom) using the same primer set as the qPCR. In brief, lyophilized RPA was resuspended in rehydration buffer and mixed with 480 nM of each primer. Then, 14 mM of magnesium acetate (final concentration) and 1 μL of extracted DNA were added to the reaction mixture. The lipL32 gene was amplified by incubating at 39°C for 40 min, followed by heat inactivation at 75°C for 5 min.
CRISPR RNA preparation
We designed the CRISPR RNA (crRNA) using in-silico analysis and the basic local alignment search tool (BLAST) to specifically detect the lipL32 gene sequence adjacent to the TTTN protospacer adjacent motif (PAM) site. The lipL32 crRNA sequence was 5′-UAAUUUCUACUAAGUGUAGAUUUCUGAGCGAGGACACAAUC-3’, consisting of a scaffold 5′-UAAUUUCUACUAAGUGUAGAU-3’ and a guide RNA 5′-UUCUGAGCGAGGACACAAUC-3’, both of which were synthesized using the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, UK). For the preparation of crRNA, synthetic oligonucleotides were ordered as ultramer DNA (Macrogen, South Korea) with an appended T7 promoter sequence. Oligonucleotides for crRNA (1 μM) were annealed to a short T7 primer (final concentration of 10 μM each) and incubated with T7 polymerase at 37°C for 2 h. The crRNA was then purified using a Monarch RNA Cleanup Kit (New England Biolabs, UK). The concentration of purified crRNA product was measured using a Qubit miRNA assay kit and Qubit 4 Fluorometer (Thermo Scientific, USA) and stored at -80°C until further use.
CRISPR/Cas12a-fluorescent based detection assay (FBDA)
The CRISPR/Cas12a-FBDA performed as described previously with minor modifications.[23,24] The CRISPR/Cas12a reaction was composed of 30 nM of crRNA, 330 nM of EnGen Lba Cas12a (Cpf1) (New England Biolabs, USA), 600 nM of fluorescent probe (5′-FAM-TTATTATT-BHQ1-3′), 1X of NEBuffer 2.0 (New England Biolabs, USA), and 1 μL of RPA amplicons in a total reaction volume of 15 μL. The CRISPR/Cas12a reaction was incubated at 39°C for 20 min. The fluorescent signal was then observed by naked eye using a BluePAD Dual LED Blue/White Light Transilluminator (BIO-HELIX, Taiwan) at 470 nm wavelength. Each test was observed by three certified laboratory technicians who were instructed to identify the qualitative test outcome as positive or negative. The tests were considered positive if at least two of the three technicians read the results as positive. Both investigators and the technicians were unaware of the outcome.
Limit of detection (LOD) and cross-reactivity testing
The analytic sensitivity of the assay was determined using genomic DNA isolated from Leptospira interrogans in EMJH cultures. Serial dilutions of genomic DNA were made from 106 cells/mL down to 1 cell/mL [25]. The LOD was determined by detecting the fluorescent signal in the tube containing the lowest cells. The specimens obtained from patients with an acute febrile illness, including acute viral hepatitis, cellulitis, scrub typhus, systemic bacterial infection, acute cystitis, influenza, Escherichia coli septicemia, and dengue hemorrhagic fever, were tested to establish the analytical specificity of the RPA-CRISPR/Cas12a-FBDA.
RPA-CRISPR/Cas12a combined with a lateral flow detection assay (LFDA) pilot study
A lateral flow test strip was developed to improve the RPA-CRISPR/Cas12a test and make it easier to use and read. The FITC-biotin reporter molecule and lateral flow strips were designed to capture labeled nucleic acids. The DNA probe (5′- FITC-AGGACCCGTATTCCCA-BIOTIN -3′) was used at 12 nM instead of the fluorescence probe at 600 nM under the otherwise same condition as the FBDA above. The reaction was incubated at 39°C for 30 min. The reaction was then mixed with 100 μL of running buffer and pipetted into the commercial lateral flow strip test (Kestrelbioscience, Thailand). Uncleaved reporter molecules were captured at the first detection line (test line), whereas the indiscriminate ssDNA cleavage activity of CRISPR/Cas12a did not generate a signal at the first detection line, but only a signal at the second line (control line).
Rapid diagnostic testing
The analytic sensitivity of the RPA-CRISPR/Cas12a detection system was compared with a commercial rapid diagnostic test (RDT). A total of 96 blood samples were tested with the RDT from the Medical Science Public Health (Department of Medical Sciences, Ministry of Public Health, Thailand). The RDT kit was designed to detect anti-Leptospira IgM antibodies and was used according to the manufacturer’s instructions. First, the blood sample was thawed at room temperature and added to the sample well without air bubbles. Next, the assay diluent was added to the diluent well. The results were read at the end of 15 min by three trained technicians. The tests were considered positive if at least two of three technicians read the results as positive.
