Abstract
Chemosensitivity assays are commonly used for preclinical drug discovery and clinical trial optimization. However, data from independent assays are often discordant, largely attributed to uncharacterized variation in the experimental materials and protocols. We report here the launching of Minimal Information for Chemosensitivity Assays (MICHA), accessed via https://micha-protocol.org. Distinguished from existing efforts that are often lacking support from data integration tools, MICHA can automatically extract publicly available information to facilitate the assay annotation including: 1) compounds, 2) samples, 3) reagents and 4) data processing methods. For example, MICHA provides an integrative web server and database to obtain compound annotation including chemical structures, targets and disease indications. In addition, the annotation of cell line samples, assay protocols and literature references can be greatly eased by retrieving manually curated catalogues. Once the annotation is complete, MICHA can export a report that conforms to the FAIR principle (Findable, Accessible, Interoperable and Reusable) of drug screening studies. To consolidate the utility of MICHA, we provide FAIRified protocols from five major cancer drug screening studies as well as six recently conducted COVID-19 studies. With the MICHA web server and database, we envisage a wider adoption of a community-driven effort to improve the open access of drug sensitivity assays.
Keywords: drug discovery, drug sensitivity assays, data integration tools, FAIR research data
Introduction
Drug sensitivity or chemosensitivity assay is an important tool to measure cellular response to drug perturbation, which has been increasingly used for preclinical drug discovery and clinical trial optimization. However, poor inter- and intralaboratory reproducibility has been reported when comparing batches that differ at assay conditions [1–3]. Central to improving the data reproducibility is the standardization of material and method descriptions, summarized as protocols, which should be sufficiently annotated and easily comparable. To make the assay protocols FAIR (Findable, Accessible, Interoperable and Reusable), a large variety of efforts to define the minimal information (MI) for specific assay types have been developed. In total, Minimum Information for Biological and Biomedical Investigations has reported 40 MI-based initiatives [4]. Among these, protocols for common omics assays include Minimal Information About Microarray Experiment, Minimum Information About a Next-generation Sequencing Experiment, Metabolomics Standards Initiative and Minimum Information About a Proteomics Experiment. For bioactivity assays in general, Minimum Information About Bioactive Entity (MIABE) has been widely used [5]. However, MIABE does not include specific guidelines for annotating drug sensitivity assays. Furthermore, like many other MI efforts, there is a lack of data integration tools to facilitate its implementation.
With an increasing number of drug sensitivity studies, efficient experimental annotation is critically needed to ensure the accessibility and reuse of the data. The solution we present here, MICHA (https://micha-protocol.org/), includes a guideline to annotate the MI for four major components of a drug sensitivity assay, including 1) compounds, 2) samples, 3) reagents and 4) data analysis. Furthermore, to make the annotation as efficient as possible, MICHA provides an integrative web tool that allows a user to retrieve the information about these assay components from public databases by standardized identifiers and ontologies. Without MICHA, a user would need to annotate a drug sensitivity experiment by retrieving multiple databases separately, which is often time-consuming and error prone. With the help of MICHA, we have catalogued the major drug sensitivity screening protocols in cancer and COVID-19 that may help users assess the FAIRness of existing experiments as well as inform the design of new experiments.
