Abstract
Disparate occurrence of breast cancer remains an intriguing question since only a subset of women with known risk factors develop cancer. Recent studies suggest an active role of local and distant microbiota in breast cancer initiation, progression, and overall prognosis. A dysbiotic microbiota predisposes the body to develop cancer by inducing genetic instability, initiating DNA damage and proliferation of the damaged progeny, eliciting favorable immune response, metabolic dysregulation and altered response to therapy. In this review, we present our analyses of the existing datasets and discuss the local dysbiosis observed in breast cancer patients and different aspects of breast carcinogenesis that can be potentially influenced by local breast microbiota. Striking differences between microbial community compositions in breast of cancer patients compared to healthy individuals were noted. Differences in microbiome were also apparent between benign and malignant disease and between nipple aspirate fluid of healthy individuals and breast survivors. We also discuss the identification of distinct bacterial, fungal, viral as well as parasite signatures for breast cancer. These microbes are capable of producing numerous secondary metabolites that can act as signaling mediators effecting breast cancer progression. We review how microbes potentially alter response to therapy affecting drug metabolism, pharmacokinetics, anti-tumor effects and toxicity. In conclusion, breast harbors a community of microbes that can communicate with the host cells inducing downstream signaling pathways and modulating various aspects of breast cancer growth and metastatic progression and an improved understanding of microbial dysbiosis can potentially reduce breast cancer risk and improve outcomes of breast cancer patients.
The human microbiome, now referred to as, the “forgotten organ” contains a metagenome that is 100-fold more diverse compared to the human genome, thereby, is critically associated with human health [1,2]. With the revelations of the human microbiome project and advent of deep sequencing techniques, a plethora of information has been acquired in recent years. Body sites like stomach, bladder and lungs, once thought to be sterile, are now known to harbor millions of indigenous microbial species. Approximately 80% of the healthy microbiome consists of Firmicutes and Bacteroidetes accompanied by Verrucomicrobia, Actinobacteria, Proteobacteria, Tenericutes and Cyanobacteria [2–7]. The role of microbiome in diabetes, obesity and even neurodegenerative diseases was greatly appreciated in the last decade [1,7–14] and now it has been established that microbiome significantly contributes to many organ specific cancers [1,15,16].
1. Microbiome in homeostasis and dysbiosis
Each anatomical site of the body harbors a distinct community of microorganisms and between individuals, higher taxonomic levels and functional groups are generally conserved but enormous differences exist in the genus, species and strain levels [1,2,7]. Human cells and the cells of the microbiota compete and cooperate with each other; the metagenome has both genetic and functional plasticity [2]. The complex dynamic interaction of the microbes with the host cells affects cellular metabolism, inflammatory, immunologic and neurological functions. The host microbiome is shaped by a co-evolution of various microbial species with host cells starting at birth leading to beneficial or harmful phenotypes [2]. Emerging evidences assert that interactions between the host and microbiome are not locally confined but have remote effects via hormonal intermediates, metabolites and immunological mediators in organs like liver, pancreas and breast. Liver is not known to harbor an active microbial population, but is extremely susceptible to alterations in gut microbiome due to microorganism-associated molecular patterns (MAMPs) and bacterial metabolites through close anatomical associations [1,2,11,17]. One such inflammatory MAMP, lipopolysaccharide (LPS), a conserved component of gram-negative bacteria, and its receptor Toll like receptor 4 (TLR4), are also known to promote pancreatic cancer [1,2]. Majority of hepatic cancer associated deaths are due to secondary hepatic cancers. The gut microbiota plays a major role in the development of secondary liver cancers through bile acid metabolism and production of secondary bile acids which in turn regulate the Natural killer T cell population of the liver via the hepatic CXCR6/CXCL16 axis [17]. The process is primarily regulated by the Clostridium species [17].
Most of our knowledge regarding microbiome’s role in human cancers is derived from gut studies since it harbors 99% of the microbial mass which contributes to gross metabolic functions of the body. In recent years, studies using germ free animals revealed tumor-promoting as well as tumor suppressing effects of the microbiota in spontaneous and carcinogen-induced models of colon, liver, breast and lung cancers [1,2,9]. The microbiota has also been identified in skin and the urinogenital tract. There is also a known association of the oral microbiome and periodontitis with breast [18–21] and pancreatic cancer [2,22]; though the mechanism remains elusive. Organ specific carcinogenesis by microbes is asserted by the fact that depletion of the intestinal bacteria, with antibiotics, reduces the development of primary and secondary liver cancer [17] and colon cancer [23]. A number of studies also indicated antitumor effects of the microbiota [24–27], eg., bacterial infection or injection of heat-killed bacteria or Coley’s toxin regressed tumors in Sarcoma patients [2]. Subsequently, it was revealed that conserved bacterial components in Coley’s toxin can activate Toll-like receptor (TLR) and NOD-like receptor (NLR) by acting as receptor agonists, eliciting anti-tumor effects [2].
In most cases, a community of microbes as opposed to a single pathogen dictates the effect of microbiota in cancer progression. One such example is H. pylori, a well know carcinogenic pathogen. Despite being the master regulator, H. pylori mono-associated mice develop fewer tumors in hypergastrinemic transgenic mouse model. Gastric atrophy and hypochlorhydria induced by chronic H. pylori infection promotes bacterial overgrowth thus increasing the conversion of dietary nitrates to carcinogens [1,2,28,29]. However, H. pylori lowers oesophageal adenocarcinoma risk, emphasizing the organ-specificity of bacterial microbiota in carcinogenesis [2]. Similarly, chronic infection with Salmonella enterica subsp. enterica serovar Typhi and Salmonella enterica subsp. enterica serovar Paratyphi induces adaptive immune response which promotes gall bladder cancer and mucosa-associated lymphoid tissue (MALT) lymphomas [1,2]. Gastric MALT lymphoma elicits activation of immune cells reactive to H. pylori-derived antigens. H. pylori clearance reduces the burden of gastric MALT lymphoma. Campylobacter jejuni, Borrelia burgdorferi and Chlamydia psittaci are also known to trigger certain lymphomas which can be regressed by antibiotic administration [1,2].
The general principles of microbiome driven malignancies have been extensively described by Plottel and Blaser [2]. They categorized microbiome-induced cancer into three classes. Class A involves an immune reaction, e.g. colonization by certain microbes recruits innate and adaptive immune effectors to the lamina propria inducing epithelial cell hyperproliferation while microbial adjuvancy induces immune surveillance restricting tumor progression [2]. Class B involves direct interaction with the parenchymal cells; for e.g. H. pylori, the highly motile, microaerophilic, gram-negative bacteria colonizing the human stomach. The bacterium physically attaches and injects a 128 kDa secretory protein, CagA via type IV secretion system into the host cell [2,29–31]. Scenarios where microbes residing in certain organs exert a remote effect on another organ are categorized as Class C interactions, for example, protective effect of H. pylori in esophageal adenocarcinoma and its precursors, Barret’s esophagus and gastrointestinal reflux disease. It corroborates with the fact that decrease in H. pylori infection in populations over decades positively correlates with a steady rise in incidence of esophageal adenocarcinoma [2,29,31]. It has been proposed that CagA induced changes in gastric physiology affecting acid secretion, hormone interactions and T cell populations [2,30]. Many organs including lungs, skin female genital tract and oral cavity harbor a rich and metabolically active bacterial population but its role in oncogenic transformation in these sites remains to be explored.
2. The microbiome and its association with breast cancer
Breast cancer remains the leading cause of cancer-related mortality in women worldwide. Approximately 12.4% women in USA confront invasive breast cancer and it is estimated that 266,120 women in USA will be diagnosed with invasive breast cancer while 63,960 will be diagnosed with carcinoma in situ in 2018 (Breast Cancer Facts & Figs. 2017–2018 - American Cancer Society). In addition, it is projected that 2550 men in US will succumb to invasive breast cancer in 2018 (Breast Cancer Facts & Figs. 2017–2018 - American Cancer Society). Breast cancer is a multifactorial disease attributable to multiple closely interdependent risk factors however 70% of women who develop breast cancer in their life time do not carry familial risk factors. The etiology of breast cancer therefore remains elusive; probability of cancer development, therapeutic outcomes and survival are still not accurately predictable. Clearly there are additional risk factors pertaining to breast cancer development and one such factor that has attracted a lot of attention in recent years is the ‘Human Microbiome’. Human microbiome has been implicated as a major contributor to incidence as well as progression for many cancer types. Breast tissue was originally considered a sterile site with no microbial population but multiple studies have revealed the presence of a distinct local microbiota in the breast and a dysbiosis in this community structure is thought to induce and aid carcinogenesis. Existence of the breast microbiota was first proposed by Urbaniak et al., in 2014 when they observed and characterized a distinct microbial population which persisted beyond lactation [32]. Thereafter, a number of studies have confirmed their claims using deep sequencing techniques and sparked further interest in the functional significance of microbiota in breast carcinogenesis. Although all these studies confirmed the presence of microbiota in breast tissue, they focused on different cohorts spanning normal breast vs. breast cancer, healthy controls vs. breast cancer survivors and benign vs. malignant disease. As a result, each study reported some findings unique to their cohort albeit the sample sizes were relatively small. We analyzed all existing data from these studies to define common trends among their results as well as unique aspects using One Codex (a bioinformatics platform for microbial genomics) (Fig. 1). In this section, we will discuss our analyses of all existing data sets and the functional significance of some important results.
Fig. 1.
A schematic representation of the planned analyses. Patient data from previous studies were analyzed to define common trends among their results as well as unique aspects using One Codex (a bioinformatics platform for microbial genomics).
2.1. Evidence from clinical studies: comparing microbiome of normal breast and breast cancer
In 2014, Urbaniak and group screened tissue from multiple sites around the breast of 18 to 90-year-old Canadian and Irish women. Both cohorts harbored rich microbial communities independent of lactation status [32].
