Abstract
Reliable and sufficiently discriminative methods are needed for differentiating individual strains of Salmonella enterica serotype Enteritidis beyond the phenotypic level; however, a consensus has not been reached as to which molecular method is best suited for this purpose. In addition, data are lacking on the molecular fingerprinting of serotype Enteritidis from poultry environments in the United Kingdom. This study evaluated the combined use of classical methods (phage typing) with three well-established molecular methods (ribotyping, macrorestriction analysis of genomic DNA, and plasmid profiling) in the assessment of diversity within 104 isolates of serotype Enteritidis from eight unaffiliated poultry farms in England. The most sensitive technique for identifying polymorphism was PstI-SphI ribotyping, distinguishing a total of 22 patterns, 10 of which were found among phage type 4 isolates. Pulsed-field gel electrophoresis of XbaI-digested genomic DNA segregated the isolates into only six types with minor differences between them. In addition, 14 plasmid profiles were found among this population. When all of the typing methods were combined, 54 types of strains were differentiated, and most of the poultry farms presented a variety of strains, which suggests that serotype Enteritidis organisms representing different genomic groups are circulating in England. In conclusion, geographical and animal origins of Salmonella serotype Enteritidis isolates may have a considerable influence on selecting the best typing strategy for individual programs, and a single method cannot be relied on for discriminating between strains.
Salmonella enterica serotype Enteritidis remains the most common Salmonella serotype isolated from humans in England and Wales, with 10,596 cases reported by the Public Health Laboratory Service (Colindale, United Kingdom) in 1999. However, the number of reported incidents associated with serotype Enteritidis in poultry is declining rapidly, with only 4.3% of the Salmonella incidents during 1999 caused by this serotype (Ministry of Agriculture, Fisheries and Food, London, United Kingdom). The implementation of European Directive 92/117 (on prevention and control of zoonoses) in breeding flocks and the establishment by major egg producers of a voluntary quality scheme (called the Lion code) involving vaccination and improved sanitation and farming practices for commercial layer flocks, may have accounted for this reduction in the number of serotype Enteritidis isolations. However, serotype Enteritidis is still considered to be the main serotype infecting humans and poultry worldwide. In order to maintain and improve the current situation in the United Kingdom, effective epidemiological surveillance and control programs are needed. Accurate means of subtyping isolates and an extensive investigation of the diversity and sources of genotypes within animal strains of serotype Enteritidis in the United Kingdom are urgently required.
Traditionally, epidemiological investigations for Salmonella spp. have been based on phenotypic characteristics; however, reliable and sufficiently discriminative methods of differentiating individual strains beyond the phenotypic level are required. The predominance of certain phenotypes (phage types [PTs]) of serotype Enteritidis within certain geographical locations makes further epidemiological subgrouping necessary. In Western Europe, the predominant PT is PT4 (14), whereas PT8 has most often been seen in the United States (12).
DNA typing techniques are now frequently used for epidemiological investigations, and the combination of conventional disease-tracing investigations and molecular epidemiology is yielding important insights into the epidemiology of many infectious diseases. Molecular epidemiology has been used to track specific strains of pathogens and to identify outbreaks of salmonellosis (4, 6, 10, 20, 23, 34). The possibility for identification of sources of infection and routes of transmission strongly suggests that these new tools should be important components of surveillance programs.
The degree of genomic polymorphism in a population is a crucial factor for molecular epidemiology. For serotype Enteritidis, one problem common to molecular methods is the supposed highly clonal nature of some of the PTs (11, 31). As a consequence, very powerful techniques are necessary to detect minor differences in the genotype of the isolates. At present, there is not a consensus as to which method is best suited for differentiation of serotype Enteritidis strains. Comparison of antibiotic resistance patterns (19), plasmid profiles (32, 38), IS200 restriction fragment length polymorphism (31), ribotyping (16–18), and pulsed-field gel electrophoresis (PFGE) (20, 27, 34, 35) have been used to differentiate strains for epidemiological purposes with various degrees of success. Also several PCR-based methods, such as random amplification of polymorphic DNA, have been used for this purpose, but these methods may lack sensitivity and reproducibility for routine use, and they do not appear to be suitably discriminatory due to the inability to separate artefactual variation and true polymorphism (18, 36).
rRNA operons are highly conserved, and they are present in several copies on the bacterial chromosome. The stable regions of the gene can act as molecular chronometers of the phylogenetic relationship of organisms, with variable rRNA gene and flanking regions allowing discrimination between strains (41). The number and location of rRNA operon copies and restriction sites within the genes and in their flanking regions differed in different bacterial clones. Ribotyping allows determination of these differences, although there is no consensus yet on the best restriction enzyme for digestion of chromosomal DNA. PFGE is a well-established procedure for the analysis of large DNA fragments and has been successfully applied to epidemiological studies for several Salmonella serotypes. Plasmid profile analysis has also been shown to be of some value in some epidemiological investigations within this genus. This study evaluated the combined use of classical methods with three well-established molecular methods (ribotyping, macrorestriction analysis of genomic DNA, and plasmid profiling) in the assessment of diversity within 104 isolates of serotype Enteritidis from eight unaffiliated poultry farms in England.
MATERIALS AND METHODS
Salmonella isolates.
One hundred and four Salmonella serotype Enteritidis isolates originating from samples taken at eight poultry farms located in different geographical areas of England (Lancashire, Herefordshire, Northampton, Suffolk, Hertfordshire, Wiltshire, Somerset, and Surrey) were examined in this study. Samples (litter, feces, and environmental swabs) were processed according to methods previously described (8). The Salmonella cultures were serotyped following a microagglutination method (29) and phage typed (39) at the Veterinary Laboratories Agency (Weybridge).
Chromosomal DNA isolation.