Materials and methods
Workflow
Using MICHA, users can upload their compounds, samples and experimental design information (Figure 1). To start, users need to upload the names and InChiKeys for the compounds, after which MICHA will automatically extract primary and secondary target information, physiochemical properties and disease indications. This information will help users annotate the mechanisms of action of the compounds. After obtaining the compound annotations, users may continue filling in the other experimental details, such as sample (cell lines or patient-derived samples) information and assay conditions. For cell lines, only the names of the cell lines are required, as the other information will be retrieved automatically from internal databases. For annotating assay protocols, we derive a consensus on the MI that is needed, including assay format, detection technology, end point mode of action, experimental medium, plate type, cell density, time for treatment, dilution fold, vehicle of compound, dispensation method and volume per well. These terms are defined in Supplementary File 1 as well as in the ‘Glossary’ tab at the MICHA website. Most of the terms are linked with the BioAssay ontology [6], which is commonly used for high-throughput chemical biology experiments [7]. Next, users are directed to a web form to report data processing information, including minimal and maximal concentrations of the compounds, publication references and drug response metric types such as IC50 or area under the dose–response curve (AUC). Finally, a tabular report can be generated according to the user’s input augmented with information retrieved from public resources (Supplementary File 2). In addition, MICHA provides a checklist of annotation items (Supplementary File 3). When preparing a manuscript, it is recommended to use the checklist to confirm the MICHA compliance, so that journals and reviewers can evaluate the FAIRness of the experiment more easily and more systematically. The Supplementary File 4 shows example template for FAIRified data that should be requested from the authors by the journals or reviewers.
Figure 1.
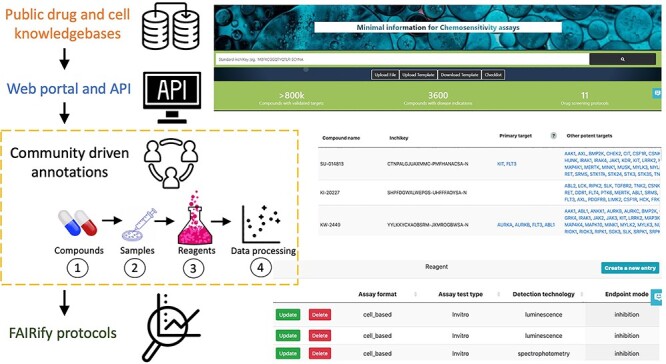
User interface and workflow of MICHA. Users start with compound annotation by uploading a list of compounds with names and standard InChiKeys. MICHA will return the pharmacological (drug targets, disease indications) and physiochemical properties of the compounds via an integrative web server and database, available under the ‘Compounds’ tab. Users then may click on Samples, Reagents or Data processing tabs to annotate their drug screening protocols. Auto suggestions are provided to avoid spelling mistakes or terminology conflicts. Finally, users can download the summary reports containing input data as well as integrated information provided by MICHA.
Data integration tools
Three types of datasets are retrievable via data integration tools in MICHA:
FAIRified protocols
A prime objective for MICHA is to provide a pipeline for the FAIRification of drug sensitivity assays, such that these established protocols can be well documented with enhanced visibility to the research community. To initiate such an effort, we have FAIRified drug screening protocols from major cancer studies including GDSC (345 compounds and 987 cell lines) [8], CCLE (24 compounds and 504 cell lines) [9] and CTRPv2 (203 compounds and 242 cell lines) [10]. Furthermore, we have provided drug sensitivity screening protocols extracted from six recent COVID-19 antiviral studies (5525 compounds and 2 cell lines) [11–16]. On the other hand, we have provided an example of protocols established at the research institution level (528 compounds and 4 cell lines utilized at the high-throughput Drug-Screening Unit at the Institute for Molecular Medicine Finland, University of Helsinki). These FAIRified protocols can be freely obtained at http://micha-protocol.org/protocols/. With more protocols annotated via MICHA, the drug discovery and translational medicine community shall be better informed on the variations on the experimental condition across different studies and institutions. Table 1 shows an overview of the FAIRified protocols by MICHA.
Table 1.
