Abstract
Cas13 nucleases are a class of programmable RNA-targeting CRISPR effector proteins that are capable of silencing target gene expression in mammalian cells. Here, we demonstrate that RfxCas13d, a Cas13 ortholog with favorable characteristics to other family members, can be delivered to the mouse spinal cord and brain to silence neurodegeneration-associated genes. Intrathecally delivering an adeno-associated virus vector encoding an RfxCas13d variant programmed to target superoxide dismutase 1 (SOD1), a protein whose mutation can cause amyotrophic lateral sclerosis, reduced SOD1 mRNA and protein in the spinal cord by >50% and improved outcomes in a mouse model of the disorder. We further show that intrastriatally delivering an RfxCas13d variant programmed to target huntingtin (HTT), a protein whose mutation is causative for Huntington’s disease, led to a ~50% reduction in HTT protein in the mouse brain. Our results establish RfxCas13d as a versatile platform for knocking down gene expression in the nervous system.
RfxCas13d, an RNA-targeting CRISPR effector, can be delivered to the nervous system to silence neurodegeneration-linked genes.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated (Cas) proteins constitute a diverse family of adaptive bacterial immune systems that, in addition to serving as valuable tools for DNA editing (1, 2), have emerged as promising platforms for transcriptome engineering. Among the CRISPR-based technologies capable of perturbing RNA is Cas13, a programmable class 2 type VI CRISPR effector protein that naturally binds and cleaves single-stranded RNA (3–7) and, as such, can be co-opted for RNA targeting in prokaryotic (3) and eukaryotic cells (5, 8). Cas13 proteins can be directed to a unique RNA sequence via an engineered CRISPR RNA (crRNA) guide molecule that encodes a programmable spacer sequence that mediates target engagement by RNA-RNA base complementarity. Upon engaging with a target sequence, Cas13 undergoes a conformational change that activates its intrinsic ribonuclease (RNase) activity and results in the cleavage of the complexed RNA (Fig. 1A) (9).
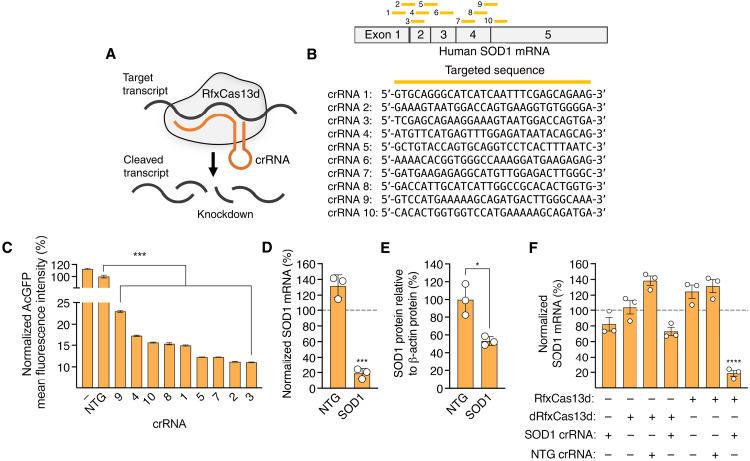
Fig. 1. RfxCas13d can be programmed to target hSOD1.
(A) Cartoon of RfxCas13d targeting. (B) (Top) Mature human SOD1 (hSOD1) mRNA and locations of the crRNA binding sites (yellow bars). (Bottom) crRNA target sequences. (C) Mean fluorescence intensity (MFI) of AcGFP in HEK293T cells transfected with SOD1 reporter plasmid and RfxCas13d with crRNAs. Data were normalized to cells transfected with RfxCas13d and nontargeted crRNA, denoted NTG. “−” indicates cells transfected with reporter plasmid only (n = 3). (D) Normalized SOD1 mRNA in HEK293T cells transfected with RfxCas13d and crRNAs. Data were normalized to relative SOD1 mRNA in untreated cells (n = 3). (E) hSOD1 protein in cell lysate from HEK293T cells transfected with RfxCas13d and crRNAs. SOD1 protein was normalized to β-actin protein (n = 3). (F) SOD1 mRNA in HEK293T transfected with RfxCas13d, catalytically dead RfxCas13d (dRfxCas13d), or no RfxCas13d with crRNAs. Data are normalized to relative SOD1 mRNA in untreated cells. (D and F) Dashed lines indicate relative SOD1 mRNA in untreated cells (n = 3). All analyses were conducted 72 hours after transfection. Error bars indicate SEM. *P < 0.05; ***P < 0.001. (C, D, and F) One-way ANOVA and (E) two-tailed unpaired t test.
To date, several different Cas13 subtypes have been identified and repurposed to silence target gene expression in mammalian cells (5, 8, 10, 11). Among these is the Cas13d nuclease from Ruminococcus flavefaciens XPD3002 (RfxCas13d) (5), a Cas13 ortholog that has favorable targeting characteristics to other family members (5, 12, 13) and is small enough to fit within a single adeno-associated virus (AAV) vector with a crRNA expression cassette for in vivo gene transfer (14, 15). Given these features, we sought to determine whether RfxCas13d could suppress gene function in vivo. More specifically, we sought to establish whether RfxCas13d could be delivered to the central nervous system (CNS) to silence the expression of genes whose mutations are associated with debilitating gain-of-function phenotypes.
In the present study, we demonstrate that RfxCas13d can be deployed to the mouse spinal cord and brain to knock down the genes causative for two progressive neurodegenerative disorders: amyotrophic lateral sclerosis (ALS) and Huntington’s disease (HD). We find that intrathecally delivering an AAV vector encoding RfxCas13d with a crRNA programmed to target superoxide dismutase 1 (SOD1), a protein whose mutation can cause ALS, reduced SOD1 mRNA and protein in the spinal cord by >50%, an outcome that we show results in decreased muscle atrophy, improved neuromuscular function, and a slowing in the overall progression of the disease in a mouse model of the disorder. Furthermore, we demonstrate that RfxCas13d can be programmed to target huntingtin (HTT), a protein that, when mutated to carry an abnormal expansion of a polyglutamine (polyQ)–encoding tract, causes HD. Intrastriatally delivering RfxCas13d to a mouse model of the disease led to a potent reduction in HTT protein and its aggregates. Our results thus establish RfxCas13d as a versatile platform for knocking down gene expression in the CNS.
RESULTS
Silencing SOD1 with RfxCas13d
We sought to determine whether CRISPR-Cas13 could be delivered to the CNS to silence neurodegeneration-linked genes. More specifically, we first asked whether RfxCas13d could be used to target SOD1, a ubiquitously expressed metalloenzyme whose mutation can cause ALS (16), a rapidly progressive and paralytic disorder characterized by the selective loss of motor neurons in the spinal cord and brain (17). Given its causative role in ALS, lowering the expression of mutant SOD1 by gene silencing has emerged as a promising strategy for treating the disease (18, 19).
To this end, we designed 10 crRNAs with 30-nucleotide (nt) protospacers targeting the mature human SOD1 (hSOD1) mRNA (Fig. 1B). These crRNAs do not overlap with any common mutations in SOD1, ensuring that they could be used in a mutation-independent manner to target the majority of the >100 different SOD1 mutations identified in ALS patients to date. Because Cas13d variants with a nuclear localization signal (NLS) sequence target RNA with greater efficiency than the native protein (5), we used an RfxCas13d variant with NLS sequences at its N and C termini for our studies.
To identify crRNAs that could target SOD1, we transfected human embryonic kidney (HEK) 293T cells with expression vector encoding RfxCas13d and a candidate crRNA, as well as a reporter plasmid expressing wild-type SOD1 fused to the green fluorescent protein from Aequorea coerulescens (AcGFP), thereby linking SOD1 expression to AcGFP fluorescence. Using flow cytometry, which was conducted 3 days after transfection, we found that all 10 of the SOD1-targeting crRNAs decreased AcGFP fluorescence by >75% relative to cells transfected with RfxCas13d and a nontargeted crRNA (P < 0.001), with the most active crRNAs found to reduce AcGFP fluorescence by >90% (P < 0.001) (Fig. 1C).
As the protospacer length of a crRNA can influence RfxCas13d targeting, we next analyzed the activities of crRNAs with variously sized targeting sequences (18, 20, 22, 24, 26, 28, 30, and 32 nt) for their ability to silence the SOD1 reporter in HEK293T cells. Flow cytometry revealed that, while spacer sequences equal to or less than 24 nt displayed modest or no activity, crRNAs with protospacers exceeding 28 nt all had virtually identical targeting activities, decreasing AcGFP fluorescence by ~95% (fig. S1). Given the lack of clear enhancement in targeting with alternatively sized crRNAs, we used a 30-nt protospacer for all subsequent studies.
We next evaluated the ability of RfxCas13d to target endogenous SOD1 mRNA. We therefore transfected HEK293T cells with RfxCas13d and the most active crRNA identified from our reporter screen (crRNA 3). Compared to untreated cells, we measured a ~80% decrease in SOD1 mRNA in cells transfected with RfxCas13d and the SOD1-targeting crRNA (P < 0.001) (Fig. 1D). Western blot corroborated this, showing a twofold reduction in total SOD1 protein at the same time point in cells transfected with RfxCas13d and the crRNA for SOD1 (Fig. 1E and fig. S2). Thus, RfxCas13d can lower SOD1 mRNA and protein in a human cell line.