A single colony of serotype Enteritidis was grown overnight at 37°C in 3 ml of Luria Bertani (LB) broth (10 g of tryptone per liter, 5 g of yeast extract per liter, and 10 g of NaCl per liter [pH 7.5]). Bacterial cells were pelleted by centrifugation at 6,000 rpm in a Centaur 2 MSE centrifuge, and the DNA was extracted from approximately 200 mg wet weight as previously described (7).
Restriction fragment length polymorphism. (i) Preparation of the rrn probe.
An Escherichia coli strain harboring plasmid pKK3535 (1) was grown at 37°C on LB agar supplemented with 50 μg of ampicillin per ml. Plasmid pKK3535 carrying the rrnB ribosomal RNA operon from E. coli was extracted using a QIAfilter plasmid Midi purification kit (Qiagen, Crawley, United Kingdom) and labeled with digoxigenin-11-dUTP by a random primed DNA labeling technique using the DIG-High prime kit (Roche Molecular Biochemicals, Lewes, United Kingdom).
(ii) Southern blotting hybridization.
Restriction enzyme digests of Salmonella DNAs were prepared using 4 μg of extracted DNA, 20 U of PstI, and 10 U of SphI, and these mixtures were incubated for 16 h at 37°C in incubation buffer (MULTI-CORE) as recommended by the manufacturer (Promega, Southampton, United Kingdom). Digested DNA (2 μg) was fractionated by electrophoresis on a 25-cm-long gel of 0.8% agarose type II (Sigma, Poole, United Kingdom) for 20 h at 45 V using TAE buffer (40 mM Tris-acetate [pH 8.3], 1 mM EDTA) with recirculaiton at 14°C. A DNA molecular weight marker that had been II-digoxigenin labeled (Roche Molecular Biochemicals) was used as the size standard in three wells of each gel. Fractionated DNA was transferred to positively charged nylon membranes (Roche Molecular Biochemicals) using 0.4 mM NaOH in a vacuum blotting apparatus (Pharmacia Biotech, Herts, United Kingdom) connected to a variable pump set at 40 mbar for 1 h. Membranes were rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and air dried before DNA was fixed to the membranes by cross-linking under UV light. Membranes were prehybridized for 4 h at 42°C in 20 ml of DIG Easy Hyb (Roche Molecular Biochemicals). Probes were denatured by boiling and added to fresh hybridization fluid at 20 ng/ml, and hybridizations were performed overnight at 42°C in a Hybaid oven. Following hybridization, excess probe was removed by washing twice for 5 min in 2× SSC–0.1% sodium dodecyl sulfate at room temperature and twice for 15 min in 0.1× SSC–0.1% sodium dodecyl sulfate at 68°C. The presence of the labeled probe was detected using the alkaline phosphatase-conjugated antibody DNA detection kit (Roche Molecular Biochemicals) and the chemiluminescent substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13.7]decan}-4-yl)phenyl phosphate (CSPD) as recommended by the supplier. The images produced on X-ray film were computer analyzed using the GelCompar II 1.01 software (Applied Maths, Kortrijk, Belgium). Molecular weights of the probed fragments were calculated by comparison with the external markers, and images from different gels were normalized accordingly. For the purposes of this study different PstI/SphI ribotypes (PS types) were allocated to strains when a genetic difference could be detected.
(iii) Pulsed-field gel electrophoresis.
Single colonies of Salmonella isolates were incubated overnight at 37°C in 3-ml amounts of LB broth with moderate shaking. One-milliliter aliquots of the cultures were transferred into microfuge tubes and washed twice with 1 ml of saline solution (0.85% [wt/vol] NaCl); finally cells were resuspended in 0.8 ml of saline solution and equilibrated at 40°C. This suspension was mixed in equal parts with molten 2% agarose (CleanCut; Bio-Rad, Hempstead, United Kingdom) and pipetted into disposable molds. Three of these agarose plugs were incubated overnight at 56°C in 2 ml of ES lysis buffer (0.5 M EDTA, 1% N-laurylsarcosine [Sigma]) with proteinase K (Sigma) at a final concentration of 250 μg/ml. The next morning, the lysis buffer was replaced with fresh ES buffer–proteinase K solution followed by a second overnight incubation at 56°C. Thereafter DNA-containing-plugs were thoroughly washed in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]) and stored at 4°C. Chromosomal DNA was digested with 30 U of XbaI (Promega), and PFGE was performed with a CHEF DRII system (Bio-Rad) in 0.5× Tris-borate-EDTA extended-range buffer (Bio-Rad) (130 mM Tris, 45 mM boric acid, 2.5 mM EDTA) with recirculation at 14°C. DNA macrorestriction fragments were resolved on 1% agarose gels (PFGE-certified agarose [Bio-Rad]), and a lambda Ladder pulsed-field gel marker (New England BioLabs, Hitchin, United Kingdom) was used as a size standard. Pulse times were ramped from 5 to 60 s during a 48-h run at 5.4 V/cm. The preparation and digestion of DNA from a proportion of the strains were repeated, and samples were electrophoresed under the same conditions to assess the reproducibility of the method. Macrorestriction patterns were compared with the use of GelCompar II software. The molecular weights of the restriction fragments were calculated by comparison with the external markers, and images were normalized accordingly. Different profiles were assigned to types in accordance with differences in the restriction patterns.
Plasmid analysis.
Plasmid DNA was isolated by the alkaline lysis method as described before (15). Samples were analyzed by electrophoresis in 1× Tris-borate-EDTA buffer at 150 V for 4 h on 0.8% and 1.5% agarose gels. Plasmid-containing strain E. coli 39R861 (42) and a supercoiled DNA ladder (Gibco BRL, Paisley, United Kingdom) were used to estimate plasmid sizes.
Discriminatory power of the methods.