FAIRified protocols by MICHA
| Protocol name | Type | Comp-ounds | Cell lines | Detection technology | Dilution fold | Plate type | Min concentration (nM) | Max concentration (nM) | Metric | References |
|---|---|---|---|---|---|---|---|---|---|---|
| GDSC | Cancer | 345 | 987 | Fluorescence | 2 | 384 and 96 | 0.03 | 4000 000 | IC50 | [10] |
| CCLE | Cancer | 24 | 504 | Luminescence | 3.16 | 1536 | 2.5 | 8000 | IC50, EC50 | [9] |
| CTRPv2 | Cancer | 203 | 242 | Luminescence | 2 | 384 | 0.56 | 592,000 | IC50 | [8] |
| FIMM | Cancer | 528 | 4 | Fluorescence | 10 | 384 | 0.0 | 1000 000 | DSS | [17] |
| Mario Negri | Cancer | 1 | 16 | Spectrophotometry, luminescence | 10 | 96 | 0.0 | 10 000 | AUC | [18] |
| NCATS | COVID-19 | 5430 | 1 | Luminescence | 2–5 | 384 | 0.0 | 120 000 | AC50 | [15] |
| Ellinger et al. | COVID-19 | 103 | 1 | Label free | 3.33 | 384 | 20 | 20 000 | IC50, CC50 | [16] |
| Gordon et al. | COVID-19 | 73 | 1 | Spectrophotometry | 96 | 1 | 10 000 | IC50 | [12] | |
| Jeon et al. | COVID-19 | 43 | 1 | Microscopy | 2 | 384 | 50 | 50 000 | IC50 | [14] |
| Touret et al. | COVID-19 | 83 | 1 | qPCR | 2 | 96 | 600 | 40 000 | EC50 | [13] |
| Weston et al. | COVID-19 | 43 | 1 | Luminescence | 2 | 96 | 50 | 5000 | IC50 | [11] |
Compound target profiles
Compound–target profiles are integrated from the most comprehensive drug–target databases including DrugTargetCommons (DTC) [19] [20], BindingDB [21], ChEMBL [22], GtopDB [23], DGiDB [24] and DrugBank [25]. The first four databases (DTC, BindingDB, ChEMBL and GtopDB) contain quantitative bioactivity data, whereas DGiDB and DrugBank contain unary drug–target information. We have focused on the primary and secondary targets of a compound, defined as those displaying binding affinities ≤ 1000 nM from the bioactivity databases, or those that are recorded in the unary databases. We have integrated drug targets for 277K chemicals from DTC, 513K from ChEMBL, 258K from Binding DB, 4.8K from GtopDB, 7.6K from DGiDB and 6.8K from DrugBank. Furthermore, we have merged overlapping targets across these databases to avoid duplications, resulting in high-quality target profiles for >800K chemicals. Such a data integration provides one of the most comprehensive compounds collection along with their potent primary and secondary targets.
Compound properties, cell line and assay information
Compound physiochemical properties and structures for 1.9 million compounds are obtained from the ChEMBL database. Furthermore, we have integrated disease indications and clinical phase information for 3600 clinical drugs from the DTC database. This information together with the drug–target profiles will be retrieved for user-uploaded compound list. When users annotate the cell lines, the majority of cell line information can be retrieved automatically from Cellosaurus [26], which is a comprehensive knowledge database on cell lines. For assay annotation, commonly used techniques will be provided for users to choose from to ease the burden of manual editing.
Added values by MICHA
Comprehensive compound–target profiles
For annotating the mechanisms of action of compounds, MICHA integrates compound–target profiles from various databases, ranging from quantitative bioactivity values to unary drug–target hits. For instance, DrugBank, GtopDB and DGiDB are mainly focused on approved compounds with putative target information, whereas ChEMBL, BindingDB and DTC include bioactivity values for more versatile investigational and preclinical chemicals. In MICHA, we have improved target coverage across the druggable genome by integrating nonoverlapping data points from the latest releases of these databases. As shown in Figure 2, the average number of targets for 2993 approved drugs (and salts) in MICHA is 7.33, as compared with that from ChEMBL (5.5), DGiDB (4.71), DrugBank (3.56), BindingDB (2.74) and GtopDB (0.96). Similarly, for 1992 investigational compounds (defined as those in clinical trials), the average number of targets per chemical is higher in MICHA as compared with other databases. Secondly, MICHA provides efficient Application Programming Interface (API) for retrieving comprehensive target profiles, available at: https://api.micha-protocol.org (Supplementary File 5). We believe that the API for compound–target information will further boost the usability of MICHA by programmatically integrating compound–target profiles with other related tools and shall open new applications for drug discovery researchers for training their compound–target machine learning models [27–29] as well as providing more insights on the network modeling of mechanisms of action [30, 31].
