ABSTRACT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged into a world of maturing pathogen genomics, with more than 2 million genomes sequenced at the time of writing. The rise of more transmissible variants of concern that impact vaccine and therapeutic effectiveness has led to widespread interest in SARS-CoV-2 evolution. Clinicians are also eager to take advantage of the information provided by SARS-CoV-2 genotyping beyond surveillance purposes. Here, we review the potential role of SARS-CoV-2 genotyping in clinical care. The review covers clinical use cases for SARS-CoV-2 genotyping, methods of SARS-CoV-2 genotyping, assay validation and regulatory requirements, and clinical reporting for laboratories, as well as emerging issues in clinical SARS-CoV-2 sequencing. While clinical uses of SARS-CoV-2 genotyping are currently limited, rapid technological change along with a growing ability to interpret variants in real time foretells a growing role for SARS-CoV-2 genotyping in clinical care as continuing data emerge on vaccine and therapeutic efficacy.
KEYWORDS: ASM, IDSA, RT-qPCR, SARS-CoV-2, allele specific, clinical applications, consensus, genomics, genotyping, sequencing
TEXT
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has coincided with growing abilities in infectious disease genomics, resulting in an explosion of data. With more than 2 million SARS-CoV-2 genomes available as of summer 2021, SARS-CoV-2 is now defined by a multitude of different lineages. To put this in context, there are currently fewer than 500 publicly available genomes for the four seasonal human coronaviruses combined. While SARS-CoV-2 variants may be associated with multiple clinical outcomes, the clinical utility of providing SARS-CoV-2 genotype results remains unclear (1). Here, we describe the potential roles of SARS-CoV-2 sequencing for clinical care and the challenges faced by laboratories endeavoring to implement this process.
SARS-CoV-2 is a member of the family Coronaviridae and has a large RNA genome at around 30 kb. Coronaviruses are able to propagate large genomes in part due to mutation rates substantially lower than those of other RNA viruses, achieved by means of viral accessory proteins compensating for the lack of intrinsic proofreading among RNA polymerases. As a result, the observed evolutionary rate for SARS-CoV-2 is roughly one mutation per genome per 2 weeks (corresponding to approximately two generations of host infection) (2). Coronavirus evolutionary rates are much slower than those of other common RNA viruses, such as influenza virus or HIV, leading to a lower observed genotype sequence variation among SARS-CoV-2 isolates on a per-nucleotide basis (3).
For SARS-CoV-2, we are still learning which nucleotide sequence differences convey clinically significant information. The short time frame of study has been matched by a plethora of clinical studies and incipient abilities to profile mutation function at scale. At this point, a handful of known SARS-CoV-2 mutations have been associated with substantial in vivo effects.
OF INTEREST, CONCERN, AND HIGH CONSEQUENCE
SARS-CoV-2 has accrued a number of mutations that enhance its ongoing adaptation to spread in humans and to circumvent the adaptive immune system. These variants are classified by the World Health Organization (WHO) and the U.S. Centers for Disease Control and Prevention (CDC) based on specific attributes demonstrated by a lineage in viral culture and/or in people. Variants of interest (VOI) are defined by changes to receptor binding, neutralization activity, therapeutic efficacy, or diagnostics, while variants of concern (VOC) are marked by evidence of an increase in transmissibility or disease severity or a greater reduction in neutralization, therapeutic efficacy, and/or diagnostic detection. Variants of high consequence are defined by reduced effectiveness of prevention measures or medical countermeasures; none have yet been described. To date, many of the mutations of interest have occurred in select locations of the SARS-CoV-2 spike protein, such as the receptor binding domain, enabling more focused approaches to SARS-CoV-2 genotyping. Interpretation has also been greatly assisted by high-throughput assays that allow for characterization of thousands of potential variants in parallel.
