Abstract
Hepatotropic viruses naturally have narrow host and tissue tropisms, challenging the development of robust experimental models. The advent of organoid technology provides a unique opportunity for moving the field forward. Here, we demonstrate that three-dimensional cultured organoids from fetal and adult human liver with cholangiocyte or hepatocyte phenotype support hepatitis E virus (HEV) replication. Inoculation with infectious HEV particles demonstrates that human liver–derived organoids support the full life cycle of HEV infection. By directing organoids toward polarized monolayers in a transwell system, we observed predominantly apical secretion of HEV particles. Genome-wide transcriptomic and tRNAome analyses revealed robust host responses triggered by viral replication. Drug screening in organoids identified brequinar and homoharringtonine as potent HEV inhibitors, which are also effective against the ribavirin resistance variant harboring G1634R mutation. Thus, successful recapitulation of HEV infection in liver-derived organoids shall facilitate the study of virus-host interactions and development of antiviral therapies.
Human liver–derived organoids support HEV infection and enable the study of virus-host interaction and antiviral drug development.
INTRODUCTION
Hepatitis E virus (HEV) is a positive-sense, single-stranded RNA virus with approximately 7.2-kb genome encoding three open reading frames (ORFs). It is the most common causative agent of acute hepatitis (1) and is responsible for substantial morbidity and mortality in specific populations, including immunocompromised patients and pregnant women (2, 3). Among the eight classified HEV genotypes (GTs), GT1 imposes a heavy clinical burden on pregnant women in resource-limited regions (4), and occurrence of maternal-fetal transmission can seriously affect fetal/neonatal outcomes (5). GT3 is widely circulating in many countries and causes chronic hepatitis E in organ transplantation patients (6).
Because currently no approved medication is available, ribavirin monotherapy has been used as off-label treatment in some chronic hepatitis E cases, showing 70 to 80% sustained virological response rate (7, 8). As an RNA virus, the HEV genome has a high mutation rate, in particular, under selection pressure. Emerging mutations, for example, the G1634R mutant in polymerase, have been implicated in ribavirin resistance and treatment failure of patients with chronic hepatitis E (9, 10). Collectively, suboptimal efficacy, poor tolerability, and contraindication during pregnancy have limited the wide application of ribavirin for treating hepatitis E. Thus, there is a clear need of further advancing HEV biology and developing new therapeutics. This, in turn, requires the establishment of innovative and robust experimental models.
Hepatotropic viruses intrinsically have narrow tropism, challenging the development of experimental models. Although human liver cancer cell lines (e.g., Huh7, HepaRG, and HepG2) have been used to model hepatitis virus infections, these models have fundamental limitations. Because of extensive and long-term culturing, these two-dimensional (2D) cultured cancer cell lines harbor enormous genetic, epigenetic, and functional alterations (11, 12). Thus, many important host factors and antiviral targets are likely dysregulated, compromising the authenticity in recapitulating virus-host interactions and antiviral drug assessment. The development of organoid technology provides a unique opportunity to circumvent these contemporary challenges (13). Primary organoids are tissue stem cell–derived “mini organs” cultured in 3D structure. They are much better in recapitulating the architecture, composition, diversity, organization, and functionality of cell types of the original tissue/organ (14).
Fetal and adult human livers harbor residential stem cell populations that can be cultured into 3D organoids (15). These stem cells are located in the intrahepatic biliary compartment and give rise to organoids with a cholangiocyte phenotype, which can be differentiated toward a hepatocyte-like phenotype by specific culture conditions (16). Notably, HEV infects both hepatocytes and cholangiocytes in the liver of patients with hepatitis E (17, 18). Epithelial cells, including hepatocytes and cholangiocytes, are columnar polarized epithelium with functional tight junctions, which allows directional trafficking and secretion of molecules (19). Transwell system has been explored for generating polarized hepatocyte-like cells in 2D cell culture (20). This study aims to establish innovative HEV models using 2D and 3D cultured human liver–derived organoids for mapping virus-host interactions and discovering new therapeutics.
RESULTS
HEV replication in fetal and adult human liver–derived organoids
Considering the clinical relevance that maternal-fetal transmission can occur in HEV-infected pregnant women (21), we cultured organoids from both fetal and adult human liver. Because these organoids are originated from the intrahepatic biliary compartment, they are also termed as intrahepatic cholangiocyte organoids (ICOs) (22). Although HEV primarily infects hepatocytes, direct infection of the bile duct epithelia (cholangiocytes) has been reported in patients with chronic hepatitis E who developed cholangitis (18). Here, we have stained liver biopsies from 11 patients with HEV and found positive staining of HEV ORF2 protein in bile duct of 4 patients (fig. S1A). Furthermore, inoculating the human cholangiocyte-like cell line TFK-1 with HEV particles resulted in clear expression of ORF2 protein shown by immunostaining (fig. S1B). These results collectively indicate that cholangiocyte is also the target of HEV infection.
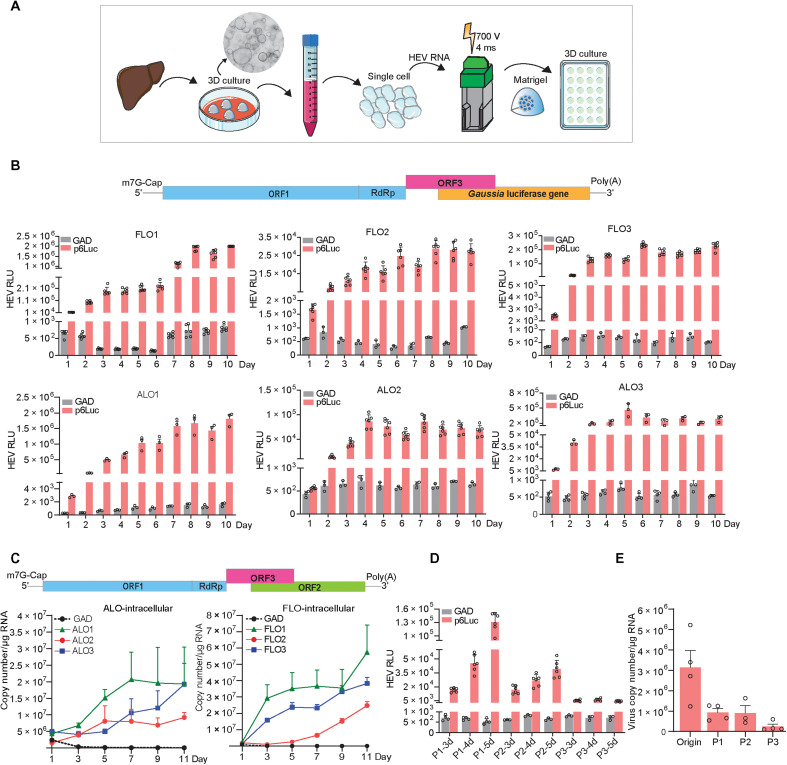
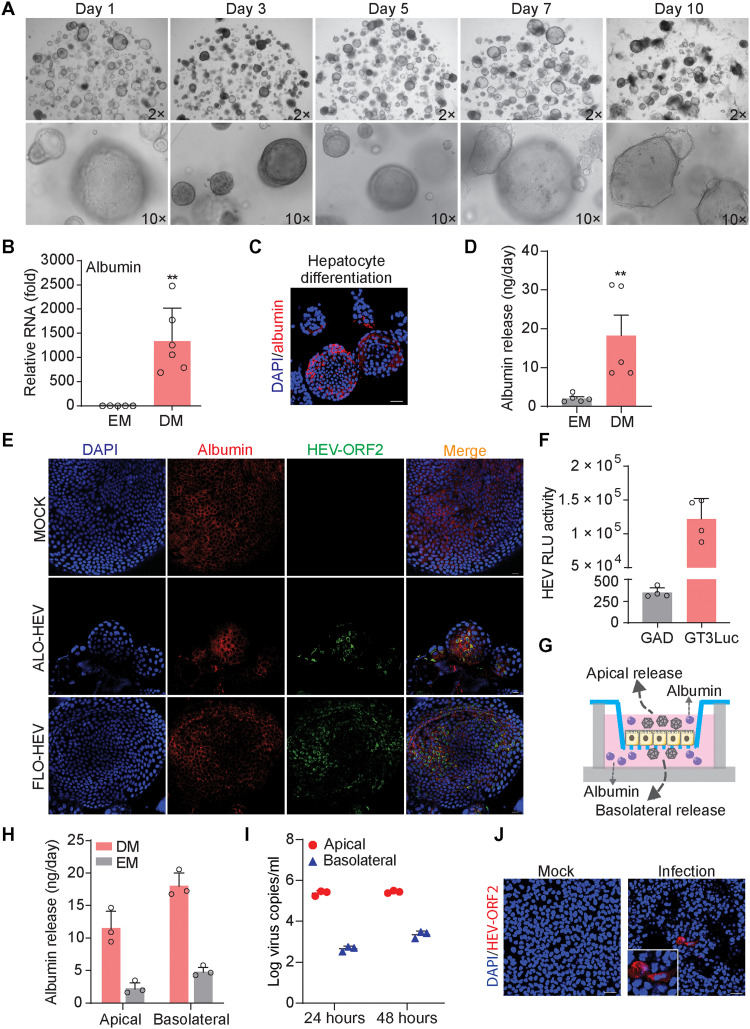
To first investigate whether human ICOs are permissive to HEV replication, we generated subgenomic and full-length GT3 (Kernow-C1 p6 strain) HEV RNA, namely, p6Luc and p6, based on previously established reverse genetics protocols (23, 24). The in vitro–transcribed genomic RNA was delivered into individual organoid cells by electroporation (Fig. 1A). Notably, this system generates not only functional but also defective RNA. Thus, the defective RNA would rapidly degrade in the cells, whereas the functional HEV genomic RNA sustains viral replication. In organoids harboring the p6Luc replicon that secretes Gaussia luciferase as an indicator of viral replication, we observed a steady increase of luciferase activity within the first 5 days after electroporation. This robust luciferase activity was stably maintained for at least 10 days. The maximal luciferase signal (104 to 106 U) varied between the three fetal and the three adult ICO lines, which were derived from six different donors (Fig. 1B). In contrast, organoids harboring the p6Luc replicon with a GAD mutation in the polymerase, only produced basal luciferase activity around 102 U (Fig. 1B).
Fig. 1. Human liver–derived organoids are highly permissive for HEV replication.
(A) Schematic representation of the experimental layout. (B) Luciferase activity in adult (ALOs) and fetal (FLOs) ICOs from days 1 to 10 after electroporation (n = 3 to 6). (C) Dynamics of HEV replication (intracellular) in adult and fetal ICOs from days 1 to 11 after electroporation. (D) Dynamic changes of p6Luc HEV replication–related luciferase activity within three passages (FLO1, n = 3 to 6). (E) Dynamic changes of p6 HEV copy numbers within three passages (FLO1, n = 3 to 4). RdRp, RNA-dependent RNA polymerase; RLU, relative luminescence units.