Figure 2.
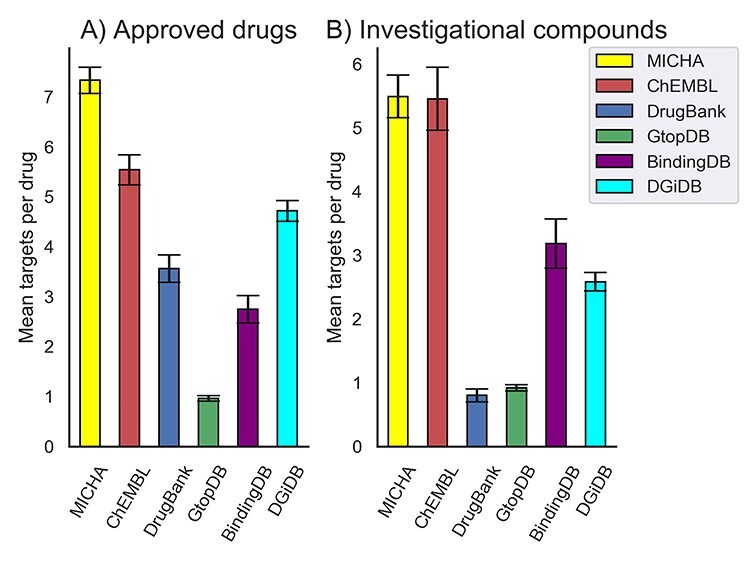
Average number of targets for compounds in multiple databases. (A) Approved drugs. (B) Investigational compounds.
Systematic comparison of drug sensitivity screening protocols
We have FAIRified the screening protocols for three major cancer drug studies including CCLE, GDSC and CTRPv2 (Table 1). FAIRification of these protocols is performed using MI-based information as mentioned in checklist available at the home page of MICHA (as well as in Supplementary File 3). These drug screening studies share similar objectives of linking genetic features of cancer cell lines to small-molecule sensitivity to accelerate drug discovery. Note that MICHA focuses on the annotation of drug screening protocols while the actual data points are available in their corresponding databases. Here we report the comparison of the major components in the assay protocols (Table 2).
Table 2.
Comparison between protocols of CCLE, GDSC and CTRPv2
| CCLE | GDSC | CTRPv2 | |
|---|---|---|---|
| Plate types | 1536 | 384, 96 | 384-Opaque white |
| Experimental mediums | NA | DMEM, RPMI | ALPHAMEM, DMEM, DMEMF, EMEM, HAMSF, IMDM, L15, McCoys5A, MCDB, MEM, RPMI, Waymouth, WilliamsE |
| Detection technology | Luminescence | Fluorescence | Luminescence |
| Cell density | 250 | NA | 500, 1000, 2000, 3000, 5000, 10 000 |
| Treatment time (h) | 72–84 | 72 | 72 |
| Analysis metrics | IC50, EC50 | IC50 | IC50 |
| Compounds | 24 | 345 | 203 |
| Number of cell lines | 504 | 987 | 242 |
NA indicates that the information is unavailable.
Both GDSC and CTRPv2 used common experimental plate type i.e. 384 wells, whereas CCLE compounds were tested on 1536 well plates. In GDSC, two different experimental mediums including DMEM and RPMI were tested for the 987 cancer cell lines, whereas the CTRPv2 cell lines were tested for many more different mediums as listed in Table 2. In contrast, we could not find experimental medium information for CCLE. On the other hand, both CCLE and CTRPv2 have used Cell-Titer-Glo (Promega), a luminescence-based assay to measure the levels of ATP as a surrogate to cell viability, whereas GDSC has used based nucleic acid staining syto60 (Invitrogen) for adherent cells and resazurin (Sigma) for suspension cells. All the three screening studies have used at least 72 h of treatment, after which the IC50 or EC50 concentrations were determined from the dose–response curves.