GENOTYPING APPROACHES
SARS-CoV-2 genotyping can be performed using allele-specific reverse transcription-quantitative PCR (RT-qPCR), targeted/Sanger sequencing, or whole-genome sequencing (WGS) (Table 1). No assay is currently authorized by the FDA for SARS-CoV-2 genotyping, so any assays developed for clinical use in the United States will require validation as a laboratory-developed test to be performed in a CLIA (Clinical Laboratory Improvement Amendments) high-complexity laboratory (4). The majority of SARS-CoV-2 genotyping is currently being performed outside a CLIA-regulated environment, with results reporting to public health allowed under CMS (Centers for Medicare and Medicaid Services) enforcement discretion. In addition, it should be noted that not all clinical laboratories can provide SARS-CoV-2 genotyping, as current assays are not as automated as other molecular microbiological tests.
TABLE 1.
Comparison of different approaches to SARS-CoV-2 genotypinga
| Parameter | Allele-specific RT-qPCR | Targeted/Sanger sequencing | WGS |
|---|---|---|---|
| Cost | $ | $$ | $$$ (depends on batch size) |
| Real-world TAT | 0−2 days | 2–7 days | 3−10 days |
| Advantages | Rapid TAT to impact MAb choice; widely available; easy to define targets | Potentially faster TAT than that of WGS; potentially more widely available | Outbreak investigation; novel mutation identification; no need to redevelop assay to identify new variants |
| Disadvantages | Limited targets; need for continuous updates to include new variants | Limited targets | Greater informatics expertise, cost, TAT |
TAT, turnaround time; MAb, monoclonal antibody. Reagent costs for WGS can be as low as $30 to $40 per sample if sufficient batch size is obtained. Given that none of these tests are highly automated, labor costs comprise a significant proportion of the total cost.
Allele-specific RT-qPCR.
Due to the desire for rapid turnaround times and the ability to focus on mutational hot spots associated with viral function, allele-specific RT-qPCR has emerged as a potential solution for identifying SARS-CoV-2 variants with potential therapeutic implications. Utilization of RT-qPCR by frontline laboratories can mitigate the demand for WGS, in which capacity may currently be limited. This approach also serves as a solution to test samples with relatively small amounts of viral RNA, as indicated by higher PCR cycle threshold (Ct) values (e.g., >30). Testing by WGS often yields lower depth of genome coverage in these situations (5). For identification of specific mutations, amplification primers are designed to carry a nucleotide sequence complementary to the sequence of the mutation; this leads to preferential binding to mutant genomes and causes the wild-type genome to have a mismatch at the mutation site. Viral genomes bearing the mutation of interest are selectively amplified in RT-qPCRs and then detected with a fluorescence-labeled oligonucleotide probe. These reactions can be multiplexed, allowing multiple mutations to be analyzed simultaneously (6). The primary drawback of this method is that the focus on a select group of mutations may not specifically define the correct lineage. Allele-specific RT-qPCR assay design requires continuous updating as new variants emerge.
SARS-CoV-2 whole-genome sequencing.
The alternative approach to genotypic analysis is to determine the full nucleotide sequence, either for the whole viral genome or for a specific region containing most of the known medically relevant mutation sites (such as the spike glycoprotein [S] gene). SARS-CoV-2 WGS is generally performed via amplicon tiling approaches, which involve hundreds of small overlapping RT-PCRs to cover the entire genome. Amplicon panels are relatively sensitive and can generally recover viral genomes for specimens for which the Ct is <30 (i.e., approximately 50,000 viral copies/mL). Numerous commercial and lab-developed wet-lab sequencing protocols are available, including ARTIC, Swift, Illumina COVID-Seq (a derivation of ARTIC), and the Ion AmpliSeq SARS-CoV-2 research panel (7, 8). While specific amplicons may drop out occasionally due to mutations at primer binding sites, most protocols are robust enough to recover >99% of the SARS-CoV-2 genome. Informatics requirements for validation will depend on what is reported. To enable data sharing, consensus SARS-CoV-2 genomes should be deposited in the GISAID and/or GenBank database.