We next profiled the kinetics of viral replication in organoids electroporated with the p6 full-length HEV genome. We quantified intracellular viral RNA levels by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and calculated as copy numbers (table S1 and fig. S2). We observed a continuous increase of viral RNA levels from days 1 to 11 after electroporation, to a maximum of 1 × 107 to 5 × 107 copies/μg of total RNA across the six organoid lines (Fig. 1C). As expected, the control HEV RNA having GAD mutation in the polymerase that prevents viral replication rapidly degraded within the first 3 days of electroporation (Fig. 1C). Notably, passaging organoids in culture gradually attenuated viral replication in both p6Luc and p6 models (Fig. 1, D and E).
Although this reverse genetics system based on patient-derived GT3 HEV (Kernow-C1 p6 strain) has been widely used in the field, there are also other established but less commonly used reverse genetics systems available, including GT1-, GT3- (swine origin), GT4-, GT5-, and GT7-based systems (25–27) (fig. S3A). We thus further assessed these systems by electroporation of produced genomic RNA into ICOs. Immunofluorescence staining of viral double-stranded RNA (dsRNA), the replicating intermediate, indicated active viral replication, but the replication level varied among these different systems (fig. S3B). Quantification of HEV RNA from the intracellular and supernatant compartments at 1, 5, and 11 days after electroporation indicated variable levels of sustained viral replication in the organoids but showed limited viral secretion (fig. S3, C to H). Overall, HEV is capable to replicate in fetal and adult human liver–derived organoids, but the level varies among different reverse genetics systems established from different HEV GTs/strains. Because the GT3 Kernow-C1 p6 system is most robust, this study mainly used this system to establish HEV liver organoid models.
Liver-derived organoids support the full life cycle of HEV infection
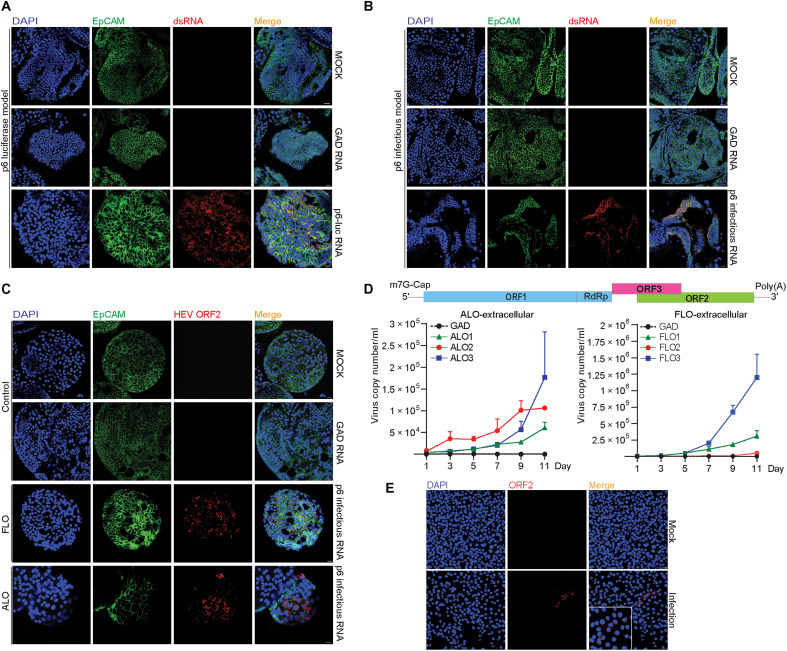
To dissect the different steps of HEV infection in ICOs using the GT3 Kernow-C1 p6 system, we performed immunofluorescence staining of viral replicating dsRNA. We found that dsRNA was widely present in the p6- or p6Luc-electroporated organoid cells (Fig. 2, A and B). High expression of HEV ORF2 protein was also observed in both fetal and adult ICOs electroporated with the p6 genome (Fig. 2C). Because p6 efficiently replicates in organoids, we quantified the release of viruses into supernatant by qRT-PCR. The amount of released viruses increased gradually from day 3 after electroporation up to day 11. It finally reached the range of 5 × 104 to 1 × 106 copies/ml supernatant in the six organoid lines (Fig. 2D). Previous studies in cell culture models have shown that secreted extracellular HEV particles are predominately quasi-enveloped viruses with limited infectivity, whereas intracellular HEV particles are more infectious (28). By inoculating a human liver cell line with extracellular HEV from supernatant of infected ICOs, we observed evidence of viral replication based on increase of viral RNA level, suggesting that infectious virus particles are also present in organoid supernatant (fig. S4A). Therefore, for producing liver organoid–derived HEV particles, we harvested viruses from both the supernatant and intracellular compartments. We demonstrated that these HEV particles are able to infect the human liver cancer cell line Huh7, as shown by immunofluorescence staining of HEV ORF2 protein (Fig. 2E).
Fig. 2. Human liver–derived organoids support HEV replication and production of infectious virus.
(A and B) Immunofluorescence staining for viral dsRNA in liver organoids. DAPI, 4′,6-diamidino-2-phenylindole. (C) Immunofluorescence staining for HEV ORF2 expression in organoids. (D) Dynamics of HEV production (extracellular) in adult and fetal ICOs from days 1 to 11 after electroporation. (E) Immunofluorescence staining of HEV ORF2 in Huh7 cells. HEV genomic RNA with GAD mutation (defect in viral replication) was used as a negative control. Data are presented as means ± SD.
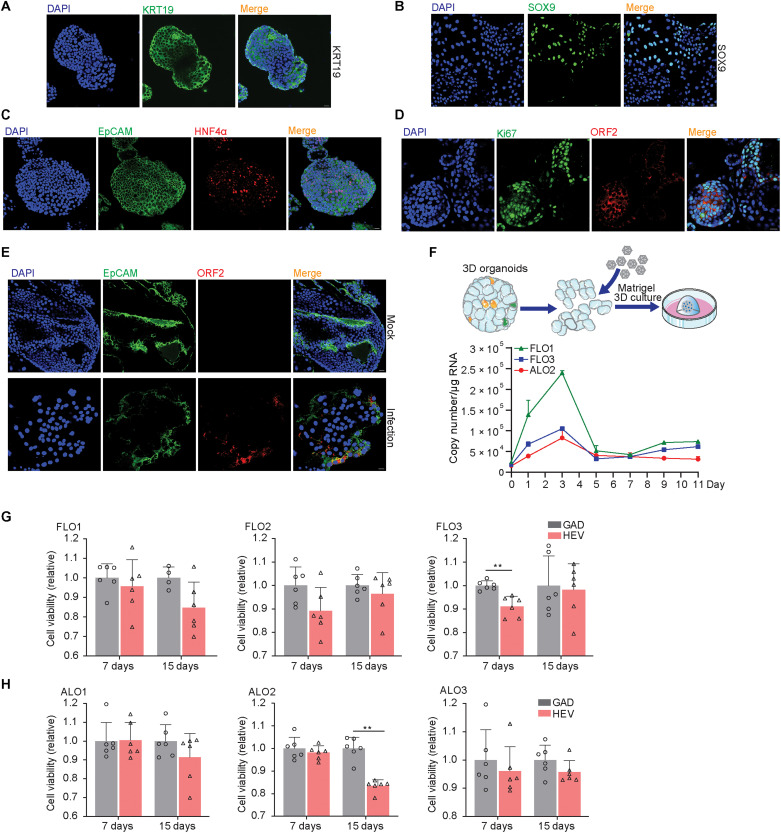
In addition to the general ductal markers KRT19 (keratin 19) and SOX9 (SRY-box transcription factor 9), hepatocyte marker HNF4α (hepatocyte nuclear factor 4 alpha), and epithelial surface marker EpCAM (epithelial cellular adhesion molecule) (Fig. 3, A to C), we specifically included proliferation marker Ki67 for better characterization. We found that HEV ORF2 was expressed in both Ki67-positive and Ki67-negative cells (Fig. 3D), suggesting that HEV replication is not restricted to proliferating cells only. Last, we performed direct inoculation of cell culture–produced HEV particles in organoids and confirmed successful infection by staining ORF2 expression at day 3 after inoculation (Fig. 3E). Intracellular and secreted viral RNA levels were further analyzed by qRT-PCR after inoculation. Intracellular HEV peaked at 3 days after inoculation and then decreased but stabilized during days 5 to 11 at lower levels of viral replication (Fig. 3F). Extracellular HEV persistently increased, peaked at 5 or 7 days after inoculation, and then decreased at variable degrees among different organoid lines (fig. S4B). In summary, 3D cultured human liver–derived organoids support the full life cycle of HEV infection.
Fig. 3. Immunostaining of relevant markers, HEV infection in polarized organoid cells with cholangiocyte phenotype, and the effects of HEV replication on organoid growth.
(A) Immunofluorescence staining of KRT19 in ICOs. (B) Immunofluorescence staining of SOX9 in organoids. (C) Immunofluorescence staining of hepatic marker HNF4α in organoids. (D) Immunofluorescence staining of proliferation marker Ki67. (E) Immunofluorescence staining for viral ORF2 protein in ICOs on day 3 after inoculation of infectious HEV particles. (F) Virus growth curve by inoculation of organoids with HEV particles. Virus titers of 1 hour after inoculation was set as day 0 as starting point. (G and H) Cell viability of organoids electroporated with p6 RNA and HEV RNA with GAD mutation. Cell viability is tested on days 7 and 15 after electroporation (n = 4 to 6).
Mild effects on organoid growth by HEV infection
We recently have shown that rotavirus infection in human liver–derived organoids induced severe cytopathogenesis (15). To evaluate whether these effects also occur during HEV infection, we measured cell viability of the three adult donor–derived and three fetal liver–derived organoids on days 7 and 15 after electroporation with p6 HEV RNA. Significantly reduced growth was observed in one fetal ICO line [fetal liver organoid 3 (FLO3)] on day 7 and one adult ICO line [adult liver organoid 2 (ALO2)] on day 15. Trends of affecting organoid growth were also observed in FLO2 (days 7 and 15), ALO1 (day 15), and ALO3 (days 7 and 15), but they are not statistically significant (Fig. 3, G and H). On the basis of morphological characteristics, there was no major impact on the appearance of the organoids (fig. S4, C and D). Overall, the effects of HEV replication on organoid growth are mild.