Statistical analysis
Continuous variables are shown as the mean ± one standard deviation (SD) in case of a normal distribution and as a median and interquartile range (IQR) in case of non-normally distributed variables. The Student’s t-test or Mann-Whitney test was used to analyze the differences between two continuous variables. Categorical variables were presented as numbers with percentages and were compared using the Chi-square test. The performance of the RPA-CRISPR/Cas12a targeting the lipL32 gene detection system was expressed by calculating the sensitivity, specificity, accuracy, and positive and negative predictive values compared to the qPCR analysis of the same samples. All statistical analyses were performed using the SPSS Version 22 software (SPSS, Chicago, IL).
Analysis of the crRNA target sequence using bioinformatics
The lipL32 sequences of Leptospira spp. were acquired from the National Center for Biotechnology Information (NCBI) to identify the extent of homology in the crRNA target sequences of Leptospira spp.
Results
The LOD and cross-reactivity of the RPA-CRISPR/Cas12a-FBDA
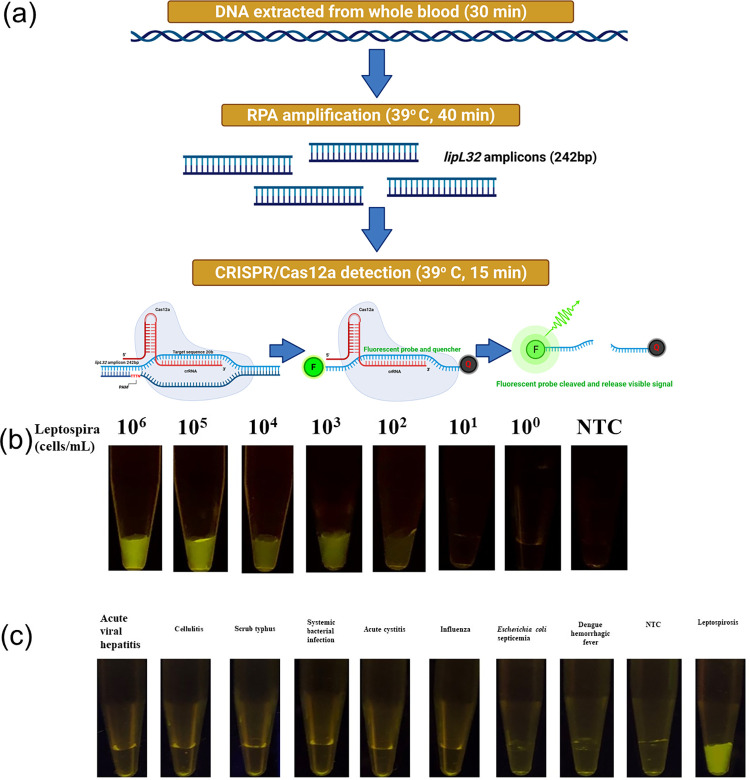
The LOD of the RPA-CRISPR/Cas12a-FBDA was tested by extracting DNA from a Leptospira interrogans culture that was serially diluted from 106 to 1 cell/mL. The diluted DNA was amplified using RPA, followed by the CRISPR/Cas12a-FBDA. The workflow of the assay is summarized in Fig 1A. The LOD was found to be 100 cells/mL (Fig 1B). In addition, eight specimens from patients with other acute febrile illnesses, including acute viral hepatitis, cellulitis, scrub typhus, systemic bacterial infection, acute cystitis, influenza, Escherichia coli septicemia, and dengue hemorrhagic fever, were tested to explore potential cross-reactivity. The results showed no cross-reactivity (Fig 1C).
Fig 1. Detection of leptospirosis using the RPA-CRISPR/Cas12a-FBDA.
(A) Schematic representation of the RPA-CRISPR/Cas12-FBDA’s workflow (Created with BioRender.com). (B, C) The (B) LOD and (C) cross-reactivity of the RPA-CRISPR/Cas12a targeting lipL32 gene against several infectious diseases with similar clinical manifestations as leptospirosis.
