Abstract
RNA structural elements called pseudoknots are involved in various biological phenomena including ribosomal frameshifts. Because it is infeasible to construct an efficiently computable secondary structure model including pseudoknots, secondary structure prediction methods considering pseudoknots are not yet widely available. We developed IPknot, which uses heuristics to speed up computations, but it has remained difficult to apply it to long sequences, such as messenger RNA and viral RNA, because it requires cubic computational time with respect to sequence length and has threshold parameters that need to be manually adjusted. Here, we propose an improvement of IPknot that enables calculation in linear time by employing the LinearPartition model and automatically selects the optimal threshold parameters based on the pseudo-expected accuracy. In addition, IPknot showed favorable prediction accuracy across a wide range of conditions in our exhaustive benchmarking, not only for single sequences but also for multiple alignments.
Keywords: RNA secondary structure prediction, pseudoknots, integer programming
Introduction
Genetic information recorded in DNA is transcribed into RNA, which is then translated into protein to fulfill its function. In other words, RNA is merely an intermediate product for the transmission of genetic information. This type of RNA is called messenger RNA (mRNA). However, many RNAs that do not fit into this framework have been discovered more recently. For example, transfer RNA and ribosomal RNA, which play central roles in the translation mechanism, nucleolar small RNA, which guides the modification sites of other RNAs, and microRNA, which regulates gene expression, have been discovered. Thus, it has become clear that RNAs other than mRNAs are involved in various biological phenomena. Because these RNAs do not encode proteins, they are called non-coding RNAs. In contrast to DNA, which forms a double-stranded structure in vivo, RNA is often single-stranded and is thus unstable when intact. In the case of mRNA, the cap structure at the 5′ end and the poly-A strand at the 3′ end protect it from degradation. On the other hand, for other RNAs that do not have such structures, single-stranded RNA molecules bind to themselves to form three-dimensional structures and ensure their stability. Also, as in the case of proteins, RNAs with similar functions have similar three-dimensional structures, and it is known that there is a strong association between function and structure. The determination of RNA three-dimensional (3D) structure can be performed by X-ray crystallography, nuclear magnetic resonance, cryo-electron microscopy, and other techniques. However, it is difficult to apply these methods on a large scale owing to difficulties associated with sequence lengths, resolution and cost. Therefore, RNA secondary structure, which is easier to model, is often computationally predicted instead. RNA secondary structure refers to the set of base pairs consisting of Watson–Crick base pairs (A–U, G–C) and wobble base pairs (G–U) that form the backbone of the 3D structure.
RNA secondary structure prediction is conventionally based on thermodynamic models, which predict the secondary structure with the minimum free energy (MFE) among all possible secondary structures. Popular methods based on thermodynamic models include mfold [1], RNAfold [2], and RNAstructure [3]. Recently, RNA secondary structure prediction methods based on machine learning have also been developed. These methods train alternative parameters to the thermodynamic parameters by taking a large number of pairs of RNA sequences and their reference secondary structures as training data. The following methods fall under the category of methods that use machine learning: CONTRAfold [4], ContextFold [5], SPOT-RNA [6] and MXfold2 [7]. However, from the viewpoint of computational complexity, most approaches do not support the prediction of secondary structures that include pseudoknot substructures.
Pseudoknots are one of the key topologies occurring in RNA secondary structures. The pseudoknot structure is a structure in which some bases inside of a loop structure form base pairs with bases outside of the loop (e.g. Figure 1A). In other words, it is said to have a pseudoknot structure if there exist base pairs that are crossing each other by connecting bases of base pairs with arcs, as shown in Figure 1B. The pseudoknot structure is known to be involved in the regulation of translation and splicing, and ribosomal frameshifts [8–10]. The results of sequence analysis suggest that the hairpin loops, which are essential building blocks of the pseudoknots, first appeared in the evolutionary timescale [11], and then the pseudoknots were configured, resulting in gaining those functions. We therefore conclude that pseudoknots should not be excluded from the modeling of RNA secondary structures.
Figure 1 .
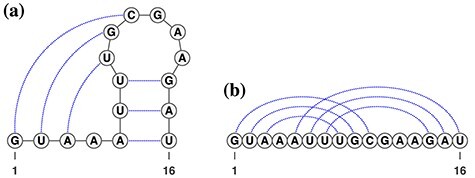
(A) A typical psudoknot structure. The dotted lines represent base pairs. (B) A linear presentation of the pseudoknot.
The computational complexity required for MFE predictions of an arbitrary pseudoknot structure has been proven to be NP-hard [12, 13]. To address this, dynamic programming-based methods that require polynomial time (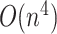 –
–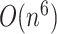 for sequence length
for sequence length 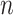 ) to exactly compute the restricted complexity of pseudoknot structures [12–16] and heuristics-based fast computation methods [17–20] have been developed.
) to exactly compute the restricted complexity of pseudoknot structures [12–16] and heuristics-based fast computation methods [17–20] have been developed.
We previously developed IPknot [21], a fast heuristic-based method for predicting RNA secondary structures including pseudoknots. IPknot decomposes a secondary structure with pseudoknots into several pseudoknot-free substructures and predicts the optimal secondary structure using integer programming (IP) based on maximization of expected accuracy (MEA) under the constraints that each substructure must satisfy. The threshold cut technique, which is naturally derived from MEA, enables IPknot to perform much faster calculations with nearly comparable prediction accuracy relative to other methods. However, because the MEA-based score uses base pairing probability without considering pseudoknots, which requires a calculation time that increases cubically with sequence length, it is difficult to use for secondary structure prediction of sequences that exceed 1000 bases, even when applying a threshold cut technique. Furthermore, as the prediction accuracy can drastically change depending on the thresholds determined in advance for each pseudoknot-free substructure, thresholds must be carefully determined.
To address the limitations of IPknot, we implemented the following two improvements to the method. The first is the use of LinearPartition [22] to calculate base pairing probabilities. LinearPartition can calculate the base pairing probability, with linear computational complexity with respect to sequence length, using the beam search technique. By employing the LinearPartition model, IPknot is able to predict secondary structures while considering pseudoknots for long sequences, including mRNA, lncRNA and viral RNA. The other improvement is the selection of thresholds based on pseudo-expected accuracy, which was originally developed by Hamada et al. [23]. We show that the pseudo-expected accuracy is correlated with the ‘true’ accuracy, and by choosing thresholds for each sequence based on the pseudo-expected accuracy, we can select a nearly optimal secondary structure prediction.