Predominately apical release of HEV particles in polarized organoid cells
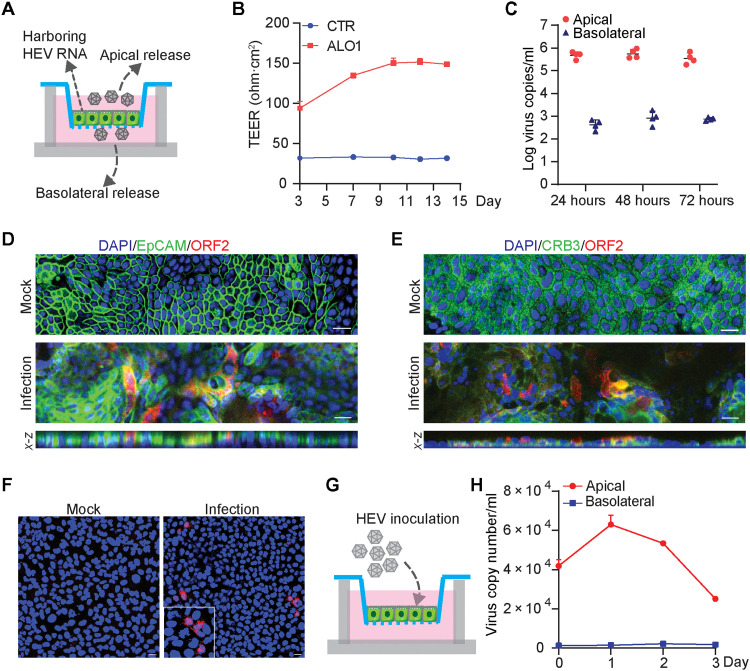
We cultured ICOs harboring p6 HEV RNA in a transwell system to generate columnar polarized monolayers mimicking the structure of bile duct (Fig. 4A and fig. S5A). The transepithelial electrical resistance (TEER) of monolayers in a transwell membrane was stably maintained from 10 days after seeding, showing favorable integrity and permeability (Fig. 4B). As a collagen-coated transwell filter is semipermeable, this permits secretion of viruses and molecules from both apical and basolateral sides of the cells. Overall, the apical supernatant contained over 99% (median, 5 × 105 copies), but the basolateral supernatant only contained less than 1% (median, 7 × 102 copies) of the total secreted viral RNA (Fig. 4C). Confocal microscopy demonstrated robust expression of HEV ORF2 protein in a proportion of polarized cholangiocyte-like organoid cells (Fig. 4, D and E). Consistently, x-z confocal microscopy showed that ORF2 protein localized at the apical side of infected cells by colocalization with EpCAM and apical marker CRB3 (Fig. 4, D and E). Apically released HEV particles are infectious when inoculating naïve Huh7 cells shown by immunostaining ORF2 protein (Fig. 4F).
Fig. 4. Recapitulating HEV infection in polarized organoid cells in transwell system.
(A) Schematic representation of virus release from polarized organoid cells with cholangiocyte phenotype in a transwell system. (B) TEER value of monolayers in transwell membrane tested at 3, 7, 10, 12, and 14 days after seeding. (C) Polarized release of HEV RNA into apical and basolateral supernatant at 24, 48, and 72 hours (n = 4). (D) Immunofluorescence staining for HEV ORF2 and EpCAM and x-z sections for viral ORF2 and EpCAM colocalizaiton. (E) Immunofluorescence staining for HEV ORF2 and apical marker CRB3 and x-z sections for viral ORF2 and CRB3 colocalization. (F) Immunofluorescence staining of HEV ORF2 in Huh7 cells at 48 hours after inoculation with apical released viruses. (G) Schematic representation of HEV virus inoculation with organoid cells in a transwell system. (H) Quantification of released viruses from polarized cells inoculated with cell culture–produced HEV particles. Scale bar, 20 μM. Data are presented as means ± SD.
Next, we performed direct inoculation of cell culture–produced HEV particles from the apical side of polarized ICO cells (Fig. 4G). We further confirmed apical release of HEV particles (Fig. 4H). Our results showing predominately apical release of infectious HEV particles from polarized ICO cells suggest that HEV-infected bile ducts may contribute to fecal dissemination of HEV.
HEV replication and apical release from hepatocyte-differentiated organoids
As HEV infection was primarily demonstrated in liver-derived organoids that exhibit mostly cholangiocyte features (Figs. 1 to 3), we next evaluated whether hepatocyte-differentiated organoids also support HEV replication. ICOs harboring p6Luc replicon or p6 infectious RNA were cultured in previously described hepatocyte differentiation medium (DM) (29). Upon 10 days of differentiation, organoids acquired hepatocyte morphology of polygonal cell shapes (Fig. 5A). There was a marked increase in the expression of hepatic markers including albumin (1300-fold increase), HNF4α (4-fold), and hepatic metabolic marker G6PC (300-fold) compared with undifferentiated organoids. In contrast, the expression of stem cell marker LGR5 (leucine-rich repeat-containing G-protein coupled receptor 5), cytokeratin marker KRT19 and ductal marker SOX9 was significantly down-regulated in hepatocyte-like organoids (Fig. 5B and fig. S5, B to F). Albumin protein expression and secretion were visualized by immunostaining (Fig. 5C) and quantified by enzyme-linked immunosorbent assay (ELISA) assay (Fig. 5D), respectively. HEV ORF2 protein was expressed in albumin-positive organoid cells (Fig. 5E). In apparent agreement, high luciferase activity was detected in organoids harboring p6Luc replicon after hepatocyte differentiation (Fig. 5F). Collectively, hepatocyte-like organoids remain permissive to HEV replication.
Fig. 5. Hepatocyte-differentiated organoids support HEV replication.
(A) Morphology of hepatocyte-differentiated organoids in different time points. (B) Gene expression level of albumin upon hepatocyte differentiation (n = 5 to 6). EM, expansion medium. (C) Immunofluorescence staining for mature hepatic marker albumin in hepatocyte-differentiated organoids. Scale bar, 40 μM. (D) The secretion of albumin into supernatant quantified by ELISA (n = 5). (E) Immunofluorescence staining of albumin and HEV ORF2 in hepatocyte-differentiated organoids (FLO1 and ALO1 lines were used for differentiation). Scale bar, 20 μM. (F) HEV replication–related luciferase activity in hepatocyte-differentiated organoids (ALO1, n = 4). (G) Illustrating secretion of albumin and HEV from polarized hepatocyte-like cells in a transwell system. (H) Release of albumin into apical and basolateral compartments from polarized cells cultured in EM with cholangiocyte phenotype or DM with hepatocyte phenotype (n = 3). (I) Polarized release of HEV from hepatocyte-like cells differentiated from ICOs harboring p6 HEV genome (ALO1, n = 3). (J) Immunofluorescence staining of HEV ORF2 in Huh7 cells at 48 hours after inoculation of apical released virus from hepatocyte-like cells. Scale bar, 40 μM. Data are presented as means ± SD, **P < 0.01.
Next, we seeded ICOs harboring HEV genomic RNA in the transwell system, which were then subjected to hepatocyte differentiation (Fig. 5G and fig. S5G). As expected, hepatocyte-differentiated compared to undifferentiated organoid cells secrete much more albumin (Fig. 5H). Albumin secretion preferentially directs to the basolateral compartment (Fig. 5H and fig. S5H), whereas HEV particles are exclusively (over 99%) released into the apical side (Fig. 5I). The infectivity of apical released HEV particles was confirmed by inoculating naïve Huh7 cells (Fig. 5J). Thus, hepatocyte-differentiated organoids also support HEV infection and release infectious virus particles from the apical side.
HEV replication triggers robust host response in liver-derived organoids
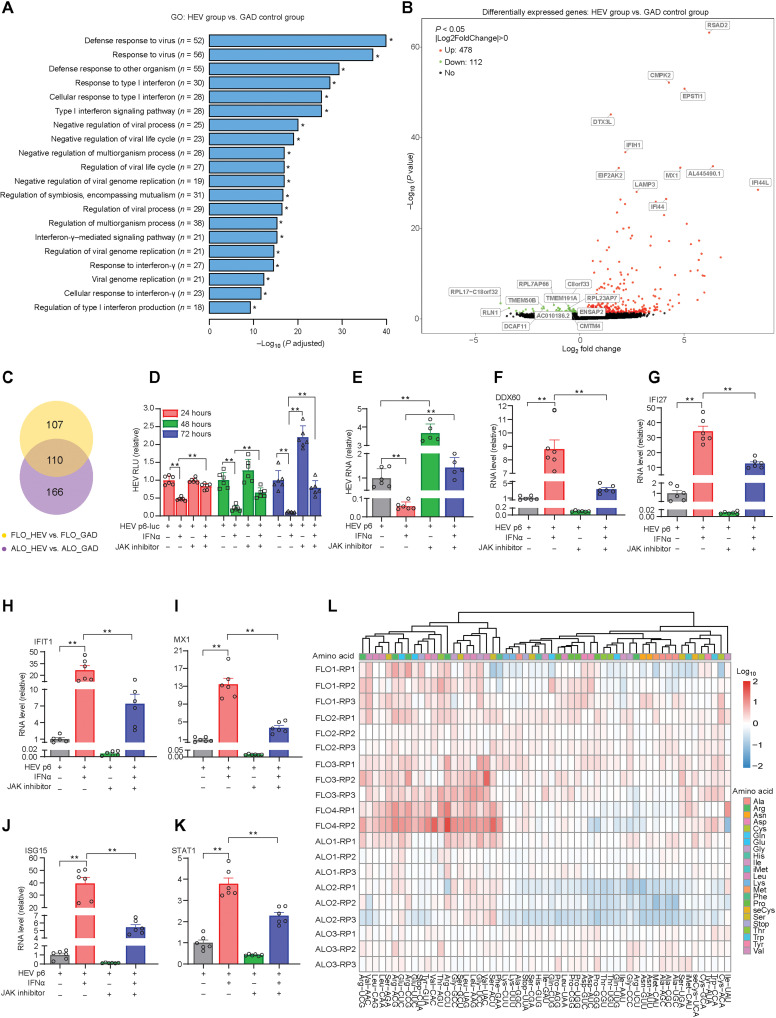
To specially map host responses of liver-derived organoids to HEV replication, we electroporated fetal and adult ICOs with p6 RNA and control HEV RNA having the GAD mutation. We harvested the samples on day 7 after electroporation and performed a genome-wide transcriptomic analysis. Gene ontology enrichment analysis of differentially expressed genes revealed that the major regulated pathways were related to viral infection and innate immunity, including type I interferon (IFN) signaling pathway, response to virus pathway, and IFNγ mediated signaling pathway (Fig. 6A). These responses were similar in fetal and adult liver–derived organoids (fig. S6, A and B). Notably, a series of metabolic processes with respect to glycolysis were remarkably down-regulated in HEV-infected adult ICOs, which was not the case in fetal ICOs (fig. S6, C and D). Volcano plots showed significant down-regulation of 112 genes and up-regulation of 478 genes in response to HEV infection. The major highly up-regulated genes such as RSAD2, IFIH1, MX1, EIF2AK2, CMPK2, and IFI44 were interferon-stimulated genes (ISGs) (Fig. 6B). Subgroup analysis of fetal and adult ICOs revealed similar gene expression patterns (fig. S6, E and F). In total, 110 differentially expressed genes were common in fetal and adult organoids, but 107 and 166 genes were specifically regulated in fetal and adult organoids, respectively (Fig. 6C).
Fig. 6. Genome-wide transcriptomic analysis in liver-derived organoids upon HEV replication.
(A) Top 20 significantly enriched pathways by gene ontology (GO) analysis in ICOs. (B) Differential gene expression analysis in ICOs. (C) Venn diagram of overlapped differentially expressed genes in fetal ICOs compared to adult ICOs. (D) Treatment of IFNα and JAK inhibitor in organoids with p6Luc HEV replicon model (FLO1, n = 6). (E) Treatment of IFNα and JAK inhibitor in organoids harboring p6 HEV infectious model (n = 5 to 6). (F to K) Expression of ISGs in organoids by IFNα or JAK inhibitor treatment (FLO1, n = 5 to 6). (L) Cluster analysis of the tRNAome profiles upon HEV replication in organoids. Results are normalized with value from organoids electroporated with HEV RNA of the GAD mutation. Data are presented as means ± SD, *P < 0.05 and **P < 0.01.