Study population
The performance of the RPA-CRISPR/Cas12a-FBDA was validated with 110 blood samples from clinically suspected leptospirosis patients. Fifty-four samples (49.1%) were leptospirosis confirmed cases (positive by qPCR) and 56 (50.9%) were non-leptospirosis confirmed cases (negative by qPCR). The clinical characteristics of the enrolled patients are shown in Table 1. Compared with non-leptospirosis, leptospirosis patients had significantly higher serum levels of white blood cells, creatinine, total bilirubin, direct bilirubin, and potassium, but a lower systolic blood pressure (p<05). In addition, there was a significant difference in terms of days of fever until enrollment between the groups (p = .01). Other relevant laboratory investigations were not found to be significantly different between the two groups.
Table 1. Characteristics of the qPCR positive and negative groups with patient clinical and laboratory data.
| Characteristic | Leptospirosis (N = 54) | Non-leptospirosis (N = 56) | Total (N = 110) | P-value |
|---|---|---|---|---|
| Male gender, n (%) | 44 (81.48%) | 45 (80.36%) | 89 (80.91%) | 0.88 |
| Age, years, Mean (SD) | 50.78 (16.71) | 51.75 (15.93) | 51.25 (16.26) | 0.82 |
| Days of fever until enrollment, Median (IQR) | 3 (3, 5) | 3 (2, 4) | 3 (2, 4) | *0.01 |
| Exposure to flood waters, n (%) | 47 (85.45%) | 40 (81.63%) | 87 (79.09%) | 0.18 |
| Exposure to animals, n (%) | 8 (14.55%) | 9 (18.37%) | 17 (15.45%) | 0.73 |
| Body temperature, Mean (SD) | 38.14 (1.20) | 38.16 (1.24) | 38.15 (1.21) | 0.97 |
| SBP, mm HG, Median (IQR) | 109.00 (96.00, 121.50) | 120.00 (101.00, 129.50) | 111.00 (100.00, 126.00) | *0.02 |
| DBP, mm Hg, Median (IQR) | 61.00 (58.00, 73.25) | 68.00 (60.00, 76.75) | 64.00 (60.00, 74.00) | 0.10 |
| Platelet (x 103/μL), Median (IQR) | 94500.00 (59500.00, 213250.00) | 132000.00 (68500, 194750.00) | 118500.00 (63000.00, 204000.00) | 0.70 |
| *WBC (x 103/μL), Median (IQR) | 10950.00 (8525.00, 14025.00) | 8600.00 (5375.00, 12250.00) | 10500.00 (6350.00, 13375.00) | *0.02 |
| Creatinine, mg/dL, Median (IQR) | 1.33 (1.00, 2.90) | 1.10 (0.86, 1.27) | 1.12 (0.94, 1.96) | *0.01 |
| *TB, g/dL, Median (IQR) | 1.40 (0.82, 3.30) | 0.90 (0.50, 2.35) | 1.18 (0.70, 2.90) | *0.02 |
| *DB, g/dL, Median (IQR) | 0.90 (0.46, 1.97) | 0.50 (0.24, 1.55) | 0.70 (0.30, 1.80) | *0.03 |
| *SGOT, U/L, Median (IQR) | 63.00 (41.50, 147.00) | 64.50 (39.50, 170.00) | 63.00 (41.00, 164.00) | 0.91 |
| *SGPT, U/L, Median (IQR) | 59.00 (31.50, 103.50) | 60.00 (33.00, 84.75) | 60.00 (32.50, 96.00) | 0.77 |
| Na, mEq/L, Median (IQR) | 135.00 (132.00, 138.00) | 135.00 (131.45, 139.00) | 135.00 (131.70, 139.00) | 0.71 |
| K, mEq/L, Median (IQR) | 3.77 (3.40, 4.26) | 3.50 (3.09, 3.90) | 3.63 (3.26, 4.01) | *0.01 |
| HCO3, mEq/L, Median (IQR) | 24.00 (20.15, 25.75) | 25.00 (22.25, 26.45) | 24.55 (21.73, 26.00) | 0.11 |
Abbreviations: WBC: white blood cell; TB: total bilirubin; DB: direct bilirubin; SGOT: serum glutamic oxaloacetic transaminase; SGPT: serum glutamic; pyruvic transaminase, Na: sodium; K: potassium; HCO: bicarbonate; DBP: diastolic blood pressure; SBP: systolic blood pressure. Continuous data are expressed as the mean, standard deviation (SD) or median shown with interquartile range (IQR). Categorical variables are expressed as numbers (%)
* represents P < 0.05.
