ABSTRACT
This multicenter study evaluated the IMMY Aspergillus Galactomannan Lateral Flow Assay (LFA) with automated reader for diagnosis of pulmonary aspergillosis in patients with COVID-19-associated acute respiratory failure (ARF) requiring intensive care unit (ICU) admission between 03/2020 and 04/2021. A total of 196 respiratory samples and 148 serum samples (n = 344) from 238 patients were retrospectively included, with a maximum of one of each sample type per patient. Cases were retrospectively classified for COVID-19-associated pulmonary aspergillosis (CAPA) status following the 2020 consensus criteria, with the exclusion of LFA results as a mycological criterion. At the 1.0 cutoff, sensitivity of LFA for CAPA (proven/probable/possible) was 52%, 80% and 81%, and specificity was 98%, 88% and 67%, for bronchoalveolar lavage fluid (BALF), nondirected bronchoalveolar lavage (NBL), and tracheal aspiration (TA), respectively. At the 0.5 manufacturer’s cutoff, sensitivity was 72%, 90% and 100%, and specificity was 79%, 83% and 44%, for BALF, NBL and TA, respectively. When combining all respiratory samples, the receiver operating characteristic (ROC) area under the curve (AUC) was 0.823, versus 0.754, 0.890 and 0.814 for BALF, NBL and TA, respectively. Sensitivity and specificity of serum LFA were 20% and 93%, respectively, at the 0.5 ODI cutoff. Overall, the Aspergillus Galactomannan LFA showed good performances for CAPA diagnosis, when used from respiratory samples at the 1.0 cutoff, while sensitivity from serum was limited, linked to weak invasiveness during CAPA. As some false-positive results can occur, isolated results slightly above the recommended cutoff should lead to further mycological investigations.
KEYWORDS: Aspergillus galactomannan lateral flow assay (LFA), serum, bronchoalveolar lavage, nondirected bronchial lavage, tracheal aspirate, galactomannan, COVID-19, SARS-CoV-2
INTRODUCTION
As of 28 July 2021, the COVID-19 pandemic that emerged in 2020 has been responsible for more than 190,000,000 confirmed cases worldwide (https://covid19.who.int/). This respiratory infection, resulting in pulmonary damages and systemic inflammatory burst, may promote superinfections (1). Among these, pulmonary aspergillosis has been rapidly recognized as a specific disease, termed COVID-19-associated pulmonary aspergillosis (CAPA), favored by the classical risk factors for aspergillosis in Intensive Care Units (ICU), such as mechanical ventilation and frequent use of steroids (2, 3).
After several months using diagnostic criteria partially adapted to CAPA, such as the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) criteria, the AspICU and the modified AspICU criteria (4–6), consensus CAPA case definitions were published on the main behalf of the European Confederation of Medical Mycology (ECMM) and the International Society for Human and Animal Mycology (ISHAM) (7), allowing standardization of the clinical studies. Utilizing an approach similar to other case definitions for fungal infections (4–6, 8), these were based on the combination of a host factor (positive SARS-CoV-2 RT-PCR), clinical signs evocative for the disease (refractory fever, pleural rub, chest pain and/or haemoptysis) associated with pulmonary and/or cavitary infiltrates on imaging, and mycological evidence specific for Aspergillus (microscopic detection, culture, PCR or galactomannan antigen) in a deep respiratory sample. The classification as “proven,” “probable,” and “possible” relied on positive mycology in a sterile aspiration/biopsy specimen, bronchoalveolar lavage fluid (BALF), and nonbronchoscopic lavage (NBL), respectively. Additionally, patients with a positive Aspergillus PCR and/or galactomannan immunoassay (GM) in serum are also classified as “probable” CAPA. However, as these criteria were compiled to answer the urgent need for consensus recommendations, some mycological factors were included based on test performance in non-COVID-19 ICU patients at risk of pulmonary aspergillosis, accepting there was limited evidence available specifically for the diagnosis of CAPA (7).
The IMMY sōna Aspergillus Galactomannan Lateral Flow Assay (LFA) (IMMY, Norman, OK, USA) is a point-of-care diagnostic test that has been recently released to supplement some weaknesses of the most frequently used Aspergillus biomarker, the galactomannan immunoassay (GM) (9). The value of the GM assay for the diagnosis of invasive aspergillosis has been clearly established, initially in serum from neutropenic patients (10), but then in BALF also for both neutropenic and nonneutropenic patients (11). However, the laborious nature of the ELISA format and requirement for significant sample numbers to meet batch testing requirements can become an obstacle for the patient management of this life-threatening infection. The Aspergillus LFA has several advantages, including simple use, single sample testing, results after minimal processing (around 45 min), and good concordance with GM ELISA (12, 13). In this multicenter study, we evaluated the performance of the IMMY Aspergillus LFA, for diagnosis of CAPA in COVID-19 patients admitted to ICUs.
MATERIALS AND METHODS
This retrospective multicenter study was conducted as part of the ECMM Aspergillus in COVID-19 study. All included patients and controls had COVID-19-associated ARF and were admitted to the ICU between March 2020 and April 2021. Participating centers were the University Hospital Center of Rennes (France), the University Hospital of Graz (Austria), the Public Health Wales Mycology Reference Laboratory (Cardiff, United-Kingdom), and the Hospital General Universitario Gregorio Marañón of Madrid (Spain). Patients were retrospectively included following two different conditions: (i) CAPA patients were those classified with possible, probable or proven CAPA according to the ECMM/ISHAM criteria (7), with the exclusion of Aspergillus LFA as a mycological criterion, and (ii) the “no CAPA” patients were randomly selected among COVID-19 patients admitted in ICU but not fulfilling the ECMM/ISHAM criteria (7), thus without CAPA. Each center included all their CAPA patients and selected “no CAPA” patients in order to obtain balanced cohort sizes for each sample type. The study protocol and all study-related procedures were approved by the Human Research Protections Program at the Medical University of Graz (#32-296 ex 19/20), the Ethics Committee of the Rennes University Hospital (approval N 20–56 obtained the 30 April 2020), and the Research Ethics Committee of Hospital General Universitario Gregorio Marañón (study MICRO.HGUGM.2020‐027). In Cardiff, the study was as a retrospective, anonymous, case/control performance evaluation of the LFA, using surplus clinical samples, with no impact on patient management and not requiring ethical approval.