The biological and therapeutic implications of antiviral interferon response
Interferon signaling provides an early innate defense against many viral infections. Secreted interferons bind to their receptors to initiate the Janus kinase signal transducer and activator of transcription (JAK-STAT) cascade to activate ISG transcription and establish antiviral state (30, 31). We have previously shown evidence of activating interferon response in the liver of patients with hepatitis E (17), and now, we also demonstrated this in human liver–derived organoids (Fig. 6A). As a first step to better understand the biological implications of this observation, we used pharmacological inhibition of interferon signaling by JAK inhibitor 1. This resulted in a significant promotion of viral replication in organoids harboring p6Luc or p6 HEV (Fig. 6, D and E) and, consistently, inhibition of ISG expression (Fig. 6, F to K). These results suggest that HEV-induced interferon response functionally restricts HEV replication but is not sufficient to immediately clear the infection.
Recombinant IFNα has been widely used for treating viral infections in the clinic. We previously demonstrated the anti-HEV activity of IFNα in cell culture models and in treated patients with chronic HEV (32). Here, we confirmed significant inhibition of HEV replication by IFNα treatment in both p6Luc- and p6-based organoid models, and this antiviral effect was attenuated by JAK inhibitor 1 (Fig. 6, D and E). As expected, therapeutic IFNα effectively activated the transcription of ISGs including DDX60, IFI27, IFIT1, MX1, ISG15, and STAT1, and the induction was largely blocked by JAK inhibitor 1 treatment (Fig. 6, F to K). Thus, HEV-triggered activation of interferon signaling functionally restricts viral replication but is not sufficient to completely defend the infection, whereas therapeutic IFNα treatment harnesses this innate defense mechanism.
HEV remodels the tRNAome landscape in liver-derived organoids
The robust induction of ISGs was characterized at gene expression (mRNA) level (Fig. 6B). Proteins synthesized from mRNA perform the eventual antiviral functions. A key step in protein synthesis is the recognition of codons by mature transfer RNAs (tRNAs) charged with their corresponding amino acids (33). We have recently developed a qRT-PCR–based assay for quantifying 57 human mature tRNA species that collectively constitute the tRNAome (34). We previously found that HEV infection in the Huh7 cell line profoundly reprogrammed tRNAome with most of the tRNA species that were up-regulated (35). To validate these results, we profiled the tRNAome in four fetal and three adult ICO lines at day 7 after electroporation of p6 HEV RNA. The heatmap depicts the global up-regulation of tRNA expression, although there were variations among the different donors. For example, one adult ICO line appears to have more down-regulated tRNAs (Fig. 6L). Thus, the results in ICOs are able to confirm our previous findings in human liver cancer cell lines that HEV replication remodels the host tRNAome.
Drug screening in organoids identified brequinar and homoharringtonine as potent HEV inhibitors
To explore the application of HEV organoid models for assessing antiviral drugs, we first tested the well-known HEV inhibitors ribavirin and mycophenolic acid (MPA). We demonstrated the antiviral activity of both agents in six ICO lines harboring p6Luc replicon. However, the potency varied substantially among different donors, suggesting that both drugs likely exert antiviral effects at least partially through host factors (fig. S7, A to F). The anti-HEV effects were further confirmed in the p6-based organoid model (fig. S7, G and H).
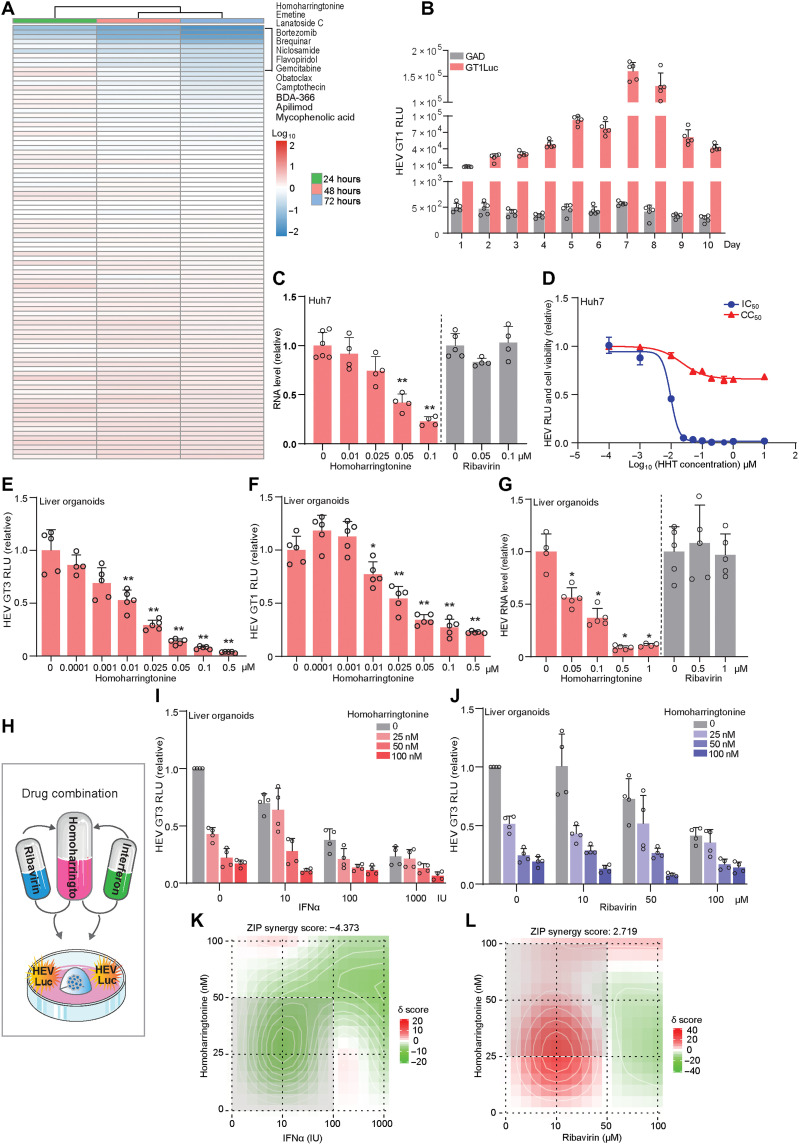
Because we have established the organoid model containing the luciferase-based HEV reporter (Fig. 1A), it is attractive to explore whether this can be used for small- to medium-scale drug screening. To provide proof of concept, we now screened a library of 94 known safe-in-human broad-spectrum antiviral agents in p6Luc organoids (table S2). We previously have screened this library in cancer line–based viral infection models (36, 37). In this study, we used a low concentration (2.5 μM) to minimize nonspecific effects. Ribavirin does not exert anti-HEV activity at this concentration, so that we may identify candidates with higher potency than ribavirin. HEV replication–related luciferase activity was determined at 24, 48, and 72 hours. Among the 13 identified compounds with over 50% inhibition on HEV luciferase activity (Fig. 7A), we prioritized two leading candidates, brequinar and homoharringtonine, for further validation.
Fig. 7. Antiviral drug discovery in liver-derived organoids harboring HEV.
(A) Screening a library of 94 antiviral agents in GT3 p6Luc organoids (ICOs). (B) HEV GT1 luciferase activity in organoids harboring GT1Luc replicon (FLO3, n = 5). (C) The inhibitory effect of homoharringtonine (HHT) and ribavirin in p6 Huh7 cells (n = 4 to 6). (D) IC50 and CC50 of homoharringtonine in p6Luc Huh7 cells (n = 5 to 6). (E) The inhibitory effect of homoharringtonine in p6Luc organoids (FLO1, n = 5). (F) The inhibitory effect of homoharringtonine in GT1Luc organoids (FLO1, n = 4 to 5). (G) The inhibitory effect of homoharingtonine and ribavirin in p6 organoids (FLO1, n = 4 to 5). (H) Schematic representation of drug combination treatment on organoids harboring p6Luc model. (I) The antiviral effects of homoharringtonine in combination with IFNα (FLO1, n = 4). (J) The antiviral effects of homoharringtonine in combination with ribavirin (FLO1, n = 4). (K) Synergy distribution of pairwise combination of IFNα and homoharringtonine (n = 4). (L) Synergy distribution of pairwise combination of ribavirin and homoharringtonine (n = 4). ZIP, zero interaction potency. Data are presented as means ± SD, *P < 0.05 and **P < 0.01.
The anti-HEV effect of brequinar was indicated in our previous study on nucleotide synthesis pathway in HEV cell line models (38). We now comparatively assessed the effects of brequinar in Huh7 and organoid models. Significant inhibition was observed even at very low concentrations (0.05 and 0.5 μM) of brequinar in Huh7-based GT3 and GT1 HEV subgenomic replicon models, respectively (fig. S8, A to C). The 50% inhibition and cytotoxicity concentrations (IC50 and CC50) were 0.0359 and 0.1475 μM in a Huh7-p6Luc model (fig. S8D). Similar results were observed in organoid-based p6Luc HEV replicon model (fig. S8, E and F). We next electroplated ICOs with GT1 HEV replicon. The viral replication–related luciferase signal peaked on day 7 and stably persisted for 10 days (Fig. 7B). Treatment with 2.5 μM brequinar significantly inhibited GT1 HEV replication in organoids (fig. S8G).
To our knowledge, the anti-HEV effect of homoharringtonine has never been described earlier. We thus comprehensively validated its effects in both Huh7- and organoid-based HEV models. Homoharringtonine inhibited viral replication–related luciferase activity in a dose-dependent manner in Huh7 cells (fig. S9A). This was confirmed at the viral RNA level in Huh7 p6 cells (Fig. 7C). The IC50 and CC50 were 0.0097 and 0.0218 μM in Huh7 cells (Fig. 7D and fig. S9, B and C). Similarly, the potent anti-HEV effect of homoharringtonine was further verified in ICO-based p6Luc and GT1Luc replicon models, as well as significant inhibition of viral RNA in organoids harboring p6 infectious HEV (Fig. 7, E to G). Notably, at these low concentrations, ribavirin failed to exert anti-HEV activity in our models (Fig. 7, C and G).
Because drug combination is a common strategy for enhancing antiviral efficacy, we tested combinations of homoharringtonine with IFNα and ribavirin, respectively (Fig. 7H). Combing homoharringtonine with IFNα hardly has a synergistic effect (Fig. 7, I and K, and fig. S10, A and C). In contrast, combination with ribavirin exerted a clear synergistic effect, in particular, at the concentrations ranging 0 to 50 nM homoharringtonine with 0 to 50 μM ribavirin (Fig. 7, J and L, and fig. S10, B and D). Thus, we have demonstrated a proof of concept of applying our organoid models for anti-HEV drug discovery.