Diagnostic performance of the RPA-CRISPR/Cas12a-FBDA
To evaluate the diagnostic performance of the RPA-CRISPR/Cas12a-FBDA, 110 DNA samples from leptospirosis and non-leptospirosis confirmed cases were tested and results were compared to the qPCR results. The RPA-CRISPR/Cas12 assay yielded 100% specificity, 85.2% sensitivity, and 92.7% accuracy, with a positive predictive value (PPV) and a negative predictive value (NPV) of 100% and 87.50%, respectively (Table 2).
Table 2. Performance of the RPA-CRISPR/Cas12a-FBDA relative to qPCR detection.
| Parameter | Performance of RPA-CRISPR/Cas12a |
|---|---|
| Total samples | 110 |
| True positive (TP) | 46 |
| True negative (TN) | 56 |
| False positive (FP) | 0 |
| False negative (FN) | 8 |
| Sensitivity | 85.19% |
| Specificity | 100% |
| Positive predictive value (PPV) | 100% |
| Negative predictive value (NPV) | 87.50% |
| Accuracy | 92.73% |
Diagnosis accuracy at different days after the fever onset
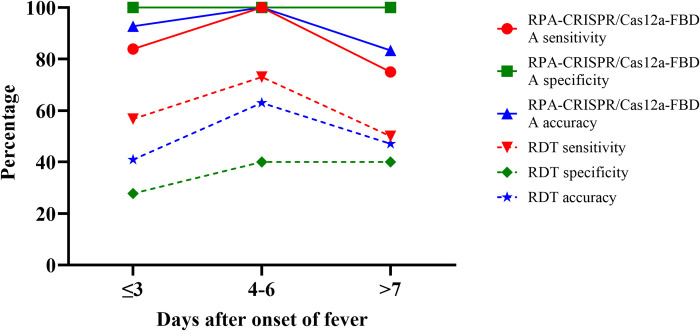
To evaluate the change in sensitivity and specificity of the assay with time after fever onset, the patients were categorized into three groups based on the time since the onset of fever (at the first day of enrollment):1) within 3 d after fever onset (n = 69), 2) within 4–6 d from fever onset (n = 19), and 3) 7 d or longer after the onset of fever (n = 18). We found that the sensitivity and accuracy of RPA-CRISPR/Cas12a-FBDA targeting lipL32 increased on days 4–6 and decreased after day 7. In contrast, the specificity was consistent for every day after the onset of fever (Fig 2).
Fig 2.
Sensitivity, specificity, and accuracy of the RPA-CRISPR/Cas12a-FBDA (solid line) and RDT (dashed line) at 3 d (red), 4–6 d (green), and ≥ 7 d (blue) after the onset of fever.
We also compared the diagnostic accuracy of our assay with a commercial RDT based on detection of anti-Leptospira IgM antibodies. We found that the commercial RDT assay yielded a lower sensitivity, specificity, and accuracy than the RPA-CRISPR/Cas12a-FBDA every day after the onset of fever (Fig 2).
Inter-observer variability
The sensitivity, specificity, and accuracy were calculated separately to investigate each observer’s variability, with the results summarized in S1 Table. The data revealed no significant difference between observers in the ability to identify the fluorescent signal.
The RPA-CRISPR/Cas12a LFDA
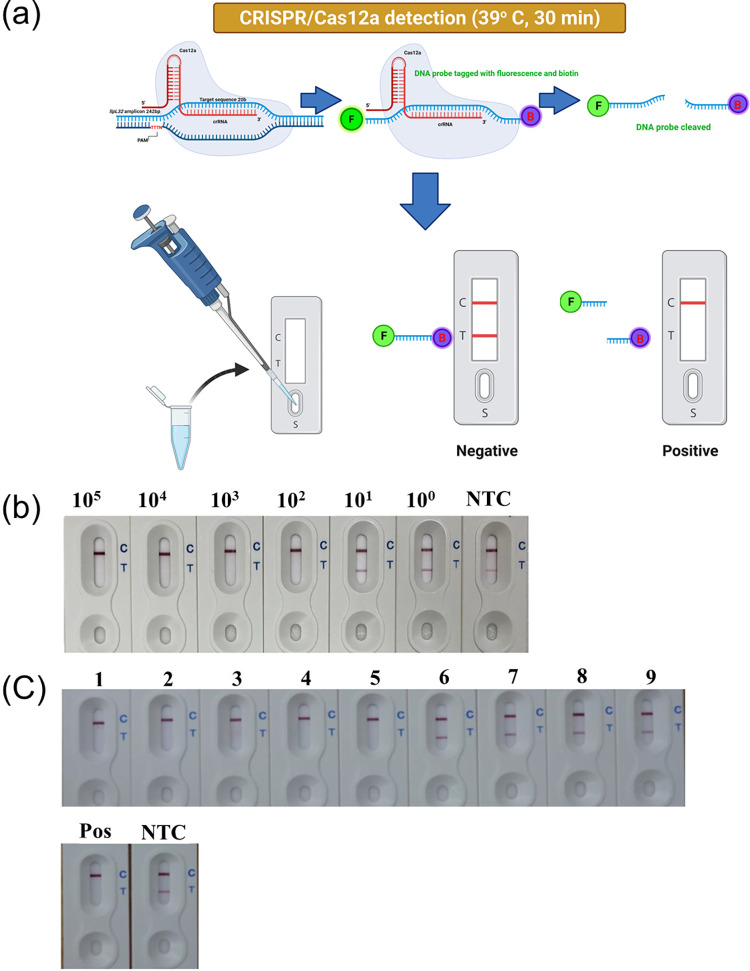
We also developed an RPA-CRISPR/Cas12a-LFDA (Fig 3A) to improve this test and make it more accessible for general use and easier to read. The LOD of this LFDA was 100 cells/ml similar to the FBDA (Fig 3B). Nine DNA samples from leptospirosis confirmed cases (N = 5) at a Ct between 27–37 and non-leptospirosis cases (N = 4) were tested in the pilot study. The results showed that the LFDA could reliably distinguish between the known positive and negative clinical samples (Fig 3C).
Fig 3. The RPA-CRISPR/Cas12a-LFDA.