The discriminatory power is the average probability that a typing system will assign a different type to two unrelated strains randomly sampled from a population and can be measured by the Simpson's index of diversity (D):
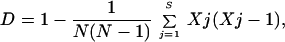 |
where S is the number of types recognized by a particular technique, Xj is the number of isolates identical to the jth strain, and N is the total number of unrelated strains tested.
RESULTS
Phage typing of Salmonella serotype Enteritidis isolates.
Phage typing differentiated the 104 isolates into 10 types (Table 1). The most common was PT4, which was found in 54 (51.9%) of the isolates from seven of the eight farms studied. Some types (PT1, PT4, and PT7) were found on more than one farm (common types) while other phage types (PT6, PT8, PT21, PT24, PT29, PT35, and PT36) appeared only on specific farms. The Simpson's index of diversity (D) for this typing method was 0.66.
TABLE 1.
Results of phage typing and DNA fingerprinting (ribotyping, PFGE, plasmid profile) of 104 serotype Enteritidis isolates from eight poultry farms
| Farm | No. of isolates | PT | Ribotype (PS type) | PFGE (X type) | Plasmid profileb | Overall typec |
|---|---|---|---|---|---|---|
| I | 6 | PT4 | PS17 | X1 | 1A | 1a |
| 1 | PT4 | PS17 | X1 | 0 | 2 | |
| 3 | PT4 | PS16 | X1 | 1A | 3 | |
| 1 | PT7 | PS16 | X2 | 0 | 4 | |
| 1 | NT | PS17 | X1 | 1A | 5 | |
| 1 | PT1 | PS17 | X1 | 0 | 6 | |
| 2 | PT1 | PS17 | X2 | 0 | 7 | |
| II | 10 | PT4 | PS2 | X1 | 1A | 8 |
| 3 | PT4 | PS1 | X1 | 1A | 9 | |
| 2 | PT4 | PS1 | X2 | 0 | 10 | |
| 1 | PT7 | PS1 | X1 | 1A | 11 | |
| 2 | NT | PS5 | X1 | 1A | 12 | |
| 1 | NT | PS3 | X1 | 2B | 13 | |
| 1 | NT | PS2 | X1 | 2B | 14 | |
| 1 | NT | PS3 | X1 | 3D | 15 | |
| 2 | PT1 | PS2 | X1 | 1A | 16 | |
| 1 | PT7 | PS2 | X1 | 1A | 17 | |
| 1 | PT7 | PS1 | X3 | 0 | 18 | |
| 1 | PT24 | PS2 | X1 | 2F | 19 | |
| III | 3 | PT4 | PS16 | X1 | 1A | 3 |
| 1 | PT4 | PS16 | X1 | 2G | 20 | |
| 2 | PT4 | PS16 | X2 | 0 | 21 | |
| 1 | PT7 | PS16 | X1 | 1A | 22 | |
| 1 | PT8 | PS22 | X5 | 1A | 23 | |
| IV | 2 | PT29 | PS12 | X1 | 3A | 24 |
| 1 | PT29 | PS8 | X2 | 3A | 25 | |
| 3 | PT29 | PS8 | X1 | 3A | 26 | |
| 2 | PT29 | PS10 | X1 | 3A | 27 | |
| 1 | PT29 | PS13 | X1 | 5A | 28 | |
| 1 | PT29 | PS13 | X1 | 3A | 29 | |
| 1 | PT29 | PS8 | X1 | 4A | 30 | |
| 1 | PT29 | PS11 | X6 | 4A | 31 | |
| 1 | PT29 | PS12 | X1 | 2E | 32 | |
| 1 | PT36 | PS9 | X1 | 3A | 33 | |
| V | 1 | PT21 | PS20 | X1 | 3B | 34 |
| 1 | PT21 | PS20 | X2 | 3B | 35 | |
| 1 | PT21 | PS20 | X2 | 2D | 36 | |
| 2 | PT21 | PS20 | X1 | 2D | 37 | |
| 1 | PT21 | PS20 | X1 | 3C | 38 | |
| 1 | PT35 | PS20 | X1 | 2D | 39 | |
| 1 | PT35 | PS20 | X1 | 2C | 40 | |
| 2 | PT6 | PS20 | X1 | 2D | 41 | |
| 1 | PT6 | PS16 | X1 | 2D | 42 | |
| 1 | PT6 | PS20 | X2 | 2D | 43 | |
| 1 | PT6 | PS20 | X1 | 3C | 44 | |
| 1 | PT6 | PS21 | X1 | 2D | 45 | |
| 1 | PT6 | PS17 | X1 | 3B | 46 | |
| 1 | PT4 | PS7 | X1 | 1A | 47 | |
| VI | 1 | PT4 | PS19 | X1 | 1A | 48 |
| 3 | PT4 | PS2 | X1 | 1A | 8 | |
| 1 | PT7 | PS2 | X1 | 1A | 17 | |
| 1 | PT7 | PS4 | X2 | 0 | 49 | |
| 1 | PT7 | PS1 | X1 | 1A | 11 | |
| VII | 1 | PT4 | PS14 | X1 | 1A | 50 |
| 2 | PT4 | PS16 | X1 | 1A | 3 | |
| 6 | PT4 | PS6 | X1 | 1A | 51 | |
| VIII | 5 | PT4 | PS16 | X1 | 1A | 3 |
| 1 | PT4 | PS16 | X2 | 0 | 21 | |
| 1 | PT4 | PS16 | X4 | 1A | 52 | |
| 1 | PT4 | PS15 | X1 | 1A | 53 | |
| 1 | PT4 | PS18 | X1 | 1A | 54 |
Types in boldface type were found only on one farm (specific types).
The numeral in the plasmid profile represents the identification number of the plasmid in the strain.