Figure 3 shows the overlapping chemicals and cell lines tested across CCLE, GDSC and CTRPv2 studies, after excluding those chemicals for which proper chemical names or identifiers were missing to assure high-quality data in MICHA. Only two chemicals are shared across the three studies including selumetinib and tanespimycin (Figure 3A). Selumetinib (AZD6244) is a MEK (kinase) inhibitor used for treating neurofibromatosis type I in children [32], whereas tanespimycin is a Hsp90 inhibitor [33] that has been studied for the treatment of leukemia or solid tumors, especially kidney tumors. In contrast, more overlap was found for the cell lines, with 112 cell lines in common across the three studies (Figure 3B).
Figure 3.
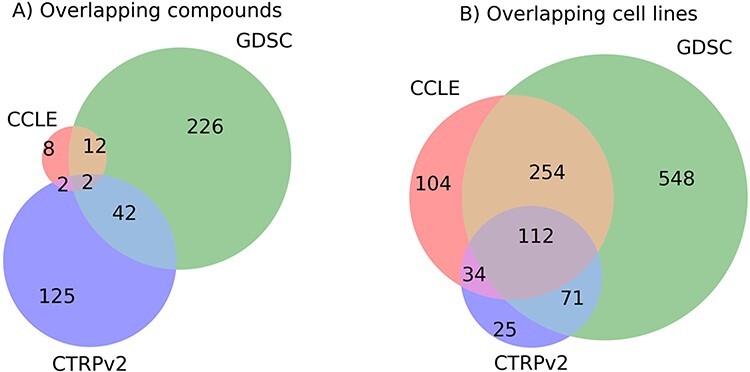
Overlapping data between CCLE, GDSC and CTRPv2 database. (A) Overlapping compounds. (B) Overlapping cell lines.
MICHA has not only FAIRified cancer-related drug screening studies but also annotated six recent studies on COVID-19, a virus that causes ongoing pandemic with limited drug treatment options. From these studies, we have identified 5525 chemicals tested across two cell lines including Vero E6 and Caco-2. The annotations of these compounds, cell lines as well as the experimental information and data analysis methods can be easily retrieved at http://micha-protocol.org/covid19.
In total, Figure 4A shows the clinical phases of the compounds FAIRified by MICHA, whereas Figure 4B shows the distribution of FAIRified cell lines from different tissue types. These statistics show a broad coverage of cell lines and compounds. We believe that with the FAIRification of more protocols, MICHA has the potential to become a standard workflow for annotating and cataloguing chemosensitivity experiments.
Figure 4.
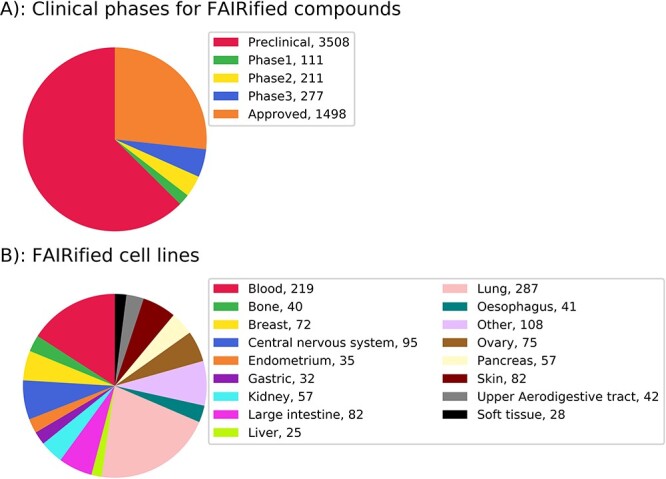
(A) Clinical phase of FAIRified compounds. (B) Tissue types for FAIRified cell lines.