Modeling the G1634R HEV variant in organoids and assessing antiviral drugs
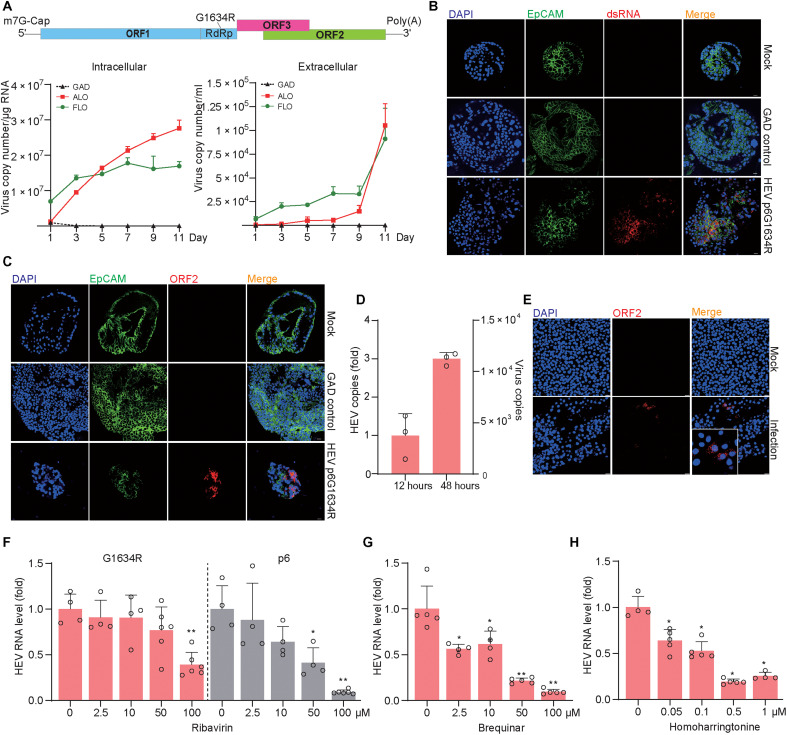
A mutation in the RNA-dependent RNA polymerase (G1634R) of GT3 HEV has been shown to confer ribavirin resistance in patients with chronic hepatitis E (39). Given the clinical relevance, we introduced the p6G1634R variant into both fetal and adult ICOs and observed efficient replication and virus secretion (Fig. 8A). Immunofluorescence staining further confirmed robust replication of viral RNA and expression of capsid protein (Fig. 8, B and C). By inoculating a human liver cell line with supernatant from G1634R mutant HEV-infected ICOs, a marked increase of intracellular viral RNA was observed when comparing a later (48 hours) to an earlier (12 hours) time point after inoculation (Fig. 8D). This suggests that the supernatant compartment also contains infectious viruses. We next produced liver organoid–derived G1634R variants by harvesting both the supernatant and intracellular compartments and performed a reinfection assay by inoculating Huh 7 cells. Successful infection in Huh7 cells was detected by immunostaining of ORF2 protein (Fig. 8E).
Fig. 8. Modeling infection of the p6G1634R HEV variant and testing antiviral drugs in liver-derived organoids.
(A) Replication (intracellular) and production (extracellular) of the HEV variant in fetal (FLO3) and adult (ALO2) ICOs. (B) Immunofluorescence staining of virus dsRNA in organoids electroporated with p6G1634R. (C) Immunofluorescence staining of HEV ORF2 protein in organoids electroporated with p6G1634R. (D) Quantification of viral RNA in HepG2 cells inoculated with extracellular HEV from organoid supernatant (n = 3). (E) Immunofluorescence staining of HEV ORF2 in Huh7 cells at 48 hours after inoculation. (F) The inhibitory effect of ribavirin in p6G1634R organoids or p6 organoids (FLO1, n = 4 to 6). (G) The inhibitory effect of brequinar in p6G1634R organoids (FLO1, n = 4 to 5). (H) The inhibitory effect of homoharringtonine in p6G1634R organoids (FLO1, n = 4 to 5). Data are presented as means ± SD, *P < 0.05 and **P < 0.01.
IFNα treatment significantly inhibited HEV replication, which was largely abrogated by JAK inhibitor 1 (fig. S11A). This was consistent with the effects on ISG induction (fig. S11, B to G). Consistent with previous reports in cell culture models (39), we observed that the p6G1634R variant is also less sensitive to ribavirin treatment in organoids. For example, significant anti-HEV effect was only observed at high concentration (100 μM) of ribavirin treatment, resulting in 61 ± 13% inhibition (means ± SD, n = 6; P = 0.0095), whereas significant inhibition of the parental p6 strain starts at 50 μM concentration (P = 0.0286), and treatment by 100 μM concentration resulted in over 90% inhibition (Fig. 8F).
Brequinar at 2.5 μM has exerted significant inhibitory effect on the p6G1634R variant, whereas 50 μM resulted in 78 ± 3% inhibition (means ± SD, n = 5; P = 0.0079). Most impressively, homoharringtonine at 0.05 μM already significantly inhibited HEV replication, whereas 0.5 μM resulted in 81 ± 3% inhibition (means ± SD, n = 5; P = 0.0159) (Fig. 8, G and H). These results demonstrated the feasibility of searching alternative antivirals using HEV organoid models for treating ribavirin resistance strains.
DISCUSSION
Hepatitis viruses naturally have narrow host and tissue tropisms, which hampers the development of robust experimental models. Compared to hepatitis B virus and hepatitis C virus (HCV), HEV has a relatively broader tropism, and in particular the availability of cell culture–adapted HEV strains has enabled viral replication in multiple cell lines (24, 40, 41). However, these immortalized (cancer) cell lines have been propagated in vitro for decades; harbor numerous genetic, epigenetic, and functional alterations; and thus are suboptimal in recapitulating the infection as seen in patients.
The development of organoid technology provides a unique opportunity for moving the field forward. These 3D-grown organoids were first successfully cultured from LGR5-marked intestinal stem cells, which can authentically recapitulate the cellular heterogeneity and physiological environment of the original tissue (42). Indications for potential usefulness of this technology for studying HEV infection came from observations that intestinal organoids were capable of sustaining human norovirus infection, the first time for more than 40 years that human norovirus can finally be cultured in vitro (43). Human intestinal organoids were also successfully used for modeling rotavirus infection and antiviral drug development (15). In the present study, we further extend the notion that the organoid technology is suitable for investigating viral biology by comprehensively demonstrating that fetal and adult human liver–derived organoids are permissive to HEV replication and support the full life cycle of infection. We found that both undifferentiated cholangiocyte lineage organoids and hepatocyte-differentiated organoids are susceptible to HEV, which is consistent with the clinical observation that both cholangiocytes and hepatocytes can be infected by HEV in the patient liver (17, 18). We further illustrated their applications in studying virus-host interactions and discovering antiviral drugs.
Identifying HEV strains that can efficiently replicate in vitro is another essential aspect for establishing robust culture models. An extensive lesson learned from the HCV field (44) is that direct inoculation with patient-derived samples is usually not efficient for establishing cell culture systems. Similar experience has been reported for culturing clinical HEV isolates in cell lines (45). An alternative and successful approach is to identify and adapt peculiar strains in cell culture and then to construct reverse genetics systems. Viral genomic RNA can be generated from these systems in vitro and delivered into host cells. However, these systems generate competent but also a proportion of defective RNA. However, the defective RNA would rapidly degrade, whereas the functional RNA sustains viral replication. Consistently, we found that HEV genomes with a GAD mutation that prevents viral replication rapidly disappear within 3 days after electroporation. In contrast, a steady increase of intracellular and extracellular viral RNA levels after electroporation was observed when using the GT3 p6 strain, indicating efficient viral replication and secretion. The other strains including GT1-, GT3- (swine origin), GT4-, GT5-, and GT7-based reverse genetics systems sustained variable level of viral replication but are much less efficient compared with the p6 model. Furthermore, they hardly secrete virus particles. Our findings are consistent with previously published studies in which only the p6 system is widely used for establishing robust HEV cell culture models. We encourage the field to further identify new strains from both animal and human origins that can be adapted and propagated in vitro. By constructing reverse genetics systems of these new strains would further enrich the availability and diversity of cell line– and organoid-based HEV models.
It is well recognized that HEV particles of fecal dissemination are more infectious. These viruses are thought to be originally produced by hepatocytes, transported across the apical membrane into the bile canaliculi, and lastly released through the biliary tract into feces. Previous studies have shown apical release of HEV particles in cell culture models (20, 46). Using the transwell system, we now demonstrated that release of HEV particles from hepatocyte-differentiated organoid cells is indeed predominately from the apical side. Recent clinical evidence indicates that HEV also infects cholangiocytes, which is associated with the development of cholangitis in patients with chronic hepatitis E (18). In the transwell system, we again observed predominately apical secretion of HEV particles from monolayers of cholangiocyte-like cells cultured from ICOs. Thus, we speculate that a proportion of infectious HEV particles in feces could be derived from infected cholangiocytes in bile ducts through apical dissemination.
Viruses solely rely on the host to survive and propagate, but host cells have powerful defense mechanisms in controlling the infection. The battles between cell autonomous host immunity and the viruses determine infection outcome. Here, we specifically investigated host response to HEV replication in liver-derived organoids by delivering full-length HEV genomic RNA compared with a mutated genome that could not replicate. We found that HEV replication provoked robust host response, in particular, type I interferon response with most of the top up-regulated genes as ISGs. This bears important biological and therapeutic implications, as inhibiting basal interferon activation by JAK inhibitor further promoted HEV replication, whereas therapeutic treatment with recombinant IFNα inhibited viral replication. PEGylated IFNα has been administered to a small number of chronic HEV patients with about 85% response rate, but this treatment has a high risk of causing acute transplant rejection (47). HEV replication in cell lines has been shown to barely activate type I but mildly type III interferon response (48). We have previously shown that type III interferons do not have anti-HEV activity (32). The alterations and defects of innate immune machinery may explain persisting HEV infection in immortalized cell lines. In contrast, de novo infection in liver-derived organoids by inoculation of infectious HEV particles peaked viral replication on day 3 and rapidly declined by day 5 after inoculation. This highly resembles acute HEV infection in healthy individuals (49), and the “self-limiting outcome” in organoids is likely to be attributed to the robust host antiviral response. Obviously, useful model systems should allow for replication to be maintained long enough to support experimentation. We thus adopted an approach of electroporating sufficient HEV RNA generated on the basis of the reverse genetics system into organoids. High levels of viral replication were stably maintained for at least 10 days and hence are sufficient for various applications. HEV replication, however, gradually diminishes following subsequent passaging of culture, showing the efficacy of cell-autonomous host responses against the virus.
We finally demonstrated a proof of concept for using organoid models in the identification and validation of novel anti-HEV drugs. Consistent with previous studies in cell culture models (50), we confirmed the anti-HEV effects of ribavirin and MPA in organoids. However, the potency varied substantially among different donor-derived organoids, suggesting that both drugs likely exert antiviral effects at least partially through host factors. This points to a potential application of personalized medicine, but taking liver biopsy from patients with HEV for organoids culture might be unsafe and logistically challenging. Nevertheless, the clinical benefit may overweigh risks and challenges.