Respiratory (BALF, NBL, tracheal aspirate [TA]), as well as serum samples were included, with a maximum of one each sample type per patient. Samples were taken as part of routine microbiological investigations, including the usual mycological examinations for CAPA diagnosis: direct microscopic examination and culture on Sabouraud medium for respiratory samples, and GM assay (Platelia Aspergillus Ag ELISA; Bio-Rad Laboratories, Marnes-la-Coquette, France) and Aspergillus qPCR on serum and respiratory samples (30). Routine investigations were prospectively performed at each participating center, before samples were stored at −70°C or lower for less than a year, which is known to limit any deleterious storage effects for the detection of GM (31). For patients with CAPA, the samples included in the study were either positive for mycological evidence or sampled within ±3 days from the positive result.
Between March 2020 and April 2021 these samples where thawed, vortexed, and tested with the LFA according to the manufacturer’s instructions, as previously described (14). Briefly, 300 μl of sample was pretreated with 100 μl of provided EDTA buffer, heated at 120°C for 5–8 min and centrifuged at 10,000 × g, 80 μl of supernatant was mixed with 40 μl running buffer before LFA strips were inserted. After 30 min, test lines intensities were read by an automated cube reader provided (sōna LFA Cube Reader, IMMY, Norman, OK, USA) and displayed in ODI (14). Invalid tests due to the absence of a control line were excluded from the study (n = 3).
Statistical analyses were performed using SPSS 25 (SPSS Inc., Chicago, IL, USA). The ODI cutoffs of 0.5 and 1.0 were compared by calculating sensitivity and specificity of the LFA assay for diagnosis of proven/probable/possible CAPA versus no CAPA and for proven/probable CAPA versus no CAPA, respectively. The calculations were made for each sample type separately, as well as BALF and NBL specimens combined (utilizing only 1 sample per patient and prioritizing BALF), and all respiratory specimens combined (utilizing only 1 sample per patient prioritizing first BALF and second NBL). Characteristics of the cohorts were compared by Student's t test for quantitative data (age) or Fisher’s exact test for qualitative data. Sensitivities and specificities were compared between BALF, NBL and TA by Chi-square test. For continuous data, including LFA and GM ELISA ODIs, receiver operating characteristic (ROC) curve analyses were performed and area under the curve (AUC) values were presented, including 95% confidence intervals (95% CI) for the outcomes proven/probable/possible and proven/probable disease (versus no aspergillosis), in the overall study cohort and subcohorts. Correlation between serum GM ELISA and LFA ODIs was calculated using Spearman rho correlation analysis due to the nonnormal distributions. A two-sided P value ≤ 0.05 was considered significant.
RESULTS
A total of 196 respiratory samples and 148 serum samples (N = 344) from 239 patients with proven CAPA (n = 1), probable CAPA (n = 47), possible CAPA (n = 19) and patients not fulfilling CAPA criteria (n = 172) were included into this study. Among the 239 patients, 63, 20, 55, and 101 were included in the Graz, Madrid, Rennes, and Cardiff centers, respectively. A detailed breakdown of patients per center, sample type, and CAPA classification is available in Table 1. Besides COVID-19 acute respiratory failure other underlying diseases were present in the following proportions: 42% (101/238) had underlying cardiovascular disease, 22% (52/238) pulmonary disease, 21% (50/238) diabetes, 10% (23/238) hematological malignancies, 5% (13/238) prior solid organ transplantation, 4% (10/238) oncological malignancies, 2% (5/238) autoimmune disease and each 1% (3/238) HIV or advanced liver cirrhosis. The 344 samples included 90 BALF samples from 90 patients, 72 NBL samples from 72 patients, 34 TA samples from 34 patients and 148 serum samples from 148 patients. Of note, all NBL samples came from Cardiff, which did not provide TA. The majority of patients were male (163/238, 68% versus 75/238, 32% female) and median age was 61 years (range 21–88). The most common mycological evidence that allowed CAPA diagnosis was GM ELISA in BALF (39%, 36/67), followed by BALF culture (34%, 23/67), serum GM ELISA and BALF PCR (both 24%, 16/67). The detailed characteristics of the CAPA and no CAPA patients are available in Table 2. No difference was observed in the characteristics of the CAPA and no CAPA groups.
TABLE 1.
Number of patients per sample type, participating center, and CAPA classificationa
| Center | Sample type | Proven CAPA | Probable CAPA | Possible CAPA | No CAPA | All patients |
|---|---|---|---|---|---|---|
| Graz | Serum | 0 | 3 | 0 | 54 | 57 |
| Tracheal aspirate | 0 | 2 | 0 | 1 | 3 | |
| NBL | 0 | 0 | 0 | 0 | 0 | |
| BALF | 0 | 3 | 0 | 29 | 32 | |
| All patients | 0 | 5 | 0 | 58 | 63 | |
| Madrid | Serum | 0 | 10 | 0 | 0 | 13 |
| Tracheal aspirate | 1 | 5 | 0 | 0 | 8 | |
| NBL | 0 | 0 | 0 | 3 | 0 | |
| BALF | 0 | 5 | 0 | 2 | 5 | |
| All patients | 1 | 14 | 0 | 5 | 20 | |
| Rennes | Serum | 0 | 13 | 0 | 28 | 41 |
| Tracheal aspirate | 0 | 8 | 0 | 15 | 23 | |
| NBL | 0 | 0 | 0 | 0 | 0 | |
| BALF | 0 | 9 | 0 | 18 | 27 | |
| All patients | 0 | 14 | 0 | 41 | 55 | |
| Cardiff | Serum | 0 | 11 | 9 | 17 | 37 |
| Tracheal aspirate | 0 | 0 | 0 | 0 | 0 | |
| NBL | 0 | 4 | 16 | 52 | 72 | |
| BALF | 0 | 9 | 3 | 14 | 26 | |
| All patients | 0 | 14 | 19 | 68 | 101 | |
| Total | Serum | 0 | 37 | 9 | 102 | 148 |
| Tracheal aspirate | 1 | 15 | 0 | 18 | 34 | |
| NBL | 0 | 4 | 16 | 52 | 72 | |
| BALF | 0 | 26 | 3 | 61 | 90 | |
| All patients | 1 | 47 | 19 | 172 | 239 |
NBL, nondirected bronchial lavage; BALF, bronchoalveolar lavage fluid.