The overall type assigned to the strains represents a combination of results from the different typing methods.
PstI-SphI ribotyping.
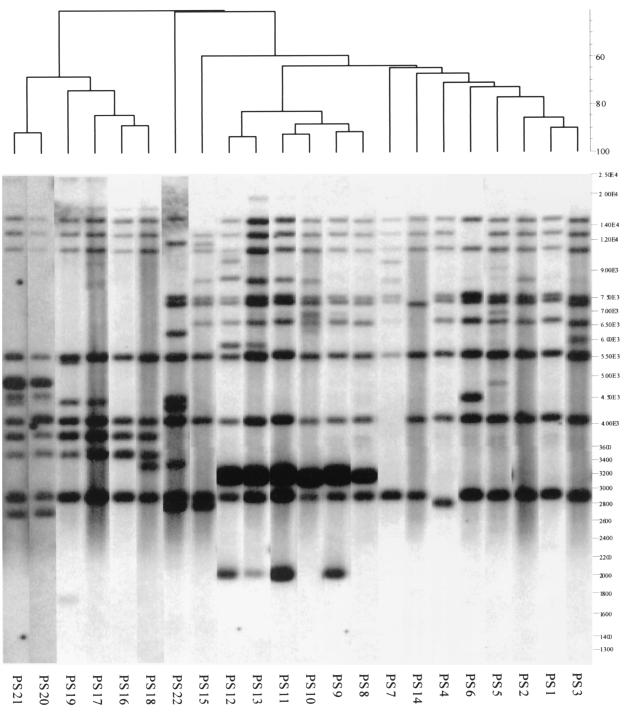
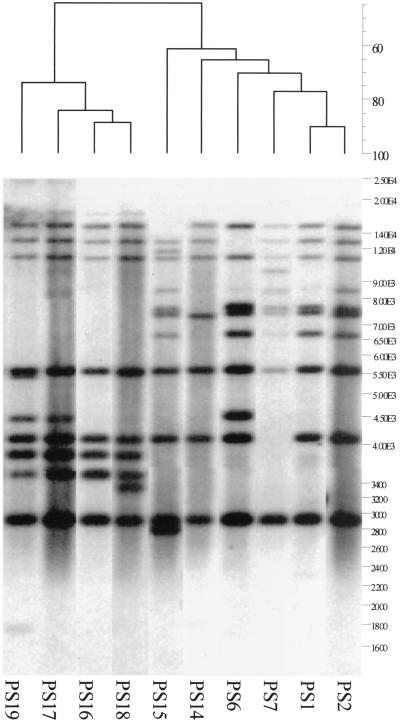
PstI-SphI ribotyping differentiated the serotype Enteritidis isolates into 22 different ribotypes (Table 1). The patterns produced with this method showed between 7 and 15 hybridizing fragments with only one conserved fragment of 5.7 kb among all isolates tested. The ribotypes were clustered into six main clusters (inter-cluster similarity percentage, < 65%) by GelCompar II. The similarity percentages between strains are shown in the dendrogram presented in Fig. 1. The most prevalent type (PS16) was found in 21 (20.2%) of the 104 isolates from five of the eight farms. Ribotypes PS1, PS2, PS16, and PS17 were found on more than one farm, while the remaining ribotypes were present only on specific farms. Figure 2 shows a dendrogram with relationships between the 10 ribotypes found among PT4 isolates. The D value for this typing method was 0.89.
FIG. 1.
Dendrogram generated by the Gelcompar II software, showing the relationship of 22 representative fingerprints (PstI-SphI ribotypes or PS types) for 104 isolates of serotype Enteritidis from England. The bands generated were analyzed using the Dice coefficient and unweighted-pair-group method with arithmetic averages (UPGMA).
FIG. 2.
Dendrogram generated by the Gelcompar II software showing the relationship of 10 representative fingerprints (PstI-SphI ribotypes or PS types) for 54 isolates of serotype Enteritidis PT4. The analysis of the bands generated was performed using the Dice coefficient and UPGMA.
PFGE.
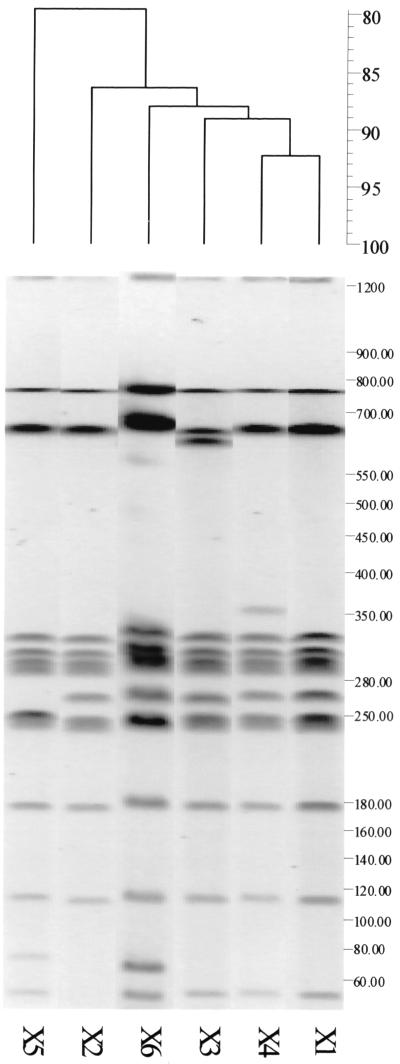
Electrophoresis of XbaI-digested genomic DNAs from the 104 isolates showed six different macrorestriction profiles (Fig. 3). XbaI profiles typically had 11 to 13 restriction fragments between 40 to 800 kb. The predominant type (X1) was found in 87 (83.6%) of the isolates analyzed and from all eight farms. Second in prevalence to type X1, type X2 was found in 13 (12.5%) isolates from seven of the eight farms. All of the remaining types were found only in single isolates from specific farms. The D value for this typing method was 0.28.