Materials and Methods
Given an RNA sequence 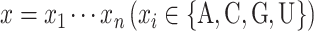 , its secondary structure is represented by a binary matrix
, its secondary structure is represented by a binary matrix 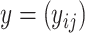 , where
, where 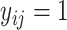 if
if 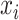 and
and 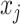 form a base pair and otherwise
form a base pair and otherwise 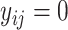 . Let
. Let 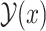 be a set of all possible secondary structures of
be a set of all possible secondary structures of 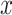 including pseudoknots. We assume that
including pseudoknots. We assume that 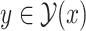 can be decomposed into a set of pseudoknot-free substructures
can be decomposed into a set of pseudoknot-free substructures 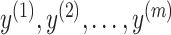 , such that
, such that 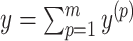 . In order to guarantee the uniqueness of the decomposition, the following conditions should be satisfied: (i)
. In order to guarantee the uniqueness of the decomposition, the following conditions should be satisfied: (i) 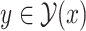 should be decomposed into mutually exclusive sets; that is, for all
should be decomposed into mutually exclusive sets; that is, for all 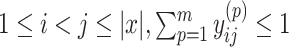 ; (ii) every base pair in
; (ii) every base pair in 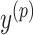 should be pseudoknotted with at least one base pair in
should be pseudoknotted with at least one base pair in 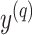 for
for 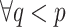 .
.
Maximizing expected accuracy
One of the most promising techniques for predicting RNA secondary structures is the MEA-based approach [4, 24]. First, we define a gain function of prediction 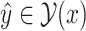 with regard to the correct secondary structure
with regard to the correct secondary structure 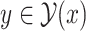 as
as
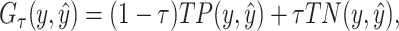 |
(1) |
where 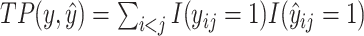 is the number of true positive base pairs,
is the number of true positive base pairs, 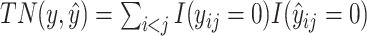 is the number of true negative base pairs, and
is the number of true negative base pairs, and 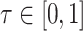 is a balancing parameter between true positives and true negatives. Here,
is a balancing parameter between true positives and true negatives. Here, 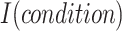 is the indicator function that takes a value of 1 or 0 depending on whether the
is the indicator function that takes a value of 1 or 0 depending on whether the 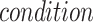 is true or false.
is true or false.
Our objective is to find a secondary structure that maximizes the expectation of the gain function (1) under a given probability distribution over the space 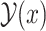 of pseudoknotted secondary structures, as follows:
of pseudoknotted secondary structures, as follows:
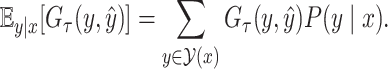 |
(2) |
Here, 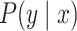 is a probability distribution of RNA secondary structures including pseudoknots.
is a probability distribution of RNA secondary structures including pseudoknots.
Because the calculation of the expected gain function (2) is intractable for arbitrary pseudoknots, we approximate Eq. (2) by the sum of the expected gain function for decomposed pseudoknot-free substructures 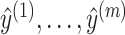 for
for 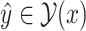 such that
such that 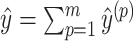 , and thus, we find a pseudoknotted structure
, and thus, we find a pseudoknotted structure 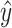 and its decomposition
and its decomposition 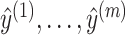 that maximize
that maximize
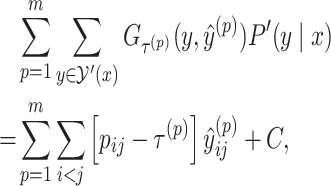 |
(3) |
where 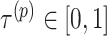 is a balancing parameter between true positives and true negatives for a level
is a balancing parameter between true positives and true negatives for a level 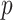 , and
, and 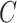 is a constant independent of
is a constant independent of 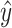 . The base pairing probability
. The base pairing probability 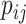 is the probability that the bases
is the probability that the bases 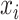 and
and 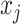 form a base pair, which is defined as
form a base pair, which is defined as
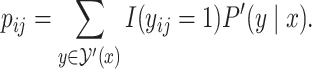 |
(4) |
See Section S1 in Supplementary Information for the derivation. Notably, it is no longer necessary to consider the base pairs whose probabilities are at most the threshold 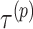 , which we refer to as the threshold cut.
, which we refer to as the threshold cut.
We can choose 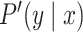 , a probability distribution over a set
, a probability distribution over a set 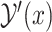 of secondary structures without pseudoknots, from among several options. Instead of using a probability distribution with pseudoknots, we can employ a probability distribution without pseudoknots, such as the McCaskill model [25] and the CONTRAfold model [4], whose computational complexity is
of secondary structures without pseudoknots, from among several options. Instead of using a probability distribution with pseudoknots, we can employ a probability distribution without pseudoknots, such as the McCaskill model [25] and the CONTRAfold model [4], whose computational complexity is 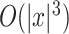 for time and
for time and 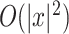 for space. Alternatively, the LinearPartition model [22], which is
for space. Alternatively, the LinearPartition model [22], which is 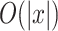 in both time and space, enables us to predict the secondary structure of sequences much longer than 1000 bases.
in both time and space, enables us to predict the secondary structure of sequences much longer than 1000 bases.