No women in either of the cohort exhibited any sign of infection and no differences based on the site of sample collection and surgeon collecting the samples were observed. However, clear differences in breast microbial communities of the two ethnicities were apparent. From their analyses, Canadian women appeared to harbor a richer diversity of microbes compared to Irish women. Bacillus (11.4%), Acinetobacter (10%), Comamonadaceae (5.7%), Gammaproteobacteria (5%) and Prevotella (5%) were some of the major taxa in Canadian cohort not detected in Irish women. Listeria welshimeri (12.1%) was detected in Irish women but not in Canadian women. Pseudomonas, Staphylococcus and Propionibacterium were detected in both cohorts with nearly similar relative abundance. Enterobacteriaceae was one of the major taxa found in both cohorts but the relative abundance varied with Canadian women exhibiting only 8.3% relative abundance whereas Irish women presented with 30.8% relative abundance. These differences are intriguing and could be contributing factors to disparity in breast cancer incidence and prognosis. Since a number of bacteria such as Bacillus cereus, are capable of metabolizing sex hormones, local microbiota can also determine breast cancer subtypes [32]. The female breast has a nutrient rich environment composed of fatty tissue with widespread vasculature, lymphatics and diffusely located lobules and ducts leading from nipple, conducible for growth of diverse microbial communities. Proteobacteria and Firmicutes comprised majority of the breast bacterial community [32,33]. Higher abundance of Proteobacteria and Firmicutes in the breast tissue could be due to the fatty acid rich environment of the breast; these phyla are positively associated with adiposity [34].
We obtained the raw data from sequence read archive (SRP076038, NCBI-SRA) for a successive study by Urbaniak and group and examined abundant taxa in each group and their relative abundance. This analysis showed that breast cancer patients have a relative abundance of Bacillus and Staphylococcus (Phylum Firmicutes) and Enterobacteriacae (Phylum Proteobacteria) (SRP076038) [35] (Fig. 2A, B). Healthy individuals show considerable higher abundance of Lactobacillus, Thermoanaerobacterium thermosaccharolyticum, Candidatus Aquiluna sp. IMCC13023, Anoxybacillus, Leuconostoc, Lactococcus, Geobacillus, Methylobacterium, and Turicella otitidis compared to cancer patients where there was an abundance of Thermus scotoductus, E. coli, Bacillus cereus, Shewanella, and Corynbacterium (Fig. 2D). Interestingly, there was no difference in community composition based on invasiveness and stage of cancer but the microbial composition in normal tissue adjacent to the tumor was very similar to that obtained from the actual tumor but markedly different from the breast of healthy women [35]. Upon species level analysis, we observed that the lactic acid bacteria, Lactobacillus, comprised, of an average 2.2% of the total bacterial count in control samples compared to 1.4% in cancer cases (Fig. 2C). A number of recent studies have emphasized the role of Lactobacillus species in treating and preventing breast cancer growth and metastasis [36,37]. The possible mechanisms of Lactobacillus induced protection include immune activation, competitive inhibition of pathogenic strains and synthesis of signaling intermediates.
Fig. 2.
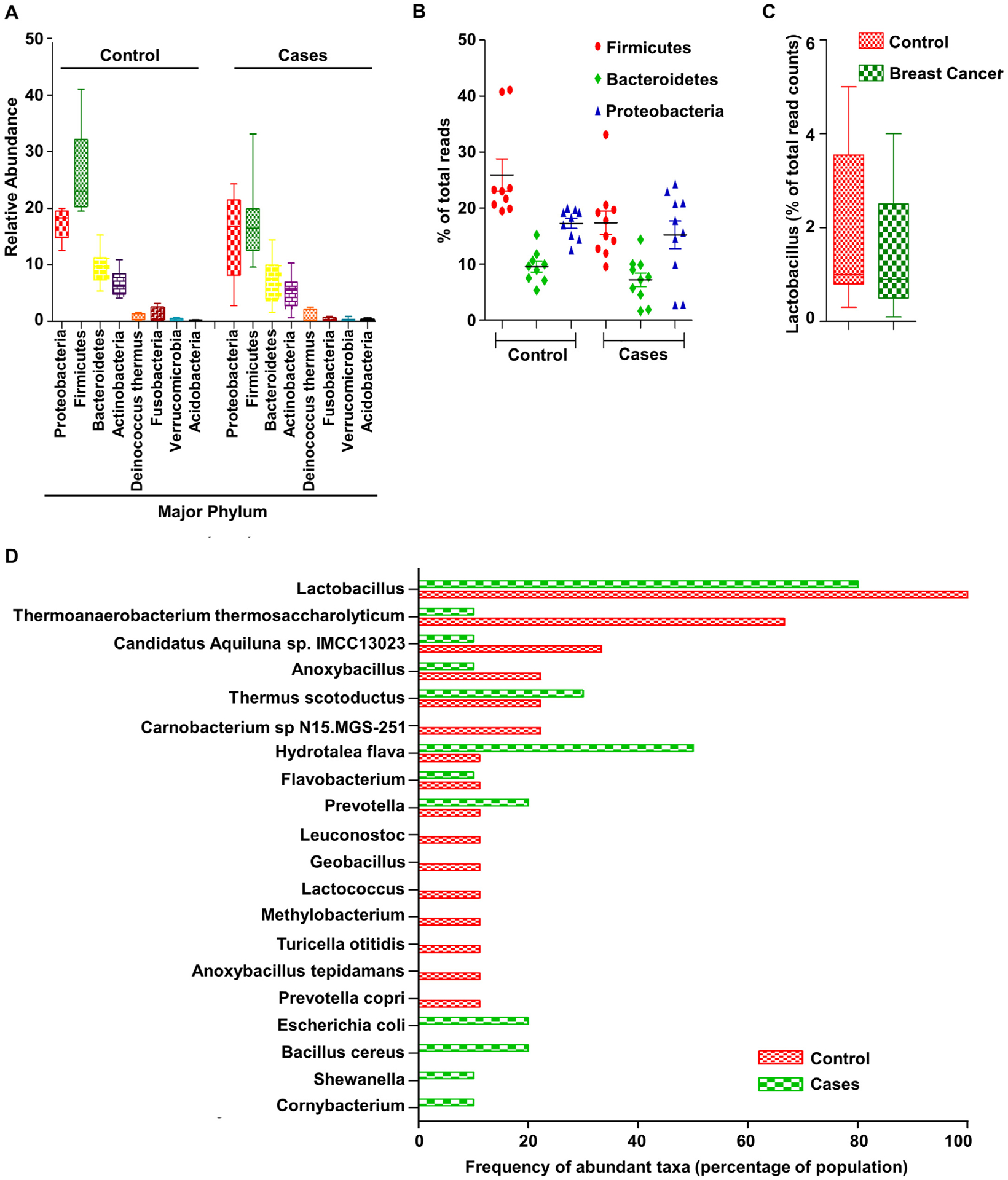
(A) Relative abundance of major phyla. (B) Percentage of three major phyla, Firmicutes, Bacteroides and Proteobacteria. (C) Relative percentage of lactobacillus species in breast cancer compared to control. (D) Frequency of occurrence of abundant taxa in control and cancer tissue samples (SRP076038, Analysis: One Codex).
To investigate the functional importance, Escherichia coli and Staphylococcus epidermis isolated from breast cancer samples were tested for their ability to induce double stranded breaks in HeLa cells [32,35]. Breast cancer samples showed a higher abundance of family Enterobacteriacae. Notably, B2 phylotype E coli (a member of the Enterobacteriacae family), possess pks pathogenicity islands encoding genotoxin colibactin [35] that can induce double stranded DNA breaks and has been implicated in colon cancer. In vitro, E coli isolated from breast cancer patients induced double stranded breaks in DNA in Hela cells as determined by ϒ-H2AX phosphorylation [35]. Similar breaks were also induced by Staphylococcus epidermis isolated from breast cancer samples. Additionally, lactic acid bacteria known to be associated with tumor suppression were less abundant [35]. Double-Strand break repair by non-homologous end joining is an extremely error prone mechanism and accumulation of these misrepairs over time leads to genomic instability, and eventually cancer. In colon, double-strand breaks induced by E coli as well as H pylori are known to induce genomic instability over long exposures [35]. Bacteria-induced DNA damage may not be sufficient to induce carcinogenesis on its own but combined with other risk factors, this may be detrimental. Bacillus cereus, elevated in breast cancer tissue (Fig. 2D), does not damage DNA but might be involved in other pro-carcinogenic effects. B. cereus possesses 5α steroid hydrogenase, 3β, 17β and 20α hydroxysteroid dehydrogenase enzyme activities which can metabolize steroid hormones progesterone and testosterone [38]. It is known to metabolize progesterone to 5-alpha-3,20-dione(5αP) which is higher in breast tumors compared to normal breast; and induces breast cancer cell proliferation in vitro [35,39]. Prevotella copri was prevalent in healthy breast tissue and it is known to synthesize short chain fatty acid, propionate [35]. It has been found to be associated with arthritis [40] and to have contrasting role on colon and oral cancers warranting further study in breast cancer [35].
In another unique study conducted by Xuan and group, genomic DNA was extracted from formalin fixed paraffin-embedded estrogen-receptor (ER) positive breast tumors and paired normal samples and 16 s pyrosequencing was performed [33]. Gene expression analyses was also performed using RNA extracted from fresh frozen breast tissue samples from healthy individuals and breast cancer patients. We obtained the raw data from public database PRJEB4755, EBI-ENA and reanalyzed using OneCodex. Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Verrucomicrobia formed the majority of microbial population in all samples (Fig. 3A, B). There was no significant difference in species richness, but evenness of communities varied significantly. Genus Sphingomonas (Proteobacteria) was abundant in 50% of normal tissue while 66.7% of tumor samples were abundant in genus Methylobacterium (Proteobacteria) [33]. Sphingomonas yanoikuyae, a benzopyrene degrading bacterium, was most significantly enriched and most prevalent in 95% of normal tissue while Methylobacterium radiotolerance, a radiation tolerant rare pathogen, was significantly enriched in 100% of tumor samples. Health implications of either species is not well known. Staphylococcus and Corynbacterium, the most common species of skin bacteria, also showed significant difference between the tumor and paired normal tissue [33]. Shift in ratio of the major phyla in cancer patients compared to control is reflected in Fig. 3C.Veillonella, Spiroplasma, Acinetobacter and Streptococcus pneumonia were some of the species limited to cancerous tissue in contrast to paired normal (Fig. 3D) [33].