FIG. 3.
Dendrogram generated by the Gelcompar II software showing the relationship of six representative fingerprints (XbaI-PFGE or X types) for 104 isolates of serotype Enteritidis from England. The analysis of the bands generated was performed using the Dice coefficient and UPGMA.
Plasmid profiles.
Fourteen different plasmid profiles were found in the 104 serotype Enteritidis isolates. Fifty-eight (55.7%) of the isolates examined in this study harbored a single plasmid of approximately 57 kb. Table 2 shows the distribution of plasmid types and the molecular weights of the different plasmids for the eight farms included in the study. It is interesting that a correlation was found between PT and the presence of plasmids of unusual size. Plasmids of approximately 45 kb were found only in three isolates that were nontypeable by phage typing from farm II; a plasmid of 40 kb was found only in a single PT24 isolate from farm II; plasmids of sizes 6.5 and 3.1 kb were present only in PT29 and PT36 strains from farm IV; and a plasmid of 8.5 kb was found only in a PT29 strain from the same property. Plasmids of 4.8 and 4.2 kb were present only in PT6, PT21, and PT35 strains from farm V. The vast majority of PT4 (98%) and PT7 (100%) isolates were of plasmid types 1A or 0, thus limiting the potential of this technique for epidemiological studies in these PTs. The D value for this typing method was 0.66.
TABLE 2.
Results of plasmid profiles of 104 serotype Enteritidis isolates from eight poultry farms
| Farm | No. of isolates | Plasmid type | Size(s) of plasmids (kb) |
|---|---|---|---|
| I | 5 | 0 | |
| 10 | 1A | 57 | |
| II | 3 | 0 | |
| 19 | 1A | 57 | |
| 2 | 2Ba | 57, 45a | |
| 1 | 3D | 57, 45, 5.8 | |
| 1 | 2F | 57, 40 | |
| III | 2 | 0 | |
| 5 | 1A | 57 | |
| 1 | 2G | 83, 57 | |
| IV | 1 | 2E | 6.5, 3.1 |
| 10 | 3A | 57, 6.5, 3.1 | |
| 2 | 4A | 57, 6.5, 5.8, 3.1 | |
| 1 | 5A | 57, 8.5, 6.5, 5.8, 3.1 | |
| V | 1 | 1A | 57 |
| 1 | 2C | 4.8, 4.2 | |
| 9 | 2D | 57, 4.8 | |
| 3 | 3B | 57, 4.8, 4.2 | |
| 2 | 3C | 83, 57, 4.8 | |
| VI | 1 | 0 | |
| 6 | 1A | 57 | |
| VII | 8 | 1A | 57 |
| 1 | 1A | 57 | |
| VIII | 1 | 0 | |
| 8 | 1A | 57 |
Types or plasmid sizes in bold text were found only on one farm.
Combination of fingerprinting profiles.
The use of various typing methods identified different groups of clones. Therefore, their results could be combined to obtain a detailed overall fingerprint type. With the combination of results described above, we were able to identify a total of 54 strains (Table 1). Of these strains, 40 (74%) were farm specific and were identified by at least one specific result (that appeared only within a unique farm) with some of the fingerprinting methods applied. The remaining nine (17.4%) specific strains were identified from a unique combination of more common types. Strains PT4/PS16/X1/1A, PT4/PS2/X1/1A, PT7/PS2/X1/1A, PT7/PS1/X1/1A, and PT4/PS16/X2/0 were found in samples from several properties which suggests a common source of contamination.
DISCUSSION
In the present report, we describe the typing of 104 Salmonella serotype Enteritidis isolates using four commonly accepted methods. The different techniques identified different degrees of polymorphism in the following order: PS ribotyping (22 types, D = 0.89) > plasmid profiling (14 types, D = 0.66) > phage typing (10 types, D = 0.66) > PFGE (6 types, D = 0.28). The types obtained with each of the methods did not coincide, and a combination of these methods allowed for better discrimination.
Phage typing has been used traditionally as a first means of subdivision within serotype Enteritidis. However, this system presents some limitations. Not all organisms can be ascribed to recognized types, phage conversions are possible (3, 26), and the method requires access to special reagents available only in reference laboratories. Ribotype data from our study suggest a close relationship between PT1, PT4, and PT7. Strains from each of these PTs were found in combination with a fingerprint PS2/X1/1A; also strains belonging to PT4 and PT7 were found in combination with fingerprints PS16/X1/1A, PS16/X2/0, and PS1/X1/1A. Finally, strains of PT1 and PT4 were found in combination with fingerprint PS17/X1/0. The relationship found among PT1, PT4, and PT7 is in agreement with the results of a recent study using similar methodology for the analysis of serotype Enteritidis strains (16). In our study, a total of six PS types (PS8, PS9, PS10, PS11, PS12, PS13) were found among 13 PT29 strains and 1 PT36 strain from farm IV. For all but one of these (PS16), the banding patterns produced were very similar, and the similarity percentage shown in the dendrogram generated by GelCompar was > 80%. Finally, a correlation was found between PT6, PT21, and PT35; isolates from each of the three PTs were found in combination with fingerprint PS20/X1/2D; PT6 and PT21 isolates were found in combination with fingerprints PS20/X2/2D and PS20/X1/3C. The only PT8 isolate included in our study presented a clearly distinct ribotype (PS22) and PFGE type (X5), and a similar observation was made in a study by Landeras et al. (16).
PT4 strains have been considered as a rather homogeneous group (11). However, in our study, PT4 isolates could be differentiated into 10 PS types, and a dendrogram showed that these types were distributed in two main clusters with a similarity percentage of <60%. Four of the 10 types were present in isolates other than PT4, and within these four fingerprints, three ribotypes (PS2, PS16, and PS17) appeared in PTs other than PT1 or PT7.