IP formulation
We can formulate our problem described in the previous section as the following IP problem:
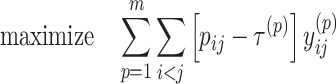 |
(5) |
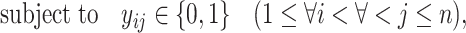 |
(6) |
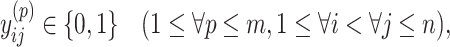 |
(7) |
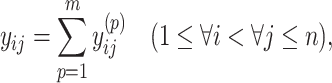 |
(8) |
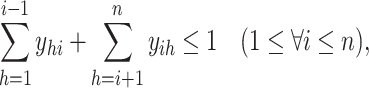 |
(9) |
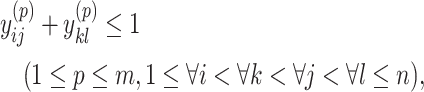 |
(10) |
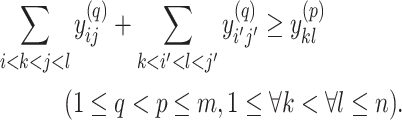 |
(11) |
Because Equation (5) is an instantiation of the approximate estimator (3) and the threshold cut technique is applicable to Eq. (3), the base pairs 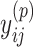 whose base pairing probabilities
whose base pairing probabilities 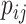 are larger than
are larger than 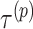 need to be considered. The number of variables
need to be considered. The number of variables 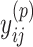 that should be considered is at most
that should be considered is at most 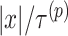 because
because 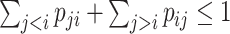 for
for 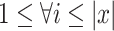 . Constraint (9) means that each base
. Constraint (9) means that each base 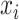 is paired with at most one base. Constraint (10) disallows pseudoknots within the same level
is paired with at most one base. Constraint (10) disallows pseudoknots within the same level 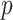 . Constraint (11) ensures that each base pair at level
. Constraint (11) ensures that each base pair at level 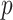 is pseudoknotted with at least one base pair at every lower level
is pseudoknotted with at least one base pair at every lower level 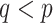 to guarantee the uniqueness of the decomposition
to guarantee the uniqueness of the decomposition 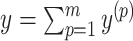 .
.
Pseudo-expected accuracy
To solve the IP problem (5)–(11), we are required to choose the set of thresholds for each level 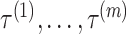 , each of which is a balancing parameter between true positives and true negatives. However, it is not easy to obtain the best set of
, each of which is a balancing parameter between true positives and true negatives. However, it is not easy to obtain the best set of 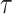 values for any sequence beforehand. Therefore, we employ an approach originally proposed by Hamada et al. [23], which chooses a parameter set for each sequence among several parameter sets that predicts the best secondary structure in terms of an approximation of the expected accuracy (called pseudo-expected accuracy) and makes the prediction by the best parameter set the final prediction.
values for any sequence beforehand. Therefore, we employ an approach originally proposed by Hamada et al. [23], which chooses a parameter set for each sequence among several parameter sets that predicts the best secondary structure in terms of an approximation of the expected accuracy (called pseudo-expected accuracy) and makes the prediction by the best parameter set the final prediction.
The accuracy of a predicted RNA secondary structure 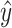 against a reference structure
against a reference structure 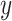 is evaluated using the following measures:
is evaluated using the following measures:
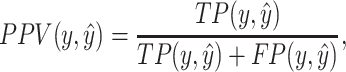 |
(12) |
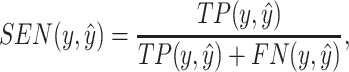 |
(13) |
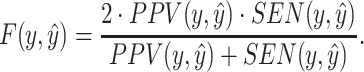 |
(14) |
Here, 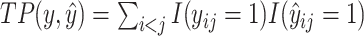 ,
, 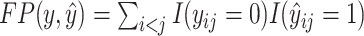 and
and 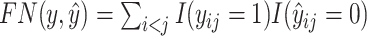 . To estimate the accuracy of the predicted secondary structure
. To estimate the accuracy of the predicted secondary structure 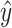 without knowing the true secondary structure
without knowing the true secondary structure 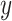 , we take an expectation of
, we take an expectation of 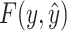 over the distribution of
over the distribution of 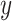 :
:
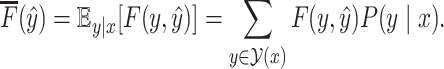 |
(15) |
However, this calculation is intractable because the number of  increases exponentially with the length of sequence
increases exponentially with the length of sequence  . Alternatively, we first calculate expected
. Alternatively, we first calculate expected  and
and 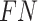 as follows:
as follows:
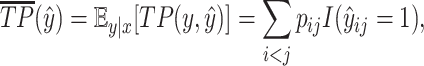 |
(16) |
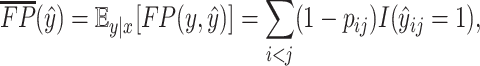 |
(17) |
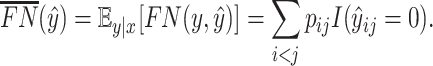 |
(18) |
Then, we approximate 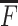 by calculating Equation (14) using
by calculating Equation (14) using 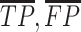 , and
, and 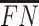 instead of
instead of 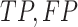 and
and 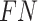 , respectively.
, respectively.
In addition to the original pseudo-expected accuracy described above, we introduce the pseudo-expected accuracy for crossing base pairs to predict pseudoknotted structures. Prediction of secondary structures including pseudoknots depends on both the conventional prediction accuracy of base pairs described above and the accuracy of crossing base pairs. A crossing base pair is a base pair 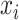 and
and 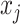 such that there exists another base pair
such that there exists another base pair 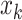 and
and 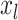 that is crossing the base pair
that is crossing the base pair 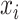 and
and 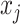 ; that is,
; that is, 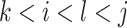 or
or 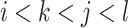 . We define the expectations of true positives, false positives and false negatives for crossing base pairs as follows:
. We define the expectations of true positives, false positives and false negatives for crossing base pairs as follows:
 |
(19) |
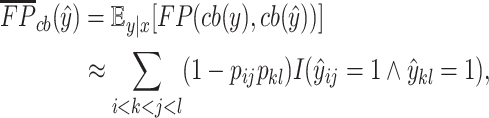 |
(20) |
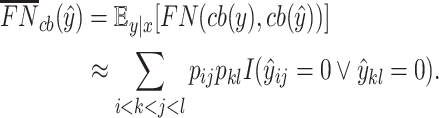 |
(21) |
Here, 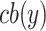 is an
is an 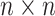 binary matrix, whose
binary matrix, whose 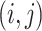 -element is
-element is 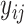 itself if there exists
itself if there exists 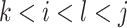 or
or 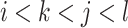 such that
such that 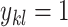 , and 0 otherwise. Then, we calculate the pseudo-expected
, and 0 otherwise. Then, we calculate the pseudo-expected  -value for crossing base pairs
-value for crossing base pairs 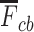 using Equation (14) with
using Equation (14) with 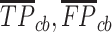 and
and 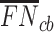 instead of
instead of 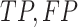 and
and 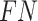 , respectively. Equations (19)–(21) require
, respectively. Equations (19)–(21) require 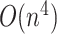 for naive calculations, but can be reduced to acceptable computational time by utilizing the threshold cut technique.
for naive calculations, but can be reduced to acceptable computational time by utilizing the threshold cut technique.