An alternative strategy to SARS-CoV-2 WGS would be to use RT-PCR to amplify the region of interest (i.e., S gene), followed by “traditional” Sanger/dideoxy sequencing to derive the nucleotide sequence for this smaller region of the viral genome. Compared to WGS, this approach is less expensive and more widely available, with faster turnaround times due to small batch sizes as the number of positive samples decreases. For example, with typical read lengths of 500 to 600 nucleotides, a single Sanger sequence could cover the region of the S gene, including most of the known consequential mutations: K417N/T, L452R, E484K/Q, and N501Y. The limitation of this strategy is that it will not cover all potentially important mutation sites, including those not yet appreciated as relevant.
REPORTING SARS-CoV-2 GENOTYPING RESULTS
What to report, and how to report it.
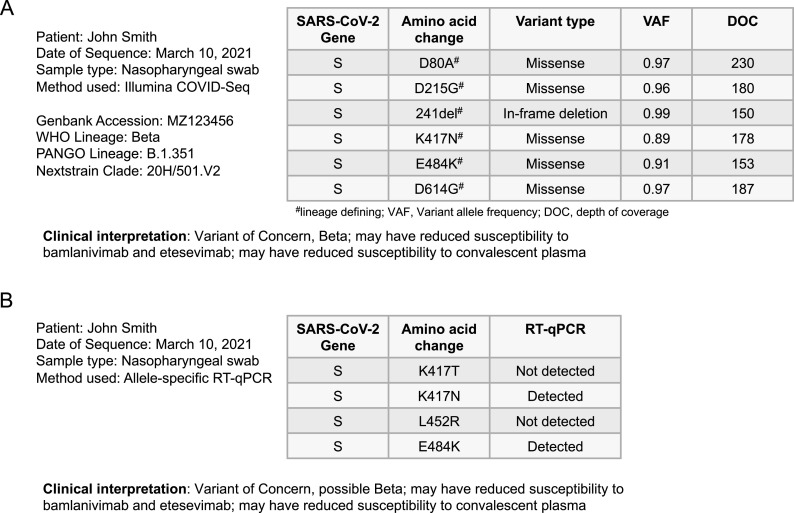
As most clinicians are now familiar with the predominant circulating VOI and VOC, it is reasonable to consider including variant lineages and clades in clinical reports, including if they were derived from the PANGO or Nextclade classification systems, as well as the genotyping method used (9). Laboratories may also elect to include the WHO naming convention (based on the Greek alphabet), but this should not be the only identification used. Updated listings can be found at https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Since new lineages continue to be identified and updated, it is prudent also to specifically report coding mutations from at least the spike protein. This allows clinical reports to be robust for future lineage classifications or changes to VOI/VOC groupings, though such analysis is unlikely to impact the individual patient whose virus has already been sequenced. Reporting coding mutations is also consistent with existing therapeutic resistance reports for other viruses and allows for the specific interrogation of mutations as new genotypic-phenotypic data become available. Due to the large size of coronavirus genomes, it may be impractical to report all coding mutations across the genome. To allow for future genome-wide interrogation should it become clinically relevant, SARS-CoV-2 genotyping reports may also include accession numbers from public databases, such as GISAID, GenBank, or even the Sequence Read Archive. Accession numbers allow end users to see the full genome, which also could be requested by the patient, but obtaining accession numbers may delay reporting. Accession numbers may also enable data uses that have not been specifically interrogated or validated during clinical testing. Finally, interpretation of the clinical implications of detecting a specific variant may be summarized in the report, including potential therapeutic impact (Fig. 1).
FIG 1.
(A) Hypothetical patient report for WGS including lineage/clade designations, coding mutations, variant allele frequency, and depth of coverage or fold coverage, as well as clinical interpretation; (B) example report for the same specimen tested by allele-specific qRT-PCR.