Results of other studies suggest that the value of ribotyping depends on the restriction enzyme used for digestion, the gene probe, and the origin of the serotype Enteritidis isolates studied (40). Such a variety of methods makes the results obtained difficult to compare. Martinetti and Altwegg (21) demonstrated that serotype Enteritidis strains from different clusters of patients presented different SphI ribotypes. Usera et al. (37) were able to segregate 30 PT8 strains into 7 ribotypes using a combination of AccI and SmaI. In a more comprehensive study, PstI and SphI gave maximum sensitivity for ribotyping (PS ribotyping) of serotype Enteritidis strains (16), and 76 unrelated strains of human origin were differentiated into 22 types (17). In a further study, 90 isolates from environmental sources were separated into 20 PS ribotypes (18). Gruner et al. (10) found six ribotypes among 20 PT4 isolates and four ribotypes among 10 PT8 unrelated strains. In their study, a high association between SphI ribotype and phage type was reported.
In contrast with these results from other studies and the results from our work, several studies (11, 22) concluded that ribotyping is not a suitable technique to differentiate serotype Enteritidis strains. These studies suggested that there is a highly clonal structure within some PTs (PT4, PT8). We argue however that although there is a degree of uniformity within these strains, they can be effectively subdivided by means of a combination of methods. Ribotyping is a technique that requires considerable laboratory expertise, although the method does present some important advantages such as a high degree of typeability, stability, good sensitivity, and the possibility of normalization of data. With adequate software it becomes possible to compare results from different laboratories. In addition there is the potential for automation of the technique by using the riboprinter (13, 25) although this approach would require more flexibility in the use of enzymes and more entries in the database of types. One of the limitations of this technique is the occurrence of both weak and very strong hybridizing bands in the same pattern. The difference in band intensity may be due to a variety of factors such as the number of rRNA operon copies within the restriction fragments or internal rRNA SphI-PstI sites nicking off the gene, resulting in poor hybridization of the probe. We have tested the problematic strains several times and have found that some of the faint bands are not always reproducible in all of the runs; therefore, we did not include them for the clustering analysis.
XbaI was selected for this study as the most discriminative enzyme for the analysis of S. enteritidis strains by PFGE based on previous reports (20, 27, 34, 35, 40). Some studies have suggested that increasing the number of enzymes used for the digestion of DNA would allow for a better discrimination of strains. However the limitation of running 48-h electrophoresis in separate gels for each enzyme makes this approach rather impractical for routine use. In our study three XbaI-PFGE types (X1, X2, and X4) were found within the 54 PT4 isolates. Within these, X1 and X2 types accounted for the vast majority of isolates (89 and 9.3%, respectively) and appeared broadly distributed among other phage types, whereas type X4 was found in a single PT4 isolate. The results presented suggest that PFGE is not sensitive enough to distinguish effectively among these PT4 isolates. Furthermore, PFGE was not effective in distinguishing between PT1, PT4, PT6, PT7, PT21, PT24, PT29, PT35, and PT36 since isolates of these PTs showed identical PFGE patterns. The strains with different PFGE subtypes had a difference of only one or two fragments in their banding patterns; the literature suggests that recent point mutations may account for these minor differences (33), although a study by Murase et al. (23) concluded that XbaI-PFGE profiles are not affected by point mutations. Some other studies have also reported the limitation of PFGE for differentiation of serotype Enteritidis; for example, Thong et al. (34) found that 29 of 32 S. enteritidis isolates from sporadic unrelated cases were indistinguishable by PFGE with three different restriction enzymes.
The analysis of plasmid profiles has been found to be a useful tool for typing of some Salmonella serotypes; however, this method presents serious limitations. The present study, in agreement with previous experiments (11), found that the vast majority of serovar Enteritidis strains carry just the serospecific virulence plasmid. Extrachromosomal DNA is regarded as an unstable genetic marker (2, 5). Also, it is recognized that strains with the same chromosomal features may show different plasmid restriction patterns (28) and that the same plasmid profile may be present in strains which are different at a chromosomal level (24). Finally, open circular or linear plasmid forms display different electrophoretic migration patterns to confuse the interpretation of banding patterns. In our study we found a similarity of plasmid sizes on specific farms and this could be due to preferential carrying of plasmids by specific PTs (predominant on those farms) or to transmission of these elements within bacteria on the farms. Several other studies have shown that multiple plasmid profiles can be seen among isolates of the same phage type and conversely that isolates assigned to different phage types can exhibit the same plasmid profile (30, 40). This observations would suggest the possibility of plasmid transmission within isolates of different phage types. However, there is also evidence suggesting a link between specific plasmids and specific PTs (9). Further studies involving plasmid curing and plasmid transmission are necessary to assess these relationships.
The present study has shown that most of the poultry farms presented a variety of strains, which may be indicative of several sources of contamination or of genetic diversification during prolonged residence on the same premises. Several explanations can be given for this finding; one possible source of contamination is the existence of reservoirs (wild birds and rodents) that contribute to the maintenance and spread of Salmonella spp.; a second source of contamination comes from the difficulty of eliminating persistent site contamination of poultry units which results in maintenance of “in-house” strains; and finally, there is also the possibility of acquiring new strains by introduction of new animals falsely diagnosed as Salmonella free.