Fig. 3.

(A) Phylum distribution of local breast microbiota in cancer and nearby normal tissue samples. (B) Mean percentages of major phyla in cancer and nearby normal breast tissue samples. (C) Percentage of three major phyla, Firmicutes, Bacteroides and Proteobacteria. (D) Frequency of occurrence of abundant taxa in breast cancer and nearby normal tissue samples (PRJEB4755, Analysis: One Codex).
In a recent study, Wang et al. extensively examined the microbiome profiles in the breast tissue, urine and oral rinse of breast cancer patients [22] and compared cancerous and histologically normal tissue of cancer patients as well as cancerous and non-cancerous samples (from women undergoing cosmetic surgery). Tumors, adjacent normal and non-cancerous samples did not significantly vary in community richness (p = .12) [22]. Unlike other cohort studies, significant variation was observed neither in overall diversity (estimated by Shannon Diversity index, H) nor number of observed species (microbial richness indicated by number of observed OTUs, N) between cancerous and non-cancerous samples (H = 3.5 ± 0.7, N = 20.3) [22]. Similarly, differences were non-significant between tumor and adjacent normal tissue from cancer patients (H = 3.3 ± 0.8, N = 20.4 ± 7.7) (p = .32, 0.98) [22]. Interestingly, differences in diversity and richness were significant when compared based on alcohol consumption; regular intake vs. no intake, and infrequent intake vs. no intake [22]. Also, diversity was higher in case of hormone positive breast cancers (H = 3.6 ± 0.8) compared to hormone negative cancers (H = 2.9 ± 0.6); p = .03 [22], possibly due to the ability of multiple microbes to metabolize and regulate bioavailability of steroid hormones. Species richness also trended significantly towards hormone receptor positive as compared to hormone receptor negative breast cancer [22]. No differences were observed based on racial identitiy, premenopausal/postmenopausal status, smoking/nonsmoking, use of antibiotic, last liquid/solid food intake, pathologic classification, histological evaluation, HER2-amplification, invasion to lymphovascular system, or node-positivity [22]. Samples could be differentiated significantly based on alcohol-intake (p < .01, R2 = 0.06; principal component analysis) [22] but not by other demographic factors. They did not observe significant clustering in principal component analysis between cancerous tissue and nearby histologically normal tissue, p = .92 [22] but clustering was observed by histologic-grade (p = .02, R2 = 0.05) [22], status of invasion to lymphovascular system (p = .02, R2 = 0.08) [22], and amplification of HER2 (p = .02, R2 = 0.05) [22]. Among cancer cases, increase in 165 taxa, 6 genera and two classes were observed while 5 genera showed a decreasing trend. In contrast to study by Xuan et al [33], genera Methylobacterium was less abundant in cancer patients [22] relative to non-cancer samples. Adding to the complexity, Methylobacterium was less abundant in tumor tissue compared to nearby normal tissue but more abundant relative to tissues from non-cancer persons [22]. Alcaligenaceae was abundant in non-cancer patient samples compared to cancer patients [22]. Significant differences based on BMI, age or any other demographic features were not observed. Hormone receptor positive tumors exhibited significantly lower levels of Methylobacterium in comparison to hormone receptor-negative samples [22], opposed to previously discussed study where Methylobacterium radiotolerance was abundant in 100% of hormone positive breast cancers. No difference was observed in terms of richness or diversity from oral rinse of all the subjects. Even in urine samples, cancer bearing (median 125,037) and non-cancer bearing (median 203,842) subjects exhibited similar read counts (p = .15). Subjects with cancer showed higher diversity compared to non-cancer as indicated by Shannon diversity index; however, they were influenced mostly by menstrual status [22]. Steroid metabolism by microbes, important in hormone positive breast cancer, is understood to some extent but crosstalk between human and microbial cells within the tumor microenvironment remains to be fully elucidated. Richer diversity in hormone positive breast tumors, via metabolic pathways, might have prognostic significance, thus, warrants further investigation.
2.2. Evidence from clinical studies: comparing microbiome of healthy controls vs breast cancer survivors
In a uniquely designed study, Chan et al. investigated the microbial population of the nipple aspirate fluid (NAF) of healthy volunteers and breast cancer survivors using 16S rRNA gene amplicon sequencing [41]. We obtained sequencing data from NCBI-SRA, accession number SRP071608, and analyzed using OneCodex. The NAF from the two groups was composed of significantly different microbial communities (Fig. 4A–C). The major phyla Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria formed 89.1% and 82.9% of total microbial population in healthy volunteers and breast cancer survivors respectively (Fig. 4B). Higher Firmicutes to Bacteroidetes ratio was observed in healthy volunteers compared to breast cancer survivors (Fig. 4C). Neisseria, Ruminococci, Lachnospiraceae, Faecalibacterium prausnitzii, Dolosigranulum pigrum, Corynbacterium durum, Mycobacterium kansasii, Clostridium baratii, and Rothia mucilaginosa were dominantly found only in the survivors whereas others like Lactobacillus salivarius were abundant in healthy women (Fig. 4D) [41]. The microbes associated with breast cancer are enriched in β-Glucuronidase, an enzyme that regulates the bioavailability of estrogens, thus may promote breast cancer [41]. Analysis of the ductal source DNA is important since breast ducts are the only passage of translocation of microbes from skin or other sources like infant’s oral cavity to the breast. Microbial community composition of NAF was similar to breast tissue dominated by Firmicutes, Proteobacteria and Bacteroidetes; differences between healthy women and women with history of breast cancer were apparent [41].
Fig. 4.

(A) Phylum distribution of breast ductal microbiota in Nipple aspirate fluid of breast cancer survivors and healthy volunteers. (B) Mean percentages of major phyla in nipple aspirate fluid of breast cancer survivors and healthy volunteers. (C) Percentage of three major phyla, Firmicutes, Bacteroides and Proteobacteria. (D) Frequency of occurrence of abundant taxa in NAF of breast cancer survivors and healthy volunteers (SRP071608, Analysis: One Codex).
Breast milk is a ductal secretion similar to NAF. Microbes of the duct, via the breast milk, transmit to the infant during breastfeeding and help with digestion of the complex polysaccharides of the milk and also establish the infant’s gut flora and prime the immune system. Found in NAF samples, Clostridium, Prevotella, Corynebacterium, Lactobacillus and Actinobacteria (Fig. 4D), overlap with breast milk. Both NAF and breast milk were noted to be rich in Bacteroidetes (Prevotella), Proteobacteria (Ralstonia, Sphingomonas, Pseudomonas, Bradyrhizobiaccea), Actinobacteria (Propionibacterium, Corynebacterium) and Firmicutes (Staphylococcus, Clostridium, Lactobacillus) [41]. Chan et al. report that the genus Alistipes was abundant in aspirate of cancer survivor’s breast but not in healthy individuals [41], though our analysis showed an abundance of only 0.01%. Alistipes abundance is associated with good gastrointestinal health and a decrease in Alistipes population has been reported in obesity and IBD patients [42–44]. Chan et al., also report an abundance of an unclassified genus from Sphingomonadaceae family in NAF of healthy individuals. In the earlier study classifying microbiota of normal breast and breast tumor, Xuan et al., reported Sphingobium yanoikuyae in 95% on healthy breasts [33]. Though health implications of this bacterium is not well known, some Sphingomonadaceae family members breakdown polycyclic aromatic hydrocarbons and aromatic hydrocarbons related with breast cancer [41]. Duct epithelial cells express TLRs and NLRs, which are stimulated by microbial signals. Sphingobium yanoikuyae, can activate the TLR5 pathway since its flagellum is a ligand for the TLR receptor, hence, inhibit cancer progression [41]. Other classes including Prevotella, capable of metabolizing hormones, have also been reported. Among metabolites, they report higher Beta-Glucuronidase levels in NAF of breast cancer survivors, an enzyme that regulates estrogen bioavailability [41].
2.3. Evidence from clinical studies: comparing microbiome of benign vs malignant disease
In a study by Hieken et al., cancer-associated differences in microbiota were explored in breast tissue of 13 benign and 15 invasive breast cancer (ER+/PR+ and ER+/PR+/Her2+) patients and compared to respective adjacent normal tissue [45]. Significant differences between microbiota composition of the normal breast tissue contiguous with invasive cancer and benign disease were observed [45]. We obtained the raw data from sequence read archive (PRJNA335375, EBI-ENA) and examined the abundant taxa in each group and their relative abundance (Fig. 5A–C). The microbial community in both benign and malignant breast cancer appeared similar with majority formed by Bacteroidetes and Firmicutes. Women with invasive breast cancer carried a relative abundance of low abundant genera Fusobacterium, Atopobium, and Lactobacillus (Fig. 5A). The major phyla Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria formed 84.5% and 83.5% of total microbial population in benign and malignant breast cancer cases, respectively (Fig. 5B). Higher Firmicutes to Bacteroidetes ratio was observed in malignant compared to benign breast cancer patients (Fig. 5C). There was a higher abundance of Amycolatopsis orientalis, Oscillibacter sp., Lactobacillus sp N15.MGS-260, Porphyromonas macacae, Staphylococcus epidermis, Prevotella timonensis, Clostridium sp ASF 356 and Atopobium vaginae and lower abundance of Carnobacterium sp., Alistipes sp., Bordetella bronchiseptica, and Delftia tsuruhatensis in malignant disease as compared to benign disease (Fig. 5D). These differences suggest a drastic change in community composition between benign and malignant tissue, though larger cohort studies are indispensable for moving forward. The variations did not change with age and menopausal status adjustment [45]. Compared to overlaying skin, breast microbiota exhibited more species richness [45]. It can be attributed to microenvironmental factors within the breast such as pH, oxygen availability, and fat density. Malignant breast tissue was enriched with Lactobacillus, Hydrogenophaga, Gluconacterobacter, Atopobium and Fusobacterium. Among these Fusobacterium was found to be associated with multiple other cancers including colon carcinogenesis, probably via secretion of virulence factors capable of inducing a pro-carcinogenic, pro-inflammatory environment. Analysis of metabolic pathways suggested a depletion of cystine and methionine metabolism, C5-branched dibasic acid metabolism, fatty acid biosynthesis and glycosyltransferases in malignant disease [45]. A number of cancers are known to be methionine dependent [46,47], hence, impaired methionine metabolism can accelerate cancer progression.