We predict secondary structures 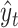 (
(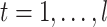 ) for several threshold parameters
) for several threshold parameters 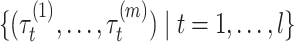 . Then, we calculate their pseudo-expected accuracy
. Then, we calculate their pseudo-expected accuracy 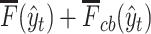 and choose the secondary structure
and choose the secondary structure 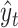 that maximizes the pseudo-expected accuracy as the final prediction.
that maximizes the pseudo-expected accuracy as the final prediction.
Common secondary structure prediction
The average of the base pairing probability matrices for each sequence in an alignment has been used to predict the common secondary structure for the alignment [26, 27]. Let 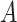 be an alignment of RNA sequences that contains
be an alignment of RNA sequences that contains 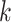 sequences and
sequences and 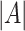 denote the number of columns in
denote the number of columns in 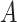 . We calculate the base pairing probabilities of an individual sequence
. We calculate the base pairing probabilities of an individual sequence 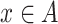 as
as
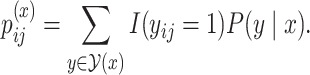 |
(22) |
The averaged base pairing probability matrix is defined as
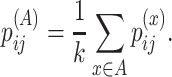 |
(23) |
The common secondary structure of the alignment 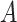 can be calculated in the same way by replacing
can be calculated in the same way by replacing 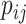 in Equations (5) with
in Equations (5) with 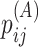 . While the common secondary structure prediction based on the average base pairing probability matrix has been implemented in the previous version of IPknot [21], the present version employs the LinearPartition model, which enables the calculation linearly with respect to the alignment length.
. While the common secondary structure prediction based on the average base pairing probability matrix has been implemented in the previous version of IPknot [21], the present version employs the LinearPartition model, which enables the calculation linearly with respect to the alignment length.
Implementation
Our method has been implemented as the newest version of a program called IPknot. In addition to the McCaskil model [25] and CONTRAfold model [4], which were already integrated into the previous version of IPknot, the LinearPartition model [22] is also supported as a probability distribution for secondary structures. To solve IP problems, the GNU Linear Programming Kit (GLPK; http://www.gnu.org/software/glpk/), Gurobi Optimizer (http://gurobi.com/) or IBM CPLEX Optimizer (https://www.ibm.com/analytics/cplex-optimizer) can be employed.
Datasets
To evaluate our algorithm, we performed computational experiments on several datasets. We employed RNA sequences extracted from the bpRNA-1m dataset [28], which is based on Rfam 12.2 [29], and the comparative RNA web dataset [30] with 2588 families. In addition, we built a dataset that includes families from the most recent Rfam database, Rfam 14.5 [31]. Since the release of Rfam 12.2, the Rfam project has actively collected about 1400 RNA families, including families detected by newly developed techniques. We extracted these newly discovered families. To limit bias in the training data, sequences with higher than 80% sequence identity with the sequence subsets S-Processed-TRA from RNA STRAND [32] and TR0 from bpRNA-1m [28], which are the training datasets for CONTRAfold and SPOT-RNA, respectively, were removed using CD-HIT-EST-2D [33]. We then removed redundant sequences using CD-HIT-EST [33], with a cutoff threshold of 80% sequence identity.
For the prediction of common secondary structures, the sequence selected by the above method was used as a seed, and 1–9 sequences of the same Rfam family and with high sequence identity (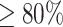 ) with the seed sequence were randomly selected to create an alignment. Common secondary structure prediction was performed on the reference alignments from Rfam and the alignments calculated by MAFFT [34]. Because there are sequences from bpRNA-1m that do not have Rfam reference alignments, only sequences from Rfam 14.5 were tested for common secondary structure prediction. To capture the accuracy of the common secondary structure prediction, the accuracy for the seed sequence is shown.
) with the seed sequence were randomly selected to create an alignment. Common secondary structure prediction was performed on the reference alignments from Rfam and the alignments calculated by MAFFT [34]. Because there are sequences from bpRNA-1m that do not have Rfam reference alignments, only sequences from Rfam 14.5 were tested for common secondary structure prediction. To capture the accuracy of the common secondary structure prediction, the accuracy for the seed sequence is shown.
A summary of the dataset created and utilized is shown in Table 1.
Table 1.
Datasets used in our experiments. Each element of the table shows the number of sequences
| Pseudoknot-free | Pseudoknotted | |||||
|---|---|---|---|---|---|---|
| Short | Medium | Long | Short | Medium | Long | |
| Length (nt) | (12–150) | (151–500) | (501–4381) | (12–150) | (151–500) | (501–4381) |
| (Single) | ||||||
| bpRNA-1m | 1971 | 514 | 420 | 125 | 162 | 245 |
| Rfam 14.5 | 6299 | 723 | 9 | 1692 | 477 | 151 |
| (Multiple) | ||||||
| Rfam 14.5 | 5118 | 554 | 4 | 1692 | 477 | 151 |
Results
Effectiveness of pseudo-expected accuracy
First, to show the effectiveness of the automatic selection from among thresholds 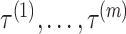 based on the pseudo-expected accuracy, Figure 2 and Table S1 in Supplementary Information show the prediction accuracy on the dataset of short sequences (
based on the pseudo-expected accuracy, Figure 2 and Table S1 in Supplementary Information show the prediction accuracy on the dataset of short sequences (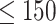 nt) using automatic selection and manual selection of the threshold
nt) using automatic selection and manual selection of the threshold  values. For IPknot, we fixed the number of decomposed sets of secondary substructures
values. For IPknot, we fixed the number of decomposed sets of secondary substructures 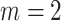 , and varied threshold parameters
, and varied threshold parameters 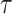 values for base pairing probability in such a way that
values for base pairing probability in such a way that 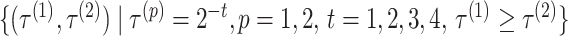 . In IPknot with pseudo-expected accuracy, the best secondary structure in the sense of pseudo-expected
. In IPknot with pseudo-expected accuracy, the best secondary structure in the sense of pseudo-expected 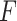 is selected from the same range of
is selected from the same range of 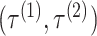 for each sequence. For these variants of IPknot, the LinearPartition model with CONTRAfold parameters (LinearPartition-C) was used to calculate base pairing probabilities. In addition, we compared the prediction accuracy of IPknot with that of ThreshKnot [35], which also calculates base pairing probabilities using LinearPartition-C. We used
for each sequence. For these variants of IPknot, the LinearPartition model with CONTRAfold parameters (LinearPartition-C) was used to calculate base pairing probabilities. In addition, we compared the prediction accuracy of IPknot with that of ThreshKnot [35], which also calculates base pairing probabilities using LinearPartition-C. We used 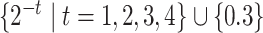 as the threshold parameter
as the threshold parameter 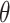 for ThreshKnot because the default threshold parameter of ThreshKnot is
for ThreshKnot because the default threshold parameter of ThreshKnot is 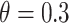 . IPknot with threshold parameters of
. IPknot with threshold parameters of 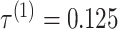 and
and 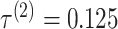 had the highest prediction accuracy of
had the highest prediction accuracy of 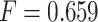 . IPknot with pseudo-expected accuracy has a prediction accuracy of
. IPknot with pseudo-expected accuracy has a prediction accuracy of 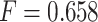 , which is comparable to the highest accuracy obtained. ThreshKnot with a threshold of 0.25 has an accuracy of
, which is comparable to the highest accuracy obtained. ThreshKnot with a threshold of 0.25 has an accuracy of 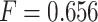 , which is also comparable to the best accuracy obtained.