The finding of a genetic difference between two strains by the use of any particular marker would imply that they are different strains presumably originating from different sources. Confidence in this assumption would be increased if more than one typing system detected a difference, since every method detects polymorphism in different regions. One example of this would be strains PT4/PS16/X2/0 and PT8/PS22/X5/1A from farm III. Each of the typing techniques identified a difference between these two isolates, suggesting that they were not closely related genetically and that contamination was introduced from different sources. However, it must be remembered that some genetic change must account for the development of different strains and that one method could identify an initial change earlier than another technique. Also because the information provided by the different methods may be unrelated, changes may be found in only one of the markers. How frequently a strain of Salmonella will alter its genetic makeup is an unknown variable, and more studies are required to specifically assess the frequency of genetic changes that could result in modification of the fingerprint type.
Based on many studies carried out for serotype Enteritidis, it is possible to conclude that the geographical and animal origins of the isolates may have a considerable influence in deciding the best typing strategy for individual programs and that a single method cannot be relied upon for discriminating between strains. GelCompar II software was used in this study to analyze data from ribotyping and PFGE. Ideally, molecular-typing laboratories would use standardized methodology and computerized pattern recognition systems that permit data sharing. At present there is an urgent need for such an approach within Salmonella fingerprinting. Also, it is essential to determine the true extent of genetic diversity among serotype Enteritidis isolates at an international level and to determine whether a limited number of clones are associated with human disease and the molecular basis for the virulence of these strains (34). In summary, we have found that, within isolates from the United Kingdom, the most sensitive technique for identifying polymorphism was PstI-SphI ribotyping. XbaI PFGE of genomic DNA was of only limited use in differentiation of these isolates, demonstrating only minor differences between them. The combined use of molecular and phenotypic methods allowed more accurate discrimination within strains.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Agriculture, Fisheries and Food (United Kingdom).
We gratefully acknowledge M. Altwegg, who provided plasmid pKK3535.
REFERENCES
- 1.Brosious J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 2.Brown D J, Threlfall E J, Rowe B. Instability of multiple drug resistance plasmids in Salmonella typhimurium isolated from poultry. Epidemiol Infect. 1991;106:247–257. doi: 10.1017/s0950268800048391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chart H, Rowe B, Threlfall E J, Ward L R. Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves the loss of lipopolysaccharide with concomitant loss of virulence. FEMS Microbiol Lett. 1989;60:37–40. doi: 10.1016/0378-1097(89)90073-6. [DOI] [PubMed] [Google Scholar]
- 4.Chisholm S A, Crichton P B, Knight H I, Old D C. Molecular typing of Salmonella serotype Thompson strains isolated from human and animal sources. Epidemiol Infect. 1999;122:33–39. doi: 10.1017/s0950268898001836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrichton P B, Old D C, Taylor A, Rankin S C. Characterization of strains of Salmonella serotype Livingstone by multiple typing. J Med Microbiol. 1996;44:325–331. doi: 10.1099/00222615-44-5-325. [DOI] [PubMed] [Google Scholar]
- 6.Corbett-Feeney G, Riain U N. The use of pulsed field gel electrophoresis for subdivision of Salmonella typhimurium in an outbreak situation. J Infect. 1998;36:175–177. doi: 10.1016/s0163-4453(98)80009-1. [DOI] [PubMed] [Google Scholar]
- 7.Cousins D V, Williams S N, Ross B C, Ellis T M. Use of a repetitive element isolated from Mycobacterium tuberculosis in hybridisation studies with Mycobacterium bovis: a new tool for epidemiological studies of bovine tuberculosis. Vet Microbiol. 1993;37:1–17. doi: 10.1016/0378-1135(93)90178-a. [DOI] [PubMed] [Google Scholar]
- 8.Davies R H, Wray C. Use of antibody-coated cellulose sponges for enhanced isolation of Salmonella. Lett Appl Microbiol. 1997;24:246–248. doi: 10.1046/j.1472-765x.1997.00211.x. [DOI] [PubMed] [Google Scholar]
- 9.Frost J A, Ward L R, Rowe B. Acquisition of a drug resistance plasmid converts Salmonella enteritidis phage type 4 to phage type 24. Epidemiol Infect. 1989;103:243–248. doi: 10.1017/s0950268800030594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruner E, Martinetti Lucchini G, Hoop R K, Altwegg M. Molecular epidemiology of Salmonella enteritidis. Eur J Epidemiol. 1994;10:85–89. doi: 10.1007/BF01717458. [DOI] [PubMed] [Google Scholar]
- 11.Helmuth R, Schroeter A. Molecular typing methods for S. enteritidis. Int J Food Microbiol. 1994;21:69–77. doi: 10.1016/0168-1605(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 12.Hickman-Brenner F W, Stubbs A D, Farmer J J. Phage typing of Salmonella enteritidis in the United States. J Clin Microbiol. 1991;29:2817–2823. doi: 10.1128/jcm.29.12.2817-2823.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton A C, Penn C W. Comparison of ribotyping and arbitrarily primed PCR for molecular typing of Salmonella enterica and relationships between strains on the basis of these molecular markers. J Appl Microbiol. 1998;85:933–940. doi: 10.1111/j.1365-2672.1998.tb05256.x. [DOI] [PubMed] [Google Scholar]