Although 2D cultures of (cancer) cell lines are convenient and widely used for high-throughput drug screening, their deviant genetic, epigenetic, and functional makeup makes this approach prone to obtaining false-positive and false-negative signals. The luciferase reporter–containing HEV liver organoids may provide a more suitable strategy for performing antiviral drug screening. In this study, we successfully screened a safe-in-human broad-spectrum antiviral drug library composed of 94 agents, representing a small- to medium-throughput screening. However, for high-throughput drug screening (e.g., from thousand to million compounds), large-scale organoid production is required. Fortunately, such technology has recently been established for liver organoid propagation (51). Among the identified leading candidates, we prioritized the validation of brequinar and homoharringtonine, which are more potent than ribavirin in our experimental settings. The G1634R HEV mutant, which has been shown to resist ribavirin (39), is very sensitive to brequinar and even more to homoharringtonine in liver-derived organoid models.
Brequinar is a potent and selective inhibitor of the enzyme dihydroorotate dehydrogenase, blocking pyrimidine nucleotide biosynthesis. This aligns well with our previous findings that nucleotide biosynthesis pathways can be therapeutically targeted to inhibit HEV replication (38). Homoharringtonine is a U.S. Food and Drug Administration–approved medication for treating chronic myeloid leukemia (CML) (52). The curative treatment for patients with CML and acute myeloid leukemia (AML) is bone marrow transplant or allogeneic stem cell transplant (53). These patients are generally treated with immunosuppressive medication after transplantation, therefore conferring a high risk of developing chronic hepatitis E (54, 55). Thus, repurposing homoharringtonine is attractive for treating HEV-infected patients with leukemia and it simultaneously inhibits malignant cells and the virus, whereas animal studies are recommended to further validate the anti-HEV efficacy before clinical application.
In summary, fetal and adult human liver–derived organoids are highly permissive to HEV replication and support the full life cycle of infection. By establishing these models, we have comprehensively mapped HEV-host interactions and successfully performed antiviral drug screening and validation. We expect that our models could serve as innovative tools to further advance HEV research, but we do not object to the use of conventional cell culture models. Notably, the infection dynamics in human liver–derived organoids appears to resemble acute HEV infection in healthy individuals. Because severe infections in pregnant women and organ transplant patients inevitably involve immune cells, coculture systems of organoids with immune cells deserve to be explored in future to further advance our models. Last, human liver–derived organoids shall also be explored for modeling infection of other hepatitis viruses, such as the neglected hepatitis D virus for which innovative experimental models are urgently needed.
MATERIALS AND METHODS
Viruses and HEV models
Plasmid constructs containing the full-length HEV genome (Kernow-C1 p6 clone; GenBank accession number JQ679013), variant harboring an RNA-dependent RNA polymerase mutation G1634R (p6G1634R), subgenomic GT3 HEV replicon coupled with a Gaussia luciferase reporter gene (p6Luc), the full-length GT1 HEV genome (Sar55 strain, GenBank accession number M80581), GT1 HEV subgenomic that contained Gaussia luciferase reporter gene (GT1Luc), the full-length GT3 HEV genome (swine origin, GenBank accession number AY575859), the full-length GT4 HEV genome (human origin, GenBank accession number HQ634346), the full-length GT5 HEV genome (wild boar origin, GenBank accession number AB573435), the full-length GT7 HEV genome (dromedary camel origin, GenBank accession number KJ496144), and HEV replication–defective replicon (the GDD motif mutated to GAD) were transcribed into genomic RNA in vitro from corresponding enzyme-digested and linearized plasmid DNA using mMessage mMachine T7 RNA kits (Invitrogen) (24–27). Kernow-C1 p6 full-length genomic RNA was delivered into Huh7 cells by electroporation to produce infectious HEV viruses in vitro. For HEV-related Gaussia luciferase analysis (p6Luc and GT1Luc), the activity of secreted luciferase in the culture supernatant was measured by the BioLux Gaussia Luciferase Flex Assay Kit (New England Biolabs, Ipswich, MA, USA). In parallel, GAD RNA was electroporated into organoids as control.
Reagents
Ribavirin, MPA, brequanir, and homoharringtonine were purchased from Sigma-Aldrich and dissolved in dimethyl sulfoxide (DMSO). Human IFNα (Sigma-Aldrich, H6166) was dissolved in phosphate-buffered saline (PBS). Stock of JAK inhibitor 1 (SC-204021, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was dissolved in DMSO with a final concentration of 5 mg/ml. A library of 94 safe-in-human broad-spectrum antiviral agents (https://drugvirus.info) was dissolved in DMSO with a stock concentration of 10 mM.
Cell lines and human liver–derived organoids
Human hepatoma Huh7 and HepG2 cell lines and bile duct carcinoma cell line TFK-1 were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza Biowhittaker, Verviers, Belgium) complemented with 10% (v/v) fetal calf serum (FCS) (Hyclone, Logan, UT, USA), penicillin (100 IU/ml), and streptomycin (100 mg/ml). Tissue samples (≤0.5 cm3) of donor liver biopsies (n > 40) used for organoids isolation and culture were collected during liver transplantation at the Erasmus Medical Center Rotterdam. Use of liver tissues for research purposes was approved by the Medical Ethical Council of the Erasmus MC, and informed consent was given (MEC2006-202 for fetal liver tissues and MEC-2014-060 for adult liver tissues).
Human fetal ICOs and adult ICOs from different donors were isolated and cultured as previously described (16). Human organoids were cultured in organoid expansion medium (EM), based on advanced DMEM/F12 (Invitrogen), supplemented with 1% penicillin/streptomycin (Life Technologies), 1 M Hepes (Life Technologies), 200 mM ultraglutamine (Life Technologies), 1% (v/v) of N2 (Gibco), 2% (v/v) of B27 (Gibco), 1 mM N-acetylcysteine (Sigma-Aldrich), 10 mM nicotinamide (Sigma-Aldrich), 5 μM A83.01 (Tocris), 10 μM forskolin (Tocris), 10 nM gastrin (Sigma-Aldrich), epidermal growth factor (EGF) (50 ng/ml; PeproTech), 10% (v/v) of R-spondin-1 (conditioned medium), fibroblast growth factor 10 (FGF10) (100 ng/ml; PeproTech), hepatocyte growth factor (HGF) (25 ng/ml; PeproTech), and 10 μM Y27632 (Sigma-Aldrich). To ensure the quality (genetically stable) and the replicability (cryopreservable), early-passage organoids (up to 15 passages) were used in this study.
Hepatocyte differentiation of organoids
When ICOs cultured in EM were in 80 to 90% density, organoids were mechanically dissociated into small fragments and cultured in EM complemented with bone morphogenetic protein 7 (BMP7) (25 ng/ml; PeproTech) for 5 days. Subsequently, medium was changed into organoid DM: AdDMEM/F12 medium supplemented with 1% penicillin/streptomycin (Life Technologies), 1 M Hepes (Life Technologies), 200 mM ultraglutamine (Life Technologies), 1% N2 and 1% B27 (with vitamin A) and containing EGF (50 ng/ml; PeproTech), 10 nM gastrin (Sigma-Aldrich), HGF (25 ng/ml; PeproTech), FGF19 (100 ng/ml; PeproTech), 500 nM A8301 (Tocris), 10 μM DAPT (Sigma-Aldrich), BMP7 (25 ng/ml; PeproTech), and 30 μM dexamethasone (Sigma-Aldrich). DM was refreshed every 2 to 3 days for a period of 10 days.
Electroporation of HEV genomic RNA into organoids
When human liver–derived organoids that were cultured in EM become confluent enough (over 80% of basement matrix), organoids were collected in cold AdDMEM/F12 medium to dissolve and remove base matrix (Matrigel). Next, organoids were trypsinized with TrypLE-Express (Gibco) for 5 min. When trypsinization was completed, 2 ml of Opti-MEM was added followed by pipetting up and down by a 1000-μl pipette tip 15 times to dissociate large organoids into single cells. The digested organoids were then washed by Opti-MEM three times. Next, the cell suspensions were filtered through a 20-μM cell strainer to eliminate large clusters of cells. Approximately 1 × 106 cells were suspended in Opti-MEM with 3-μg genome RNA, forming a final volume of 200 μl. Then, the mixture was transferred into a 4-mm electroporation curve (Cellprojects), and electroporation was performed according to the following configured program: voltage, 700 V; pulse length, 4 ms; pulse interval, 0 s; number of pulses, 1. After electroporation, the cells were placed into a new low-binding 15-ml tube and kept at room temperature for 30 min. After the standing period, the cells were suspended in 5 ml of Opti-MEM and centrifuged at 200g for 5 min. The cells were washed by Opti-MEM three times to remove left RNA. Last, organoids were embedded in Matrigel and cultured in EM at 37°C with 5% CO2.
Inoculation of liver-derived organoids with HEV particles
ICOs were mechanically dissociated into small fragments and inoculated with Huh7 cell culture–derived HEV particles (5 × 108 copy numbers/ml) for 6 hours at 37°C. During the inoculation period, organoids were resuspended every 30 min. After inoculation for 6 hours, organoids were centrifuged at 300g for 5 min at 4°C, and the supernatant was discarded. Next, organoids were washed by PBS three times to thoroughly remove unabsorbed virus. Last, organoids were embedded in Matrigel and maintained in EM: advanced DMEM/F12 at a 1:3 ratio at 37°C with 5% CO2. Organoid samples were harvested to quantify HEV RNA at 1 hour and 1, 3, 5, 7, 9, and 11 days after inoculation.
Transwell culture
Dissected organoid cells were seeded on semipermeable transwell inserts (Corning BV) that were precoated with collagen. For maintaining cholangiocyte phenotype, the upper insert and lower compartment were supplemented with organoid EM. For hepatocyte differentiation, EM complemented with BMP7 (25 ng/ml) was added for the first 7 days, and then, medium was changed into organoid DM and cultured for another 14 days. For inoculating polarized cells, HEV (1 × 106 RNA copies per 105 cells) was inoculated through apical sides for 6 hours. Then, the apical sides were washed by PBS two times. Last, both apical and basolateral compartments were maintained with EM at 37°C with 5% CO2. Supernatant from the apical and basolateral sides was harvested at 1 hour and 1, 2, and 3 days after inoculation. The TEER of monolayers on transwell inserts was measured by using an epithelial voltohmmeter (EVOM2; World Precision Instruments).
Quantification of HEV genome copy numbers
Plasmids containing HEV full-length genome were used as a template for quantifying HEV genome copy number. A series of dilutions of plasmid from 10−1 to 10−9 were prepared and then were amplified and quantified by qRT-PCR to generate a standard curve. Standard curve was generated by plotting the log copy number versus the cycle threshold (CT) value (fig. S2). HEV copy numbers were calculated as following equation: Copy number (molecules/μl) = [concentration (ng/μl) × 6.022 × 1023 (molecules/mol)]/[length of amplicon × 660 (g/mol) × 109 (ng/g)].
Quantification of gene expression
Total RNA was isolated using the Macherey-Nagel NucleoSpin RNA II Kit (Bioke, Leiden, Netherlands) and quantified by Nanodrop ND-1000 (Wilmington, DE, USA). RNA expression levels were quantified by SYBR Green–based qRT-PCR (Applied Biosystems SYBR Green PCR Master Mix; Thermo Fisher Scientific Life Sciences) with the StepOnePlus System (Thermo Fisher Scientific Life Sciences). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene served as housekeeping gene. Relative gene expression was normalized to GAPDH using the formula 2−∆∆CT (∆∆CT = ∆CTsample − ∆CTcontrol). Template control and reverse transcriptase control were included in all qRT-PCR experiments. All qRT-PCR primers are listed in table S1.