Given the ability to use SARS-CoV-2 whole-genome sequencing data to track transmission or reinfection, including the pairwise distance of sequences derived from multiple specimens may also be informative. Reporting of such data to infer transmission, covered further below, requires careful consideration of the associated epidemiologic data and therefore should likely be reported only to institutional infection prevention teams rather than in the patient’s electronic health record. Reinfection, however, could be reported within the individual patient’s electronic health record if the pairwise analysis meets the validated cutoff criteria. Such reporting could contain the numerical pairwise distance between the prior and current sequences with an interpretive comment that indicates whether that value supports reinfection, associated quality metrics, such as genome coverage, and limitations. As with any laboratory test result, the need for clinical and epidemiological correlation is required.
Reporting method considerations.
Reporting of the relevant data obtained by sequencing should be included as part of the validation process and in the United States must include a mechanism to report to the appropriate public health partner following CDC and CMS guidance (10). Several of the sequencing platform and kit manufacturers offer end-to-end products that include consensus sequence generation, variant calling, and relevant quality control metrics. Illumina COVIDSeq has a component of their DRAGEN COVIDSeq pipeline for genome calling that could be validated under CLIA/CAP (College of American Pathology) guidelines for clinical use. The automated functions of this pipeline will call variants with a coverage depth greater than 10-fold and with at least 50% variant allele frequency (11). Similarly, the Thermo Fisher Scientific Ion AmpliSeq SARS-CoV-2 research assay is compatible with the GeneXus software plug-ins that will generate consensus sequences and annotated lists of variants (12). The data generated from these basic programs require additional analysis to produce PANGO lineage and Nextclade assignment as well as potential clinical interpretation of identified variants. Several additional plug-ins for downstream analysis as well as direct submission to public databases are available via Illumina’s BaseSpace SARS-CoV-2 next-generation sequencing (NGS) toolkit and Thermo Fisher’s Ion Torrent suite software. Though less automated than the prior examples, Oxford Nanopore Technologies offers the cloud-based pipeline EPI2ME with a “point and click” access to the ARTIC and Nextclade pipelines (13). Third-party companies like CosmosID, One Codex, and IDbyDNA offer Web-based pipelines intended to make complete analysis and reporting simple for those laboratories lacking expertise. Laboratories with bioinformatics expertise may elect to validate more complex or custom pipelines. Numerous custom approaches have been published, and detailed comparisons are beyond the scope of this paper. A select list of SARS-CoV-2 sequencing resources, including protocols, curated by the CDC, can be found at https://github.com/CDCgov/SARS-CoV-2_Sequencing#bioinformatics. It is important to note that there may be differences or limitations to each pipeline as well as a need to revalidate under CLIA/CAP guidelines with any modifications.
Crafting of the final clinical report that enters the electronic medical record must be compatible with and validated via each institution’s laboratory information system (LIS). This will require some type of intermediate data file to convert reportable data to a format compatible with the LIS. Many commercially available LIS systems have sequence variant reporting functionality, though the features and capabilities of reporting may differ between vendors. For example, a table format to list variants may not be compatible and might require free text, adding additional complexity and the potential for error to the reporting process. Many systems can include prescripted clinical interpretation comments linked to the detection of specific lineages or variants. Reporting sequence data to public health can also be coordinated through the LIS, though much of this infrastructure remains to be built. Furthermore, building such a complex interpretive workflow requires tremendous resources and expertise that may not be available in most clinical laboratories.
CLINICAL USE CASES OF SARS-CoV-2 GENOTYPING
SARS-CoV-2 genotyping to inform MAb therapy.