Because crRNAs have been reported as capable of knocking down genes even in the absence of a Cas13 protein (20), we also tested whether the SOD1-targeting crRNA could lower SOD1 either by itself (that is, without RfxCas13d) or with a catalytically inactivated RfxCas13d variant (dRfxCas13d). According to quantitative polymerase chain reaction (qPCR) and relative to untreated cells, we measured a ~18% decrease in SOD1 mRNA in cells transfected with just the SOD1-targeting crRNA (P = 0.36) and a ~28% decrease in SOD1 mRNA in cells transfected with dRfxCas13d and the SOD1-targeting crRNA (P = 0.08; Fig. 1F). As additional controls, we evaluated the ability for RfxCas13d alone, dRfxCas13d alone, RfxCas13d with a nontargeted crRNA, and dRfxCas13d with a nontargeted crRNA to affect SOD1. No significant changes in SOD1 mRNA were observed for any of these conditions (P > 0.05; Fig. 1F). Thus, our results demonstrate that RfxCas13d can be programmed to target SOD1. In addition, we find evidence that non-RNase mechanism(s) can potentially contribute to knockdown.
RfxCas13d silencing of SOD1 improves outcomes in a rodent model of ALS
Next, we sought to determine whether RfxCas13d could provide a therapeutic benefit to a mouse model of SOD1-linked ALS following in vivo delivery to the spinal cord by an AAV vector. While motor neurons are selectively lost in SOD1-ALS, lowering the expression of mutant SOD1 within these cells only delays the onset of the disease (21, 22), whereas decreasing its expression in astrocytes, a type of glial cell involved in the neuroinflammatory response, markedly slows the overall progression of the disorder (23–26). Thus, we sought to deliver RfxCas13d to spinal cord astrocytes, a cell type that can be efficiently targeted in adult mice by AAV9 (26–28). To this end, we injected 60- to 65-day-old G93A-SOD1 mice—which carry ~25 copies of the human mutant SOD1G93A transgene and develop an aggressive disease that phenocopies hallmarks of ALS, including motor neuron degeneration, muscle wasting, and paralysis (29, 30)—with AAV9 vector encoding RfxCas13d with either the SOD1-targeting crRNA (AAV9-RfxCas13d-hSOD1) or a nontargeted crRNA (AAV9-RfxCas13d-NTG) via the cerebrospinal fluid of the lumbar spine (Fig. 2, A and B).
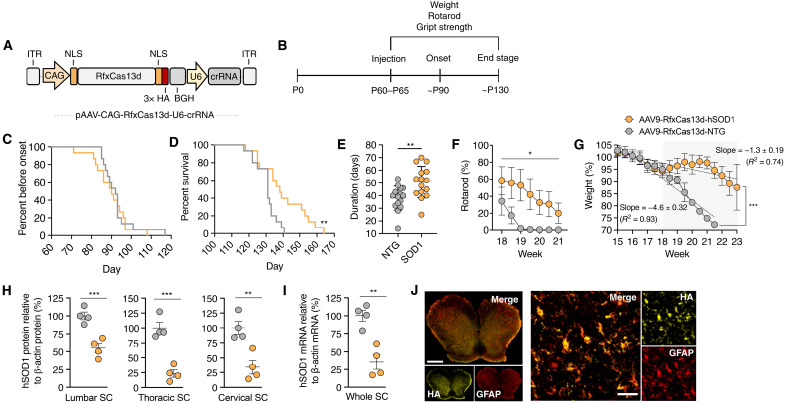
Fig. 2. RfxCas13d improves outcomes in G93A-SOD1 mice.
(A) AAV vector schematic. ITR, inverted terminal repeat; CAG, cytomegalovirus early enhancer/chicken β-actin promoter. (B) Timeline. (C) Percent before disease onset, (D) percent survival, (E) disease duration, (F) rotarod, and (G) weight of G93A-SOD1 mice injected with 2 × 1011 vg of AAV9-RfxCas13d-hSOD1 or AAV9-RfxCas13d-NTG (n = 15 for both groups for all measurements). (G) Gray indicates late-stage disease. (F and G) For NTG, weeks 19 to 19.5 had no error, whereas week 20 involved only a single animal. (H) Relative hSOD1 protein from lumbar, thoracic, and cervical spinal cord (SC) (n = 4). hSOD1 protein was normalized to β-actin protein. (I) Relative hSOD1 mRNA from whole spinal cord (n = 4). (J) Immunofluorescence of the lumbar spinal cord of injected G93A-SOD1 mice. Scale bars, 500 μm (left) and 30 μm (anterior horn). (F and G) Mean values for each mouse normalized to week 9 values for the same mouse. (G) Rate determined by linear regression analysis. Values represent means, and error bars indicate (F and G) SD or (H and I) SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (C and D) Log-rank Mantel-Cox test; (E) one-tailed unpaired t test; (F) two-way ANOVA; (H and I) one-tailed unpaired t test.
As expected, given the preference that intrathecally administered AAV9 has for spinal cord astrocytes (26–28) and the fact that disease onset in SOD1-ALS mice is modulated by the expression of mutant SOD1 in motor neurons (21, 31), we observed no delay in the onset of the disease (measured as peak weight) in mice treated by RfxCas13d versus the controls (P > 0.05; Fig. 2C). However, as compared to control animals, we observed that mice infused with AAV9-RfxCas13d-hSOD1 had both significantly increased survival (P < 0.01; Fig. 2D) and a considerably slower course of disease, evidenced by a ~33% increase in the duration of disease, a measure defined as the length of time between disease onset and end stage (hSOD1: 50.7 ± 12.4 days; NTG: 38.1 ± 9.6 days; P < 0.001; Fig. 2E). Furthermore, we measured that G93A-SOD1 mice infused with AAV9-RfxCas13d-hSOD1 had (i) significantly improved rotarod times during the late stage of the disease (the period from when animals lose 10% of their peak weight to end stage; P < 0.05) (Fig. 2F), (ii) a twofold slower rate of decline in hindlimb grip strength (hSOD1: −6.0 ± 0.36 normalized strength % per measurement; NTG: −11.0 ± 0.91 normalized strength % per measurement; P < 0.001) (fig. S3), and (iii) a nearly threefold slower rate of muscle atrophy over the course of the entire disease (hSOD1: −1.3 ± 0.19 weight % per measurement; NTG: −4.6 ± 0.32 weight % per measurement; P < 0.001) (Fig. 2G). Thus, our results demonstrate that RfxCas13d can improve therapeutic outcomes in a rodent model of a neurodegenerative disorder.
We next quantified SOD1 silencing in tissue harvested from end-stage animals. According to Western blot, G93A-SOD1 mice injected with AAV9-RfxCas13d-hSOD1 had reduced human mutant SOD1 protein in each region of the spinal cord compared to controls (Fig. 2H and fig. S4), including a 65 to 75% reduction in the cervical and thoracic regions (P < 0.001) and a ~45% reduction in the lumbar region (P < 0.01). Consistent with this finding, we also observed a ~65% decrease in the relative amount of hSOD1 mRNA in the whole spinal cord of animals treated by RfxCas13d compared to the controls (P < 0.001) (Fig. 2I), demonstrating that RfxCas13d lowered SOD1 in vivo. To evaluate off-target silencing by RfxCas13d, we analyzed from these tissues the expression of 10 candidate off-target transcripts, which each contained a potential pseudo-crRNA binding site with at least 13 nt of homology to the SOD1 crRNA (fig. S5A). Compared to control mice, no significant differences in expression (P > 0.05 for all) were measured by qPCR for any of the candidate genes in mice injected with AAV9-RfxCas13d-hSOD1 (fig. S5B).
We also analyzed the distribution of the RfxCas13d protein in G93A-SOD1 mice. Immunofluorescence analysis of end-stage tissue revealed that ~90% of glial fibrillary acidic protein–positive (GFAP+) astrocytes in the anterior horn of the lumbar, thoracic, and cervical spinal cord were positive for RfxCas13d, which we detected via its genetically fused hemagglutinin (HA) epitope tag (Fig. 2J and fig. S6). Consistent with the preference that intrathecally delivered AAV9 has for spinal cord astrocytes (22, 26), we observed limited RfxCas13d expression in other cell types within the spinal cord, including Iba1+ microglia and NeuN+ neurons (fig. S7).
Next, we determined whether RfxCas13d reduced the accumulation of SOD1 immunoreactive inclusions in the spinal cord, an outcome that we and others have observed can occur in SOD1-ALS mice after mutant SOD1 expression is ablated in spinal cord astrocytes (26, 32). According to immunofluorescence, mice injected with AAV9-RfxCas13d-hSOD1 had, on average, ~40% fewer SOD1 immunoreactive inclusions in the spinal cord white matter and ~29% fewer immunoreactive inclusions in the spinal cord anterior horn (P < 0.05 for all; fig. S8).