RNA sequencing analysis
Total RNA of ICOs was isolated on day 7 after electroporation (GT3 p6 versus GAD as mock). The quality of RNA was measured by Bioanalyzer RNA 6000 Picochip as a quality-control step, followed by RNA sequencing performed by Novogene with paired-end 150–base pair (PE 150) sequencing strategy. The identification of differentially expressed genes is based on P < 0.05 and absolute values of logFc > 1.5. Data are publically available at https://doi.org/10.17026/dans-zgz-hbgk.
AlamarBlue assay
First, the culture supernatant of organoids was discarded. Then, the organoids were incubated with alamarBlue (Invitrogen, DAL1100; 1:20 dilution in organoid expansion medium) for 2 hours (37°C). Next, all the medium was collected for analyzing the metabolic activity of the organoids. Absorbance was determined by using a fluorescence plate reader (CytoFluor Series 4000, PerSeptive Biosystems) at the excitation of 530/25 nm and emission of 590/35 nm.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
Supernatants of cells seeded in a 96-well plate were supplemented with 10-μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich) and then incubated at 37°C with 5% CO2 for 3 hours. Next, the medium was removed, and 100 μl of DMSO was added to each well. The absorbance of each well was read on a microplate absorbance reader (Bio-Rad, Hercules, CA, USA) at a wavelength of 490 nm.
Immunofluorescence assay
Organoids were washed in cold AdDMEM/F12 medium three times to remove all basal matrix. Subsequently, organoids were added into the CytoSpin II Cytocentrifuge (Shandon Scientifi Ltd., Runcorn, England) and spun down into slides at 1000 rpm for 5 min. Next, the organoids were fixed in 4% paraformaldehyde solution at 4°C for 10 min. The slides containing organoids were then rinsed three times with PBS for 5 min each time, followed by permeabilizing with PBS containing 0.2% (v/v) Triton X-100 for 5 min. Then, the slides were twice rinsed with PBS for 5 min, followed by incubation with blocking solution (5% donkey serum, 1% bovine serum albumin, and 0.2% Triton X-100 in PBS) at room temperature for 1 hour. Next, the slides were incubated in a humidity chamber with primary antibody diluted in blocking solution at 4°C overnight. Primary antibodies used in this study are as follows: Anti–HEV ORF2 antibody (1:250, mouse monoclonal antibody (mAb); Millipore, MAB8002), anti-EpCAM antibody (1:1000, rabbit mAb; Abcam), anti-dsRNA antibody (1:500, mouse mAb; Scicons J2), anti-SOX9 antibody (1:1000, rabbit mAb; Merck), anti-HNF4a antibody (1:1000, mouse mAb; Abcam), anti-KRT19 antibody (1:1000, rabbit mAb; Bio SB), anti-albumin antibody (1:500, goat mAb; Santa Cruz Biotechnology), anti-CRB3 antibody (1:250, rabbit polyclonal antibody, Atlas Antibodies). Slides were washed three times for 5 min each in PBS before 1-hour incubation with 1:1000 dilutions of the anti-mouse immunoglobulin G (IgG) (H + L, Alexa Fluor 594), the anti-rabbit IgG (H + L, Alexa Fluor 488), and anti-goat IgG (H + L, Alexa Fluor 594) secondary antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole (Invitrogen).
Measuring albumin secretion by ELISA
Secreted albumin was assayed with the human ALB/serum albumin ELISA Kit (RAB0603-1KT, Merck), following the manufacturer’s instructions. Organoids or polarized cells on transwell filters cultured in EM were served as control.
Infectious virus production and secondary infection
Human liver–derived organoids electroporated with p6 or p6G1634R variant RNA were cultured for 11 days. Subsequently, viruses were harvested from organoids and supernatants through repeated freezing and thawing. These viruses were used to inoculate Huh7 cells overnight (12 hours). Huh7 cells were washed by PBS three times on the second day and cultured for 48 hours coated with DMEM supplemented with 3% FCS. Naïve Huh7 cells without infection were set as mock. A similar approach was used for inoculating Huh7 cells with viruses derived from apical supernatant of polarized cells.
tRNAsome quantification
In total, four fetal and three adult ICO lines were electroporated with p6 RNA and controlled by GAD. After 7 days of culturing in EM, organoids were harvested to isolate tRNAsome following our previous protocol (35).
Statistics
The statistical significance of differences between means was assessed with the Mann-Whitney test (GraphPad Prism 8; GraphPad Software Inc., La Jolla, CA). The threshold for statistical significance was defined as P ≤ 0.05. Heatmaps was created by R Statistical software (version 4.0.2) with the “pheatmap” package. Synergistic scores of drug combinations were analyzed by SynergyFinder 2.0 (56).
Acknowledgments
We would like to thank S. U. Emerson (National Institute of Allergy and Infectious Diseases, NIH, USA) for providing the plasmids to generate subgenomic and full-length HEV genomic RNA. We thank D. Todt and E. Steinmann (Department for Molecular and Medical Virology, Ruhr University Bochum, Bochum Germany) for providing the plasmids to generate HEV G1634R-mutated full-length genomic RNA. We thank X. Meng (Center for Emerging, Zoonotic and Arthropod-borne Pathogens, Virginia Polytechnic Institute and State University, USA) for providing plasmids to generate GT3 (pSHEV-3) and GT4 (pHEV-4TW) full-length genomic RNA. We are grateful to the staff of the CASA clinics in Leiden and Rotterdam for collecting fetal tissues.
Funding: This research is supported by funding of the educational committee of Guangdong for specific program of key scientific research (2021ZDZX2016), National Natural Science Foundation of China (81802020 and 31770186), and Shenzhen government and SUSTech for talent research start-up funding (Y01416122 and Y01416222) to Yijin Wang; a VIDI grant (no. 91719300) from the Netherlands Organization for Scientific Research and the Dutch Cancer Society Young Investigator Grant (10140) to Q.P.; the European Regional Development Fund to the Mobilitas Pluss Project (MOBTT39) from Estonian Research Council to D.E.K.; and the China Scholarship Council for funding PhD fellowship to P.L. (no. 201808370170).
Author contributions: P.L., Yijin Wang, M.P.P., and Q.P. conceived and designed the research. P.L., Yunlong Li, J.L., M.L., Yang Li, R.Z., and Yining Wang performed experiments and analyzed data. P.L., Yijin Wang, and Q.P. interpreted the data and drafted the manuscript. T.-C.L., Z.M., and D.E.K. provided key reagents/research tools and discussed the project. M.M.A.V., M.J.B., R.A.d.M., and L.J.v.d.L. contributed to key research materials and discussed the project design. All authors critically revised the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 and S2
REFERENCES AND NOTES
- 1.Li P., Liu J., Li Y., Su J., Ma Z., Bramer W. M., Cao W., de Man R. A., Peppelenbosch M. P., Pan Q., The global epidemiology of hepatitis E virus infection: A systematic review and meta-analysis. Liver Int. 40, 1516–1528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Metselaar H. J., Peppelenbosch M. P., Pan Q., Chronic hepatitis E in solid-organ transplantation: The key implications of immunosuppressants. Curr. Opin. Infect. Dis. 27, 303–308 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Hakim M. S., Wang W., Bramer W. M., Geng J., Huang F., de Man R. A., Peppelenbosch M. P., Pan Q., The global burden of hepatitis E outbreaks: A systematic review. Liver Int. 37, 19–31 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Khuroo M. S., Khuroo M. S., Khuroo N. S., Hepatitis E: Discovery, global impact, control and cure. World J. Gastroenterol. 22, 7030–7045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma X. X., Ji Y., Jin L., Baloch Z., Zhang D. R., Wang Y., Pan Q., Ma Z., Prevalence and clinical features of hepatitis E virus infection in pregnant women: A large cohort study in Inner Mongolia, China. Clin. Res. Hepatol. Gastroenterol. 45, 101536 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., de Man R. A., de Knegt R. J., Metselaar H. J., Peppelenbosch M. P., Pan Q., Epidemiology and management of chronic hepatitis E infection in solid organ transplantation: A comprehensive literature review. Rev. Med. Virol. 23, 295–304 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Kamar N., Izopet J., Tripon S., Bismuth M., Hillaire S., Dumortier J., Radenne S., Coilly A., Garrigue V., D’Alteroche L., Buchler M., Couzi L., Lebray P., Dharancy S., Minello A., Hourmant M., Roque-Afonso A. M., Abravanel F., Pol S., Rostaing L., Mallet V., Ribavirin for chronic hepatitis E virus infection in transplant recipients. N. Engl. J. Med. 370, 1111–1120 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Kamar N., Abravanel F., Behrendt P., Hofmann J., Pageaux G. P., Barbet C., Moal V., Couzi L., Horvatits T., De Man R. A., Cassuto E., Elsharkawy A. M., Riezebos-Brilman A., Scemla A., Hillaire S., Donnelly M. C., Radenne S., Sayegh J., Garrouste C., Dumortier J., Glowaki F., Matignon M., Coilly A., Figueres L., Mousson C., Minello A., Dharancy S., Rerolle J. P., Lebray P., Etienne I., Perrin P., Choi M., Marion O., Izopet J., Hepatitis E Virus Ribavirin Study Group Ribavirin for hepatitis E virus infection after organ transplantation: A large european retrospective multicenter study. Clin. Infect. Dis. 71, 1204–1211 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Debing Y., Gisa A., Dallmeier K., Pischke S., Bremer B., Manns M., Wedemeyer H., Suneetha P. V., Neyts J., A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147, 1008–1011.e7; quiz e15-6 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Todt D., Gisa A., Radonic A., Nitsche A., Behrendt P., Suneetha P. V., Pischke S., Bremer B., Brown R. J., Manns M. P., Cornberg M., Bock C. T., Steinmann E., Wedemeyer H., In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut 65, 1733–1743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marion O., Lhomme S., Nayrac M., Dubois M., Pucelle M., Requena M., Migueres M., Abravanel F., Peron J. M., Carrere N., Suc B., Delobel P., Kamar N., Izopet J., Hepatitis E virus replication in human intestinal cells. Gut 69, 901–910 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Gural N., Mancio-Silva L., He J., Bhatia S. N., Engineered livers for infectious diseases. Cell. Mol. Gastroenterol. Hepatol. 