With the help of MICHA platform, all these COVID-19- and cancer-related drug screening protocols are freely accessible to the users (Findable and Accessible). Moreover, these protocols can be accessed programmatically using MICHA API, which makes it possible for in silico models to programmatically access MICHA to obtain compound information such as protein targets and physiochemical properties and use this information for novel drug–target predictions (Interoperable). The MICHA drug screening protocols can be considered as a reference for the experimental design of future drug screening studies as well as serve as a source of information to evaluate the experimental reproducibility (Reusable). MICHA is also indexed at https://fairsharing.org/ to be accepted as a potential tool for chemosensitivity data FAIRification.
Conclusion
Chemosensitivity assay screening has been increasingly utilized for preclinical drug discovery and clinical trial optimization. However, chemosensitivity assays often lack sufficient annotation to make the data FAIR, which has become a limiting factor for supporting its clinical translation. To improve the assay annotation, web portals that facilitate information retrieval from different assay components are critically needed. To address this issue, we have recently launched MICHA as a web server for the annotation of chemosensitivity screens that covers critical information including 1) compounds 2) samples 3) reagent protocols and 4) data processing methods.
The novelty of MICHA is 2-folds. First, it provides a protocol for defining MI for annotating drug sensitivity assays. Second, it provides software tools to implement such a protocol. To enable an effective data annotation pipeline, comprehensive compound–target profiles are deposited to the MICHA database for more than 800K compounds. These high-quality pharmacological data shall help improve the annotation on the mechanisms of action not only for approved drugs but also for investigational and preclinical compounds. Furthermore, the target profiles at the druggable genome scale provide more information on the polypharmacological effects, which might lead to new opportunities for drug repositioning [34]. To facilitate the data retrieval, the API in MICHA is highly optimized such that it can return target profiles for hundreds of compounds within seconds.
With the MICHA web portal, we have FAIRified major drug sensitivity screening protocols from five cancer studies and six recent COVID-19 studies, serving as the first instances of the catalogue. Comparing these deeply curated assay protocols should allow a more systematic analysis of data reproducibility. With the FAIR-compliant data resources and tools to deliver content standards and ontology services, MICHA will ensure the characterization of critical assay components, allow the FAIRification and cataloguing of drug sensitivity studies and support the downstream analysis toward clinical translation. We invite the drug discovery community to use MICHA for annotating their drug sensitivity assays to improve the knowledge sharing, which shall ultimately lead to a bigger impact in translational medicine.
Key Points
We proposed a novel workflow called MICHA (https://micha-protocol.org) for the FAIRification of drug sensitivity screening protocols.
MICHA provides an integrated platform to obtain drug screening assay annotations, drug–target profiles and other pharmacological information in an easy and fast manner.
MICHA FAIRified drug screening protocols related to cancer and COVID-19, which are made comparable and informative for designing new experiments.
Data availability statement
FAIRified protocols by MICHA are freely accessible using MICHA API as well as using a standalone file at https://micha-protocol.org/download/index. Compound–target profiles can be retrieved by the MICHA annotation pipeline or programmatically using the API. Furthermore, compound–target profiles can also be downloaded as a standalone file at https://micha-protocol.org/download/index.
Supplementary Material
Acknowledgements
We thank the CSC-IT Center for Science Finland for providing database storage and computing resources.
Ziaurrehman Tanoli is senior researcher at University of Helsinki. His research is mostly focused on computational drug repurposing. He is also developing bioinformatics tools for drug–target interactions.
Jehad Aldahdooh is a PhD student at University of Helsinki. He is developing text mining applications for drug–target interactions.
Farhan Alam is PhD student at University of Helsinki.
Yinyin Wang is a PhD student at University of Helsinki. She is developing methods for network pharmacology modeling for herb medicine.
Umair Seemab is PhD student at University of Helsinki. He is working on NGS and single-cell sequencing.
Maddalena Fratelli is head of Phamacogenomics Unit at Mario Negri Institute for Pharmacological Research. She is working on genomic and transcriptomic systems for the study of drug action and resistance.
Petr Pavlis is senior developer at Institute of Molecular and Translational Medicine. He is developing bioinformatics applications.
Marian Hajduch is founding director of the Institute of Molecular and Translational Medicine. His research interests are in molecular and translational medicine.