Last, we examined whether intrathecally administering the AAV9-RfxCas13d-hSOD1 vector induced an inflammatory response in mice. As neuroinflammation is a hallmark of disease in G93A-SOD1 mice, vector was delivered to B6SJLF1/J mice, the background strain for the G93A-SOD1 model used in this study. Compared to spinal cord sections from control animals injected with phosphate-buffered saline (PBS), we observed, at both 14 and 28 days after injection, no increase in the number of CD4+ or CD8+ T cells, NK1.1+ natural killer cells, Iba1+ and Mac2+ microglia, or GFAP+ astrocytes in mice injected with AAV9-RfxCas13d-hSOD1 (fig. S9). These findings suggest that delivering vector did not elicit a detectable inflammatory response in the spinal cord. In summary, our results demonstrate that RfxCas13d can be programmed to target SOD1 and that its AAV-mediated delivery to the spinal cord can impart a therapeutic benefit to a mouse model of SOD1-linked ALS.
Lowering HTT by RfxCas13d targeting
We next sought to determine the general applicability of RfxCas13d targeting in the CNS. To this end, we asked whether RfxCas13d could be used to knock down HTT (33), a protein with a polyQ tract near its N terminus that, when expanded beyond 37 repeats, causes HD, a progressive and ultimately fatal neurodegenerative disorder characterized by the loss of neurons in the striatum (34). Given the causal link between mutant HTT and HD, modalities capable of lowering mutant HTT gene expression hold the potential to slow the clinical progression of the disease (35, 36).
On the basis of the versatility and flexibility observed by RfxCas13d for SOD1, we designed just four crRNAs to target the sequence corresponding to exon 1 of the human HTT (hHTT) mRNA, both upstream and downstream of the polyQ-encoding repeat expansion (Fig. 3A). We then evaluated the silencing activity of the crRNAs in HEK293T cells using a previously described (37) reporter plasmid encoding exon 1 of the hHTT gene with 94Q fused to cyan fluorescent protein (CFP) (Fig. 3A). The expression of this reporter is controlled by the tetracycline response element, which is activated by the tetracycline-controlled transactivator protein. Thus, with this system, expression of HTT exon 1 is linked to CFP fluorescence.
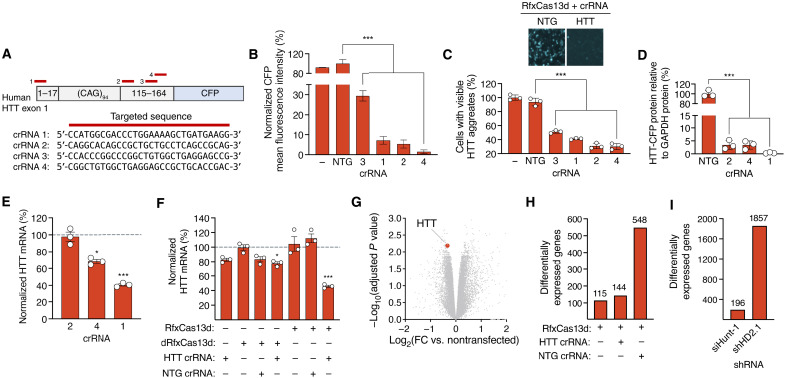
Fig. 3. RfxCas13d can target hHTT.
(A) (Top) HTT reporter and crRNA binding sites (red bars). (Bottom) crRNA target sequences. (B) CFP MFI in HEK293T cells transfected with reporter, RfxCas13d, and crRNAs. Data are normalized to cells transfected with RfxCas13d and NTG crRNA (n = 3). (C) (Top) Fluorescence image of HEK293T cells transfected with reporter, RfxCas13d, and HTT crRNA 1. (Bottom) Percentage of cells with HTT-CFP aggregates (n = 3). (B and C) “−” indicates cells transfected with reporter only. (D) HTT-CFP protein in HEK293T cells transfected with reporter, RfxCas13d, and crRNAs. CFP protein normalized to β-actin protein. (E and F) Relative HTT mRNA in HEK293T cells transfected with RfxCas13d/dRfxCas13d and crRNAs. Data are normalized to relative HTT mRNA in (E) cells transfected with RfxCas13d and NTG crRNA or (F) untreated cells, both indicated by dashed lines (n = 3). (G) Volcano plot of RNA-seq data comparing HEK293T cells transfected with RfxCas13d and HTT crRNA to untreated cells (n = 3). Red circle denotes HTT. (H and I) Number of differentially expressed genes [>1.25-fold change (FC), FDR-adjusted P < 0.01] in cells transfected with (H) RfxCas13d and crRNAs or (I) shRNAs, as compared to nontransfected cells (n = 3). All analyses are conducted 72 hours after transfection. Error bars indicate SEM. *P < 0.05; ***P < 0.001. (B to F) One-way ANOVA.
According to flow cytometry, all four of the crRNAs tested with this reporter reduced CFP expression by >70% relative to a nontargeting crRNA, with the three most active crRNAs found to decrease fluorescence by ~95% (P < 0.001; Fig. 3B). As this reporter expresses a mutant HTT protein fragment that also forms fluorescent inclusions in cells, we used fluorescence microscopy to analyze aggregate formation, observing a reduction in the number of visible inclusions in cells with RfxCas13d and the most active crRNAs (Fig. 3C). Specifically, compared to the nontargeting crRNA, we measured a ~50 to 70% decrease in the number of cells with CFP+ aggregates (P < 0.001 for all; Fig. 3C). Western blot supported this, revealing at least an 80% reduction in the total amount of mutant HTT-CFP protein for the three most active crRNAs (P < 0.001 for all; Fig. 3D and fig. S10).
We next sought to determine whether RfxCas13d could target endogenous HTT mRNA. We therefore transfected HEK293T cells with RfxCas13d and the most active HTT crRNAs identified from the reporter screen (crRNAs 1, 2, and 4). By qPCR, we measured a significant decrease in HTT mRNA for two of the crRNAs tested, with the most active crRNA, crRNA 1, observed to reduce HTT mRNA by ~60% compared to cells transfected with RfxCas13d and a nontargeting crRNA (P < 0.001; Fig. 3E). These results thus demonstrate that RfxCas13d can be programmed to target endogenous HTT mRNA in a human cell line.
Because we measured a modest decrease in SOD1 mRNA in cells transfected with just the SOD1 crRNA, we also determined whether the HTT crRNA could lower HTT mRNA either by itself or with a dRfxCas13d variant. Consistent with our findings for the SOD1 crRNA, we measured by qPCR a ~17% decrease in HTT mRNA in cells transfected with just the HTT crRNA (P = 0.15) and a ~28% decrease in HTT mRNA in cells transfected with dRfxCas13d and the HTT crRNA (P < 0.05) (Fig. 3F), with no significant changes in HTT mRNA observed in cells transfected with RfxCas13d alone, dRfxCas13d alone, RfxCas13d with a nontargeted crRNA, and dRfxCas13d with a nontargeted crRNA (P > 0.05; Fig. 3F). These results thus raise the possibility that non-RNase mechanism(s) can contribute to knockdown. Additional studies will be needed to determine whether, and potentially how, these mechanisms function alongside catalytically active RfxCas13d.
As RfxCas13d has been reported as capable of silencing nontarget transcripts (38), we analyzed on a transcriptome-wide scale the targeting specificity of RfxCas13d with the HTT-targeting crRNA in HEK293T cells. Using RNA sequencing (RNA-seq), we found that, in addition to HTT, whose down-regulation was expected, 143 genes (~0.92% of total measured transcripts) were differentially expressed in cells transfected with RfxCas13d and the HTT crRNA [>1.25-fold change; false discovery rate (FDR)–adjusted P < 0.01] (Fig. 3G). For reference, RfxCas13d alone or with a nontargeted crRNA affected the expression of ~0.73 and ~3.5% of total transcripts, respectively (Fig. 3H). Of the 143 nontarget genes that were differentially expressed in cells transfected with RfxCas13d and the HTT crRNA, 108 of them (~75%) were also affected in cells transfected with RfxCas13d and the nontargeted crRNA, while 22 of these were affected in all three groups (fig. S11 and table S1). Among the 33 nontarget genes that were uniquely affected by RfxCas13d and the HTT crRNA, similarity searches for pseudo-crRNA binding sites revealed that no gene contained >10 nt of contiguous homology to the HTT crRNA. In addition, an overrepresentation analysis of gene ontology (GO) biological process (BP) and molecular function (MF) terms revealed no significant enrichment (FDR-adjusted P > 0.05) for any themes for the 33 genes that were uniquely differentially expressed by RfxCas13d with the HTT crRNA or the 108 genes that were found to overlap between the multiple groups.
As an additional point of comparison, we measured by RNA-seq the transcriptome-wide specificities of two short hairpin RNAs (shRNAs) for HTT. By RNA-seq, we found that siHunt-1 (39), an shRNA that targets the 5′ untranslated region (UTR) of the hHTT mRNA, affected the expression of 196 genes (~1.2% of measured transcripts), while shHD2.1 (40), an shRNA that targets a sequence in exon 2 of the hHTT mRNA, affected 1857 genes (~11.7% of measured transcripts; >1.25-fold change for both; FDR-adjusted P < 0.01 for both) (Fig. 3I). Thus, our results indicate that, on a transcriptome-wide scale, RfxCas13d specificity is comparable to shRNA.
Last, we sought to determine whether RfxCas13d could lower the mutant hHTT protein in a transgenic mouse model of HD, specifically R6/2 mice, which carry exon 1 of the hHTT gene with a ~150-polyQ repeat alongside a ~1-kb fragment of the 5′UTR that drives expression of the transgene (41). R6/2 mice produce the toxic N-terminal fragment of the human mutant HTT protein and develop inclusions in striatal neurons (which are lost in HD) as early as 4 weeks of age. They are thus a useful model for evaluating the activity of HTT silencing agents.