TABLE 2.
Cohort characteristics and mycological test results for COVID-19-associated pulmonary aspergillosis (CAPA) casesa
| Characteristics | CAPA (n = 67) | No CAPA (n = 172) | Statistical significance |
|---|---|---|---|
| Demographical | |||
| Age (yrs) (mean [95% CI]) | 61 (58–64) | 61 (58–63) | ns |
| Sex ratio (male/female) | 2.53 (48/19) | 2.02 (115/57) | ns |
| Clinical | |||
| Cardiovascular disease | 39% (26/67) | 44% (75/172) | ns |
| Diabetes mellitus | 25% (17/67) | 19% (33/172) | ns |
| Pulmonary disease | 19% (13/67) | 23% (39/172) | ns |
| Haematological malignancy | 12% (8/67) | 10% (17/172) | ns |
| Solid-organ transplant recipient | 6% (4/67) | 5% (9/172) | ns |
| Oncological malignancy | 4% (3/67) | 4% (7/172) | ns |
| Auto-immune disease | 1% (1/67) | 2% (4/172) | ns |
| HIV infection | 3% (2/67) | <1% (1/172) | ns |
| Advanced liver cirrhosis | 1% (1/67) | 1% (2/172) | ns |
| Mycological | |||
| Positive BALF | 54% (36/67) | - | na |
| GM ELISA > 1.0 ODI | 39% (26/67) | - | na |
| Culture | 34% (23/67) | - | na |
| PCR < 36 Cq | 24% (16/67) | - | na |
| Positive NBL | 31% (21/67) | - | na |
| GM ELISA > 4.0 ODI | 21% (14/67) | - | na |
| GM ELISA > 1.2 ODI + PCR | 16% (11/67) | - | na |
| Culture | 16% (11/67) | - | na |
| Microscopic examination | 3% (2/67) | - | na |
| Positive serum | 27% (18/67) | - | na |
| GM ELISA > 0.5 ODI | 24% (16/67) | - | na |
| PCR | 7% (5/67) | - | na |
ns, not significant; na, not applicable.
LFA performance per sample type when using the manufacturer’s 0.5 ODI cutoff, as well as a cutoff of 1.0 ODI, for differentiating proven/probable/possible CAPA from no CAPA or proven/probable CAPA from possible/no CAPA, are displayed in Tables 3 and 4, respectively. At the 0.5 ODI cutoff, sensitivity in NBL and TA was high for proven/probable/possible (≥90%, ns) and for proven/probable CAPA (both 100%), but specificity was limited for TA compared to NBL (44% and 83%, respectively, P < 0.01). Serum and BALF samples, at the 1.0 ODI cutoff, had the highest specificity (≥98%, ns), but with limited sensitivities of 9–11% and 52–58% (P < 0.0001), respectively. Altogether, at the 1.0 ODI cutoff, combination of all respiratory sample types (90 BALF, 72 NBL and 34 TA), or BALF and NBL only (90 BALF, 72 NBL), reached sensitivities close to 65% and specificities from 83% to 94%, for both proven/probable and proven/probable/possible CAPA. With a 2.0 ODI cutoff, NBL reached 92% specificity (75% sensitivity), while for TA specificity remained unchanged at 78% and sensitivity dropped to 63%.
TABLE 3.
LFA performances per sample type for 0.5 and 1.0 cutoffs for diagnosing proven/probable/possible CAPA versus no CAPAa
| 0.5 ODI cutoff |
1.0 ODI cutoff |
|||
|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | |
| Respiratory samples | ||||
| Tracheal aspirate (TA) (NCAPA=16; NØCAPA=18) | 100% (79–100) | 44% (22–69) | 81% (54–96) | 67% (41–87) |
| Nondirected bronchial lavage (NBL) (NCAPA=20; NØCAPA=52) | 90% (68–99) | 83% (70–92) | 80% (56–94) | 88% (77–96) |
| Bronchoalveolar lavage fluid (BALF) (NCAPA=29; NØCAPA=61) | 72% (53–87) | 79% (66–88) | 52% (33–71) | 98% (91–100) |
| BALF and NBL combinedb (NCAPA=49; NØCAPA=113) | 80% (66–90) | 81% (72–87) | 63% (48–77) | 94% (88–97) |
| All combinedb (NCAPA=58; NØCAPA=127) | 83% (71–91) | 76% (67–83) | 66% (52–78) | 90% (83–94) |
| Serum samples (NCAPA=46; NØCAPA=102) | 20% (9–34) | 93% (86–97) | 9% (2–21) | 99% (95–100) |
For LFA performance for diagnosing proven/probable CAPA versus no CAPA (possible CAPA excluded) use sensitivity from Table 4 and specificity from Table 3. NCAPA, number of patients with proven/probable/possible CAPA; NØCAPA, number of patients without CAPA.
For these lines, calculations were based on patients with any of the mentioned sample types. However, only 1 sample per patient was considered, with the following order of priority: BALF, NBL, TA.
TABLE 4.