Florence Bietrix is the head of operations at EATRIS. Her research interests are to develop new therapeutic targets for atherosclerosis and nonalcoholic steatohepatitis.
Philip Gribbon is head of Discovery Research at Fraunhofer Institute for Translational Medicine and Pharmacology.
Andrea Zaliani is senior bioinformatics scientist at Fraunhofer Institute for Translational Medicine and Pharmacology. He has expertise in pharmaceutical research and development.
Matthew D. Hall is biology group leader at National Center for Advancing Translational Sciences. He optimizes both biochemical and cell-based assays for automated, small molecule, high-throughput screening in collaboration with NIH.
Min Shen has extensive experience in cheminformatics and computational chemistry through her work at NCATS.
Kyle Brimacombe is biochemist at National Center for Advancing Translational Sciences.
Evgeny Kulesskiy is senior researcher at Institute for Molecular Medicine Finland. He is working on drug sensitivity and resistance testing.
Jani Saarela is head of High Throughput Unit at Institute for Molecular Medicine Finland. He is working on drug sensitivity and resistance testing.
Krister Wennerberg is professor at Biotech Research & Innovation Centre. He is doing research on chemical systems biology applications.
Markus Vähä-Koskela is senior researcher at Institute for Molecular Medicine Finland. He is doing research in areas such as: onco-immunology, immunotherapy and translational cancer medicine.
Jing Tang is assistant professor at University of Helsinki. He is working on mathematical, statistical and informatics tools to tackle biomedical questions.
Contributor Information
Ziaurrehman Tanoli, Research Program in Systems Oncology, Faculty of medicine, University of Helsinki, Finland.
Jehad Aldahdooh, Research Program in Systems Oncology, Faculty of medicine, University of Helsinki, Finland.
Farhan Alam, Research Program in Systems Oncology, Faculty of medicine, University of Helsinki, Finland.
Yinyin Wang, Research Program in Systems Oncology, Faculty of medicine, University of Helsinki, Finland.
Umair Seemab, Research Program in Systems Oncology, Faculty of medicine, University of Helsinki, Finland.
Maddalena Fratelli, Istituto di Ricerche Farmacologiche Mario Negri IRCCS, Italy.
Petr Pavlis, Institute of Molecular and Translational Medicine, Czech.
Marian Hajduch, Institute of Molecular and Translational Medicine, Czech.
Florence Bietrix, European Infrastructure for Translational Medicine, UK.
Philip Gribbon, Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany.
Andrea Zaliani, Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany.
Matthew D Hall, National Center for Advancing Translational Sciences, USA.
Min Shen, National Center for Advancing Translational Sciences, USA.
Kyle Brimacombe, National Center for Advancing Translational Sciences, USA.
Evgeny Kulesskiy, Institute for Molecular Medicine Finland, University of Helsinki, Finland.
Jani Saarela, Institute for Molecular Medicine Finland, University of Helsinki, Finland.
Krister Wennerberg, Biotech Research & Innovation Centre (BRIC), University of Copenhagen, Denmark.
Markus Vähä-Koskela, Institute for Molecular Medicine Finland, University of Helsinki, Finland.
Jing Tang, Research Program in Systems Oncology, Faculty of medicine, University of Helsinki, Finland.
Funding
This work is supported by the European Union's Horizon 2020 research and innovation programme under EOSC-Life (Grant agreement No. 824087) and the European Research Council (ERC) (Grant agreement No. 716063), and by the Academy of Finland (Grant agreement No. 317680).