Given its ability to transduce neurons in the striatum (40, 42–45), we chose to use AAV1 to deliver RfxCas13d in vivo. Thus, we injected the striatum of 4-week-old R6/2 mice with AAV1 vector encoding RfxCas13d with either the HTT-targeting crRNA (AAV1-RfxCas13d-hHTT, left hemisphere) or a nontargeted crRNA (AAV1-RfxCas13d-NTG, right hemisphere). At 4 weeks after delivery, we quantified the abundance of the human mutant HTT protein in dissected striatal tissue. According to Western blot and compared to control tissue, we measured a ~60% decrease in human mutant HTT protein in hemispheres injected with AAV1-RfxCas13d-hHTT (P < 0.05) (Fig. 4A and fig. S12), a finding that was corroborated by qPCR, which revealed a ~45% reduction in the amount of relative hHTT mRNA in treated hemispheres compared to control hemispheres (P < 0.05) (Fig. 4B).
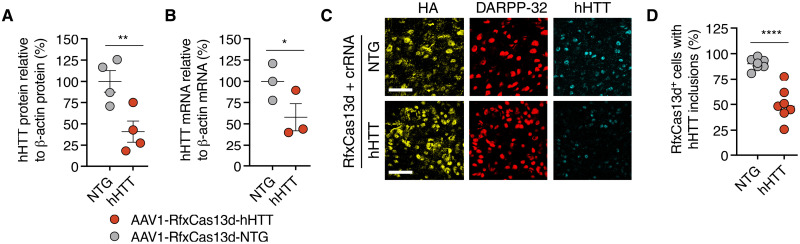
Fig. 4. RfxCas13d can reduce HTT protein in the brain of a mouse model of HD.
(A) hHTT protein (n = 4) and (B) hHTT mRNA (n = 3) from striatal tissue 4 weeks after R6/2 mice were injected with 6 × 1010 vg of AAV1-RfxCas13d-hHTT or AAV1-RfxCas13d-NTG. (A) hHTT protein in each sample was normalized to β-actin protein. (C) Representative immunofluorescence staining of striatal tissue 4 weeks after R6/2 mice were injected with either 6 × 1010 vg of AAV1-RfxCas13d-hHTT or AAV1-RfxCas13d-NTG. Scale bars, 30 μm. (D) Quantification of the percentage of RfxCas13d+ cells with measurable HTT immunoreactive inclusions, as determined by immunofluorescence staining of the striatum 4 weeks after R6/2 mice were injected with 6 × 1010 vg of AAV1-RfxCas13d-hHTT or AAV1-RfxCas13d-NTG. A total of >100 cells were counted per animal (n = 7). Values represent means, and error bars indicate SEM. *P < 0.05; **P < 0.01; ****P < 0.0001. (A, B, and D) One-tailed unpaired t test.
As R6/2 mice develop HTT immunoreactive inclusions (35, 42, 46), we next used immunohistochemistry to determine whether RfxCas13d could reduce their abundance in the brain. In addition to observing RfxCas13d expression in ~54% of cells positive for the medium spiny neuron marker DARPP-32 (Fig. 4C), the principal cell type lost in HD (34), we measured that hemispheres injected with AAV1-RfxCas13d-hHTT had ~50% fewer RfxCas13d+ cells with human mutant HTT immunoreactive inclusions as compared to RfxCas13d+ cells from the control hemisphere (P < 0.0001) (Fig. 4, C and D), demonstrating that RfxCas13d can reduce a pathology associated with HD. In conclusion, our results establish that RfxCas13d can be delivered to the mammalian spinal cord and brain to knock down target gene expression.
DISCUSSION
By virtue of their programmability and their ability to cleave target RNAs via an intrinsic RNase activity, CRISPR-Cas13 effectors can be used to knock down target gene expression in mammalian cells. In the present study, we demonstrate that RfxCas13d, a Cas13 effector that is compact enough to fit within a single AAV vector particle alongside a crRNA expression cassette, can be programmed to target (i) SOD1, a protein whose mutation has been linked to inherited forms of ALS, and (ii) HTT, a protein that, when mutated to carry an expansion of a polyQ stretch in its N terminus, causes HD. Our results thus indicate that RfxCas13d is a versatile platform for suppressing gene expression.
Gene silencing has emerged as a promising strategy for treating various CNS disorders (47–50). As evidence for this, tofersen, an antisense oligonucleotide (ASO) for SOD1 (19), has advanced to phase 3 trials (ClinicalTrials.gov identifier: NCT02623699), while microRNA (miRNA)–based approaches for SOD1-ALS (18) and HD (51) (ClinicalTrials.gov identifier: NCT04120493) are under evaluation in patients. Yet despite their broad potential, traditional gene silencing modalities have certain limitations. For instance, ASOs have a transient life cycle and will require a lifetime of administrations, which, in addition to potentially producing periods of diminished activity before a redosing, could pose a physical and financial burden on patients. Conversely, while small interfering RNAs (siRNAs), shRNAs, and miRNAs can be expressed from a viral vector to continuously engage with a target RNA, these modalities rely on endogenous pathways whose activation can trigger the silencing of off-target transcripts (52). Thus, there remains a need to explore alternate RNA targeting platforms.
To this end, in addition to evaluating its ability to mediate gene knockdown in the CNS, we determined whether RfxCas13d could be used to improve therapeutic outcomes in a mouse model of a neurodegenerative disorder, specifically the G93A-SOD1 mouse model of SOD1-linked ALS. Our results establish that RfxCas13d can impart a therapeutic benefit, as we found that G93A-SOD1 mice intrathecally injected with AAV encoding RfxCas13d and a SOD1-targeting crRNA had a significantly slowed disease, improved survival, a decreased rate of muscle atrophy, and improved rotarod and grip strength. However, despite this effect, the G93A-SOD1 mice treated by RfxCas13d did eventually succumb to disease. Although the reason for this remains unknown, one possible explanation could be a lack of delivery to microglia, a cell type that, like astrocytes, contributes to disease progression in SOD1-ALS (21). Should this be the case, the development of delivery vehicles (53) capable of targeting microglia, in addition to the other cell types involved in SOD1-ALS, could further slow disease progression.
Unexpectedly, our results also raise the possibility that RNase-independent mechanisms may contribute to knockdown, as we observed a relatively modest but nonetheless measurable decrease in SOD1 and HTT mRNA in cells transfected with either just crRNA or a catalytically dead variant of RfxCas13d with crRNA [a separate study reported a similar observation following the transfection of just crRNA (20)]. Additional experiments will be needed to fully unravel these mechanisms and to determine whether they function alongside catalytically active RfxCas13d.
Our study also examined RfxCas13d targeting specificity, a critical parameter given that the Cas13 family of enzymes has promiscuous RNase activity that can result in the cleavage of nontarget RNAs in some environments (3, 4, 6, 7, 13, 38), although this effect has not been observed for all targets in mammalian cells (5, 8). While our targeted qPCR analysis revealed no significant changes in the expression of 10 candidate off-target transcripts in whole spinal cord tissue, a whole-transcriptome survey conducted in HEK293T cells revealed that, compared to untreated cells, RfxCas13d with the HTT crRNA affected the expression of 144 genes. However, our analyses revealed that ~75% of these genes were also affected in cells transfected with RfxCas13d and a nontargeted crRNA or RfxCas13d alone, suggesting the possibility for common mechanisms in response to the RfxCas13d protein or its delivery to HEK293T cells. For additional benchmarking, we compared RfxCas13d specificity to HTT-targeting shRNAs, observing that, on a transcriptome-wide scale, both modalities could induce a comparable number of off-target effects. These results notwithstanding, RfxCas13d specificity and safety are important questions that require further study, particularly given that Cas13 proteins can collaterally cleave nontarget RNAs.
For our proof-of-concept study, we implemented non–allele-specific strategies to target both SOD1 and HTT, which holds the advantage of being applicable to a wider patient population than mutation-specific approaches for these disorders. For instance, in the case of SOD1, >100 different mutations have been identified in ALS patients. This degree of genetic heterogeneity poses considerable technical and economic challenges for the treatment of the disorder by a mutation-dependent strategy. In addition, while single-nucleotide polymorphisms (SNPs) that are associated with the mutant HTT allele exist, there remains no single allele-specific strategy that is applicable to the entire HD patient population (54–56). As a result, non–allele-specific strategies have been implemented with ASOs and shRNA/miRNAs for both SOD1-linked ALS (22, 27, 57–59) and HD (40, 43, 60). However, these approaches are expected to reduce the wild-type protein, which hold risks for inducing adverse effects. While, to date, non–allele-specific targeting for SOD1-linked ALS has resulted in no serious side effects and produced evidence of clinical improvement (19), a phase 3 clinical trial designed to evaluate the effectiveness of an intrathecally administered non–allele-specific ASO for HD was recently halted following a recommendation by an Independent Data Monitoring Committee from an assessment of its risk-benefit profile, raising questions on the potential safety of reducing the wild-type HTT protein. However, a phase 1/2 trial for an AAV-based non–allele-specific miRNA therapy for HD involving its surgical delivery to the striatum remains ongoing (ClinicalTrials.gov identifier: NCT04120493), the results of which may offer additional insight into the tolerability of lowering wild-type HTT. Along these lines, because of its programmability, RfxCas13d may be amenable to repurposing so that, if necessary, it could (i) potentially lower HTT, SOD1, or any other protein to a minimal safe but therapeutically effective threshold; (ii) selectively target mutant allele–specific SNPs [which, notably, while possible with CRISPR-Cas9 (61–63), has yet to be demonstrated as feasible for any disease target for RfxCas13d]; or (iii) participate in a two-vector–based knockdown-and-replace therapy. Such future optimizations could be aided by the emergence of design resources capable of predicting crRNA activity for targeted sequences (12).