, which is also comparable to the best accuracy obtained.
Figure 2 .
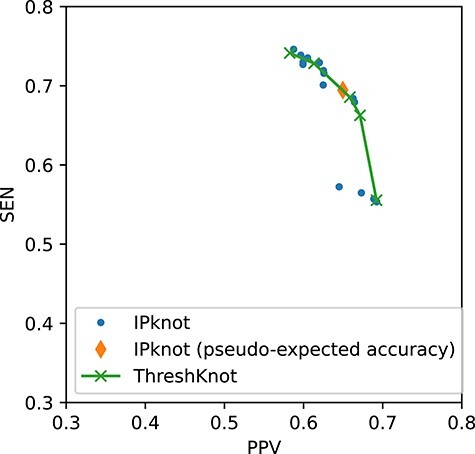
PPV–SEN plot of IPknot and ThreshKnot for short RNA sequences (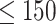 nt).
nt).
The pseudo-expected 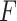 -value and “true”
-value and “true” 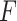 -value are relatively highly correlated (Spearman correlation coefficient
-value are relatively highly correlated (Spearman correlation coefficient 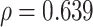 ), indicating that the selection of predicted secondary structure using pseudo-expected accuracy works well.
), indicating that the selection of predicted secondary structure using pseudo-expected accuracy works well.
While the accuracy of the prediction of the entire secondary structure has already been considered, as shown in Figure 2, for the prediction of secondary structures with pseudoknots, it is necessary to evaluate the prediction accuracy focused on the crossing base pairs. In terms of prediction accuracy limited to only crossing base pairs, IPknot with pseudo-expected accuracy yielded 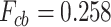 , while the highest accuracy achieved by IPknot with the threshold parameters and ThreshKnot was considerably lower at
, while the highest accuracy achieved by IPknot with the threshold parameters and ThreshKnot was considerably lower at 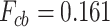 and
and 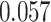 , respectively (See Table S1 in Supplementary Information). We can observe the similar tendency to the above in Figures S1 and S2, and Tables S2 and S3 in Supplementary Information for medium (151–500 nt) and long (
, respectively (See Table S1 in Supplementary Information). We can observe the similar tendency to the above in Figures S1 and S2, and Tables S2 and S3 in Supplementary Information for medium (151–500 nt) and long (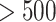 nt) sequences. These results suggest that prediction of crossing base pairs is improved by selecting the predicted secondary structure while considering both the pseudo-expected accuracy of the entire secondary structure and the pseudo-expected accuracy of the crossing base pairs.
nt) sequences. These results suggest that prediction of crossing base pairs is improved by selecting the predicted secondary structure while considering both the pseudo-expected accuracy of the entire secondary structure and the pseudo-expected accuracy of the crossing base pairs.
Comparison with previous methods for single RNA sequences
Using our dataset, we compared our algorithm with several previous methods that can predict pseudoknots, including ThreshKnot utilizing LinearPartition (committed on 17 March 2021) [22], Knotty (committed on Mar 28, 2018) [22] and SPOT-RNA (committed on 1 April 2021) [6], and those that can predict only pseudoknot-free structures, including CONTRAfold (version 2.02) [4] and RNAfold in the ViennaRNA package (version 2.4.17) [22]. IPknot has several options for the calculation model for base pairing probabilities, namely the LinearPartition model with CONTRAfold parameters (LinearPartition-C), the LinearPartition model with ViennaRNA parameters (LinearPartition-V), the CONTRAfold model and the ViennaRNA model. In addition, ThreshKnot has two possible LinearPartition models for calculating base pairing probabilities. The other existing methods were tested using the default settings.
We evaluated the prediction accuracy according to the 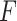 -value as defined by Equation (14) for pseudoknot-free sequences (PKF in Table 2), pseudoknotted sequences (PK in Table 2) and only crossing base pairs (CB in Table 2) by stratifying sequences by length: short (12–150 nt), medium (151–500 nt) and long (500–4381 nt).
-value as defined by Equation (14) for pseudoknot-free sequences (PKF in Table 2), pseudoknotted sequences (PK in Table 2) and only crossing base pairs (CB in Table 2) by stratifying sequences by length: short (12–150 nt), medium (151–500 nt) and long (500–4381 nt).
Table 2.