References
- 1. Hatzis C, Bedard PL, Birkbak NJ, et al. Enhancing reproducibility in cancer drug screening: how do we move forward? Cancer Res 2014;74:4016–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haverty PM, Lin E, Tan J, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature 2016;533:333–7. [DOI] [PubMed] [Google Scholar]
- 3. Larsson P, Engqvist H, Biermann J, et al. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci Rep 2020;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor CF, Field D, Sansone S-A, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol 2008;26:889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orchard S, Al-Lazikani B, Bryant S, et al. Minimum information about a bioactive entity (MIABE). Nat Rev Drug Discov 2011;10:661–9. [DOI] [PubMed] [Google Scholar]
- 6. Abeyruwan S, Vempati UD, Küçük-McGinty H, et al. Evolving BioAssay ontology (BAO): modularization, integration and applications. J Biomed Semantics 2014;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendez D, Gaulton A, Bento AP, et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 2019;47:D930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iorio F, Knijnenburg TA, Vis DJ, et al. A landscape of pharmacogenomic interactions in cancer. Cell 2016;166:740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seashore-Ludlow B, Rees MG, Cheah JH, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov 2015;5:1210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weston S, Coleman CM, Haupt R, et al. Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J Virol 2020;94:e01218–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Touret F, Gilles M, Barral K, et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep 2020;10:13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeon S, Ko M, Lee J, et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother 2020;64:e00819–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brimacombe KR, Zhao T, Eastman RT, et al. An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv 2020;2020.06.04:135046. [Google Scholar]
- 16. Ellinger B, Bojkova D, Zaliani A, et al. A SARS-CoV-2 cytopathicity dataset generated by high-content screening of a large drug repurposing collection. Sci Data 2021;8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He L, Kulesskiy E, Saarela J, et al. Methods for high-throughput drug combination screening and synergy scoring. Cancer Syst Biol 2018;351–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bolis M, Garattini E, Paroni G, et al. Network-guided modeling allows tumor-type independent prediction of sensitivity to all-trans-retinoic acid. Ann Oncol 2017;28:611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanoli Z, Alam Z, Vähä-Koskela M, et al. Drug target commons 2.0: a community platform for systematic analysis of drug–target interaction profiles. Database 2018;2018:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang J, Tanoli Z-R, Ravikumar B, et al. Drug target commons: a community effort to build a consensus Knowledge Base for drug-target interactions. Cell Chem Biol 2018;25:224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilson MK, Liu T, Baitaluk M, et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 2016;44:D1045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaulton A, Hersey A, Nowotka M, et al. The ChEMBL database in 2017. Nucleic Acids Res 2016;45:D945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexander SP, Kelly E, Marrion NV, et al. The concise guide to pharmacology 2017/18: overview. Br J Pharmacol 2017;174:S1–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner AH, Coffman AC, Ainscough BJ, et al. DGIdb 2.0: mining clinically relevant drug–gene interactions. Nucleic Acids Res 2016;44:D1036–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006;34:D668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech 2018;29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cichonska A, Pahikkala T, Szedmak S, et al. Learning with multiple pairwise kernels for drug bioactivity prediction. Bioinformatics 2018;34:i509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cichonska A, Ravikumar B, Parri E, et al. Computational-experimental approach to drug-target interaction mapping: a case study on kinase inhibitors. PLoS Comput Biol 2017;13:e1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeng X, Zhu S, Hou Y, et al. Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest. Bioinformatics 2020;36:2805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang J, Gautam P, Gupta A, et al. Network pharmacology modeling identifies synergistic Aurora B and ZAK interaction in triple-negative breast cancer. NPJ Syst Biol Appl 2019;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng F, Kovács IA, Barabási AL. Network-based prediction of drug combinations. Nat Commun [Internet]. 2019;10:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gross AM, Wolters PL, Dombi E, et al. Selumetinib in children with inoperable plexiform neurofibromas. N Engl J Med 2020;382:1430–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res 2012;18:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanoli Z, Seemab U, Scherer A, et al. Exploration of databases and methods supporting drug repurposing: a comprehensive survey. Brief Bioinform 2021;22:1656–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FAIRified protocols by MICHA are freely accessible using MICHA API as well as using a standalone file at https://micha-protocol.org/download/index. Compound–target profiles can be retrieved by the MICHA annotation pipeline or programmatically using the API. Furthermore, compound–target profiles can also be downloaded as a standalone file at https://micha-protocol.org/download/index.