Last, similar to other RNA targeting modalities, RfxCas13d must be persistently expressed in cells so that it can continuously engage with its target mRNA to maintain its effect. Cas13 effectors, however, are bacterial in origin, which could pose a challenge for their clinical implementation, as they could be recognized as foreign by the immune system. Furthermore, similar to Cas9 (64, 65), a sizable percentage of the human population may have preexisting immunity to RfxCas13d and its orthologs, a finding that could further complicate its application given its need for continuous expression. Toward the goal of beginning to determine the safety and tolerability of RfxCas13d, we measured inflammatory responses in mice intrathecally injected with RfxCas13d-encoding AAV, observing no visible evidence of activation or infiltration of CD4+ or CD8+ T cells, NK1.1+ natural killer cells, Iba1+ or Mac2+ microglia, or GFAP+ astrocytes at 14 or 28 days after injection. However, these studies probed only for inflammation; thus, additional work is needed to characterize the full spectrum of immunogenic effects that can be elicited by RfxCas13d. Longer-term studies will be also needed to determine the effects that may arise from the sustained expression of RfxCas13d. In conclusion, we have demonstrated that CRISPR-Cas13 can be delivered to the CNS to silence neurodegeneration-linked genes.
MATERIALS AND METHODS
Plasmid construction
pXR001, the plasmid encoding RfxCas13d (Addgene, 109049), and pXR003, the plasmid encoding the crRNA expression cassette (Addgene, 109053), were gifts from P. Hsu (5). To clone the pAAV plasmid pAAV-CAG-RfxCas13d-U6-crRNA, (i) RfxCas13d, (ii) three tandem copies of an HA epitope tag adjacent to a bovine growth hormone polyadenylation signal, and (iii) the human U6 promoter with a crRNA scaffold were PCR-amplified from (i) pXR001, (ii) pAAV-CAG-C-Int-CBE-U6-sgRNA (single-guide RNA) (26), and (iii) pXR003, respectively. These cassettes were then inserted between the Age I and Not I restriction sites of the plasmid pAAV-CAG-C-Int-CBE-U6-sgRNA (26) by a single-step Gibson assembly using the Gibson Assembly Master Mix [New England Biolabs (NEB)] according to the manufacturer’s instructions. Sanger sequencing (ACGT) was used to confirm the sequence of the plasmid. The full sequence of pAAV-CAG-RfxCas13d-U6-crRNA is shown in fig. S13.
To construct the SOD1-AcGFP reporter plasmid, site-directed mutagenesis was performed on pF148-pSOD1-G37R-AcGFP1 (Addgene, 26409), a gift from E. Fisher. Briefly, primers encoding nucleotides to revert the SOD1-G37R mutation back to wild type were used to amplify pF148-pSOD1-G37R-AcGFP1 using Phusion High-Fidelity DNA Polymerase (NEB). The PCR was then incubated with 1 μl of Dpn I (NEB) for 1 hour at 37°C and transformed to 5-alpha Competent Escherichia coli (NEB). Sanger sequencing (ACGT) was used to confirm the sequence of the plasmid.
Oligonucleotides encoding the crRNA targeting sequences were custom-synthesized (Integrated DNA Technologies) and then incubated with T4 polynucleotide kinase (NEB) for 30 min at 37°C, annealed at 95°C for 5 min, and then cooled to 4°C at a rate of −0.1°C/s. Annealed oligonucleotides were then ligated into the Bbs I restriction sites in pXR003 or pAAV-CAG-RfxCas13d-U6-crRNA. Sanger sequencing (ACGT) was used to confirm the sequence of the crRNA
The plasmids encoding siHunt-1 (39) and shHD2.1 (40) were custom-synthesized by Synbio Technologies. All primer sequences are shown in table S1.
Cell culture and transfections
HEK293T cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% (v/v) antibiotic-antimycotic (Thermo Fisher Scientific) in a humidified 5% CO2 incubator at 37°C. Cells were seeded onto 24-well plates at an average density of 2 × 105 cells per well and transfected with 1 μg of pAAV-CAG-RfxCas13d-U6-crRNA by Lipofectamine 3000 (Thermo Fisher Scientific), according to the manufacturer’s instructions.
For flow cytometry involving the SOD1-AcGFP reporter plasmid, cells were transfected with 800 ng of pAAV-CAG-RfxCas13d-U6-crRNA and 200 ng of SOD1-AcGFP reporter plasmid, while, for flow cytometry involving the HTT-CFP reporter plasmid, cells were transfected with 800 ng of pAAV-CAG-RfxCas13d-U6-crRNA, 100 ng of tTA/TRE-mCherry, which encoded the tetracycline-controlled transactivator, and 100 ng of pTreTight-HTT94Q-CFP (Addgene, 23966).
Flow cytometry
At 72 hours after transfection, cells were harvested, washed with PBS, and strained into single-cell suspensions using Falcon Round-Bottom Polystyrene Test Tubes with Cell Strainer Snap Caps. Fluorescence was then measured using the BD LSRFortessa Flow Cytometry Analyzer (Roy J. Carver Biotechnology Center Flow Cytometry Facility). A total of 50,000 events were recorded for each sample, and data were analyzed using FlowJo v10 (FlowJo, LLC).
Quantitative PCR
At 72 hours after transfection, RNA was extracted from cells using the PureLink RNA Mini Kit (Invitrogen) and converted to complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturers’ instructions. cDNA (50 ng) was then used per qPCR using iTaq Universal SYBR Green Supermix (Bio-Rad). qPCR measurements for each biological replicate were conducted in technical triplicates and normalized to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or mouse β-actin expression for each respective sample.
Western blot
Cells were lysed by radioimmunoprecipitation assay (RIPA) buffer (0.2% IGEPAL CA-620, 0.02% SDS with VWR Life Science Protease Inhibitor Cocktails), and protein concentration was determined by using the DC Protein Assay Kit (Bio-Rad). Protein (15 μg) was then electrophoresed by SDS–polyacrylamide gel electrophoresis and electrophoretically transferred onto a polyvinylidene fluoride (PVDF) membrane in transfer buffer [20 mM tris-HCl, 150 mM glycine, and 20% (v/v) methanol] for 1.5 hours at 100 V. Membranes were blocked with 5% (v/v) blotting-grade blocker (Bio-Rad) in tris-buffered saline (TBS) [10 mM tris-HCl and 150 mM NaCL (pH 7.5)] with 0.05% Tween 20 (TBS-T) for 1 hour and then incubated with primary antibodies in blocking solution at 4°C overnight. The following primary antibodies were used: rabbit anti-hSOD1 (1:2000; Cell Signaling Technology, 2770S), rabbit anti-GFP/CFP (1:1000; Abcam, ab6556), rabbit anti-GAPDH (1:1000; Cell Signaling Technology, 2118S), and rabbit anti–β-actin (1:1000; Cell Signaling Technology, 4970S). Membranes were then washed three times with TBS-T and incubated with goat anti-rabbit horseradish peroxidase conjugate (1:4000; Thermo Fisher Scientific, 65-6120) in blocking solution for 1 hour at room temperature (RT). Membranes were then washed again three times with TBS-T and developed using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) and visualized by automated chemiluminescence using ChemiDoc XRS+ (Bio-Rad). Band intensity was quantified using Image Lab Software (Bio-Rad) and normalized to the reference protein in each line.
To analyze the N-terminal fragment of the hHTT protein, R6/2 striatal tissue was lysed using RIPA buffer, and 20 μg of protein was electrophoresed on the 4 to 15% Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad) and transferred onto a PVDF membrane in transfer buffer [20 mM tris-HCl, 150 mM glycine, and 20% (v/v) methanol] for 1.5 hours at 100 V. Membranes were then blocked in blocking solution for 1 hour and incubated overnight at 4°C with the following primary antibodies: mouse anti-HTT (1:200; MilliporeSigma, MAB5374) and rabbit anti–β-actin (1:1000; Cell Signaling Technology, 4970S). Membranes were washed three times with TBS-T and incubated with either ready-to-use biotinylated goat anti-rabbit immunoglobulin G (IgG) (Abcam, ab64256) or ready-to-use biotinylated goat anti-mouse IgG (Abcam, ab64255) for 1 hour at RT. Membranes were then again washed three times in TBS-T and then incubated with a streptavidin–Alexa Fluor 700 conjugate (1:4000; Thermo Fisher Scientific, S21383) diluted in blocking solution (5% bovine serum albumin in TBS-T) for 1 hour at RT. Membranes were washed three more times and visualized using the Odyssey Imaging System (LI-COR). Band intensity was quantitated using LI-COR Image Studio Software (LI-COR). Human mutant HTT protein was then normalized to the β-actin reference protein in each lane.