Immunocompromised hosts are one group of patients for which there has been significant interest in using monoclonal antibody (MAb) therapy to reduce the potential for progression to severe disease. Immunocompromised patients can also be infected for prolonged periods, allowing time for the SARS-CoV-2 genotyping results to return and potentially inform patient management. Three anti-SARS-CoV-2 treatments have received FDA emergency use authorization (EUA) for the treatment of mild to moderate COVID-19 in outpatients who are at high risk for clinical progression. At the time of writing, currently authorized MAb therapies include bamlanivimab plus etesevimab, casirivimab plus imdevimab, and sotrovimab. Multiple other monoclonal antibodies are in phase III clinical trials. In laboratory studies, SARS-CoV-2 variants that contain the L452R or E484K substitution in the spike protein result in significantly reduced susceptibility to bamlanivimab, while those containing the K417T or K417N mutation have greatly reduced sensitivity to etesevimab (14). These mutations (K417N/T and E484K) are specifically combined in P.1 and B.1.351 VOC, indicating that individuals infected by these variants should not be treated with this combination monoclonal therapy (15). Indeed, the U.S. Government paused distribution of bamlanivimab plus etesevimab into multiple states, based on the high prevalence of P.1 variants in those locations (16). Because MAb therapy should ideally be started as soon as possible after the diagnosis of COVID-19, screening for these mutations for therapeutic purposes may be best accomplished using allele-specific RT-qPCR-based methods.
Genotype analysis to aid infection prevention in health care facilities.
SARS-CoV-2 WGS can be used to detect clusters of infection among patients and/or staff in health care facilities. Identical or highly related sequences may be consistent with a single-source exposure or person-to-person chain of infection, with the caveats noted due to the low rate of mutations (Text Box 1). Identification of several infections due to a rare lineage on one hospital ward over a short period may provide strong support for nosocomial transmission, whereas more detailed phylogenetic analysis may be required to make accurate inferences about a cluster of infections due to a common lineage. Decisions regarding whether to declare a SARS-CoV-2 outbreak within a health care facility should be based on epidemiologic suspicion, as results of sequencing may not be available immediately.
BOX 1: Considerations for Implementation of SARS-CoV-2 Whole-Genome Sequencing
CLINICAL/EPIDEMIOLOGIC CONSIDERATIONS
Genomic similarities/differences are defined as follows:
1. The low genetic diversity of SARS-CoV-2 (estimated mutation rate of 1.16 × 10−3 substitutions per site per year, which equates to around one mutation every 2 weeks [2]) means that SARS-CoV-2 isolates may be genomically identical even without an epidemiological link. A study from the United Kingdom reported that 22% of isolates with zero single nucleotide polymorphism (SNP) differences had no identifiable connection when further clinical and epidemiological investigations were performed (17).
2. Different studies have defined clusters based on a combination of SNP differences paired with epidemiological and clinical data (17–19).
3. Laboratories often do not have access to epidemiological information so will be able to provide only the sequencing portion of the information.
LABORATORY CONSIDERATIONS
Accuracy/Reproducibility
1. Intra-assay and inter-assay precision studies using the same positive sample should demonstrate identical variant calls relative to the reference genome. If possible, include positive SARS-CoV-2 samples from the same household, which are more likely to be identical. Also, testing of positive samples detected from within the hospital wards would increase the likelihood of identical or near identical clusters compared to testing of samples that were collected from the community setting or emergency department, which are more often phylogenetically dispersed (17).
2. Clinical laboratories can also send samples to an external laboratory that is currently offering the test as part of the validation.
Quality Control
1. Laboratories should consider restricting analysis to the consensus-level mutations and SARS-CoV-2 mutations with an allele frequency of at least 50%. Only high-quality SARS-CoV-2 genomes should be used for cluster analysis. A recent study defined this as at least a 100× coverage (number of reads aligned to a genomic position) across 97% of the genome (20).
2. The limit of detection or minimal read depth that is required at a given nucleotide position that is required to confidently make a base call needs to be evaluated and incorporated into the quality management system.
3. Robust bioinformatics and wet-lab cross-contamination checks, along with detailed clerical checks, are crucial to ensuring reliable results.
Workflow and Ordering Decisions
1. The number of positive samples sequenced within an institution may help define the approach. One option would be to have it as an orderable test placed by only the contact tracing or infection prevention and control (IPC) team when clusters are identified from the clinical and epidemiological standpoints. Phylogenetic analysis in these cases can help strengthen transmission links previously identified by the team but can also rule out suspect clusters that have been previously reported (20).