Fig. 5.
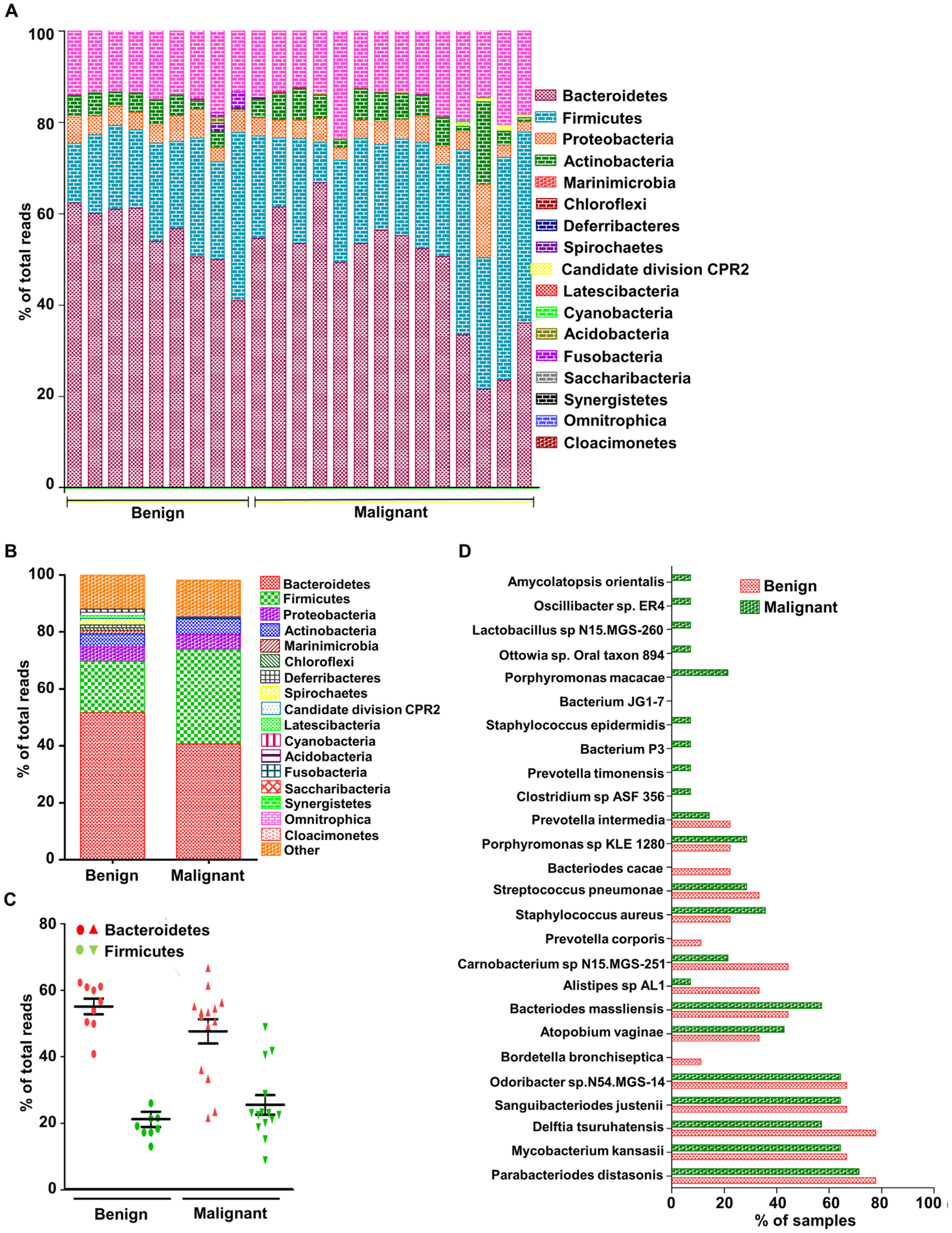
(A) Phylum distribution in local breast microbiota of breast cancer patients having benign and malignant disease. (B) Mean percentages of major phyla in benign and malignant breast cancer tissue samples. (C) Percentage of three major phyla, Firmicutes, Bacteroides and Proteobacteria. (D) Frequency of occurrence of abundant taxa in benign and malignant breast tissue samples (PRJNA335375, EBI-ENA, Analysis: OneCodex).
With growing evidence supporting the existence of a local microbiota in the breast and the ductal system, questions regarding the origin of this microbial community are becoming more important. One theory considers retrograde translocation of microbiota from the gut. This hypothesis is supported by experimental results showing that probiotics administered orally can be recovered in breast milk and could also treat lactational mastitis more efficiently than oral antibiotics [24]. An increase in mammary gland carcinogenesis has also been observed with enteral administration of some bacterial pathogens, possibly via an immune dependent mechanism [48]. These studies are however limited due to the lack of breast metagenome data. It is therefore imperative to investigate the association of gut, oral and skin microbiome with the breast microbial community.
2.4. Distinct microbiome in breast cancer subtypes
In a series of studies, Banerjee et al. have determined a microbial signature for different subtypes of breast cancer using a microarray-based approach called “PathoChip” [49,50]. They developed this unique technique for simultaneous detection of viruses, bacteria, fungi as well as parasites in the tumor microenvironment to develop a better understanding of the contribution of microbiome in carcinogenesis. The PathoChip targets viral, prokaryotic, and eukaryotic genomes with multiple DNA probes in a microarray format [51]. The chip has been optimized to detect even low copy number and fragmented genomes of microbial pathogens from fixed samples which can be further validated by PCR and sequencing methods [51]. Banerjee et al., identified distinct non-overlapping viral, bacterial, fungal as well as parasitic signatures for different breast cancer subtypes. The microbial and parasitic signatures for breast cancer subtypes using pathochip are represented in Table 1.
Table 1.
Viral, bacterial, fungal and parasitic signatures of the four major breast cancer subtypes obtained using pathochip.
| Breast cancer subtypes | Viral signatures | Bacterial signatures | Fungal signatures | Parasitic signatures |
|---|---|---|---|---|
| ER+PR+ | Hepadnaviridae | Arcanobacterium, Bifidobacterium, Cardiobacterium, Citrobacter, Escherichia | Filobasidilla, Mucor, Trichophyton | Brugia, Paragonimus |
| HER2+ | Nodaviridae | Streptococcus | Epidermophyton, Fonsecaea, Pseudallescheria | Balamuthia |
| ER+PR+HER2+ | Birnaviridae, Hepeviridae, Polyomaviridae, Hepadnaviridae | Bordetella, Campylobacter, Chlamydia, Chlamydophila, Legionella, Pasteurella | Penicillium | Ancylostoma, Angiostrongylus, Echinococcus, Sarcocystis, Trichomonas, Trichostrongylus |
| ER−PR−HER2− | Aerococcus, Arcobacter, Geobacillus, Orientia, Rothia | Alternaria, Malassezia, Piedraia, Rhizomucor | Centrocestus, Contracaecum, Leishmania, Necator, Onchocerca, Toxocara, Trichinella, Trichuris |
Using pathochip, Polyomaviridae was detected as a viral signature for ER+/PR+/HER2+ based on signal intensity, though detectable in all subtypes [50]. High intensity signals were achieved for Hepadnaviridae in ER+/PR+ and ER+/PR+/HER2+ subtype [50]. Parapoxviridae family signature was detected with highest intensity in ER+/PR+ subtype. These viruses encode homologs of human angiogenesis genes [52]. As reported by all previous studies, bacterial population was dominated by Proteobacteria followed by Firmicutes. Brevundimonas, Mobiluncus and Actinomyces (reported and discussed earlier) were abundant in all subtypes. HER2+ breast cancer was associated with Actinomyces signatures. Fungal signatures included yeasts like Candida, Geotrichum, Rhodotorula, Trichosporon, Epidermophyton and Trichophyton. Focus of current microbiome research in breast cancer focuses mostly on bacterial pathogens though some studies have shed light on viral contributors as well. Parasites have never really been studied in the context of breast cancer.
Salient feature of this series and the pathochip system is the screening and detection of subtype specific parasite signatures. Trichinella was associated with triple negative and recurrent invasive ductal carcinoma, most aggressive of the breast cancer subtypes [53]. Schistosoma was detected in both triple negative and triple positive subtypes and it has been linked to bladder cancer [54]. Ascaris was detected in HER2+, ER+/PR+ subtypes. Trichuris, another parasite detected in triple negative breast cancers in known to be associated with pediatric cancer [55]. Strongyloides detected in triple negative and HER2+ subtypes, has also been reported in adult cancer patients [56]. Leishmania (ER−/PR−/HER2−) and Plasmodium (HER2+, ER+/PR+ HER2+, ER+/PR+) inhibit apoptosis [57,58], and could be pro-carcinogenic. Some parasitic and fungal signatures in triple negative breast cancer were observed in patients with poor prognosis compared to patients who showed good response to therapy while some others trended in responsive patients. Once validated in larger cohorts, this pathochip could be a highly efficient and economic tool to assess breast cancer prognosis and suggest adjuvant therapies/interventions in addition to standard therapies to improve overall response.
3. Interactions between microbiome and cancer therapy
The microbiome affects various physiological functions of the body including metabolism, hematopoiesis, inflammation, immunity, neurological and cognitive functions [59]. Functioning as a second genome, a dysbiotic microbiota predisposes the body to develop cancers by inducing genetic instability, initiating DNA damage, proliferation of the damaged progeny and eliciting favorable immune response. It is now becoming clear that the microbiome can also alter response to therapy affecting drug metabolism, pharmacokinetics, anti-tumor effects and toxicity [59]. Absorption and bioavailability of most chemotherapy drugs require exposure to enzymes in the gut before entering the circulation. Microbes, the enzyme factories, can alter the mechanism of action and toxicity of chemotherapy drugs. Oxaliplatin and cyclophosphamide are two such widely used drugs that are modulated by gut microbes. Interestingly, germ-free mice are less susceptible to toxicity from cancer radiotherapy compared to conventional mice [60,61]. Despite multiple studies showing the influence of microbiome on the efficacy of cancer therapy, the exact mechanism regulating this phenomenon remains to be determined.