LFA performances per sample type for 0.5 and 1.0 cutoffs for diagnosing proven/probable CAPA versus possible/no CAPAa
| 0.5 ODI cutoff |
1.0 ODI cutoff |
|||
|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | |
| Respiratory samples | ||||
| Tracheal aspirate (TA) (NCAPA=16; NØCAPA=18) | 100% (79–100) | 44% (22–69) | 81% (54–96) | 67% (41–87) |
| Nondirected bronchial lavage (NBL) (NCAPA=4; NØCAPA=68) | 100% (40–100) | 66% (54–77) | 100% (40–100) | 74% (61–84) |
| Bronchoalveolar lavage fluid (BALF) (NCAPA=26; NØCAPA=64) | 77% (56–91) | 80% (68–89) | 58% (37–77) | 98% (92–100) |
| BALF and NBL combinedb (NCAPA=30; NØCAPA=132) | 80% (61–92) | 72% (63–79) | 63% (44–80) | 86% (78–91) |
| All combinedb (NCAPA=39; NØCAPA=146) | 85% (69–94) | 68% (60–76) | 67% (50–81) | 83% (76–89) |
| Serum samples (NCAPA=37; NØCAPA=111) | 22% (10–38) | 93% (86–97) | 11% (3–25) | 99% (95–100) |
For LFA performance for diagnosing proven/probable CAPA versus no CAPA (possible CAPA excluded) use sensitivity from Table 4 and specificity from Table 3. NCAPA, number of patients with proven/probable CAPA; NØCAPA, number of patients with possible CAPA or without CAPA.
For these lines, calculations were based on patients with any of the mentioned sample types. However, only 1 sample per patient was considered, with the following order of priority: BALF, NBL, TA.
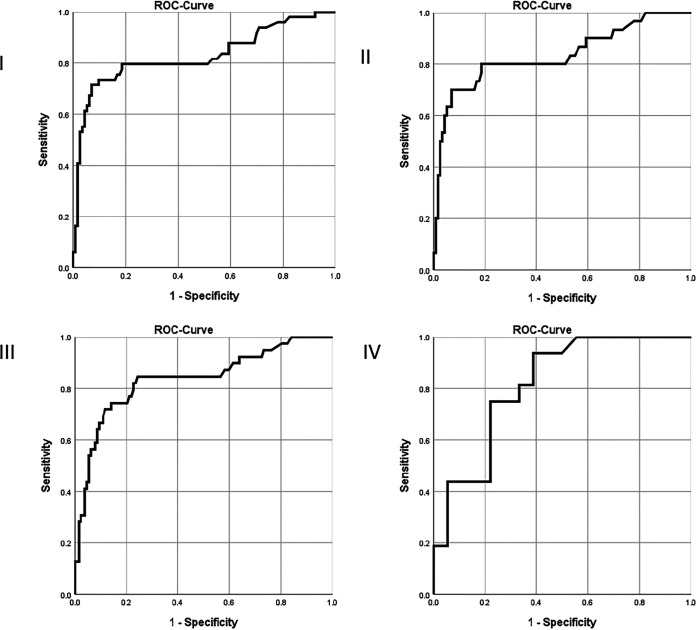
The ROC curve analysis showed an AUC of 0.829 (95%CI 0.749–0.909) for the LFA from BALF and NBL combined, for differentiating patients without CAPA (n = 113) from those with proven/probable/possible disease (n = 49) (Fig. 1). When combining all three respiratory sample types (but with only 1 sample per patient, with the following order of priority: BALF, then NBL, then TA), AUC was 0.823 (95% CI 0.750–0.896) with n = 58 CAPA versus n = 127 patients without CAPA. AUCs for each respiratory specimen considered separately were: 0.754 (95%CI 0.621–0.887) for BALF, 0.890 (95%CI 0.790–0.990) for NBL, and 0.814 (95%CI 0.671–0.958) for TA. AUCs were higher for differentiation of patients with proven/probable CAPA from those without CAPA (possible CAPA excluded): 0.833 (95% CI 0.737–0.928) for BALF and NBL combined (Fig. 1), 0.836 (95% CI 0.756–0.917) for all respiratory samples combined (Fig. 1), 0.790 (95% CI 0.658–0.921) for BALF, 0.976 (95% CI 0.973–1.000) for NBL and 0.814 (95% CI 0.671–0.958) for TA (Fig. 1). AUC for serum was indiscriminative at 0.512 (95%CI 0.402–0.622) for diagnosis of proven/probable/possible CAPA, and at 0.541 (95% CI 0.420–0.661) for diagnosis of proven/probable CAPA.
FIG 1.
Receiver operating characteristics analysis curves for lateral flow assay for diagnosing COVID-19 associated pulmonary aspergillosis versus no aspergillosis in the overall study cohort. (I) BALF/NBL combined for proven/probable/possible CAPA (n = 49) versus no CAPA (n = 113). (II) BALF/NBL combined for proven/probable CAPA (n = 30) versus no CAPA (n = 113). (III) BALF/NBL/TA combined for proven/probable CAPA (n = 39) versus no CAPA (n = 127). (IV) TA for proven/probable CAPA (n = 16) versus no CAPA (n = 18).
A total of 81/344 (23.5%) of samples were obtained from patients receiving antifungal prophylaxis. In sub analysis, AUCs generally tended to be lower in the subgroup of cases receiving mold active antifungal prophylaxis/treatment at the time of sampling. In those on antifungal prophylaxis AUC was 0.763 (95%CI 0.608–0.907) for BALF, NBL and TA combined (n = 12 with CAPA and n = 26 without; AUC was 0.841 in those not receiving prophylaxis), and 0.652 (95%CI 0.338–0.965) for BAL fluid alone (n = 6 with CAPA and n = 11 without; AUC was 0.787 in those not receiving prophylaxis).
Same day serum GM ELISA ODI, obtained with the Platelia Aspergillus Ag ELISA, were available for 98/148 samples: 12 had proven/probable/possible CAPA and positive serum GM ELISA, 17 had proven/probable/possible CAPA and negative serum GM ELISA, 68 did not have CAPA nor positive GM ELISA, and one “no CAPA” patient had a positive serum GM ELISA >0.5 ODI (but without clinical signs and without further antifungal treatment). AUC of serum GM ELISA ODI was 0.736 (95% CI 0.615–0.857) for differentiating proven/probable/possible CAPA versus no CAPA. Among those with proven/probable/possible CAPA, serum GM LFA was positive in 58% (7/12) of the GM ELISA positive samples (0.5 ODI cutoff), versus 11.8% (2/17) of the GM ELISA negative samples. Among patients without CAPA and with negative serum GM ELISA (and no other mycological evidence), serum LFA was >0.5 ODI in 7.4% of samples (5/68), while LFA was also negative in the single false-positive serum GM ELISA sample. Spearman correlation analysis showed a significant correlation between serum LFA ODI and serum GM ELISA ODI (rho 0.371, P < 0.001). Same day BALF GM ELISA ODI were available in 62/90 BALF samples (AUC 0.973, 95%CI 0.936–1.000 for differentiating proven/probable/possible CAPA versus no CAPA), and Spearman correlation analysis showed a moderate to strong correlation with BALF LFA ODI (rho 0.523, P < 0.001).