RNA sequencing
Library construction was performed by the Roy J. Carver Biotechnology Center (University of Illinois). Purified deoxyribonuclease-treated RNAs were converted into individually barcoded polyadenylated mRNA sequencing libraries using the TruSeq Stranded mRNA Sample Prep Kit (Illumina). Libraries were then barcoded with unique dual indexes to prevent index switching. Adaptor-ligated double-stranded cDNAs were then PCR-amplified for eight cycles with KAPA HiFi DNA Polymerase (Roche). Final libraries were quantitated by Qubit (Thermo Fisher Scientific), and the average cDNA fragment sizes were determined on a fragment analyzer. Libraries were diluted to 10 nM and quantitated by qPCR on a CFX Connect Real-Time qPCR system (Bio-Rad) to confirm accurate pooling of barcoded libraries and to maximize the number of clusters in the flowcell. Barcoded RNA-seq libraries were sequenced with NovaSeq 6000 (Illumina). FastQ read files were generated and demultiplexed using the bcl2fastq v2.20 Conversion Software (Illumina). The quality of the demultiplexed FastQ files was evaluated using FastQC.
RNA-seq analysis was conducted by the High-Performance Biological Computing Core (University of Illinois). Briefly, Salmon3 version 1.2.0 was used to quasi-map reads to the transcriptome and to quantify the abundance of each transcript. Transcriptomes were indexed using the decoy-aware method in Salmon with the entire genome file, and gene-level counts were estimated on the basis of transcript-level counts using the “bias corrected counts without an offset” method from the tximport package. The raw RNA-seq data are available in the Supplementary Materials. Overrepresentation analysis on differentially expressed genes was performed using NetworkAnalyst 3.0 (66) using the GO:BP and GO:MF databases (67).
AAV packaging
AAV vectors were packaged according to a previously established protocol (68). Briefly, 2 × 107 HEK293T cells were seeded onto 15-cm cell culture plates in DMEM supplemented with 10% (v/v) FBS (Thermo Fisher Scientific) and 1% (v/v) antibiotic-antimycotic (Thermo Fisher Scientific). At 16 hours after seeding, cells were transfected with 15 μg of either pAAV-CAG-RfxCas13d-U6-NTG, pAAV-CAG-RfxCas13d-U6-SOD1, or pAAV-CAG-RfxCas13d-U6-HTT; 15 μg of AAV1 or AAV9; and 15 μg of pHelper using 135 μl of polyethylamine (1 μg/μl). Cells were gently harvested after 72 hours using a cell scraper, centrifuged at 4000g for 5 min at RT, and then resuspended in lysis buffer [50 mM tris-HCl and 150 mM NaCl (pH 8.0)]. Using liquid nitrogen and a water bath set to 37°C, cells were freeze-thawed three times and then incubated with 10 U of benzonase (Sigma-Aldrich) per 1 ml of cell supernatant for 30 min at 37°C. Supernatant was then centrifuged at 18,500g for 30 min at RT, and the resulting lysate was overlayed on an iodixanol density gradient, which was centrifuged at 140,000g for 2 hours at 18°C. After centrifugation, the virus was isolated by extraction, washed three times with 15 ml of PBS with 0.001% Tween 20 using an Ultra-15 centrifugal filter unit (Amicon), and concentrated to <150 μl. Viral genomic titer was determined by qPCR using iTaq Universal SYBR Green Supermix (Bio-Rad) and stored at 4°C.
Injections
All animal procedures were approved by the Illinois Institutional Animal Care and Use Committee at the University of Illinois and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Treatment and control groups for all studies were sex-balanced and litter-matched.
Following the procedures previously described by our laboratory (26), genotyped P60-P65 G93A-SOD1 mice bred from male G93A-SOD1 mice [B6SJL-Tg(SOD1*G93A)1Gur/J; The Jackson Laboratory, stock no. 002726] and female B6SJLF1/J mice (The Jackson Laboratory, stock no. 100012) were injected with 2 × 1011 vector genomes (vg) of AAV9-RfxCas13d-hSOD1 or AAV9-RfxCas13d-NTG in 10 μl of PBS with 0.001% Tween 20 into the mouse lumbar subarachnoid space between the L5 and L6 vertebrae using a Hamilton syringe with a 29-gauge 1.5-inch needle. The crRNA used in the animal study corresponded to SOD1 crRNA 3 from our initial screen. Identical injection procedures (26) were used to inject B6SJLF1/J mice with AAV vector for inflammation studies.
Genotyped P28 R6/2 mice bred from male R6/2 mice [B6CBA-Tg(HDexon1)62Gpb/3 J; The Jackson Laboratory, stock no. 006494] and female B6CBAF1/J mice (The Jackson Laboratory, stock no. 100011) were injected with a total of 6 × 1010 vg of AAV1-RfxCas13d-HTT (left hemisphere) and AAV1-RfxCas13d-NTG (right hemisphere) in 3 μl of PBS with 0.001% Tween 20 at stereotaxic coordinates anterior-posterior = 0.50 mm; medial-lateral = ±1.65 mm; and dorsal-ventral = −3.5, −3.0, and − 2.5 mm using a 25-μl syringe with a 30-gauge Point Style 4 needle with a 30° angle (Hamilton). Brain injections were performed using a drill and injection robot (NeuroStar). The crRNA used in this animal study corresponded to HTT crRNA 1 from our initial screen.
Behavior
All behavior measurements were conducted by a blinded investigator. Beginning 1 week after injection, motor coordination of G93A-SOD1 mice was measured using a Rotamex-5 rotarod (Columbus Instruments). Mice were placed onto an apparatus programmed to accelerate from 4 to 40 rpm in 180 s (26). The latency to fall was recorded, and each session was composed of three trials (26).
Hindlimb strength was measured using a grip strength meter (Harvard Apparatus), as described (26). Mice were scruffed and allowed to firmly latch onto a pull bar with their hindlimbs and then pulled in the opposite direction (26). The maximum force exerted before the release of the bar was recorded for each animal. Each session consisted of at least three measurements (26).
Following our previous procedures (26), the weights of each mouse were recorded twice per week using an electronic scale. Disease onset and late-stage disease onset were then calculated as the day at which animals reached peak weight and lost 10% of their peak weight, respectively (26).
As previously described (26), end stage was determined as the point when the animal could no longer turn themselves over within 10 s of being placed on their back, lost more than 20% of their peak weight, or had complete paralysis. Mice were provided with wet, mashed food in their cages at the first sign of hindlimb paralysis and were monitored daily thereafter (26). All behavior measurements were normalized to the starting value at day 63.
Immunohistochemistry
Immunohistochemistry was performed according to our previous procedures (26). After transcardially perfusing mice with PBS, tissues were harvested and fixed overnight with 4% paraformaldehyde in PBS at 4°C and then cut into 40-μm sagittal or coronal sections using a CM3050 S cryostat (Leica). Sections of spinal cord were then transferred to a 48-well plate, while sections of brain were transferred to a 24-well plate and then stored in cryoprotectant at −20°C. Sections were washed three times with PBS and incubated with blocking solution [PBS with 10% (v/v) donkey serum (Abcam) and 1% Triton X-100] for 2 hours at RT and stained with primary antibodies in blocking solution for 72 hours at 4°C. Sections were then again washed three times with PBS and incubated with secondary antibodies in blocking solution for 2 hours at RT. Following incubation with the secondary antibodies, sections were washed three final times with PBS and then mounted onto slides using VECTASHIELD HardSet Antifade Mounting Medium (Vector Laboratories). Slides were imaged using a Leica TCS SP8 confocal microscope and a Zeiss Observer Z1 microscope (Beckman Institute Imaging Technology Microscopy Suite, University of Illinois). All image analyses were performed using ImageJ software.
The following primary antibodies were used: rabbit anti-hSOD1 (1:250; Cell Signaling Technology, 2770S), goat anti-choline acetyltransferase (ChAT) (1:25; EMD Millipore, AB144P), goat anti-HA (1:250; GenScript, A00168), rabbit anti-HA (1:500; Cell Signaling Technology, 3724S), chicken anti-HA (1:500; Abcam, ab9111), rabbit anti-NeuN (1:500; Abcam, ab177487), rabbit anti-Iba1 (1:500; Wako Pure Chemical Industries, 019-19741), mouse anti–β3-tubulin (1:1000; Sigma-Aldrich, T8578), chicken anti-GFAP (1:1000; Abcam, ab4674), rabbit anti–DARPP-32 (1:100; Cell Signaling Technology, 2306S), mouse anti-HTT (1:50; MilliporeSigma, MAB5374), rabbit anti-CD4 (1:200; Abcam, ab183685), rat anti-CD8a (1:200; Thermo Fisher Scientific, 14-0081-82), mouse anti-NK1.1 (1:200; Thermo Fisher Scientific, MA1-70100), and rat anti-Mac2 (1:500; Cedarlane, CL8942AP).
The following secondary antibodies were used: donkey anti-rabbit Cy3 (Jackson ImmunoResearch, 711-165-152), donkey anti-rabbit Alexa Fluor 488 (Jackson ImmunoResearch, 711-545-152), donkey anti-goat Cy3 (Jackson ImmunoResearch, 705-165-147), donkey anti-goat Alexa Fluor 488 (Thermo Fisher Scientific, A-11055), donkey anti-goat Alexa Fluor 647 (Jackson ImmunoResearch, 705- 605-147), donkey anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch, 715-545-150), donkey anti-chicken Alexa Fluor 647 (Jackson ImmunoResearch, 703-605-155), donkey anti-chicken Cy3 (Jackson ImmunoResearch, 703-165-155), donkey anti-rat Cy3 (Jackson ImmunoResearch, 712-165-153), and donkey anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch, 715-545-150).