(A) The RPA-CRISPR/Cas12a-LFDA’s workflow (Created with BioRender.com), (B) LOD, and (C) clinical sample validation [1–5 and 6–9 are known positive and negative samples, respectively, while NTC and Pos are the no-template negative and positive control, respectively].
Bioinformatics analysis of the crRNA target sequence
The lipL32 sequences of Leptospira spp. were obtained from the National Center for Biotechnology Information (NCBI) in order to determine the similarity between Leptospira spp. crRNA target sequences. Fig 4 and S2 Table illustrate that the majority of the sequences were found identical except for L. borgpetersenii strain M10 and FMAS_AP2, L. santarosai serovar Shermani and L. weilii serovar Manhao II.
Fig 4. The Bioinformatics analysis of lipL32 multiple sequence alignment between pathogenic Leptospira.
Sequence accession numbers are as follows, CP048830.1, CP043891.1, EU526391.1, AY609326.1, AY461918.1, AY461920.1, AY609333.1, AY568680.1, NZ_CP072630.1, AY461917.1, AY461912.1, AY461911.1, AY609331.1, and CP006694.1.
Discussion
The RPA-CRISPR/Cas12a-FBDA is a new nucleic acid detection platform able to diagnose many infectious diseases [12,23,24,26–29]. This study is the first report for Leptospira detection using the RPA-CRISPR/Cas12aFBDA assay targeting the lipL32 gene. The assay demonstrated acceptable sensitivity (85%) and excellent specificity (100%) of Leptospira detection compared to qPCR as the reference test. The RPA is an isothermal nucleic acid amplification platform, which is less time-consuming than conventional PCR and qPCR [11,30]. Due to the absence of lipL32 in nonpathogenic or intermediate Leptospira spp., this newly developed test identified exclusively pathogenic Leptospira [16,17,31] and without cross-reactivity when tested against blood from non-leptospirosis patients.
A previous study revealed that PCR inhibitors in clinical samples can affect qPCR performance in detecting lipL32 [32]. The RPA has the advantage of being more tolerant of PCR inhibitors making it an ideal amplification platform for this study [33]. The qPCR system provides a highly specific and sensitive tool for detecting and quantifying Leptospira [34]. However, due to its high cost compared to other diagnostic methods and its requirements for specialized instruments, it has not been widely used as an early diagnostic tool at the point of care.
Results from our study support usage of the RPA-CRISPR/Cas12a-based system to detect Leptospira with an acceptable sensitivity and high specificity. To address the sensitivity and specificity issues raised by prior studies, point-of-care testing for leptospirosis such as LAMP and RDT were created [9,10]. A previous study reported that having more than 1,000 Leptospira cells/mL was associated with severe leptospirosis [35]. It is notable that our assay was sensitive enough to detect Leptospira in the patient’s blood and administer treatment before disease symptoms became severe.