3.1. Influence of microbiome on drug metabolism
The gut microbiota has been shown to metabolize over 40 drugs and might contribute to the therapeutic efficacy of many more [62]. Studies using meta-transcriptomic, 16srRNA sequencing and flow cytometry have demonstrated that the response to drug treatments involves activation of drug metabolism and stress response pathways across diverse phyla, especially Firmicutes [62]. Gut commensals catalyze multiple chemical reactions including removal of functional groups, cleavage of N-oxide, proteolytic degradation, isoxazole scission, denitration, deconjugation, acetylation/deacetylation, amine formation and/or hydrolysis, and opening of thiazole ring, thus can biotransform chemical drugs [59,62]. Certain microbes physically adhere to the drugs, e.g. H. pylori interacts with L-DOPA via surface adhesins, reducing their absorption and bioavailability in the body [62]. Radio-sensitizer Misonidazole undergoes nitroreduction while anti-metabolite methotrexate is hydrolyzed by the gut microflora. Studies also describe the deconjugation of topoisomerase I inhibitor irinotecan (CPT-11) by gut microbes after hepatic detoxification [59,62]. RNA sequencing studies demonstrate constitutive overexpression of xenobiotic-sensing genes including Car or Nr1i3, Por, Ahr, PPAR α and Nrf2 in germ free mice [59,63,64] resulting in faster metabolism of xenobiotics. Colonization with gut microbiota from conventional mice restores conventional gene expression but administration of a probiotic preparation only restores a portion of the altered genes indicating that the difference in response to chemotherapy in cancer patients, at least partially, can be explained by differences in microbiome composition [59,64]. Intravenous chemotherapeutic drug, Irinotecan, is esterified into SN-38, the active form, in liver and small intestine. It then undergoes hepatic detoxification by UDP glucuronosyltransferases to form inactive SN-38-G prior to getting transported to gut [65]. Gut bacteria possessing ß-glucuronidases that can re-activate it to SN-38, resulting in serious intestinal toxicity. In experimental rodents, Irinotecan treatment increased Clostridium cluster XI and Enterobacteriaceae abundance in proximal colon thereby increasing the bacterial species expressing ß-glucuronidase (mostly firmicutes within Clostridium clusture XIVa and IV) [59,64]. Clostridium cluster XI is also known to regulate bile acid metabolism and liver immune activity, thereby affecting secondary tumor development in liver [17]. Administration of antibiotics inhibiting these species or bacterial ß-glucuronidase inhibitors effectively reduced Irinotecan-induced inflammation and systemic toxicity. In experimental animals, administration of probiotics also reduced Irinotecan induced diarrhea [59,64]. These studies warrant further investigations regarding the influence of microbiota on chemotherapeutic drugs and development of effective interventions to maximize the benefits and limit undesirable side effects using antibiotics and/or probiotics.
3.2. Influence of microbiome on drug efficacy
Lehouritis et al., tested the direct interaction of non-pathogenic Gram-negative Escherichia coli and Gram-positive Listeria welshimeri with 30 commonly used chemotherapeutics in vitro and in vivo. Efficacy of ten drugs was markedly reduced by either one or both of the species while six drugs showed enhanced efficacy [66]. Mass spectrometry and HPLC confirmed biotransformation of these drugs upon direct interaction with the bacteria [59]. Antitumor efficiency of gemcitabine was inhibited while that of prodrug CB1954 was enhanced by bacterial contact and was confirmed in vivo. Doxorubicin, one of the most commonly used chemotherapeutic drug, retains its therapeutic potential in microbiota depleted mice, however, colonization by Parabacteroides distasonis in microbiota depleted mice has been demonstrated to inhibit its activity [59,67]. Another important class of drugs, the platinum compounds, oxaliplatin and cisplatin, act via formation of intra-strand platinum– DNA adducts leading to DNA double-strand breaks, thus hindering DNA replication. They are therefore extremely toxic to the rapidly dividing cells of intestinal mucosa. Disruption of the barrier functions allows commensal microbes to leak to the lymphatic circulation causing a situation of septic shock and systemic inflammation [67–69]. Germ-free and broad-spectrum antibiotic treated mice bearing tumor do not respond to antineoplastic drugs oxaliplatin or cisplatin [67]. In experimental setting, absence of an active microbiota resulted in a lower inflammatory response upon oxaliplatin treatment. DNA damage was minimal despite the fact that an active microbiota is not required for penetration of the drug into the tumor or for formation of platinum-DNA adducts [67,70,71]. Platinum drug-induced cellular damage is brought about by reactive oxygen species or ROS [67,72,73]. ROS induces DNA damage and apoptotic response in tumor cells and interaction with the platinum drugs was thought to induce the cancer cells to generate ROS [73], however, depletion of microbiota in experimental mice prevented the paracrine ROS generation by tumor-in-filtrating myeloid cells engaging NADPH oxidase 2 (NOX2) [67,70,71]. This is supported by the fact that efficacy of oxaliplatin is reduced in CYBB (encoding gp91phox chain of NOX2)-deficient mice or myeloid cells depleted mice.
In Lewis lung carcinoma model, antibiotic treatment depleting most of the commensals resulted in inferior response to cisplatin treatment resulting in poor survival rate [74]. Co-administration of probiotic bacteria, Lactobacillus acidophilus, resulted in restoration of cisplatin induced gene expression and resulted in superior cisplatin response [74]. In multiple mice models bearing subcutaneous tumors, clearance of microflora resulted in diminished response to CpG oligonucleotide immunotherapy as well as platinum chemotherapy [67]. Depletion of commensals downregulated expression of genes related to antigen presentation, inflammation, phagocytosis and adaptive immunity and upregulation of genes related to cancer progression and metabolism [67]. Iida et al. describe that the tumor associated myeloid cells are primed by the commensals to induce cytokine production, which is impaired in absence of an active microbiome [67]. Microbiota induced myeloid cell priming is dependent on TLR4 expression and can be restored by oral administration of consistent doses of LPS. Gram-negative genera Alistipes is thought to be important in priming of cells and TNF production; however, other genera and receptors are likely to have important roles. In contrast to cisplatin, Oxaliplatin induces immunogenic cell death involving adoptive T cell response. Early autophagy uncovers signals associated with cell surface and soluble immunostimulatory signals which elicits T cell activation via antigen-presenting cells [74], resulting in long-term tumor regression [67,75,76]. The adoptive T cell activation depends on interleukin-1β (IL-1β) induction mediated innate immune receptor Toll-like receptor 4 (TLR4) and NLRP3 inflammasome [77]. DAMPs induce inflammation and immune response by reacting with germline-encoded PRRs [78]. Anthracycline agents on the other hand, require activation of FPR1 on dendritic cells via endogenous ligand annexin A1 from dying cancer cells [79]. Despite the fact that cisplatin induces a non-immunogenic cell death by direct DNA damage which results in rapid tumor clearance followed by a rapid recurrence [67], both early impact of cisplatin and long-lasting effects of oxaliplatin are compromised in microbiota-depleted mice [67], suggesting a role of microbiota in rapid antitumor response as well as the adaptive immune response however the underlying mechanisms might be comlpletely independent.
Alkylating agent Cyclophosphamide (CTX) is an important chemotherapeutic drug capable of stimulating antitumor immune response and at least partially dependent on intestinal flora. Viaud et al. in 2013 demonstrated that CTX alters the microbial community composition in the small intestine and promotes translocation of certain gram-positive bacteria to mesenteric lymph nodes where they induce generation of T helper 17 cells (pTH17 cells) and memory TH1 cells initiating long term anti-cancer immune response [67,70,78]. Germfree mice exhibited depleted pTH17 population and lower response to CTX treatment. Adoptive transfer of pTH17 cells restored CTX response in mice models [67]. Further studies demonstrated that bacterial species Enterococcus hirae and Barnesiella intestinihominis can restore and enhance CTX anticancer immune response in germ free mice [70,80]. While Enterococcus hirae translocated to secondary lymph nodes and increased intra-tumoral CD8+ T cell/T regulatory (Treg) cell ratio, Barnesiella intestinihominis accumulated in the colon and promoted the infiltration of interferon-γ (IFNγ)-producing γδ T cells in cancer lesions of CTX-treated mice. Interestingly, E. hirae-and B. intestinihominis-specific TH1 cells also correlated with better response to chemo-immunotherapy in lung and ovarian cancer cases [80].
In cervical cancer patients undergoing cisplatin chemotherapy and pelvic radiotherapy, L. acidophilus and Bifidobacterium bifidum probiotic administration prevented radiation induced intestinal toxicity [74,81]. Pharmacological inhibition of NADH oxidase using acetovanillone prevents cisplatin induced nephrotoxicity by inhibiting ROS generation in damaged tubular cells of the nephrons as well as the tissue myeloid cells [82] in addition to improving the response to cisplatin. Chemotherapy induced intestinal toxicity is regulated by the microbial sensors, TLRs. TLR4 activated by conserved microbial products or DAMPs mediates methotrexate induced mucosal damage while TLR2 activation prevents the same by inducing multidrug resistance factor, pglycoprotein overexpression [83–85]. Doxorubicin is known to induce severe organ toxicities including cardiovascular and mucosal damage and is also associated with significant changes in oral and gut microbial population. Interestingly, Rigby et al. elegantly demonstrated that microbial activity is essential for doxorubicin induced intestinal toxicity but not drug induced apoptosis [86–88]. However, the mechanistic details of microbial regulation of innate immune activation and regulation of chemotherapy related organ toxicities remains to be fully understood [86–88]. In the intestinal crypts, stem cells constitutively overexpress the innate immune receptor NOD2 that is activated by Muramyl peptide, common in most bacterial species. It promotes stem cell survival and prevents oxidative stress mediated cell death [89]. Gut commensals are known to play important role in fat metabolism, improving energy efficiency, adipose tissue inflammation, adipokine production and lipid uptake by cells. A dysbiotic microbiota is known to promote diet induced obesity via the farnesoid X receptor(FXR) pathway [90,91]. Cachexia and impaired fat metabolism are common observation in cancer patients undergoing platinum-chemotherapy [92–95] which enhances overall toxicity of cancer treatment in patients, partly due to altered drug pharmacokinetics [96,97]. Though further research regarding mechanism of cachexia in these conditions is needed but microbiota could be involved since energy metabolism and gut microbiota are closely related [95,98,99]. It has been shown that E. coli strain O21:H+ gut colonization prevents cachexia induced by Salmonella or Burkholderia infection via a sustained activation of PI3K/Akt activation in skeletal muscles which was dependent on NLRCF inflammasome formation [100].