Inclusion area was quantified as previously described (26). Briefly, a region of interest was first highlighted in ImageJ. A pixel intensity threshold was then applied to identify regions covered by inclusions, and the area of the respective region was determined by the measure function in ImageJ. The measured area was then normalized to the total area initially highlighted to derive the percentage of the area occupied by immunoreactive inclusions. All measurements were performed by a blinded investigator.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8. For in vitro studies, SOD1-AcGFP and HTT-CFP fluorescence intensity and protein, and hSOD1 and hHTT mRNA were compared using one-way analysis of variance (ANOVA). hSOD1 protein was compared using an unpaired two-tailed t test. For in vivo studies, mRNA, protein, disease onset, and late-stage disease onset were compared using an unpaired one-way t test. Survival was analyzed by Kaplan-Meier analyses using the Mantel-Cox test. Rotarod and grip strength were analyzed using a two-way ANOVA followed by a Bonferroni post hoc test. Weight loss and hind-limb grip strength were analyzed using a linear regression analysis. Reactive inclusion data were compared using a one-tailed unpaired t test.
Acknowledgments
Funding: C.K.W.L. was supported by a Roy J. Carver Fellowship in Engineering. M.A.Z. was supported by the NIH/NIBIB (T32EB019944), the Mavis Future Faculty Fellows Program, and a University of Illinois Aspire Fellowship. This work was supported by the Muscular Dystrophy Association (MDA602798), the Judith & Jean Pape Adams Foundation, and the NIH/NIGMS (1R01GM141296).
Author contributions: T.G. conceived the study. J.E.P. designed and cloned the plasmids. J.E.P., C.S.-M., and R.K. screened crRNAs by flow cytometry. J.E.P., C.S.-M., and R.K. analyzed knockdown in HEK293T cells by RT-qPCR and Western blot. J.E.P. packaged AAV. C.K.W.L. and J.E.P. bred and genotyped animals. C.K.W.L. performed intrathecal injections. C.K.W.L., M.A.Z., and T.X.M. performed stereotaxic injections. J.E.P. conducted behavior measurements as a blinded investigator. C.K.W.L., G.D.M., and J.E.P. performed immunohistochemistry. J.E.P. analyzed imaging data. J.E.P. and C.K.W.L. analyzed knockdown in spinal cord and brain tissue by qPCR and Western blot. T.G., C.K.W.L., and J.E.P. wrote the manuscript with input from all authors.
Competing interests: The Board of Trustees of the University of Illinois have filed a provisional patent application related to the research in this study (application no. 63/220,385, filed on 9 July 2021). T.G., J.E.P., and C.K.W.L. are listed as co-inventors on this application. T.G. has consulted for Sarepta Therapeutics. The other authors declare no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data files S1 and S2
REFERENCES AND NOTES
- 1.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E., A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abudayyeh O. O., Gootenberg J. S., Konermann S., Joung J., Slaymaker I. M., Cox D. B. T., Shmakov S., Makarova K. S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E. S., Koonin E. V., Zhang F., C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smargon A. A., Cox D. B. T., Pyzocha N. K., Zheng K., Slaymaker I. M., Gootenberg J. S., Abudayyeh O. A., Essletzbichler P., Shmakov S., Makarova K. S., Koonin E. V., Zhang F., Cas13b is a type VI-B CRISPR-associated RNA-guided rnase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konermann S., Lotfy P., Brideau N. J., Oki J., Shokhirev M. N., Hsu P. D., Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan W. X., Chong S., Zhang H., Makarova K. S., Koonin E. V., Cheng D. R., Scott D. A., Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.East-Seletsky A., O’Connell M. R., Knight S. C., Burstein D., Cate J. H., Tjian R., Doudna J. A., Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abudayyeh O. O., Gootenberg J. S., Essletzbichler P., Han S., Joung J., Belanto J. J., Verdine V., Cox D. B. T., Kellner M. J., Regev A., Lander E. S., Voytas D. F., Ting A. Y., Zhang F., RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slaymaker I. M., Mesa P., Kellner M. J., Kannan S., Brignole E., Koob J., Feliciano P. R., Stella S., Abudayyeh O. O., Gootenberg J. S., Strecker J., Montoya G., Zhang F., High-resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Rep. 26, 3741–3751.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox D. B. T., Gootenberg J. S., Abudayyeh O. O., Franklin B., Kellner M. J., Joung J., Zhang F., RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C., Zhou Y., Xiao Q., He B., Geng G., Wang Z., Cao B., Dong X., Bai W., Wang Y., Wang X., Zhou D., Yuan T., Huo X., Lai J., Yang H., Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat. Methods 18, 499–506 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Wessels H.-H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N. E., Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 38, 722–727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynh N., Depner N., Larson R., King-Jones K., A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 21, 279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He B., Peng W., Huang J., Zhang H., Zhou Y., Yang X., Liu J., Li Z., Xu C., Xue M., Yang H., Huang P., Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell 11, 518–524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C., Hu X., Tang C., Liu W., Wang S., Zhou Y., Zhao Q., Bo Q., Shi L., Sun X., Zhou H., Yang H., CasRx-mediated RNA targeting prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Natl. Sci. Rev. 7, 835–837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J. P., Deng H.-X., Rahmani Z., Krizus A., McKenna-Yasek D., Cayabyab A., Gaston S. M., Berger R., Tanzi R. E., Halperin J. J., Herzfeldt B., Van den Bergh R., Hung W.-Y., Bird T., Deng G., Mulder D. W., Smyth C., Laing N. G., Soriano E., Pericak-Vance M. A., Haines J., Rouleau G. A., Gusella J. S., Horvitz H. R., Brown R. H., Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Brown R. H., Al-Chalabi A., Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Mueller C., Berry J. D., McKenna-Yasek D. M., Gernoux G., Owegi M. A., Pothier L. M., Douthwright C. L., Gelevski D., Luppino S. D., Blackwood M., Wightman N. S., Oakley D. H., Frosch M. P., Flotte T. R., Cudkowicz M. E., Brown R. H., SOD1 suppression with adeno-associated virus and MicroRNA in familial ALS. N. Engl. J. Med. 383, 151–158 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Miller T., Cudkowicz M., Shaw P. J., Andersen P. M., Atassi N., Bucelli R. C., Genge A., Glass J., Ladha S., Ludolph A. L., Maragakis N. J., McDermott C. J., Pestronk A., Ravits J., Salachas F., Trudell R., Van Damme P., Zinman L., Bennett C. F., Lane R., Sandrock A., Runz H., Graham D., Houshyar H., McCampbell A., Nestorov I., Chang I., McNeill M., Fanning L., Fradette S., Ferguson T. A., Phase 1–2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med. 383, 109–119 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Blanchard E. L., Vanover D., Bawage S. S., Tiwari P. M., Rotolo L., Beyersdorf J., Peck H. E., Bruno N. C., Hincapie R., Michel F., Murray J., Sadhwani H., Vanderheyden B., Finn M. G., Brinton M. A., Lafontaine E. R., Hogan R. J., Zurla C., Santangelo P. J., Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. 39, 717–726 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Boillée S., Yamanaka K., Lobsiger C. S., Copeland N. G., Jenkins N. A., Kassiotis G., Kollias G., Cleveland D. W., Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312, 1389–1392 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Foust K. D., Salazar D. L., Likhite S., Ferraiuolo L., Ditsworth D., Ilieva H., Meyer K., Schmelzer L., Braun L., Cleveland D. W., Kaspar B. K., Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol. Ther. 21, 2148–2159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai M., Re D. B., Nagata T., Chalazonitis A., Jessell T. M., Wichterle H., Przedborski S., Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 10, 615–622 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamanaka K., Chun S. J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D. H., Takahashi R., Misawa H., Cleveland D. W., Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 11, 251–253 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haidet-Phillips A. M., Hester M. E., Miranda C. J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M. J., Foust K. D., Rao M., Eagle A., Kammesheidt A., Christensen A., Mendell J. R., Burghes A. H. M., Kaspar B. K., Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 29, 824–828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim C. K. W., Gapinske M., Brooks A. K., Woods W. S., Powell J. E., Zeballos M. A., Winter C. J., Perez-Pinera P., Gaj T., Treatment of a mouse model of ALS by in vivo base editing. Mol. Ther. 28, 1177–1189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Yang B., Qiu L., Yang C., Kramer J., Su Q., Guo Y., Brown R. H. Jr., Gao G., Xu Z., Widespread spinal cord transduction by intrathecal injection of rAAV delivers efficacious RNAi therapy for amyotrophic lateral sclerosis. Hum. Mol. Genet. 23, 668–681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bey K., Ciron C., Dubreil L., Deniaud J., Ledevin M., Cristini J., Blouin V., Aubourg P., Colle M. A., Efficient CNS targeting in adult mice by intrathecal infusion of single-stranded AAV9-GFP for gene therapy of neurological disorders. Gene Ther. 24, 325–332 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Gurney M., Pu H., Chiu A., Dal Canto M., Polchow C., Alexander D., Caliendo J., Hentati A., Kwon Y., Deng H., Chen W., Zhai P., Sufit R. L., Siddique T., Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Vinsant S., Mansfield C., Jimenez-Moreno R., Del Gaizo Moore V., Yoshikawa M., Hampton T. G., Prevette D., Caress J., Oppenheim R. W., Milligan C., Characterization of early pathogenesis in the SOD1(G93A) mouse model of ALS: Part II, results and discussion. Brain Behav. 