A comparison of prediction accuracies (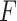 -values) by sequence length for each method
-values) by sequence length for each method
| Length | Short (12–150 nt) | Medium (151–500 nt) | Long (501–4381 nt) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PKF | PK | CB | PKF | PK | CB | PKF | PK | CB | |
| IPknot | |||||||||
| (LinearPartition-C) | 0.681 | 0.552 | 0.258 | 0.492 | 0.482 | 0.128 | 0.433 | 0.428 | 0.061 |
| (LinearPartition-V) | 0.669 | 0.499 | 0.143 | 0.478 | 0.461 | 0.091 | 0.380 | 0.370 | 0.038 |
| (CONTRAfold) | 0.678 | 0.550 | 0.259 | 0.495 | 0.505 | 0.154 | 0.426 | 0.413 | 0.066 |
| (ViennaRNA) | 0.669 | 0.500 | 0.144 | 0.480 | 0.461 | 0.091 | 0.212 | 0.317 | 0.041 |
| ThreshKnot | |||||||||
| (LinearPartition-C) | 0.681 | 0.501 | 0.027 | 0.493 | 0.475 | 0.019 | 0.439 | 0.431 | 0.008 |
| (LinearPartition-V) | 0.669 | 0.484 | 0.033 | 0.481 | 0.456 | 0.026 | 0.383 | 0.372 | 0.014 |
| Knotty | 0.641 | 0.550 | 0.315 | — | — | — | — | — | — |
| SPOT-RNA | 0.658 | 0.621 | 0.322 | 0.462 | 0.479 | 0.127 | — | — | — |
| CONTRAfold | 0.682 | 0.519 | 0.000 | 0.500 | 0.497 | 0.000 | 0.425 | 0.415 | 0.000 |
| RNAfold | 0.668 | 0.472 | 0.000 | 0.474 | 0.442 | 0.000 | 0.361 | 0.347 | 0.000 |
PKF, 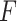 -value for pseudoknot-free sequences; PK,
-value for pseudoknot-free sequences; PK, 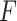 -value for pseudoknotted sequences; CB,
-value for pseudoknotted sequences; CB, 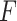 -value of crossing base pairs.
-value of crossing base pairs.
For short sequences, SPOT-RNA archived high accuracy, especially for pseudoknotted sequences. However, a large difference in accuracy between the bpRNA-1m-derived and Rfam 14.5-derived sequences can be observed for SPOT-RNA compared with the other methods (See Tables S4–S9 in Supplementary Information). Notably, bpRNA-1m contains many sequences in the same family as the SPOT-RNA training data, and although we performed filtering based on sequence identity, there is still a concern of overfitting. Knotty can predict structures including pseudoknots with an accuracy comparable to that of SPOT-RNA, but as shown in Figure 3, it can perform secondary structure prediction for only short sequences, owing to its huge computational complexity. Comparing IPknot using the LinearPartition-C and -V models with its counterparts, the original CONTRAfold model and ViennaRNA model achieved comparable accuracy. However, because the computational complexity of the original models is cubic with respect to sequence length, the computational time of the original models increases rapidly as the sequence length exceeds 1500 bases. On the other hand, the computational complexity of the LinearPartition models is linear with respect to sequence length, so the base pairing probabilities can be quickly calculated even when the sequence length exceeds 4000 bases. In addition to calculating the base pairing probabilities, IP calculations are required, but because the number of variables and constraints to be considered can be greatly reduced using the threshold cut technique, the overall execution time is not significantly affected if the sequence length is several thousand bases. Because ThreshKnot, like IPknot, uses the LinearPartition model, it is able to perform fast secondary structure prediction even for long sequences. However, for the prediction accuracy of crossing base pairs, ThreshKnot is even less accurate.
Figure 3 .
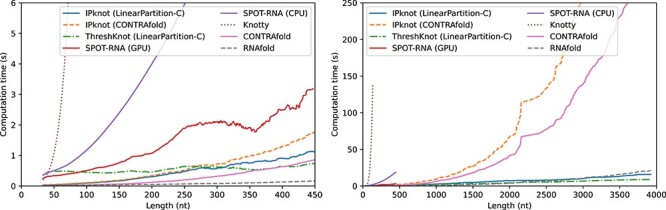
Computational time of each method as a function of sequence length. For SPOT-RNA with GPGPU, we used a Linux workstation with Intel Xeon Gold 6136 and NVIDIA Tesla V100. All other computations were performed on Linux workstations with AMD EPYC 7702. For IPknot, we employed IBM CPLEX Optimizer as the IP solver.
Pseudoknots are found not only in cellular RNAs but also in viral RNAs, performing a variety of functions [8]. Tables S10–S11 in Supplementary Information show the results of the secondary structure prediction by separating the datasets into cellular RNAs and viral RNAs, indicating that there is no significant difference in the prediction accuracy between cellular RNAs and viral RNAs.
Prediction of common secondary structures with pseudoknots
Few methods exist that can perform prediction of common secondary structures including pseudoknots for sequence alignments longer than 1000 bases. Table 3 and Tables S12–S20 in Supplementary Information compare the accuracy of IPknot that employs the LinearPartition model, and RNAalifold in the ViennaRNA package. We performed common secondary structure prediction for the Rfam reference alignment and the alignment calculated by MAFFT, as well as secondary structure prediction of single sequences only for the seed sequence included in the alignment, and evaluated the prediction accuracy for the seed sequence. In most cases, the prediction accuracy improved as the quality of the alignment increased (Single < MAFFT < Reference). IPknot predicts crossing base pairs based on pseudo-expected accuracy, whereas RNAalifold is unable to predict pseudoknots.
Table 3.
A comparison of prediction accuracies (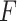 -values) of common secondary structure prediction by sequence alignments for each method
-values) of common secondary structure prediction by sequence alignments for each method
| Reference | MAFFT | Single | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PKF | PK | CB | PKF | PK | CB | PKF | PK | CB | |
| IPknot | |||||||||
| (LinearPartition-C) | 0.765 | 0.616 | 0.220 | 0.732 | 0.585 | 0.218 | 0.718 | 0.548 | 0.227 |
| (LinearPartition-V) | 0.761 | 0.565 | 0.177 | 0.729 | 0.529 | 0.165 | 0.714 | 0.494 | 0.124 |
| RNAalifold | 0.804 | 0.611 | 0.000 | 0.745 | 0.540 | 0.000 | 0.716 | 0.474 | 0.000 |
PKF, 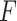 -value for pseudoknot-free sequences; PK,
-value for pseudoknot-free sequences; PK, 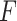 -value for pseudoknotted sequences; CB,
-value for pseudoknotted sequences; CB, 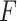 -value of crossing base pairs.
-value of crossing base pairs.