- 14.Humphrey T J, Baskerville A, Maver S, Rove B, Hopper S. Salmonella enteritidis phage type 4 from the contents of intact eggs: a study involving naturally infected hens. Epidemiol Infect. 1989;103:415–423. doi: 10.1017/s0950268800030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landeras E, Gonzalez-Hevia M A, Alzugaray R, Mendoza M C. Epidemiological differentiation of pathogenic strains of Salmonella enteritidis by ribotyping. J Clin Microbiol. 1996;34:2294–2296. doi: 10.1128/jcm.34.9.2294-2296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landeras E, Mendoza M C. Evaluation of PCR-based methods and ribotyping performed with a mixture of PstI and SphI to differentiate strains of Salmonella serotype enteritidis. J Med Microbiol. 1998;47:427–434. doi: 10.1099/00222615-47-5-427. [DOI] [PubMed] [Google Scholar]
- 18.Landeras E, Gonzalez-Hevia M A, Mendoza M C. Molecular epidemiology of Salmonella serotype Enteritidis: relationship between food, water and pathogenic strains. Int J Food Microbiol. 1998;43:81–90. doi: 10.1016/s0168-1605(98)00092-0. [DOI] [PubMed] [Google Scholar]
- 19.Ling J M, Koo I C, Kam K M, Cheng A F. Antimicrobial susceptibilities and molecular epidemiology of Salmonella enterica serotype Enteritidis strains isolated in Hong Kong from 1986 to 1996. J Clin Microbiol. 1998;36:1693–1699. doi: 10.1128/jcm.36.6.1693-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukinmaa S, Schildt R, Rinttila T, Siitonen A. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J Clin Microbiol. 1999;37:2176–2182. doi: 10.1128/jcm.37.7.2176-2182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinetti G, Altwegg M. rRNA gene restriction patterns and plasmid analysis as a tool for typing Salmonella enteritidis. Res Microbiol. 1990;141:1151–1162. doi: 10.1016/0923-2508(90)90088-8. [DOI] [PubMed] [Google Scholar]
- 22.Millemann Y, Lesage M C, Chaslus-Dancla E, Lafont J P. Value of plasmid profiling, ribotyping, and detection of IS200 for tracing avian isolates of Salmonella typhimurium and S. enteritidis. J Clin Microbiol. 1995;33:173–179. doi: 10.1128/jcm.33.1.173-179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murase T, Nakamura A, Matsushima A, Yamai S. An epidemiological study of Salmonella enteritidis by pulsed field gel electrophoresis (PFGE): several PFGE patterns observed in isolates from a food poisoning outbreak. Microbiol Immunol. 1996;11:873–875. doi: 10.1111/j.1348-0421.1996.tb01153.x. [DOI] [PubMed] [Google Scholar]
- 24.Olsen J E, Skov M N. Genomic lineage of Salmonella enterica serovar Dublin. Vet Microbiol. 1994;40:271–282. doi: 10.1016/0378-1135(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 25.Oscar T P. Identification and characterization of Salmonella isolates by automated ribotyping. J Food Prot. 1998;61:519–524. doi: 10.4315/0362-028x-61.5.519. [DOI] [PubMed] [Google Scholar]
- 26.Rankin S, Platt D J. Phage conversion in Salmonella enterica serotype Enteritidis: implications for epidemiology. Epidemiol Infect. 1995;114:227–236. doi: 10.1017/s0950268800057897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridley A M, Threlfall E J, Rowe B. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1998;36:2314–2321. doi: 10.1128/jcm.36.8.2314-2321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiaffino A, Beuzon C R, Uzzau S, et al. Strain typing with IS200 fingerprints in Salmonella abortus ovis. Appl Environ Microbiol. 1996;40:271–282. doi: 10.1128/aem.62.7.2375-2380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shipp C R, Rowe B. A mechanical microtechnique for Salmonella serotyping. J Clin Pathol. 1980;33:595–597. doi: 10.1136/jcp.33.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer J T, Opitz H M, Gerhman M, Hall M M, Muniz I G, Rao S V. Molecular characterization of Salmonella enteritidis isolates from Maine poultry and poultry farm environments. Avian Dis. 1992;36:324–333. [PubMed] [Google Scholar]
- 31.Stanley J, Jones C S, Threlfall E J. Evolutionary lines among Salmonella enteritidis phage types are identified by insertion sequence IS200 distribution. FEMS Microbiol Lett. 1991;82:83–90. doi: 10.1016/0378-1097(91)90425-a. [DOI] [PubMed] [Google Scholar]
- 32.Stubbs A D, Hickman-Brenner F W, Cameron D N, Farmer J J., III Differentiation of Salmonella enteritidis phage type 8 strains: evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns, and ribotyping. J Clin Microbiol. 1994;32:199–201. doi: 10.1128/jcm.32.1.199-201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thong K L, Ngeow Y F, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thong K L, Puthucheary S, Pang T. Outbreak of Salmonella enteritidis gastroenteritis investigation by pulsed field gel electrophoresis. Int J Infect Dis. 1998;2:159–163. doi: 10.1016/s1201-9712(98)90120-5. [DOI] [PubMed] [Google Scholar]
- 36.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usera M A, Popovic T, Bopp C A, Strockbine N A. Molecular subtyping of Salmonella enteritidis phage type 8 strains from the United States. J Clin Microbiol. 1994;32:194–198. doi: 10.1128/jcm.32.1.194-198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachsmuth I K, Kiehlbauch J A, Bopp C A, Cameron D N, Strockbine N A, Wells J G, Blake P A. The use of plasmid profiles and nucleic acid probes in epidemiologic investigations of foodborne diarrheal diseases. Int J Food Microbiol. 1991;12:77–90. doi: 10.1016/0168-1605(91)90049-u. [DOI] [PubMed] [Google Scholar]
- 39.Ward L D, De Sa J D H, Rowe B. A phage typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–304. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weide-Botjes M, Kobe B, Schwarz S. Inter- and intra-phage type differentiation of Salmonella enterica subsp. Enterica serovar Enteritidis isolates using molecular typing methods. Zentbl Bakteriol. 1998;288:181–193. doi: 10.1016/s0934-8840(98)80037-6. [DOI] [PubMed] [Google Scholar]
- 41.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wray C, McLaren I M, Jones Y E. The epidemiology of Salmonella typhimurium in cattle: plasmid profile analysis of definitive phage type (DT) 204c. J Med Microbiol. 1998;47:483–487. doi: 10.1099/00222615-47-6-483. [DOI] [PubMed] [Google Scholar]