The specificity of the RPA-CRISPR/Cas12a-FBDA was found to be consistent for all three testing periods of time after the onset of fever. We reported that the sensitivity increased to 100% on days 4–6 after fever onset and decreased after day 7, which may have reflected the fact that the serum Leptospira spp. peaked at days 4–6 after fever onset and decreased after day 7 (39). We compared the assay performance with a commercial RDT designed for detection of anti-Leptospira spp. IgM antibodies. The RDT performance was similar to that previously reported [9], but had a lower sensitivity, specificity, and accuracy than the RPA-CRISPR/Cas12a-FBDA developed in this study. The window of positivity for the RPA-CRISPR/Cas12a platform began at the first sign of infection, compared to the RDT window which started at days 6–8 [5,36]. While Leptospira can be found in blood within the first week following the beginning of symptoms, the bacterial load diminishes over time, limiting the test’s sensitivity, whereas the IgM level begins to climb to detectable levels after the first week [36]. However, both our newly developed test and qPCR were able to detect Leptospira spp. after day 7 since Leptospira spp. can still be detected in blood albeit with a decreased number [36]. Therefore, using this RPA-CRISPR/Cas12a-FBDA targeting lipL32 combined with RDTs would expand the window of positivity and enhance the accuracy of the leptospirosis diagnosis. We also developed the RPA-CRISPR/Cas12a-LFDA to improve the test by making it more accessible for use and easier to read. The preliminary result showed that the RPA-CRISPR/Cas12a-LFDA could reliably distinguish between known positive and negative Leptospira spp. from clinical samples in our pilot study.
There were a number of strengths highlighted by this study, first we compared the test sensitivity, specificity, and accuracy at the day of fever, and the results showed that the time window after the onset of infection is a vital factor in the detection of positive infections in the different types of assays. Our newly developed test is capable of rapid and early detection of pathogenic Leptospira spp., which is critical for proper treatment. Second, our study was performed using blinded assessments and with three different observers to minimize bias. Third, we developed an LFDA to further facilitate field usage, and this pilot study achieved a similar LOD as the FBDA. Fourth, we performed a bioinformatics analysis to investigate nucleotide variation that would affect test sensitivity [7,37,38]. The results showed minimum variation in the lipL32 targeted by crRNA of CRISPR/Cas12a among pathogenic Leptospira spp. As a result, there is still room for future improvement of our crRNA to ensure that it covers all pathogenic Leptospira spp. Finally, the RPA-CRISPR/Cas12a-based detection system is ideal for rural hospitals, as it has less expensive laboratory equipment requirements. The system would require only a heat block for the isothermal reaction. Additionally, the total time required to perform the RPA-CRISPR/Cas12a FBDA test, beginning with blood collection and DNA extraction, was approximately 1 hour and 35 minutes. Due to the extended reaction time required by CRISPR/Cas12a to completely cleave the probe, results from the RPA-CRISPR/Cas12a LFDA took approximately 2 hours compared to qPCR which takes 3–4 hours.
There were some limitations to our study, which must be acknowledged. The first consideration is that, due to the study design and variable timing of hospital admission, we cannot test all patients from the first day of fever. Therefore, the interpretation for the role of RPA-CRISPR/Cas12a in diagnosis leptospirosis need to understand this limitation. Second, most patients visited the hospital early after fever onset (first 3 day) and only a small number of patients presented at day 4–6 (n = 19) and more than 7 d (n = 18) after the fever onset. This time of presentation could impact test accuracy. As a result, we should interpret the results cautiously especially if the patients presented to the hospital after 7 days of fever onset. Third, the use of more than one target gene may enhance the efficiency of the test. While lipL32 may contain a highly conserved domain, it is a single-copy gene, which may limit its sensitivity and LOD [39]. As a consequence, our future efforts should be directed toward developing additional target genes, particularly those with multiple copies, in order to increase the test’s LOD and sensitivity. Fourth, we conducted only a few cross-reactivity tests (n = 8). As a result, in our next investigation, we will undertake testing for known blood-borne diseases to confirm there is no cross-reactivity as well as testing against a broad variety of different Leptospira species. Fifth, we still do not know the stability/longevity/shelf life of the LFDA from Kestrel bioscience, Thailand. As a result, we will collaborate with Kestrel bioscience, Thailand, in the future to research the stability/longevity/shelf life of LFDA. Sixth, the qPCR CT greater than 37 had an effect on the test’s accuracy. Finally, clinical samples were collected from a single province in Thailand, which limits the generalizability of the findings.