Discussion
Both IPknot and ThreshKnot use the LinearPartition model to calculate base pairing probabilities, and then perform secondary structure prediction using different strategies. ThreshKnot predicts the base pairs 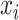 and
and 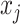 that are higher than a predetermined threshold
that are higher than a predetermined threshold 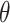 and have the largest
and have the largest 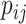 in terms of both
in terms of both 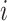 and
and 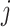 . IPknot predicts the pseudoknot structure with multiple thresholds
. IPknot predicts the pseudoknot structure with multiple thresholds 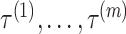 in a hierarchical manner based on IP (5)–(11), and then carefully selects from among these thresholds based on pseudo-expected accuracy. Because both the pseudo-expected accuracy of the entire secondary structure as well as the pseudo-expected accuracy of the crossing base pairs are taken into account, the prediction accuracy of the pseudoknot structure is inferred to be enhanced in IPknot.
in a hierarchical manner based on IP (5)–(11), and then carefully selects from among these thresholds based on pseudo-expected accuracy. Because both the pseudo-expected accuracy of the entire secondary structure as well as the pseudo-expected accuracy of the crossing base pairs are taken into account, the prediction accuracy of the pseudoknot structure is inferred to be enhanced in IPknot.
Because the LinearPartition model uses the same parameters as the CONTRAfold and ViennaRNA packages, there is no significant difference in accuracy between using LinearPartition-C and -V and their counterparts, the CONTRAfold and ViennaRNA models. It has been shown that LinearPartition has no significant effect on accuracy even though it ignores structures whose probability is extremely low owing to its use of beam search, which makes the calculation linear with respect to the sequence length [22]. The LinearPartition model enables IPknot to perform secondary structure prediction including pseudoknots of very long sequences, such as mRNA, lncRNA, and viral RNA.
SPOT-RNA [6], which uses deep learning, showed notable prediction accuracy in our experiments, especially in short sequences containing pseudoknots, with 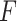 -value of 0.621, which is superior to other methods. However, SPOT-RNA requires considerable computing resources such as GPGPU and long computational time. Furthermore, SPOT-RNA showed a large difference in prediction accuracy between sequences that are close to the training data and those that are not compared with the other methods. Therefore, the situations in which SPOT-RNA can be used are considered to be limited. In contrast, IPknot uses CONTRAfold parameters, which is also based on machine learning, but we did not observe as much overfitting with IPknot as with SPOT-RNA.
-value of 0.621, which is superior to other methods. However, SPOT-RNA requires considerable computing resources such as GPGPU and long computational time. Furthermore, SPOT-RNA showed a large difference in prediction accuracy between sequences that are close to the training data and those that are not compared with the other methods. Therefore, the situations in which SPOT-RNA can be used are considered to be limited. In contrast, IPknot uses CONTRAfold parameters, which is also based on machine learning, but we did not observe as much overfitting with IPknot as with SPOT-RNA.
Approaches that provide an exact solution for limited-complexity pseudoknot structures, such as PKNOTS [14], pknotsRG [15], and Knotty [16], can predict pseudoknot structures with high accuracy but demand a huge amount of computation 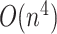 –
–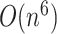 for sequence length
for sequence length 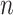 , limiting secondary structure prediction to sequences only up to about 150 bases. On the other hand, IPknot predicts the pseudoknot structure using a fast computational heuristic-based method with the linear time computation, which does not allow us to find an exact solution. Instead, IPknot improves the prediction accuracy of the pseudoknot structure by choosing the best solution from among several solutions based on the pseudo-expected accuracy.
, limiting secondary structure prediction to sequences only up to about 150 bases. On the other hand, IPknot predicts the pseudoknot structure using a fast computational heuristic-based method with the linear time computation, which does not allow us to find an exact solution. Instead, IPknot improves the prediction accuracy of the pseudoknot structure by choosing the best solution from among several solutions based on the pseudo-expected accuracy.
IPknot uses pseudoknot-free algorithms, such as CONTRAfold and ViennaRNA, to calculate base pairing probabilities, and its prediction accuracy of the resulting secondary structure strongly depends on the algorithm used to calculate base pairing probabilities. Therefore, we can expect to improve the prediction accuracy of IPknot by calculating the base pairing probabilities based on state-of-the-art pseudoknot-free secondary structure prediction methods such as MXfold2 [7].
It is well known that common secondary structure prediction from sequence alignments improves the accuracy of secondary structure prediction. However, among the algorithms for predicting common secondary structure including pseudoknots, only IPknot can deal with sequence alignments longer than several thousand bases. In the RNA virus SARS-CoV-2, programmed -1 ribosomal frameshift (-1 PRF), in which a pseudoknot structure plays an important role, has been identified and is attracting attention as a drug target [10]. Because many closely related strains of SARS-CoV-2 have been sequenced, it is expected that structural motifs including pseudoknots, such as -1 PRF, can be found by predicting the common secondary structure from the alignment.
Conclusions
We have developed an improvement to IPknot that enables calculation in linear time by employing the LinearPartition model and automatically selects the optimal threshold parameters based on the pseudo-expected accuracy. LinearPartition can calculate the base pairing probability with linear computational complexity with respect to the sequence length. By employing LinearPartition, IPknot is able to predict the secondary structure considering pseudoknots for long sequences such as mRNA, lncRNA, and viral RNA. By choosing the thresholds for each sequence based on the pseudo-expected accuracy, we can select a nearly optimal secondary structure prediction.
The LinearPartition model realized the predictiction of secondary structures considering pseudoknots for long sequences. However, the prediction accuracy is still not sufficiently high, especially for crossing base pairs. We expect that by learning parameters from long sequences [36], we can achieve high accuracy even for long sequences.
Key Points
We reduced the computational time required by IPknot from cubic to linear with respect to the sequence length by employing the LinearPartition model and enabled the secondary structure prediction including pseudoknots for long RNA sequences such as mRNA, lncRNA, and viral RNA.
We improved the accuracy of secondary structure prediction including pseudoknots by introducing pseudo-expected accuracy not only for the entire base pairs but also for crossing base pairs.
To the best of our knowledge, IPknot is the only method that can perform RNA secondary structure prediction including pseudoknot not only for very long single sequence, but also for very long sequence alignments.