2. A second option would be to perform WGS and phylogenetic analysis on all SARS-CoV-2 isolates detected by molecular testing, which may prompt further epidemiological investigation to determine if a direct transmission link can be identified. Again, the link between genomic and epidemiological data needs to be emphasized.
BILLING AND REIMBURSEMENT
At the time of writing, there is no current procedural terminology (CPT) code published by the American Medical Association for SARS-CoV-2 variant identification/genotyping. In the United States, laboratories may use a generic CPT code, such as 81479; however, it is unlikely that any of this work will be reimbursed by payers, leaving the cost to be absorbed by the performing laboratory.
While genotyping is often used to confirm findings of an outbreak investigation (21, 22), some health care institutions are conducting prospective genomic surveillance for SARS-CoV-2 infection (17, 23). Meredith et al. (17) combined results of prospective rapid SARS-CoV-2 sequencing of RT-qPCR-positive diagnostic samples with clinical and epidemiologic data. They identified 35 clusters of identical viruses infecting 159 patients at one hospital over 6 weeks. Seventy-eight percent of patients within clusters had strong or plausible epidemiologic links. Several clusters included patients who were not suspected of being linked based on epidemiologic data alone. Specifically, their results highlighted the elevated risk of infection in renal dialysis units, simultaneously ruling in and out transmission links within this ward. Results were provided weekly to clinical and infection control teams, enabling further investigation and intervention in real time. The timely availability of genotyping data has the potential to improve understanding of health care-associated transmission of SARS-CoV-2 and to expedite mitigation, control, and prevention strategies that protect patients and health care providers.
Cohorting of patients based on viral genotype may be a practical consideration, particularly for older health care facilities with limited numbers of single-bed rooms. Guidelines do not currently advise specific cohorting (24), but it has been successfully implemented in some Canadian institutions (25). Repeat infections with VOC (B.1.1.7, P.1, and B.1.351) have been reported in patients previously infected with SARS-CoV-2 (26). Cohorting may be an adjunct infection control measure considered in certain clinical situations where transmission risk is potentially higher, particularly in communities where multiple VOC are cocirculating. Health care facilities with a limited private room capacity could consider cohorting patients with the same VOC based on (i) stage of clinical presentation (early versus late; sample with a low Ct), (ii) type of VOC, and (iii) operations to minimize movement/transfer of COVID-19 patients, as has been performed in a limited fashion to date (27).
THE FUTURE IS NOW
The SARS-CoV-2 pandemic has challenged the medical system like no other but offers an opportunity to build the future clinical care-public health approach to infectious disease control. SARS-CoV-2 genotyping has definitively impacted care, not least by revoking an EUA for the therapeutic monoclonal antibody bamlanivimab due the identification of resistant variants (28). SARS-CoV-2 genotyping has been critical for multiple public health purposes, including epidemiology, vaccine efficacy monitoring, vaccine planning, therapeutic choice and design, and detection of polymorphisms causing therapeutic and/or diagnostic failure. The main current clinical use for genotyping is limited to helping determine if the presence of a mutation will impact the effectiveness of a therapeutic monoclonal antibody. It is costly to validate and implement molecular testing of this type, and clinical laboratories are generally paid only for testing deemed clinically necessary to an individual beneficiary based on CMS and insurance rules. Currently there are no specific CPT codes for SARS-CoV-2 genotyping.
As such, clinical laboratories have not typically been paid to perform testing for the public’s health. Here, contracting between public health agencies and clinical laboratories to monitor the genotypes of circulating SARS-CoV-2 could be mutually beneficial, and this cooperative approach has been successfully implemented in some areas. Clinical laboratories offer the distributed scale for viral genomic surveillance as well as potentially faster turnaround times since sentinel clinical laboratories can immediately begin the genotyping process once a positive sample is identified. Public health agencies benefit from this significantly increased scale of SARS-CoV-2 genotype information produced by clinical laboratories as well as the integration of these data at the point of clinical care, while clinical laboratories benefit from increased funding to develop and perform novel genotyping assays. In many ways, this is how clinical testing for any virus, including SARS-CoV-2, is successfully performed at scale. Building these bridges will be especially important given the numerous barriers toward SARS-CoV-2 genotyping, such as the growing dispersion of SARS-CoV-2 testing with the continued growth of antigen, direct-to-consumer, and over-the-counter tests.