If LFA ODI results had not been excluded for classification of CAPA cases, this would have resulted in reclassification of a total of 9/239 cases (3.8%), including one reclassification from possible to probable CAPA, one reclassification from no CAPA to possible CAPA (which had elevated GM ELISA ODI from NBL of 1.8 that did alone not fulfill mycological criteria), and 7 reclassifications from no CAPA to probable CAPA. While one of those 7 had also Aspergillus sp. growth in a TA culture, 5/6 of the other reclassifications from no CAPA to probable CAPA had slightly elevated serum LFA ODIs between 0.5 and 0.58 but no other mycological evidence for CAPA.
DISCUSSION
In this retrospective multicenter study, the performance of the Aspergillus galactomannan LFA, using the automated Cube Reader for quantification, was evaluated on respiratory and serum samples for diagnosis of pulmonary aspergillosis in ICU patients admitted for COVID-19-related ARF. A total of 344 samples from 239 patients were included, with a maximum of one sample of each type per patient. For respiratory samples, the 0.5 ODI cutoff allowed good sensitivity of the assay (72–100% depending on the sample type, 80–85% all types combined), but with variable specificity (i.e., from 44% for TA, to 79% for BALF, 76–81% all types combined). The 1.0 ODI cutoff improved greatly the specificity for TA and BALF (67% and 98%, respectively), at the cost of a lower sensitivity (81% and 52%, respectively). However, sensitivity was higher when considering only proven/probable CAPA cases (81% and 58% for TA and BALF, respectively). When performed on NBL, with a 1.0 ODI cutoff, the LFA presented interesting performance with 80% sensitivity and 88% specificity, but this observation relied only on samples from a single center. The ROC curves AUCs ranged from 0.754 and 0.814 for BALF and TA, respectively, to 0.890 for NBL. In serum samples, LFA had very low sensitivity (9–22%) but high specificity (≥93%), providing a low ROC AUC (0.479). For 5/102 patients without CAPA diagnosed but tested in serum, the serum LFA was between 0.5 and 0.58, which would have reclassified them as “probable CAPA.”
Several studies evaluated performance of LFA on BALF for diagnosis of aspergillosis, but mainly in mixed cohorts of patients (hematology, ICU, solid-organ transplant, etc.). Their study designs were also varied, in terms of cohort constitution, case definition, and even assay interpretation (visual versus digital read, cutoff value), generating performance that was highly variable, with sensitivity ranging from 40% to 88%, and specificity ranging from 46% to 95% (13–17). Among them, a multicenter study found a sensitivity of 74% and specificity of 83% with digital readout from BALF at 1.5 ODI cutoff, with stable test performance across centers and patient groups (13). Another recent study, which included 178 patients from Belgium and the Netherlands, used digital reader and focused on ICU patients, which by design is the closest match to our current study. A sensitivity of 87–94%, depending on the case definition used, was observed, with 81% specificity (12). The sensitivity was higher than the 72–77% we observed when testing BALF at 0.5 ODI cutoff. However, strict exclusions criteria were applied in that study (i.e., “possible” invasive fungal disease following EORTC/MSG definitions, BALF GM ELISA between 0.5 and 1.0 ODI, patients with mycological findings that survived without mold-active antifungal therapy, probable and putative IPA if autopsy did not reveal signs of IPA, patients that received more than 72h of mold-active therapy before sampling), leading to a cohort of highly probable cases and the exclusion of possible cases. This exclusion could explain the increased sensitivity in the earlier study, due to a decrease in the number of possible cases with potentially lower fungal load at greater risk of false negative results. Indeed, according to the EORTC/MSGERC definitions for IFD, possible invasive aspergillosis (IA) cases are classified due to the absence of mycological evidence (6).
Only one study has evaluated the performance of the LFA when testing TA samples, for the diagnosis of CAPA (18). The authors observed, at the 1.0 ODI cutoff, 64% sensitivity and 68% specificity, comparable to our study, except for sensitivity, which was slightly higher at this cutoff (81%), and potentially associated with the use of different case definitions, with the recently published ECMM/ISHAM consensus criteria applied in our study (7). Data are scarce concerning the value of NBL testing in the diagnosis of pulmonary aspergillosis, even if this sample type was suggested as possible alternative to the usual BAL fluid (7). Of note, in a Welsh cohort, 89% (17/19) of patients with CAPA, according to various definitions, had NBL GM ≥1.0 ODI (19). Also, a monocentric study performed in the Netherlands observed high correlation between positive Aspergillus culture and GM ELISA >1.0 ODI (78%) (20). Results from our study support this finding, but as previously emphasized, our results are based on samples that originated from a single center (Cardiff). Further studies are needed to improve the knowledge when testing NBL, if this type of sample is to provide added value for the diagnosis of aspergillosis.
In serum, the LFA with digital readout was evaluated in four studies focusing mostly on patients with hematologic malignancy. Among hematological malignancy patients one study reported a sensitivity of 49% and a specificity of 95% (21), and two reported higher sensitivities of 97% (22) and 91% (23) with specificities between 90% and 96% at an ODI of ≥0.5, reaching 100% at 0.61 ODI cutoff (22). In a recent mixed cohort study, sensitivity and specificity of the LFA were 78.6% and 80.5% a 0.5 ODI cutoff, with similar performance among patients with hematological malignancies and patients with other traditional risk factors for IA (24). Serum GM is known to provide poor sensitivity (<20%) but high specificity in CAPA diagnosis (7, 19, 25, 26), and indeed our study showed LFA sensitivity of 20–22% at the 0.5 ODI cutoff used for the classification of “probable CAPA” (7). However, at this cutoff, 7/102 patients without CAPA would have been misclassified as “probable CAPA” in our study, suggesting that classification of patients with a unique serum LFA ≥0.5 but <0.7 could be risky, when they lack any other supporting mycological evidence. This poor performance of the serum LFA and GM correlates with the fact that Aspergillus angioinvasion, which is considered to be the source of serum GM, is less frequent in CAPA than in other backgrounds of invasive pulmonary aspergillosis (27).