Supplementary Material
Funding
This work was partially supported by a Grant-in-Aid for Scientific Research (B) (No. 19H04210) and Challenging Exploratory Research (No. 19K22897) from the Japan Society for the Promotion of Science (JSPS) to K.S. and a Grant-in-Aid for Scientific Research (C) (Nos. 18K11526 and 21K12109) from JSPS to Y.K.
Acknowledgments
The supercomputer system used for this research was made available by the National Institute of Genetics, Research Organization of Information and Systems.
Kengo Sato is an assistant professor at the Department of Biosciences and Informatics at Keio University, Japan. He received his PhD in Computer Science from Keio University, Japan, in 2003. His research interests include bioinformatics, computational linguistics and machine learning.
Yuki Kato is an assistant professor at Department of RNA Biology and Neuroscience, Graduate School of Medicine, and at Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Japan. His research interests include biological sequence analysis and single-cell genomics.
Contributor Information
Kengo Sato, Department of Biosciences and Informatics, Keio University, 3–14–1 Hiyoshi, Kohoku-ku, Yokohama 223–8522, Japan.
Yuki Kato, Department of RNA Biology and Neuroscience, Graduate School of Medicine, Osaka University, Suita, Osaka 565–0871, Japan; Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Suita, Osaka 565–0871, Japan.
Availability
The IPknot source code is freely available at https://github.com/satoken/ipknot. IPknot is also available for use from a web server at http://rtips.dna.bio.keio.ac.jp/ipknot++/. The datasets used in our experiments are available at https://doi.org/10.5281/zenodo.4923158.
Author contributions statement
K.S. conceived the study, implemented the algorithm, collected the datasets, conducted experiments, and drafted the manuscript. K.S. and Y.K. discussed the algorithm and designed the experiments. All authors read, contributed to the discussion of and approved the final manuscript.
References
- 1. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31(13):3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lorenz R, Bernhart SH, Höner Zu Siederdissen C, et al. ViennaRNA package 2.0. Algorithms Mol Biol 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 2010;11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 2006;22(14):e90–8. [DOI] [PubMed] [Google Scholar]
- 5. Zakov S, Goldberg Y, Elhadad M, et al. Rich parameterization improves RNA structure prediction. J Comput Biol 2011;18(11):1525–42. [DOI] [PubMed] [Google Scholar]
- 6. Singh J, Hanson J, Paliwal K, et al. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat Commun 2019;10(1):5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sato K, Akiyama M, Sakakibara Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat Commun 2021;12(1):941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brierley I, Pennell S, Gilbert RJC. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 2007;5(8):598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol 2005;3(6):e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelly JA, Olson AN, Neupane K, et al. Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J Biol Chem 2020;295(31):10741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trifonov EN, Gabdank I, Barash D, et al. Primordia vita. deconvolution from modern sequences. Orig Life Evol Biosph December 2006;36(5–6):559–65. [DOI] [PubMed] [Google Scholar]
- 12. Akutsu T. Dynamic programming algorithms for RNA secondary structure prediction with pseudoknots. Discrete Appl Math 2000;104(1):45–62. [Google Scholar]
- 13. Lyngsø RB, Pedersen CN. RNA pseudoknot prediction in energy-based models. J Comput Biol 2000;7(3–4):409–27. [DOI] [PubMed] [Google Scholar]
- 14. Rivas E, Eddy SR. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J Mol Biol 1999;285(5):2053–68. [DOI] [PubMed] [Google Scholar]
- 15. Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics 2004;5:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jabbari H, Wark I, Montemagno C, et al. Knotty: efficient and accurate prediction of complex RNA pseudoknot structures. Bioinformatics 2018;34(22):3849–56. [DOI] [PubMed] [Google Scholar]
- 17. Ruan J, Stormo GD, Zhang W. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics 2004;20(1):58–66. [DOI] [PubMed] [Google Scholar]
- 18. Ren J, Rastegari B, Condon A, et al. HotKnots: heuristic prediction of RNA secondary structures including pseudoknots. RNA 2005;11(10):1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen X, He S-M, Bu D, et al. FlexStem: improving predictions of RNA secondary structures with pseudoknots by reducing the search space. Bioinformatics 2008;24(18):1994–2001. [DOI] [PubMed] [Google Scholar]
- 20. Bellaousov S, Mathews D. H. ProbKnot: fast prediction of RNA secondary structure including pseudoknots. RNA 2010;16(10):1870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato K, Kato Y, Hamada M, et al. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics 2011;27(13):i85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H, Zhang L, Mathews DH, et al. LinearPartition: linear-time approximation of RNA folding partition function and base-pairing probabilities. Bioinformatics 2020;36(Supplement_1):i258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamada M, Sato K, Asai K. Prediction of RNA secondary structure by maximizing pseudo-expected accuracy. BMC Bioinformatics 2010;11:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamada M, Kiryu H, Sato K, et al. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics 2009;25(4):465–73. [DOI] [PubMed] [Google Scholar]
- 25. McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990;29(6–7):1105–19. [DOI] [PubMed] [Google Scholar]
- 26. Kiryu H, Kin T, Asai K. Robust prediction of consensus secondary structures using averaged base pairing probability matrices. Bioinformatics 2007;23(4):434–41. [DOI] [PubMed] [Google Scholar]
- 27. Hamada M, Sato K, Asai K. Improving the accuracy of predicting secondary structure for aligned RNA sequences. Nucleic Acids Res 2011;39(2):393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Danaee P, Rouches M, Wiley M, et al. bpRNA: large-scale automated annotation and analysis of RNA secondary structure. Nucleic Acids Res 2018;46(11):5381–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nawrocki EP, Burge SW, Bateman A, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 2015;43(Database issue):D130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cannone JJ, Subramanian S, Schnare MN, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 2002;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalvari I, Nawrocki EP, Ontiveros-Palacios N, et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res 2021;49(D1):D192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andronescu M, Bereg V, Hoos HH, et al. RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinformatics 2008;9:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu L, Niu B, Zhu Z, et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012;28(23):3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30(4):772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Zhang H, Mathews DH, et al. ThreshKnot: Thresholded ProbKnot for improved RNA secondary structure prediction. arXiv:1912.12796v1 [q-bio.BM] 2019.
- 36. Rezaur Rahman F, Zhang H, Huang L. Learning to fold RNAs in linear time. bioRxiv. 2019. 10.1101/852871. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