Our study provides support for the use of the RPA-CRISPR/Cas12a-FBDA to detect leptospires due to its satisfactory LOD, sensitivity, and specificity. It is suitable for use in the field, especially in rural hospitals with limited resources since it is practical, portable, rapid, and simple to use. Only a heat box is required to perform the isothermal reaction. With the addition of the LFDA, we will be able to further decrease the need for equipment making the procedure even more desirable.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We thank the Critical Care Nephrology Research Unit, Faculty of Medicine, Chulalongkorn University; the Excellence Center for Critical Care Nephrology (EC-CCN), King Chulalongkorn Memorial Hospital, Tropical Medicine Cluster, Chulalongkorn University, and Sisaket Provincial Public Health Office for their collaborative effort during data collection and for participation in the study. We thank Suppalak Brameld (Section of Immunology Leptospirosis, Melioildosis, and Brucellosis, National Institute of Health, Department of Medical Sciences, Ministry of Public Health) for the MAT testing.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Sirawit Jirawannaporn received funding from the The 100th Anniversary Chulalongkorn University Fund for Doctoral Scholarship, and the 90th Anniversary of Chulalongkorn University Scholarship (Ratchadaphiseksomphot Endowment Fund). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Neglected Tropical Diseases. 2015;9(9):e0003898. doi: 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levett PN. Leptospirosis. Clinical Microbiology Reviews. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soo ZMP, Khan NA, Siddiqui R. Leptospirosis: Increasing importance in developing countries. Acta Tropica. 2019;201:105183. doi: 10.1016/j.actatropica.2019.105183 [DOI] [PubMed] [Google Scholar]
- 4.World Health O. Human leptospirosis: guidance for diagnosis, surveillance and control. Geneva: World Health Organization; 2003. [Google Scholar]
- 5.Budihal SV, Perwez K. Leptospirosis Diagnosis: Competancy of Various Laboratory Tests. Journal of Clinical and Diagnostic Research 2014;8(1):199–202. doi: 10.7860/JCDR/2014/6593.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad SN, Shah S, Ahmad FM. Laboratory diagnosis of leptospirosis. Journal of Postgraduate Medicine. 2005;51(3):195–200. [PubMed] [Google Scholar]
- 7.Narkkul U, Thaipadungpanit J, Srisawat N, Rudge JW, Thongdee M, Pawarana R, et al. Human, animal, water source interactions and leptospirosis in Thailand. Scientific Reports. 2021;11(1):3215. doi: 10.1038/s41598-021-82290-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed A, van der Linden H, Hartskeerl RA. Development of a recombinase polymerase amplification assay for the detection of pathogenic Leptospira. International Journal of Environmental Research and Public Health. 2014;11(5):4953–64. doi: 10.3390/ijerph110504953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinhuzen J, Limothai U, Tachaboon S, Krairojananan P, Laosatiankit B, Boonprasong S, et al. A prospective study to evaluate the accuracy of rapid diagnostic tests for diagnosis of human leptospirosis: Result from THAI-LEPTO AKI study. PLoS Neglected Tropical Diseases. 2021;15(2):e0009159. doi: 10.1371/journal.pntd.0009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonthayanon P, Chierakul W, Wuthiekanun V, Thaipadungpanit J, Kalambaheti T, Boonsilp S, et al. Accuracy of loop-mediated isothermal amplification for diagnosis of human leptospirosis in Thailand. American Journal of Tropical Medicine and Hygiene. 2011;84(4):614–20. doi: 10.4269/ajtmh.2011.10-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase Polymerase Amplification for Diagnostic Applications. Clinical Chemistry. 2016;62(7):947–58. doi: 10.1373/clinchem.2015.245829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–42. doi: 10.1126/science.aam9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018. doi: 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daher Ede F, de Abreu KL, da Silva Junior GB. Leptospirosis-associated acute kidney injury. J Bras Nefrol. 2010;32(4):400–7. [PubMed] [Google Scholar]
- 16.Fernandes LG, Siqueira GH, Teixeira AR, Silva LP, Figueredo JM, Cosate MR, et al. Leptospira spp.: Novel insights into host-pathogen interactions. Veterinary Immunology and Immunopathology. 2016;176:50–7. doi: 10.1016/j.vetimm.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 17.Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, Mazel M, et al. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infection and Immunity. 2000;68(4):2276–85. doi: 10.1128/IAI.68.4.2276-2285.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evangelista K, Franco R, Schwab A, Coburn J. Leptospira interrogans Binds to Cadherins. PLoS Neglected Tropical Diseases. 2014;8(1):e2672. doi: 10.1371/journal.pntd.0002672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreier S, Triampo W, Doungchawee G, Triampo D, Chadsuthi S. Leptospirosis research: fast, easy and reliable enumeration of mobile leptospires. Biological Research. 2009;42(1):5–12. doi: /S0716-97602009000100001 [PubMed] [Google Scholar]