The main limitation of this study is its retrospective, case/control design, which can lead to selection bias, and the relatively low number of TA samples. However, the multiple centers involved and the use of consensus criteria for case definition generates robust data on performance of LFA for CAPA diagnosis, with the exception of NBL testing, which originated from a single center. Overall, these results emphasize a high value of the galactomannan LFA on respiratory samples for CAPA diagnosis. Indeed, if performance seems to be partially satisfactory, especially because GM LFA was either specific in BALF or sensitive in TA, this is comparable with the well-established GM ELISA, but with an easier and faster handling. Using this assay, testing respiratory samples with a 1.0 ODI cutoff could be implemented in workflows of centers managing COVID-19 patients in ICU, for example by screening on TA, then exploring on BALF in case of positivity of a TA sample, potentially governed by the LFA GM ODI value. Finally, the study chose a more conservative approach to CAPA classification by applying the new and more restrictive ECMM/ISHAM criteria for CAPA classification instead of the modified Blot criteria. Compared to modified Blot criteria, application of these new criteria was shown to result in a reduction of the mean incidence of probable/proven CAPA cases from 19% to 11.9% among evaluable cases, bringing the prevalence of CAPA cases closer to that suggested by autopsy studies (28, 29).
In conclusion, the Aspergillus Galactomannan LFA showed good performances for CAPA diagnosis, especially on respiratory samples at the 1.0 ODI cutoff, with the benefit of a relatively short handling time (around 45 min). As some false-positive results can occur, isolated ODI slightly above the 0.5 ODI cutoff should lead to further mycological investigations.
ACKNOWLEDGMENTS
We acknowledge Marina Linhofer’s support as research coordinator.
Aspergillus galactomannan LFAs were provided by IMMY, Norman, Oklahoma, USA. They had no role in the study design, data collection, analysis, interpretation, decision to publish, in the writing of the manuscript, or in the decision to submit the manuscript for publication.
Martin Hoenigl received research funding from NIH, Gilead, Astellas, MSD, Scynexis, F2G, and Pfizer. P. Lewis White performed diagnostic evaluations for and received meeting sponsorship from Bruker, Dynamiker, and Launch Diagnostics; speaker fees, expert advice fees, and meeting sponsorship from Gilead; speaker and expert advice fees from F2G; and speaker fees from MSD and Pfizer and received speaker fees from and performed diagnostic evaluations for Associates of Cape Cod and IMMY. Jean-Pierre Gangneux received funding from Gilead and Pfizer outside the work. Maricela Valerio received speaker fees from Pfizer, MSD, and GSK. Juergen Prattes has received personal fees from Gilead Sciences and Pfizer and research funding from MSD outside of the submitted work and is a stakeholder of AbbVie and Novo Nordisk. Marina Machado received speaker fees from Pfizer and Gilead. All other authors declare no conflict of interest for this study.
Contributor Information
Martin Hoenigl, Email: hoeniglmartin@gmail.com.
Kimberly E. Hanson, University of Utah
REFERENCES
- 1.Garcia-Vidal C, Sanjuan G, Moreno-García E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, Fernandez-Pittol M, Pitart C, Inciarte A, Bodro M, Morata L, Ambrosioni J, Grafia I, Meira F, Macaya I, Cardozo C, Casals C, Tellez A, Castro P, Marco F, García F, Mensa J, Martínez JA, Soriano A, COVID-19 Researchers Group . 2021. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect 27:83–88. doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prattes J, Wauters J, Giacobbe DR, Salmanton-García J, Maertens J, Bourgeois M, Reynders M, Rutsaert L, Van Regenmortel N, Lormans P, Feys S, Reisinger AC, Cornely OA, Lahmer T, Valerio M, Delhaes L, Jabeen K, Steinmann J, Chamula M, Bassetti M, Hatzl S, Rautemaa-Richardson R, Koehler P, Lagrou K, Hoenigl M. 2021. Risk factors and outcome of pulmonary aspergillosis in critically ill coronavirus disease 2019 patients—a multinational observational study by the European Confederation of Medical Mycology. Clin Microbiol Infect doi: 10.1016/j.cmi.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangneux J-P, Bougnoux M-E, Dannaoui E, Cornet M, Zahar JR. 2020. Invasive fungal diseases during COVID-19: we should be prepared. J Mycol Med 30:100971. doi: 10.1016/j.mycmed.2020.100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blot SI, Taccone FS, Van den Abeele A-M, Bulpa P, Meersseman W, Brusselaers N, Dimopoulos G, Paiva JA, Misset B, Rello J, Vandewoude K, Vogelaers D, AspICU Study Investigators . 2012. A clinical algorithm to diagnose invasive pulmonary aspergillosis in critically ill patients. Am J Respir Crit Care Med 186:56–64. doi: 10.1164/rccm.201111-1978OC. [DOI] [PubMed] [Google Scholar]