3.3. Influence of microbiome on response to radiotherapy
Role of microbiome in regulating response to radiotherapy is poorly understood. Some studies suggest close association but the cause and effect relationship remains to be deciphered. Radiation induces non-targeted bystander effect on nearby cells leading to disruption of gap junctions dampening cell-cell interaction and release of extracellular mediators, including ROS, nitric oxide (NO), cytokines and exosomes [101–103]. It also induces abscopal effect, an antitumor immune response outside the field of radiation requiring activation of antigen-presenting dendritic cells and immune T cells [104]. Radiation-induced local and systemic toxicity may hinder anti-tumor immunity resulting in poor prognosis [105,106]. DNA damaging ionizing radiations are particularly toxic to proliferating tissue like bone marrow and epithelia [107], toxicities include oral mucositis, diarrhea, enteritis, colitis and bone marrow failure [60] and some of these effects have been attributed to changes in microbiota composition. In the intestinal crypts, ionizing radiation induces cellular damage and apoptosis [60]. Radio-tolerant pathobionts stimulate inflammatory response in the gut and alter the microbiome mediated systemic and innate immunity [60,61]. Mechanistically, TLRs known for their potential to recognize conserved microbial products like LPS and their derivatives, initiate innate immune response to ionizing radiation, for eg. TLR3 which detects dsRNA has also been shown to regulate radiation induced intestinal toxicity. TLR3 deficient mice are more sensitive to radiotherapy and show lower intestinal toxicity and increased life span [108]. However, TLRs function is complex and context-dependent stimulating a crosstalk between immune dependent and direct oncogenic pathways. Probiotic bacterium, Lactobacillus rhamnosus GG activates TLR2 which relocates COX2 expression from the cells of the villi to the base of the crypts [109], inducing ROS generation and consequently, activating the NRF cytoprotective pathway, thus protecting the intestinal mucosa from chemo and radiation induced toxicity in experimental mice [109,110]. Similarly, probiotic preparation VSL#3 (Bifidobacterium, Lactobacillus and Streptococcus spp.) has been shown to be protective against pelvic radiation induced gut toxicity [111] and Lactobacillus brevis prevents mucositis induced by chemo and radiotherapy in head and neck cancer [112]. Another association between microbiota and response to radiotherapy could possibly lie in the diurnal variation of gut microbiota composition. A number of studies have shown variation in radiation’s effect in bone marrow, blood and intestine depending on the circadian rhythm [113–115]. Microbial metabolic activities and thus the concentration of SCFAs and other immune stimulatory metabolites oscillates during the day, so does the sensitivity to radiation and post radiation tissue repair [116–120]. Studies have also indicated that conventional mice are more prone to dsDNA breaks in peripheral leukocytes in response to ionizing radiation compared to germ free and controlled microbiota bearing mice suggesting that microbes can influence the direct DNA damaging effect and bystander effect of ionizing radiation [121]. Better understanding of microbiome’s role in radiation response would help design better treatment regimens such as blockade of TLRs using natural or synthetic agonists, administration of probiotics prior to radiotherapy and timing radiotherapy sessions according to individual’s circadian rhythm. It may prove tremendously impactful in improving treatment response and preventing collateral damage.
4. Conclusions
The human metagenome is a genetic repository almost 100-folds larger than human genome and the host and microbial cells actively interact with each other. It is a dynamic system influencing every physiological process in the body while dysbiosis can alter normal physiological processes leading to cancer. However, it is highly variable across populations and no two individuals can possess exactly similar microbial signatures. Microbial composition of the body can vary depending on age, gender, ethnicity, race, BMI, dietary patterns, environmental conditions, mode of birth, exposure to antibiotics and other cultural variables. Local microbial communities have been identified in anatomical sites previously considered to be sterile with deep sequencing techniques, but microbiome research is still at its infancy. In this review, we analyzed, compared and discussed, within our scope, all available metagenomic data with relation to breast cancer. Till date, results obtained from different studies have been inconsistent due to variation in sample population in terms of age, geographical location, ethnicity, methods of sampling, sequencing techniques as well as mode of analysis. Most of the studies found statistically insignificant results making it difficult to draw definitive conclusions. However, the fact that similar trends have been observed in most, if not all, of the studies is very encouraging. Metabolic and molecular links are being discovered regularly indicating that there is a lot more to explore in this arena and all these inconsistencies are merely roadblocks appearing due to some incompletely deciphered facts.
Funding details
This work was supported by NCI NIH R01CA204555, Breast Cancer Research Foundation (BCRF) 90047965 and The Fetting Fund (to DS).
Footnotes
Disclosure statement
All authors declare that they have no conflict of interest.
References
- [1].Schwabe RF, Jobin C, The microbiome and cancer, Nat. Rev. Cancer 13 (2013) 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Plottel CS, Blaser MJ, Microbiome and malignancy, Cell Host Microbe 10 (2011) 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ohigashi S, Sudo K, Kobayashi D, et al. , Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer, Dig. Dis. Sci 58 (2013) 1717–1726. [DOI] [PubMed] [Google Scholar]
- [4].Ley RE, Obesity and the human microbiome, Curr. Opin. Gastroenterol 26 (2010) 5–11. [DOI] [PubMed] [Google Scholar]
- [5].Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI, Obesity alters gut microbial ecology, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ley RE, Knight R, Gordon JI, The human microbiome: eliminating the biomedical/environmental dichotomy in microbial ecology, Environ. Microbiol 9 (2007) 3–4. [DOI] [PubMed] [Google Scholar]
- [7].Pevsner-Fischer M, Tuganbaev T, Meijer M, Zhang SH, Zeng ZR, Chen MH, Elinav E, Role of the microbiome in non-gastrointestinal cancers, World J. Clin. Oncol 7 (2016) 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abu-Shanab A, Quigley EM, The role of the gut microbiota in nonalcoholic fatty liver disease, Nat. Rev. Gastroenterol. Hepatol 7 (2010) 691. [DOI] [PubMed] [Google Scholar]
- [9].Contreras AV, Cocom-Chan B, Hernandez-Montes G, Portillo-Bobadilla T, Resendis-Antonio O, Host-microbiome interaction and cancer: potential application in precision medicine, Front. Physiol 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Samuel BS, Shaito A, Motoike T, et al. , Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41, Proc. Natl. Acad. Sci 105 (2008) 16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF, Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4, Cancer Cell 21 (2012) 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Phelan JP, Reen FJ, Caparros-Martin JA, O’Connor R, O’Gara F, Rethinking the bile acid/gut microbiome axis in cancer, Oncotarget. 8 (2017) 115,736–115,747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vieira SM, Pagovich OE, Kriegel MA, Diet, microbiota and autoimmune diseases, Lupus. 23 (2014) 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Parracho HM, Bingham MO, Gibson GR, McCartney AL, Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children, J. Med. Microbiol 54 (2005) 987–991. [DOI] [PubMed] [Google Scholar]
- [15].Vogtmann E, Goedert JJ, Epidemiologic studies of the human microbiome and cancer, Br. J. Cancer 114 (2016) 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang J, Tan Q, Fu Q, et al. , Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications, Breast Cancer. 24 (2017) 220–228. [DOI] [PubMed] [Google Scholar]
- [17].Ma C, Han M, Heinrich B, et al. , Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells, Science (2018) 360 eaan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Freudenheim JL, Genco RJ, LaMonte MJ, et al. , Periodontal disease and breast cancer: prospective cohort study of postmenopausal women, Cancer Epidemiol. Biomark. Prev 25 (2016) 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sfreddo CS, Maier J, De David SC, Susin C, Moreira CHC, Periodontitis and breast cancer: a case-control study, Community Dent. Oral Epidemiol 45 (2017) 545–551. [DOI] [PubMed] [Google Scholar]
- [20].Nwizu NN, Marshall JR, Moysich K, et al. , Periodontal disease and incident cancer risk among postmenopausal women: results from the Women’s Health Initiative Observational Cohort, Cancer Epidemiol. Biomark. Prev 26 (2017) 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mai X, LaMonte MJ, Hovey KM, et al. , Periodontal disease severity and cancer risk in postmenopausal women: the Buffalo OsteoPerio study, Cancer Causes Control 27 (2016) 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, Grobmyer SR, Eng C, Breast tissue, oral and urinary microbiomes in breast cancer, Oncotarget. 8 (2017) 88,122–88,138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].DeStefano Shields CE, Van Meerbeke SW, Housseau F, et al. , Reduction of murine colon tumorigenesis driven by enterotoxigenic bacteroides fragilis using cefoxitin treatment, J. Infect. Dis 214 (2016) 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fernández L, Cárdenas N, Arroyo R, et al. , Prevention of infectious mastitis by oral administration of lactobacillus salivarius PS2 during late pregnancy, Clin. Infect. Dis 62 (2016) 568–573. [DOI] [PubMed] [Google Scholar]