- 20.Sukmark T, Lumlertgul N, Peerapornratana S, Khositrangsikun K, Tungsanga K, Sitprija V, et al. Thai-Lepto-on-admission probability (THAI-LEPTO) score as an early tool for initial diagnosis of leptospirosis: Result from Thai-Lepto AKI study group. PLoS Neglected Tropical Diseases. 2018;12(3):e0006319. doi: 10.1371/journal.pntd.0006319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villumsen S, Pedersen R, Borre MB, Ahrens P, Jensen JS, Krogfelt KA. Novel TaqMan(R) PCR for detection of Leptospira species in urine and blood: pit-falls of in silico validation. Journal of Microbiological Methods. 2012;91(1):184–90. doi: 10.1016/j.mimet.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 22.Srisawat N, Praditpornsilpa K, Patarakul K, Techapornrung M, Daraswang T, Sukmark T, et al. Neutrophil Gelatinase Associated Lipocalin (NGAL) in Leptospirosis Acute Kidney Injury: A Multicenter Study in Thailand. PloS One. 2015;10(12):e0143367–e. doi: 10.1371/journal.pone.0143367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. 2020;38(7):870–4. doi: 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayuramart O, Nimsamer P, Rattanaburi S, Chantaravisoot N, Khongnomnan K, Chansaenroj J, et al. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Experimental Biology and Medicine. 2021;246(4):400–5. doi: 10.1177/1535370220963793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, et al. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infectious Diseases. 2002;2(1):13. doi: 10.1186/1471-2334-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimsamer P, Mayuramart O, Rattanaburi S, Chantaravisoot N, Saengchoowong S, Puenpa J, et al. Comparative performance of CRISPR-Cas12a assays for SARS-CoV-2 detection tested with RNA extracted from clinical specimens. Journal of Virological Methods. 2021;290:114092. doi: 10.1016/j.jviromet.2021.114092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caliendo AM, Hodinka RL. A CRISPR Way to Diagnose Infectious Diseases. New England Journal of Medicine. 2017;377(17):1685–7. doi: 10.1056/NEJMcibr1704902 [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Li S, Wang J, Liu G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends in Biotechnology. 2019;37(7):730–43. doi: 10.1016/j.tibtech.2018.12.005 [DOI] [PubMed] [Google Scholar]
- 29.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nature Protocols. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abd El Wahed A, Patel P, Faye O, Thaloengsok S, Heidenreich D, Matangkasombut P, et al. Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLOS ONE. 2015;10(6):e0129682. doi: 10.1371/journal.pone.0129682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, et al. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PloS One. 2008;3(2):e1607. doi: 10.1371/journal.pone.0001607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galloway RL, Hoffmaster AR. Optimization of LipL32 PCR assay for increased sensitivity in diagnosing leptospirosis. Diagnostic Microbiology and Infectious Disease. 2015;82(3):199–200. doi: 10.1016/j.diagmicrobio.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malaria Journal. 2014;13:99. doi: 10.1186/1475-2875-13-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009;64(3):247–55. doi: 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 35.Tubiana S, Mikulski M, Becam J, Lacassin F, Lefèvre P, Gourinat AC, et al. Risk factors and predictors of severe leptospirosis in New Caledonia. PLoS Negl Trop Dis. 2013;7(1):e1991. doi: 10.1371/journal.pntd.0001991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picardeau M. Diagnosis and epidemiology of leptospirosis. Médecine et Maladies Infectieuses. 2013;43(1):1–9. doi: 10.1016/j.medmal.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 37.Murugan K, Seetharam AS, Severin AJ, Sashital DG. CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J Biol Chem. 2020;295(17):5538–53. doi: 10.1074/jbc.RA120.012933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lelu M, Muñoz-Zanzi C, Higgins B, Galloway R. Seroepidemiology of leptospirosis in dogs from rural and slum communities of Los Rios Region, Chile. BMC veterinary research. 2015;11:31-. doi: 10.1186/s12917-015-0322-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thaipadunpanit J, Chierakul W, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Boonslip S, et al. Diagnostic Accuracy of Real-Time PCR Assays Targeting 16S rRNA and lipl32 Genes for Human Leptospirosis in Thailand: A Case-Control Study. PloS One. 2011;6(1):e16236. doi: 10.1371/journal.pone.0016236 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.