- 5.Schauwvlieghe AFAD, Rijnders BJA, Philips N, Verwijs R, Vanderbeke L, Van Tienen C, Lagrou K, Verweij PE, Van de Veerdonk FL, Gommers D, Spronk P, Bergmans DCJJ, Hoedemaekers A, Andrinopoulou E-R, van den Berg CHSB, Juffermans NP, Hodiamont CJ, Vonk AG, Depuydt P, Boelens J, Wauters J, Dutch-Belgian Mycosis study group . 2018. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir Med 6:782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, Clancy CJ, Wingard JR, Lockhart SR, Groll AH, Sorrell TC, Bassetti M, Akan H, Alexander BD, Andes D, Azoulay E, Bialek R, Bradsher RW, Jr., Bretagne S, Calandra T, Caliendo AM, Castagnola E, Cruciani M, Cuenca-Estrella M, Decker CF, Desai SR, Fisher B, Harrison T, Heussel CP, Jensen HE, Kibbler CC, Kontoyiannis DP, Kullberg B-J, Lagrou K, Lamoth F, Lehrnbecher T, Loeffler J, Lortholary O, Maertens J, Marchetti O, Marr KA, Masur H, Meis JF, Morrisey CO, Nucci M, Ostrosky-Zeichner L, Pagano L, Patterson TF, Perfect JR, Racil Z, Roilides E, Ruhnke M, Prokop CS, Shoham S, et al. 2020. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis 71:1367–1376. doi: 10.1093/cid/ciz1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehler P, Bassetti M, Chakrabarti A, Chen SCA, Colombo AL, Hoenigl M, Klimko N, Lass-Flörl C, Oladele RO, Vinh DC, Zhu L-P, Böll B, Brüggemann R, Gangneux J-P, Perfect JR, Patterson TF, Persigehl T, Meis JF, Ostrosky-Zeichner L, White PL, Verweij PE, Cornely OA, European Confederation of Medical Mycology, International Society for Human Animal Mycology, Asia Fungal Working Group, INFOCUS LATAM/ISHAM Working Group, ISHAM Pan Africa Mycology Working Group, European Society for Clinical Microbiology, Infectious Diseases Fungal Infection Study Group, ESCMID Study Group for Infections in Critically Ill Patients, Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy, Medical Mycology Society of Nigeria, Medical Mycology Society of China Medicine Education Association, Infectious Diseases Working Party of the German Society for Haematology and Medical Oncology, Association of Medical Microbiology, Infectious Disease Canada . 2021. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect Dis 21:e149–e162. doi: 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verweij PE, Rijnders BJA, Brüggemann RJM, Azoulay E, Bassetti M, Blot S, Calandra T, Clancy CJ, Cornely OA, Chiller T, Depuydt P, Giacobbe DR, Janssen NAF, Kullberg B-J, Lagrou K, Lass-Flörl C, Lewis RE, Liu PW-L, Lortholary O, Maertens J, Martin-Loeches I, Nguyen MH, Patterson TF, Rogers TR, Schouten JA, Spriet I, Vanderbeke L, Wauters J, van de Veerdonk FL. 2020. Review of influenza-associated pulmonary aspergillosis in ICU patients and proposal for a case definition: an expert opinion. Intensive Care Med 46:1524–1535. doi: 10.1007/s00134-020-06091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenks JD, Mehta SR, Taplitz R, Aslam S, Reed SL, Hoenigl M. 2019. Point-of-care diagnosis of invasive aspergillosis in non-neutropenic patients: Aspergillus galactomannan lateral flow assay versus Aspergillus-specific Lateral Flow Device test in bronchoalveolar lavage. Mycoses 62:230–236. doi: 10.1111/myc.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maertens J, Theunissen K, Verbeken E, Lagrou K, Verhaegen J, Boogaerts M, Eldere JV. 2004. Prospective clinical evaluation of lower cut-offs for galactomannan detection in adult neutropenic cancer patients and haematological stem cell transplant recipients. Br J Haematol 126:852–860. doi: 10.1111/j.1365-2141.2004.05140.x. [DOI] [PubMed] [Google Scholar]
- 11.Meersseman W, Lagrou K, Maertens J, Wilmer A, Hermans G, Vanderschueren S, Spriet I, Verbeken E, Van Wijngaerden E. 2008. Galactomannan in bronchoalveolar lavage fluid. Am J Respir Crit Care Med 177:27–34. doi: 10.1164/rccm.200704-606OC. [DOI] [PubMed] [Google Scholar]
- 12.Mercier T, Dunbar A, Veldhuizen V, Holtappels M, Schauwvlieghe A, Maertens J, Rijnders B, Wauters J. 2020. Point of care Aspergillus testing in intensive care patients. Crit Care 24:642. doi: 10.1186/s13054-020-03367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenks JD, Hoenigl M. 2020. Point-of-care diagnostics for invasive aspergillosis: nearing the finish line. Expert Rev Mol Diagn 20:1009–1017. doi: 10.1080/14737159.2020.1820864. [DOI] [PubMed] [Google Scholar]
- 14.Jenks JD, Prattes J, Frank J, Spiess B, Mehta SR, Boch T, Buchheidt D, Hoenigl M. 2020. Performance of the bronchoalveolar lavage fluid Aspergillus galactomannan lateral flow assay with cube reader for diagnosis of invasive pulmonary aspergillosis: a multicenter cohort study. Clin Infect Dis:ciaa1281. doi: 10.1093/cid/ciaa1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arkell P, Mahboobani S, Wilson R, Fatania N, Coleman M, Borman AM, Johnson EM, Armstrong-James DPH, Abdolrasouli A. 2021. Bronchoalveolar lavage fluid IMMY Sona Aspergillus lateral-flow assay for the diagnosis of invasive pulmonary aspergillosis: a prospective, real life evaluation. Med Mycol 59:404–408. doi: 10.1093/mmy/myaa113. [DOI] [PubMed] [Google Scholar]
- 16.Scharmann U, Verhasselt HL, Kirchhoff L, Buer J, Rath P-M, Steinmann J, Ziegler K. 2020. Evaluation of two lateral flow assays in BAL fluids for the detection of invasive pulmonary aspergillosis: a retrospective two-centre study. Mycoses 63:1362–1367. doi: 10.1111/myc.13176. [DOI] [PubMed] [Google Scholar]