5, 131–144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Y., Bijvelds M., Dang W., Xu L., van der Eijk A. A., Knipping K., Tuysuz N., Dekkers J. F., Wang Y., de Jonge J., Sprengers D., van der Laan L. J., Beekman J. M., Ten Berge D., Metselaar H. J., de Jonge H., Koopmans M. P., Peppelenbosch M. P., Pan Q., Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res. 123, 120–131 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Dutta D., Heo I., Clevers H., Disease modeling in stem cell-derived 3d organoid systems. Trends Mol. Med. 23, 393–410 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Chen S., Li P., Wang Y., Yin Y., de Ruiter P. E., Verstegen M. M. A., Peppelenbosch M. P., van der Laan L. J. W., Pan Q., Rotavirus infection and cytopathogenesis in human biliary organoids potentially recapitulate biliary atresia development. MBio 11, e01968-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M. M., Ellis E., van Wenum M., Fuchs S. A., de Ligt J., van de Wetering M., Sasaki N., Boers S. J., Kemperman H., de Jonge J., Ijzermans J. N., Nieuwenhuis E. E., Hoekstra R., Strom S., Vries R. R., van der Laan L. J., Cuppen E., Clevers H., Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Wang Y., Qu C., Wang S., Zhou J., Cao W., Xu L., Ma B., Hakim M. S., Yin Y., Li T., Peppelenbosch M. P., Zhao J., Pan Q., The RNA genome of hepatitis E virus robustly triggers an antiviral interferon response. Hepatology 67, 2096–2112 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Beer A., Holzmann H., Pischke S., Behrendt P., Wrba F., Schlue J., Drebber U., Neudert B., Halilbasic E., Kreipe H., Lohse A., Sterneck M., Wedemeyer H., Manns M., Dienes H. P., Chronic hepatitis E is associated with cholangitis. Liver Int. 39, 1876–1883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treyer A., Musch A., Hepatocyte polarity. Compr. Physiol. 3, 243–287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capelli N., Marion O., Dubois M., Allart S., Bertrand-Michel J., Lhomme S., Abravanel F., Izopet J., Chapuy-Regaud S., Vectorial release of hepatitis E virus in polarized human hepatocytes. J. Virol. 93, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Sayed Zaki M., El Razek M. M., El Razek H. M., Maternal-fetal hepatitis E transmission: Is it underestimated? J. Clin. Transl. Hepatol. 2, 117–123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsee A., Roos F. J. M., Verstegen M. M. A., Consortium H. P. B. O., Gehart H., de Koning E., Lemaigre F., Forbes S. J., Peng W. C., Huch M., Takebe T., Vallier L., Clevers H., van der Laan L. J. W., Spee B., Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell 28, 816–832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X., Wang Y., Metselaar H. J., Janssen H. L., Peppelenbosch M. P., Pan Q., Rapamycin and everolimus facilitate hepatitis E virus replication: Revealing a basal defense mechanism of PI3K-PKB-mTOR pathway. J. Hepatol. 61, 746–754 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Shukla P., Nguyen H. T., Faulk K., Mather K., Torian U., Engle R. E., Emerson S. U., Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J. Virol. 86, 5697–5707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordoba L., Feagins A. R., Opriessnig T., Cossaboom C. M., Dryman B. A., Huang Y. W., Meng X. J., Rescue of a genotype 4 human hepatitis E virus from cloned cDNA and characterization of intergenotypic chimeric viruses in cultured human liver cells and in pigs. J. Gen. Virol. 93, 2183–2194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T. C., Zhou X., Yoshizaki S., Ami Y., Suzaki Y., Nakamura T., Takeda N., Wakita T., Production of infectious dromedary camel hepatitis E virus by a reverse genetic system: Potential for zoonotic infection. J. Hepatol. 65, 1104–1111 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Li T. C., Bai H., Yoshizaki S., Ami Y., Suzaki Y., Doan Y. H., Takahashi K., Mishiro S., Takeda N., Wakita T., Genotype 5 hepatitis E virus produced by a reverse genetics system has the potential for zoonotic infection. Hepatol. Commun. 3, 160–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin X., Ambardekar C., Lu Y., Feng Z., Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J. Virol. 90, 4232–4242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broutier L., Andersson-Rolf A., Hindley C. J., Boj S. F., Clevers H., Koo B. K., Huch M., Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Wang W., Xu L., Su J., Peppelenbosch M. P., Pan Q., Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 25, 573–584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L., Wang W., Peppelenbosch M. P., Pan Q., Noncanonical antiviral mechanisms of ISGs: Dispensability of inducible interferons. Trends Immunol. 38, 1–2 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zhou X., Xu L., Wang W., Watashi K., Wang Y., Sprengers D., de Ruiter P. E., van der Laan L. J., Metselaar H. J., Kamar N., Peppelenbosch M. P., Pan Q., Disparity of basal and therapeutically activated interferon signalling in constraining hepatitis E virus infection. J. Viral Hepat. 23, 294–304 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Ou X., Cao J., Cheng A., Peppelenbosch M. P., Pan Q., Errors in translational decoding: tRNA wobbling or misincorporation? PLOS Genet. 15, e1008017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R., Noordam L., Ou X., Ma B., Li Y., Das P., Shi S., Liu J., Wang L., Li P., Verstegen M. M. A., Reddy D. S., van der Laan L. J. W., Peppelenbosch M. P., Kwekkeboom J., Smits R., Pan Q., The biological process of lysine-tRNA charging is therapeutically targetable in liver cancer. Liver Int. 41, 206–219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou X., Ma B., Zhang R., Miao Z., Cheng A., Peppelenbosch M. P., Pan Q., A simplified qPCR method revealing tRNAome remodeling upon infection by genotype 3 hepatitis E virus. FEBS Lett. 594, 2005–2015 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Li P., Li Y., Zhang R., Yu P., Ma Z., Kainov D. E., de Man R. A., Peppelenbosch M. P., Pan Q., Drug screening identified gemcitabine inhibiting hepatitis E virus by inducing interferon-like response via activation of STAT1 phosphorylation. Antivir. Res. 184, 104967 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Chen S., Wang Y., Li P., Yin Y., Bijvelds M. J., de Jonge H. R., Peppelenbosch M. P., Kainov D. E., Pan Q., Drug screening identifies gemcitabine inhibiting rotavirus through alteration of pyrimidine nucleotide synthesis pathway. Antivir. Res. 180, 104823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Wang W., Xu L., Zhou X., Shokrollahi E., Felczak K., van der Laan L. J., Pankiewicz K. W., Sprengers D., Raat N. J., Metselaar H. J., Peppelenbosch M. P., Pan Q., Cross talk between nucleotide synthesis pathways with cellular immunity in constraining hepatitis E virus replication. Antimicrob. Agents Chemother. 60, 2834–2848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todt D., Friesland M., Moeller N., Praditya D., Kinast V., Bruggemann Y., Knegendorf L., Burkard T., Steinmann J., Burm R., Verhoye L., Wahid A., Meister T. L., Engelmann M., Pfankuche V. M., Puff C., Vondran F. W. R., Baumgartner W., Meuleman P., Behrendt P., Steinmann E., Robust hepatitis E virus infection and transcriptional response in human hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 117, 1731–1741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimgaonkar I., Ding Q., Schwartz R. E., Ploss A., Hepatitis E virus: Advances and challenges. Nat. Rev. Gastroenterol. Hepatol. 15, 96–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X., Huang F., Xu L., Lin Z., de Vrij F. M. S., Ayo-Martin A. C., van der Kroeg M., Zhao M., Yin Y., Wang W., Cao W., Wang Y., Kushner S. A., Marie Peron J., Alric L., de Man R. A., Jacobs B. C., van Eijk J. J., Aronica E. M. A., Sprengers D., Metselaar H. J., de Zeeuw C. I., Dalton H. R., Kamar N., Peppelenbosch M. P., Pan Q., Hepatitis E virus infects neurons and brains. J. Infect. Dis. 215, 1197–1206 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., Clevers H., Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Ettayebi K., Crawford S. E., Murakami K., Broughman J. R., Karandikar U., Tenge V. R., Neill F. H., Blutt S. E., Zeng X. L., Qu L., Kou B., Opekun A. R., Burrin D., Graham D. Y., Ramani S., Atmar R. L., Estes M. K., Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boonstra A., van der Laan L. J., Vanwolleghem T., Janssen H. L., Experimental models for hepatitis C viral infection. Hepatology 50, 1646–1655 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Sari G., van de Garde M. D. B., van Schoonhoven A., Voermans J. J. C., van der Eijk A. A., de Man R. A., Boonstra A., Vanwolleghem T., Pas S. D., Hepatitis E virus shows more genomic alterations in cell culture than in vivo. Pathogens 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dao Thi V. L., Wu X., Belote R. L., Andreo U., Takacs C. N., Fernandez J. P., Vale-Silva L. A., Prallet S., Decker C. C., Fu R. M., Qu B., Uryu K., Molina H., Saeed M., Steinmann E., Urban S., Singaraja R. R., Schneider W. M., Simon S. M., Rice C. M., Stem cell-derived polarized hepatocytes. Nat. Commun. 11, 1677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorris M., van der Lecq B. M., van Erpecum K. J., de Bruijne J., Treatment for chronic hepatitis E virus infection: A systematic review and meta-analysis. J. Viral Hepat. 28, 454–463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin X., Li X., Ambardekar C., Hu Z., Lhomme S., Feng Z., Hepatitis E virus persists in the presence of a type III interferon response. PLOS Pathog. 13, e1006417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamar N., Izopet J., Pavio N., Aggarwal R., Labrique A., Wedemeyer H., Dalton H. R., Hepatitis E virus infection. Nat. Rev. Dis. Primers. 3, 17086 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Wang Y., Zhou X., Debing Y., Chen K., Van Der Laan L. J., Neyts J., Janssen H. L., Metselaar H. J., Peppelenbosch M. P., Pan Q., Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology 146, 1775–1783 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Schneeberger K., Sanchez-Romero N., Ye S., van Steenbeek F. G., Oosterhoff L. A., Pla Palacin I., Chen C., van Wolferen M. E., van Tienderen G., Lieshout R., Colemonts-Vroninks H., Schene I., Hoekstra R., Verstegen M. M. A., van der Laan L. J. W., Penning L. C., Fuchs S. A., Clevers H., De Kock J., Baptista P. M., Spee B., Large-scale production of LGR5-positive bipotential human liver stem cells. Hepatology 72, 257–270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvandi F., Kwitkowski V. E., Ko C. W., Rothmann M. D., Ricci S., Saber H., Ghosh D., Brown J., Pfeiler E., Chikhale E., Grillo J., Bullock J., Kane R., Kaminskas E., Farrell A. T., Pazdur R., U.S. Food and Drug Administration approval summary: Omacetaxine mepesuccinate as treatment for chronic myeloid leukemia. Oncologist 19, 94–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Bubnoff N., Duyster J., Chronic myelogenous leukemia: Treatment and monitoring. Dtsch. Arztebl. Int. 107, 114–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Felden J., Alric L., Pischke S., Aitken C., Schlabe S., Spengler U., Giordani M. T., Schnitzler P., Bettinger D., Thimme R., Xhaard A., Binder M., Ayuk F., Lohse A. W., Cornelissen J. J., de Man R. A., Mallet V., The burden of hepatitis E among patients with haematological malignancies: A retrospective European cohort study. J. Hepatol. 71, 465–472 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Kamar N., Selves J., Mansuy J. M., Ouezzani L., Peron J. M., Guitard J., Cointault O., Esposito L., Abravanel F., Danjoux M., Durand D., Vinel J. P., Izopet J., Rostaing L., Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358, 811–817 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Ianevski A., Giri A. K., Aittokallio T., SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 48, W488–W493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 and S2