- [25].Pompei A, Cordisco L, Amaretti A, Zanoni S, Matteuzzi D, Rossi M, Folate production by bifidobacteria as a potential probiotic property, Appl. Environ. Microbiol 73 (2006) 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wollowski I, Rechkemmer G, Pool-Zobel BL, Protective role of probiotics and prebiotics in colon cancer, Am. J. Clin. Nutr 73 (2001) 451s–455s. [DOI] [PubMed] [Google Scholar]
- [27].Lakritz JR, Poutahidis T, Levkovich T, Varian BJ, Ibrahim YM, Chatzigiagkos A, Mirabal S, Alm EJ, Erdman SE, Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice, Int. J. Cancer 135 (2013) 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang W, Lu H, Graham DY, An update on helicobacter pylori as the cause of gastric cancer, Gastrointest. Tumors 1 (2014) 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ding SZ, Goldberg JB, Hatakeyama M, Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis, Future Oncol. 6 (2010) 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Backert S, Selbach M, Role of type IV secretion in Helicobacter pylori pathogenesis, Cell. Microbiol 10 (2008) 1573–1581. [DOI] [PubMed] [Google Scholar]
- [31].Lee IO, Kim JH, Choi YJ, Pillinger MH, Kim SY, Blaser MJ, Lee YC, Helicobacter pylori CagA phosphorylation status determines the gp130-activated SHP2/ERK and JAK/STAT signal transduction pathways in gastric epithelial cells, J. Biol. Chem 285 (2010) 16,042–16,050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, Scott L, O’Hanlon DM, Burton JP, Francis KP, Tangney M, Reid G, Microbiota of human breast tissue, Appl. Environ. Microbiol 80 (2014) 3007–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, Lee DJ, Microbial dysbiosis is associated with human breast cancer, PLoS ONE 9 (2014) e83744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Murphy EA, Velazquez KT, Herbert KM, Influence of high-fat diet on gut microbiota: a driving force for chronic disease risk, Curr. Opin. Clin. Nutr. Metab. Care 18 (2015) 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G, The microbiota of breast tissue and its association with breast cancer, Appl. Environ. Microbiol 82 (2016) 5039–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Aragón F, Carino S, Perdigón G, de A Moreno de LeBlanc, Inhibition of growth and metastasis of breast cancer in mice by milk fermented with lactobacillus casei CRL 431, J. Immunother 38 (2015) 185–196. [DOI] [PubMed] [Google Scholar]
- [37].Marschalek J, Farr A, Marschalek ML, Domig KJ, Kneifel W, Singer CF, Kiss H, Petricevic L, Influence of orally administered probiotic lactobacillus strains on vaginal microbiota in women with breast cancer during chemotherapy: a randomized placebo-controlled double-blinded pilot study, Breast Care (Basel) 12 (2017) 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ojanotko-Harri A, Nikkari T, Harrl M-P, Paunio K, Metabolism of progesterone and testosterone by Bacillus cereus strain Socransky 67 and Streptococcus mutans strain Ingbritt, Oral Microbiol. Immunol 5 (1990) 237–239. [DOI] [PubMed] [Google Scholar]
- [39].Wiebe JP, Muzia D, Hu J, Szwajcer D, Hill SA, Seachrist JL, The 4-pregnene and 5alpha-pregnane progesterone metabolites formed in nontumorous and tumorous breast tissue have opposite effects on breast cell proliferation and adhesion, Cancer Res. 60 (2000) 936–943. [PubMed] [Google Scholar]
- [40].Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR, Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis, Elife. 2 (2013) e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chan AA, Bashir M, Rivas MN, Duvall K, Sieling PA, Pieber TR, Vaishampayan PA, Love SM, Lee DJ, Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors, Sci. Rep 6 (2016) 28,061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Saulnier DM, Kevin R, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J, Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome, Gastroenterology. 141 (2011) 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Martínez I, Muller CE, Walter J, Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species, PLoS ONE 8 (2013) e69621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Clarke SF, Murphy EF, O’Sullivan O, Ross RP, O’Toole PW, Shanahan F, Cotter PD, Targeting the microbiota to address diet-induced obesity: a time dependent challenge, PLoS ONE 8 (2013) e65790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hieken TJ, Chen J, Hoskin TL, et al. , The microbiome of aseptically collected human breast tissue in benign and malignant disease, Sci. Rep 6 (2016) 30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hoffman RM, Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: a 40-year odyssey, Expert. Opin. Biol. Ther 15 (2015) 21–31. [DOI] [PubMed] [Google Scholar]
- [47].Cavuoto P, Fenech MF, A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension, Cancer Treat. Rev 38 (2012) 726–736. [DOI] [PubMed] [Google Scholar]
- [48].Lakritz JR, Poutahidis T, Mirabal S, Varian BJ, Levkovich T, Ibrahim YM, Ward JM, Teng EC, Fisher B, Parry N, Lesage S, Alberg N, Gourishetti S, Fox JG, Ge Z, Erdman SE, Gut bacteria require neutrophils to promote mammary tumorigenesis, Oncotarget. 6 (2015) 9387–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Banerjee S, Wei Z, Tan F, et al. , Distinct microbiological signatures associated with triple negative breast cancer, Sci. Rep 5 (2015) 15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Banerjee S, Tian T, Wei Z, et al. , Distinct Microbial Signatures Associated With Different Breast Cancer Types, (2018), p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Baldwin DA, Feldman M, Alwine JC, Robertson ES, Metagenomic Assay for Identification of Microbial Pathogens in Tumor Tissues, 5 (2014), p. e01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ueda N, Wise LM, Stacker SA, Fleming SB, Mercer AA, Pseudocowpox virus encodes a homolog of vascular endothelial growth factor, Virology. 305 (2003) 298–309. [DOI] [PubMed] [Google Scholar]
- [53].Kristek J, Marjanović K, Dmitrović B, Krajinović Z, Sakić K, Trichinella spiralis and breast carcinoma–a case report, Coll Antropol. 29 (2005) 775–777. [PubMed] [Google Scholar]
- [54].Benamrouz S, Conseil V, Creusy C, Calderon E, Dei-Cas E, Certad G, Parasites and malignancies, a review, with emphasis on digestive cancer induced by Cryptosporidium parvum (Alveolata: Apicomplexa: ), Parasite. 19 (2012) 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Menon BS, Abdullah MS, Mahamud F, Singh B, Brief report. Intestinal parasites in Malaysian children with cancer, J. Trop. Pediatr 45 (1999) 241–242. [DOI] [PubMed] [Google Scholar]
- [56].Guarner J, Matilde-Nava T, Villaseñor-Flores R, Sanchez-Mejorada G, Frequency of intestinal parasites in adult cancer patients in Mexico, Arch. Med. Res 28 (1997) 219–222. [PubMed] [Google Scholar]
- [57].Heussler VT, Küenzi P, Rottenberg S, Inhibition of apoptosis by intracellular protozoan parasites, Int. J. Parasitol 31 (2001) 1166–1176. [DOI] [PubMed] [Google Scholar]
- [58].Lowe SW, Lin AW, Apoptosis in cancer, Carcinogenesis. 21 (2000) 485–495. [DOI] [PubMed] [Google Scholar]
- [59].Roy S, Trinchieri G, Microbiota: a key orchestrator of cancer therapy, Nat. Rev. Cancer 17 (2017) 271. [DOI] [PubMed] [Google Scholar]
- [60].Touchefeu Y, Montassier E, Nieman K, et al. , Systematic Review: The Role of the Gut Microbiota in Chemotherapy- or Radiation-Induced Gastrointestinal Mucositis – Current Evidence and Potential Clinical Applications, 40 (2014), pp. 409–421. [DOI] [PubMed] [Google Scholar]
- [61].Wang A, Ling Z, Yang Z, Kiela PR, Wang T, Wang C, Cao L, Geng F, Shen M, Ran X, Su Y, Cheng T, Wang J, Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: a pilot study, PLoS ONE 10 (2015) e0126312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Haiser HJ, Turnbaugh PJ, Developing a metagenomic view of xenobiotic metabolism, Pharmacol. Res 69 (2012) 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Selwyn FP, Cheng SL, Klaassen CD, Cui JY, Regulation of Hepatic Drug-Metabolizing Enzymes in Germ-Free Mice by Conventionalization and Probiotics, 44 (2016), pp. 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Selwyn FP, Cui JY, Klaassen CD, RNA-Seq Quantification of Hepatic Drug Processing Genes in Germ-Free Mice, 43 (2015), pp. 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fujita K, Sparreboom A, Pharmacogenetics of irinotecan disposition and toxicity: a review, Curr. Clin. Pharmacol 5 (2010) 209–217. [DOI] [PubMed] [Google Scholar]
- [66].Lehouritis P, Cummins J, Stanton M, et al. , Local bacteria affect the efficacy of chemotherapeutic drugs, Sci. Rep 5 (2015) 14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Iida N, Dzutsev A, Stewart CA, et al. , Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment, 342 (2013), pp. 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hooper LV, Macpherson AJ, Immune adaptations that maintain homeostasis with the intestinal microbiota, Nat. Rev. Immunol 10 (2010) 159. [DOI] [PubMed] [Google Scholar]
- [69].Sonis ST, The pathobiology of mucositis, Nat. Rev. Cancer 4 (2004) 277. [DOI] [PubMed] [Google Scholar]
- [70].Viaud S, Saccheri F, Mignot G, et al. , The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide, 342 (2013), pp. 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Galluzzi L, Vitale I, Michels J, et al. , Systems biology of cisplatin resistance: past, present and future, Cell Death Dis. 5 (2014) e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kim S, Lee T-J, Park J-W, Kwon TK, Overexpression of cFLIPs Inhibits Oxaliplatin-Mediated Apoptosis Through Enhanced XIAP Stability and Akt Activation in Human Renal Cancer Cells, 105 (2008), pp. 971–979. [DOI] [PubMed] [Google Scholar]
- [73].Laurent A, Nicco C, Chéreau C, et al. , Controlling Tumor Growth by Modulating Endogenous Production of Reactive Oxygen Species, 65 (2005), pp. 948–956. [PubMed] [Google Scholar]
- [74].Gui QF, Lu HF, Zhang CX, Xu ZR, Yang YH, Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model, Genet. Mol. Res 14 (2015) 5642–5651. [DOI] [PubMed] [Google Scholar]