- 17.Linder KA, Kauffman CA, Miceli MH. 2020. Performance of Aspergillus galactomannan lateral flow assay on bronchoalveolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis. JoF 6:297. doi: 10.3390/jof6040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roman-Montes CM, Martinez-Gamboa A, Diaz-Lomelí P, Cervantes-Sanchez A, Rangel-Cordero A, Sifuentes-Osornio J, Ponce-de-Leon A, Gonzalez-Lara MF. 2021. Accuracy of galactomannan testing on tracheal aspirates in COVID-19-associated pulmonary aspergillosis. Mycoses 64:364–371. doi: 10.1111/myc.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White PL, Dhillon R, Cordey A, Hughes H, Faggian F, Soni S, Pandey M, Whitaker H, May A, Morgan M, Wise MP, Healy B, Blyth I, Price JS, Vale L, Posso R, Kronda J, Blackwood A, Rafferty H, Moffitt A, Tsitsopoulou A, Gaur S, Holmes T, Backx M. 2020. A national strategy to diagnose COVID-19 associated invasive fungal disease in the ICU. Clin Infect Dis:ciaa1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Biesen S, Kwa D, Bosman RJ, Juffermans NP. 2020. Detection of invasive pulmonary aspergillosis in COVID-19 with non-directed bronchoalveolar lavage. Am J Respir Crit Care Med 202:1171–1173. doi: 10.1164/rccm.202005-2018LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercier T, Dunbar A, de Kort E, Schauwvlieghe A, Reynders M, Guldentops E, Blijlevens NMA, Vonk AG, Rijnders B, Verweij PE, Lagrou K, Maertens J. 2020. Lateral flow assays for diagnosing invasive pulmonary aspergillosis in adult hematology patients: a comparative multicenter study. Med Mycol 58:444–452. doi: 10.1093/mmy/myz079. [DOI] [PubMed] [Google Scholar]
- 22.White PL, Price JS, Posso R, Cutlan-Vaughan M, Vale L, Backx M. 2020. Evaluation of the performance of the IMMY sona Aspergillus galactomannan lateral flow assay when testing serum to aid in diagnosis of invasive aspergillosis. J Clin Microbiol 58:e00053-20. doi: 10.1128/JCM.00053-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serin I, Dogu MH. 2021. Serum Aspergillus galactomannan lateral flow assay for the diagnosis of invasive aspergillosis: a single-centre study. Mycoses 64:678–683. doi: 10.1111/myc.13265. [DOI] [PubMed] [Google Scholar]
- 24.Hoenigl M, Egger M, Boyer J, Schulz E, Prattes J, Jenks JD. 2021. Serum Lateral Flow Assay with digital reader for the diagnosis of invasive pulmonary aspergillosis: a two-centre mixed cohort study. Mycoses 64:1197–1202. doi: 10.1111/myc.13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arastehfar A, Carvalho A, van de Veerdonk FL, Jenks JD, Koehler P, Krause R, Cornely OA, S Perlin D, Lass-Flörl C, Hoenigl M. 2020. COVID-19 associated pulmonary aspergillosis (CAPA): from immunology to treatment. JoF 6:91. doi: 10.3390/jof6020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prattes J, Wauters J, Giacobbe DR, Lagrou K, Hoenigl M, Koehler P, Salmanton-García J, Rautemaa-Richardson R, Hatzl S, Maertens J, Debaveye Y, Bourgeois M, Reynders M, Rutsaert L, Van Regenmortel N, Lormans P, Feys S, Reisinger AC, Cornely OA, Lahmer T, Valerio M, Delhaes L, Jabeen K, Steinmann J, Chamula M, Bassetti M, ECMM-CAPA Study Group . 2021. Diagnosis and treatment of COVID-19 associated pulmonary apergillosis in critically ill patients: results from a European confederation of medical mycology registry. Intensive Care Med 47:1158–1160. doi: 10.1007/s00134-021-06471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verweij PE, Brüggemann RJM, Azoulay E, Bassetti M, Blot S, Buil JB, Calandra T, Chiller T, Clancy CJ, Cornely OA, Depuydt P, Koehler P, Lagrou K, de Lange D, Lass-Flörl C, Lewis RE, Lortholary O, Liu P-WL, Maertens J, Nguyen MH, Patterson TF, Rijnders BJA, Rodriguez A, Rogers TR, Schouten JA, Wauters J, van de Veerdonk FL, Martin-Loeches I. 2021. Taskforce report on the diagnosis and clinical management of COVID-19 associated pulmonary aspergillosis. Intensive Care Med 47:819–834. doi: 10.1007/s00134-021-06449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fekkar A, Neofytos D, Nguyen M-H, Clancy CJ, Kontoyiannis DP, Lamoth F. 2021. COVID-19-associated pulmonary aspergillosis (CAPA): how big a problem is it? Clin Microbiol Infect 27:1376–1378. doi: 10.1016/j.cmi.2021.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prattes J, Koehler P, Hoenigl M, Wauters J, Giacobbe DR, Lagrou K, Salmanton-García J, Rautemaa-Richardson R, Hatzl S, Maertens J, Debaveye Y, Bourgeois M, Reynders M, Rutsaert L, Van Regenmortel N, Lormans P, Feys S, Reisinger AC, Cornely OA, Lahmer T, Valerio M, Delhaes L, Jabeen K, Steinmann J, Chamula M, Bassetti M, on behalf of the ECMM-CAPA Study Group . 2021. COVID-19 associated pulmonary aspergillosis: regional variation in incidence and diagnostic challenges. Intensive Care Med doi: 10.1007/s00134-021-06510-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gangneux J-P, Reizine F, Guegan H, Pinceaux K, Le Balch P, Prat E, Pelletier R, Belaz S, Le Souhaitier M, Le Tulzo Y, Seguin P, Lederlin M, Tadié J-M, Robert-Gangneux F. 2020. Is the COVID-19 pandemic a good time to include Aspergillus molecular detection to categorize aspergillosis in ICU patients? a monocentric experience. JoF 6:105. doi: 10.3390/jof6030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheat LJ, Nguyen MH, Alexander BD, Denning D, Caliendo AM, Lyon GM, Baden LR, Marty FM, Clancy C, Kirsch E, Noth P, Witt J, Sugrue M, Wingard JR. 2014. Long-term stability at −20°C of Aspergillus galactomannan in serum and bronchoalveolar lavage specimens. J Clin Microbiol 52:2108–2111. doi: 10.1128/JCM.03500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]