3, 431–457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaj T., Ojala D. S., Ekman F. K., Byrne L. C., Limsirichai P., Schaffer D. V., In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv. 3, eaar3952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Gutmann D. H., Roos R. P., Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum. Mol. Genet. 20, 286–293 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Saudou F., Humbert S., The biology of huntingtin. Neuron 89, 910–926 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Tabrizi S. J., Flower M. D., Ross C. A., Wild E. J., Huntington disease: New insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 16, 529–546 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Kordasiewicz H. B., Stanek L. M., Wancewicz E. V., Mazur C., McAlonis M. M., Pytel K. A., Artates J. W., Weiss A., Cheng S. H., Shihabuddin L. S., Hung G., Bennett C. F., Cleveland D. W., Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabrizi S. J., Leavitt B. R., Landwehrmeyer G. B., Wild E. J., Saft C., Barker R. A., Blair N. F., Craufurd D., Priller J., Rickards H., Rosser A., Kordasiewicz H. B., Czech C., Swayze E. E., Norris D. A., Baumann T., Gerlach I., Schobel S. A., Paz E., Smith A. V., Bennett C. F., Lane R. M., Targeting huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med. 380, 2307–2316 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Maynard C. J., Böttcher C., Ortega Z., Smith R., Florea B. I., Díaz-Hernández M., Brundin P., Overkleeft H. S., Li J. Y., Lucas J. J., Dantuma N. P., Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc. Natl. Acad. Sci. U.S.A. 106, 13986–13991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchman A. B., Brogan D. J., Sun R., Yang T., Hsu P. D., Akbari O. S., Programmable RNA targeting using CasRx in flies. CRISPR J. 3, 164–176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Lebron E., Denovan-Wright E. M., Nash K., Lewin A. S., Mandel R. J., Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol. Ther. 12, 618–633 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper S. Q., Staber P. D., He X., Eliason S. L., Martins I. H., Mao Q., Yang L., Kotin R. M., Paulson H. L., Davidson B. L., RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. U.S.A. 102, 5820–5825 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P., Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Ekman F. K., Ojala D. S., Adil M. M., Lopez P. A., Schaffer D. V., Gaj T., CRISPR-Cas9-mediated genome editing increases lifespan and improves motor deficits in a Huntington’s disease mouse model. Mol. Ther. Nucleic Acids 17, 829–839 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boudreau R. L., McBride J. L., Martins I., Shen S., Xing Y., Carter B. J., Davidson B. L., Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 17, 1053–1063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride J. L., Boudreau R. L., Harper S. Q., Staber P. D., Monteys A. M., Martins I., Gilmore B. L., Burstein H., Peluso R. W., Polisky B., Carter B. J., Davidson B. L., Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: Implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U.S.A. 105, 5868–5873 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieuwenhuis B., Haenzi B., Hilton S., Carnicer-Lombarte A., Hobo B., Verhaagen J., Fawcett J. W., Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: Comparison of four promoters. Gene Ther. 28, 56–74 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J. Y., Popovic N., Brundin P., The use of the R6 transgenic mouse models of Huntington’s disease in attempts to develop novel therapeutic strategies. NeuroRx 2, 447–464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett C. F., Krainer A. R., Cleveland D. W., Antisense oligonucleotide therapies for neurodegenerative diseases. Annu. Rev. Neurosci. 42, 385–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva A. C., Lobo D. D., Martins I. M., Lopes S. M., Henriques C., Duarte S. P., Dodart J.-C., Nobre R. J., Pereira de Almeida L., Antisense oligonucleotide therapeutics in neurodegenerative diseases: The case of polyglutamine disorders. Brain 143, 407–429 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Martier R., Konstantinova P., Gene therapy for neurodegenerative diseases: Slowing down the ticking clock. Front. Neurosci. 14, 580179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh R., Tabrizi S. J., Gene suppression approaches to neurodegeneration. Alzheimer’s Res. Ther. 9, 82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallès A., Evers M. M., Stam A., Sogorb-Gonzalez M., Brouwers C., Vendrell-Tornero C., Acar-Broekmans S., Paerels L., Klima J., Bohuslavova B., Pintauro R., Fodale V., Bresciani A., Liscak R., Urgosik D., Starek Z., Crha M., Blits B., Petry H., Ellederova Z., Motlik J., van Deventer S., Konstantinova P., Widespread and sustained target engagement in Huntington’s disease minipigs upon intrastriatal microRNA-based gene therapy. Sci. Transl. Med. 13, eabb8920 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Janas M. M., Schlegel M. K., Harbison C. E., Yilmaz V. O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S., Rajeev K. G., Manoharan M., Keirstead N. D., Maier M. A., Jadhav V., Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 9, 723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li C., Samulski R. J., Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 21, 255–272 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Kay C., Collins J. A., Skotte N. H., Southwell A. L., Warby S. C., Caron N. S., Doty C. N., Nguyen B., Griguoli A., Ross C. J., Squitieri F., Hayden M. R., Huntingtin haplotypes provide prioritized target panels for allele-specific silencing in huntington disease patients of european ancestry. Mol. Ther. 23, 1759–1771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao M. J., Gillis T., Atwal R. S., Mysore J. S., Arjomand J., Harold D., Holmans P., Jones L., Orth M., Myers R. H., Kwak S., Wheeler V. C., MacDonald M. E., Gusella J. F., Lee J.-M., Haplotype-based stratification of Huntington’s disease. Eur. J. Hum. Genet. 25, 1202–1209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J.-M., Kim K.-H., Shin A., Chao M. J., Elneel K. A., Gillis T., Mysore J. S., Kaye J. A., Zahed H., Kratter I. H., Daub A. C., Finkbeiner S., Li H., Roach J. C., Goodman N., Hood L., Myers R. H., MacDonald M. E., Gusella J. F., Sequence-level analysis of the major European huntington disease haplotype. Am. J. Hum. Genet. 97, 435–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borel F., Gernoux G., Cardozo B., Metterville J. P., Toro Cabreja G. C., Song L., Su Q., Gao G. P., Elmallah M. K., Brown R. H. Jr., Mueller C., Therapeutic rAAVrh10 mediated SOD1 silencing in adult SOD1(G93A) mice and nonhuman primates. Hum. Gene Ther. 27, 19–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCampbell A., Cole T., Wegener A. J., Tomassy G. S., Setnicka A., Farley B. J., Schoch K. M., Hoye M. L., Shabsovich M., Sun L., Luo Y., Zhang M., Comfort N., Wang B., Amacker J., Thankamony S., Salzman D. W., Cudkowicz M., Graham D. L., Bennett C. F., Kordasiewicz H. B., Swayze E. E., Miller T. M., Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J. Clin. Investig. 128, 3558–3567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raoul C., Abbas-Terki T., Bensadoun J.-C., Guillot S., Haase G., Szulc J., Henderson C. E., Aebischer P., Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 11, 423–428 (2005). [DOI] [PubMed] [Google Scholar]
- 60.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R. K., Rajeev K. G., Manoharan M., Sah D. W. Y., Zamore P. D., Aronin N., Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U.S.A. 104, 17204–17209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oikemus S. R., Pfister E., Sapp E., Chase K. O., Kennington L. A., Hudgens E., Miller R., Zhu L. J., Chaudhary A., Mick E. O., Sena-Esteves M., Wolfe S. A., DiFiglia M., Aronin N., Brodsky M. H., Allele-specific knockdown of mutant HTT protein via editing at coding region SNP heterozygosities. Hum. Gene Ther. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin J. W., Kim K.-H., Chao M. J., Atwal R. S., Gillis T., MacDonald M. E., Gusella J. F., Lee J.-M., Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 25, 4566–4576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monteys A. M., Ebanks S. A., Keiser M. S., Davidson B. L., CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol. Ther. 25, 12–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charlesworth C. T., Deshpande P. S., Dever D. P., Camarena J., Lemgart V. T., Cromer M. K., Vakulskas C. A., Collingwood M. A., Zhang L., Bode N. M., Behlke M. A., Dejene B., Cieniewicz B., Romano R., Lesch B. J., Gomez-Ospina N., Mantri S., Pavel-Dinu M., Weinberg K. I., Porteus M. H., Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chew W. L., Tabebordbar M., Cheng J. K. W., Mali P., Wu E. Y., Ng A. H. M., Zhu K., Wagers A. J., Church G. M., A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Med. 13, 868–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou G., Soufan O., Ewald J., Hancock R. E. W., Basu N., Xia J., NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 47, W234–W241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaj T., Schaffer D. V., Adeno-associated virus–mediated delivery of CRISPR-Cas systems for genome engineering in mammalian cells. Cold Spring Harb. Protoc. 2016, pdb.prot086868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 and S2
Data files S1 and S2