- [75].Tesniere A, Schlemmer F, Boige V, et al. , Immunogenic death of colon cancer cells treated with oxaliplatin, Oncogene. 29 (2009) 482. [DOI] [PubMed] [Google Scholar]
- [76].Michaud M, Sukkurwala AQ, Martins I, Shen S, Zitvogel L, Kroemer G, Subversion of the chemotherapy-induced anticancer immune response by the ecto-ATPase CD39, OncoImmunology. 1 (2012) 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ghiringhelli F, Apetoh L, Tesniere A, et al. , Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β–dependent adaptive immunity against tumors, Nat. Med 15 (2009) 1170. [DOI] [PubMed] [Google Scholar]
- [78].Pateras IS, Havaki S, Nikitopoulou X, Vougas K, Townsend PA, Panayiotidis MI, Georgakilas AG, Gorgoulis VG, The DNA damage response and immune signaling alliance: is it good or bad? Nature decides when and where, Pharmacol. Ther 154 (2015) 36–56. [DOI] [PubMed] [Google Scholar]
- [79].Vacchelli E, Ma Y, Baracco EE, et al. , Chemotherapy-Induced Antitumor Immunity Requires Formyl Peptide Receptor, 350 (2015), pp. 972–978. [DOI] [PubMed] [Google Scholar]
- [80].Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP, Routy B, Jacquelot N, Apetoh L, Becharef S, Rusakiewicz S, Langella P, Sokol H, Kroemer G, Enot D, Roux A, Eggermont A, Tartour E, Johannes L, Woerther PL, Chachaty E, Soria JC, Golden E, Formenti S, Plebanski M, Madondo M, Rosenstiel P, Raoult D, Cattoir V, Boneca IG, Chamaillard M, Zitvogel L, Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects, Immunity 45 (2016) 931–943. [DOI] [PubMed] [Google Scholar]
- [81].Chitapanarux I, Chitapanarux T, Traisathit P, Kudumpee S, Tharavichitkul E, Lorvidhaya V, Randomized controlled trial of live Lactobacillus Acidophilus plus Bifidobacterium Bifidum in prophylaxis of diarrhea during radiotherapy in cervical cancer patients, 5 (2010), p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang Y, Luo X, Pan H, Huang W, Wang X, Wen H, Shen K, Jin B, Pharmacological inhibition of NADPH oxidase protects against cisplatin induced nephrotoxicity in mice by two step mechanism, Food Chem. Toxicol 83 (2015) 251–260. [DOI] [PubMed] [Google Scholar]
- [83].Mercado-Lubo R, The interaction of gut microbes with host ABC transporters, Gut Microbes 1 (2010) 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cario E, Toll-like receptors in the pathogenesis of chemotherapy-induced gastrointestinal toxicity, Curr. Opin. Support. Palliat. Care 10 (2016) 157–164. [DOI] [PubMed] [Google Scholar]
- [85].Frank M, Hennenberg EM, Eyking A, et al. , TLR Signaling Modulates Side Effects of Anticancer Therapy in the Small Intestine, 194 (2015), pp. 1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Napeñas JJ, Brennan MT, Coleman S, et al. , Molecular methodology to assess the impact of cancer chemotherapy on the oral bacterial flora: a pilot study, Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod 109 (2010) 554–560. [DOI] [PubMed] [Google Scholar]
- [87].Niu QY, Li ZY, Du GH, Qin XM, (1)H NMR based metabolomic profiling revealed doxorubicin-induced systematic alterations in a rat model, J. Pharm. Biomed. Anal 118 (2016) 338–348. [DOI] [PubMed] [Google Scholar]
- [88].Rigby RJ, Carr J, Orgel K, King SL, Lund PK, Dekaney CM, Intestinal bacteria are necessary for doxorubicin-induced intestinal damage but not for doxorubicin-induced apoptosis, Gut Microbes 7 (2016) 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ, The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration, Cell Host Microbe 15 (2014) 792–798. [DOI] [PubMed] [Google Scholar]
- [90].Jiang C, Xie C, Lv Y, et al. , Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction, Nat. Commun 6 (2015) 10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Parséus A, Sommer N, Sommer F, et al. , Microbiota-Induced Obesity Requires Farnesoid X Receptor, 66 (2017), pp. 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Das SK, Eder S, Schauer S, et al. , Adipose Triglyceride Lipase Contributes to Cancer-Associated Cachexia, 333 (2011), pp. 233–238. [DOI] [PubMed] [Google Scholar]
- [93].Ruud J, Wilhelms DB, Nilsson A, et al. , Inflammation- and tumor-induced anorexia and weight loss require MyD88 in hematopoietic/myeloid cells but not in brain endothelial or neural cells, FASEB J. 27 (2013) 1973–1980. [DOI] [PubMed] [Google Scholar]
- [94].Suárez-Zamorano N, Fabbiano S, Chevalier C, et al. , Microbiota depletion promotes browning of white adipose tissue and reduces obesity, Nat. Med 21 (2015) 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].de Matos-Neto EM, Lima JD, de Pereira WO, et al. , Systemic Inflammation in Cachexia – Is Tumor Cytokine Expression Profile the Culprit? Front. Immunol 6 (2015) 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bruggeman AR, Kamal AH, LeBlanc TW, Ma JD, Baracos VE, Roeland EJ, Cancer Cachexia: Beyond Weight Loss, 12 (2016), pp. 1163–1171. [DOI] [PubMed] [Google Scholar]
- [97].Cvan Trobec K, Kerec Kos M, Trontelj J, et al. , Influence of Cancer Cachexia on Drug Liver Metabolism and Renal Elimination in Rats, 6 (2015), pp. 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bindels LB, Delzenne NM, Muscle wasting: the gut microbiota as a new therapeutic target? Int. J. Biochem. Cell Biol 45 (2013) 2186–2190. [DOI] [PubMed] [Google Scholar]
- [99].Klein GL, Petschow BW, Shaw AL, Weaver E, Gut barrier dysfunction and microbial translocation in cancer cachexia: a new therapeutic target, Curr. Opin. Support. Palliat. Care 7 (2013) 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Schieber AM, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS, Disease tolerance mediated by microbiome < em > E. coli < /em > involves inflammasome and IGF-1 signaling, 350 (2015), pp. 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Apetoh L, Ghiringhelli F, Tesniere A, et al. , Toll-like receptor 4–dependent contribution of the immune system to anticancer chemotherapy and radiotherapy, Nat. Med 13 (2007) 1050–1059. [DOI] [PubMed] [Google Scholar]
- [102].Ermolaeva MA, Segref A, Dakhovnik A, et al. , DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance, Nature. 501 (2013) 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Al-Mayah A, Bright S, Chapman K, Irons S, Luo P, Carter D, Goodwin E, Kadhim M, The non-targeted effects of radiation are perpetuated by exosomes, Mutat. Res 772 (2015) 38–45. [DOI] [PubMed] [Google Scholar]
- [104].Demaria S, Ng B, Devitt ML, et al. , Ionizing radiation inhibition of distant untreated tumors (abscopal effect) is immune mediated, Int. J. Radiat. Oncol. Biol. Phys 58 (2004) 862–870. [DOI] [PubMed] [Google Scholar]
- [105].Deng L, Liang H, Burnette B, Beckett M, Darga T, Weichselbaum RR, Fu YX, Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice, J. Clin. Invest 124 (2014) 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Baird JR, Friedman D, Cottam B, Dubensky TW, Kanne DB, Bambina S, Bahjat K, Crittenden MR, Gough MJ, Radiotherapy combined with novel STING-targeting oligonucleotides results in regression of established Tumors, Cancer Res. 76 (2015) 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Barker HE, Paget JT, Khan AA, Harrington KJ, The tumor microenvironment after radiotherapy: mechanisms of resistance and recurrence, Nat. Rev. Cancer 15 (2015) 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Vacchelli E, Eggermont A, Sautès-Fridman C, et al. , Trial watch: Toll-like receptor agonists for cancer therapy, OncoImmunology. 2 (2013) e25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Jones RM, Luo L, Ardita CS, et al. , Symbiotic Lactobacilli Stimulate Gut Epithelial Proliferation < em > via < /em > Nox-Mediated Generation Of Reactive Oxygen Species, 32 (2013), pp. 3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Jones RM, Desai C, Darby TM, Luo L, Wolfarth AA, Scharer CD, Ardita CS, Reedy AR, Keebaugh ES, Neish AS, Lactobacilli modulate epithelial Cytoprotection through the Nrf2 pathway, Cell Rep. 12 (2015) 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Blanarova C, Galovicova A, Petrasova D, Use of probiotics for prevention of radiation-induced diarrhea, Bratisl. Lek. Listy 110 (2009) 98–104. [PubMed] [Google Scholar]
- [112].Sharma A, Rath GK, Chaudhary SP, Thakar A, Mohanti BK, Bahadur S, < em > Lactobacillus brevis < /em > CD2 lozenges reduce radiation- and chemotherapy-induced mucositis in patients with head and neck cancer: A randomized double-blind placebo-controlled study, Eur. J. Cancer 48 (2012) 875–881. [DOI] [PubMed] [Google Scholar]
- [113].Duncan AM, Ronen A, Blakey DH, Diurnal variation in the response of gamma-ray-induced apoptosis in the mouse intestinal epithelium, Cancer Lett. 21 (1983) 163–166. [DOI] [PubMed] [Google Scholar]
- [114].Ishihara H, Tanaka I, Yakumaru H, et al. , Circadian transitions in radiation dose-dependent augmentation of mRNA levels for DNA damage-induced genes elicited by accurate real-time RT-PCR quantification, J. Radiat. Res 51 (2010) 265–275. [DOI] [PubMed] [Google Scholar]
- [115].Ruifrok AC, Weil MM, Thames HD, Mason KA, Diurnal variations in the expression of radiation-induced apoptosis, Radiat. Res 149 (1998) 360–365. [PubMed] [Google Scholar]
- [116].Leone V, Gibbons SM, Martinez K, et al. , Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism, Cell Host Microbe 17 (2015) 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Liang X, Bushman FD, FitzGerald GA, Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 10479–10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Mukherji A, Kobiita A, Ye T, Chambon P, Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs, Cell. 153 (2013) 812–827. [DOI] [PubMed] [Google Scholar]
- [119].Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A, Circadian Gene Bmal1 Regulates Diurnal Oscillations of Ly6Chi Inflammatory Monocytes, 341 (2013), pp. 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E, Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis, Cell. 159 (2014) 514–529. [DOI] [PubMed] [Google Scholar]
- [121].Maier I, Berry DM, Schiestl RH, Intestinal microbiota reduces genotoxic endpoints induced by high-energy protons, Radiat. Res 181 (2014) 45–53. [DOI] [PubMed] [Google Scholar]