The benchmark for implementing tests in the clinical laboratory is when a result alters patient management and impacts outcomes or has value for hospital infection control purposes. Patient management may include the use of therapeutic agents shown to be efficacious for the particular genotype in question. For SARS-CoV-2, few studies are currently available that meet this standard, though study designs are rapidly changing with the new availability of genotypic data. If met, the case may be made that precision diagnostics of viral genotypes in the clinical laboratory are required for therapeutic agents, including monoclonal antibodies, antiviral agents, and care pathways. Indeed, in the realm of cancer therapeutics, we are seeing the need for precise detection of mutations to guide chemotherapeutic choices for both blood and solid organ malignancies. Many other human viruses are significantly more genetically diverse than SARS-CoV-2 (29), indicating that genotype-specific therapies may be required. Incipient technologies, such as CRISPR-Cas9 or genotype-specific isothermal amplification, potentially offer point-of-care or same-day genotyping (30, 31). Starting now, the clinical-public health laboratory collaborative approach developed for SARS-CoV-2 genotyping could be used for many other pathogens, including genotyping for influenza virus, HIV, hepatitis C virus, Salmonella, Listeria, and any other respiratory and foodborne pathogens associated with outbreaks.
ACKNOWLEDGMENTS
D.D.R. has received funding from Cepheid, Cleveland Diagnostics, Luminex, Qiagen, and Talis Biomedical outside the submitted work. D.R.P. has a relationship with Illucidx Inc. outside the submitted work. A.L.G. has received central testing contracts from Abbott Molecular and has received funding from Gilead and Merck outside the submitted work. M.B.M. receives fees and funding from Cepheid, Luminex Molecular Diagnostics, Qiagen, Talis Biomedical, Werfen, Agena Bioscience, Sherlock Biosciences, Abbott Molecular, and ArcBio outside the submitted work. All other authors report no potential conflicts.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Contributor Information
Alexander L. Greninger, Email: agrening@uw.edu.
Alexander J. McAdam, Boston Children's Hospital
REFERENCES
- 1.Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. 2021. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ 372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duchene S, Featherstone L, Haritopoulou-Sinanidou M, Rambaut A, Lemey P, Baele G. 2020. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol 6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jo WK, Drosten C, Drexler JF. 2021. The evolutionary dynamics of endemic human coronaviruses. Virus Evol 7:veab020. doi: 10.1093/ve/veab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev 23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Govindavari JP, Davis BD, Chen SS, Kim JT, Song J, Lopategui J, Plummer JT, Vail E. 2020. Analysis of genomic characteristics and transmission routes of patients with confirmed SARS-CoV-2 in Southern California during the early stage of the US COVID-19 pandemic. JAMA Netw Open 3:e2024191. doi: 10.1001/jamanetworkopen.2020.24191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Miller JA, Verghese M, Sibai M, Solis D, Mfuh KO, Jiang B, Iwai N, Mar M, Huang C, Yamamoto F, Sahoo MK, Zehnder J, Pinsky BA. 2021. Multiplex SARS-CoV-2 genotyping RT-PCR for population-level variant screening and epidemiologic surveillance. J Clin Microbiol 59:e00859-21. doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Addetia A, Lin MJ, Peddu V, Roychoudhury P, Jerome KR, Greninger AL. 2020. Sensitive recovery of complete SARS-CoV-2 genomes from clinical samples by use of Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. J Clin Microbiol 59:e02226-20. doi: 10.1128/JCM.02226-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasir JA, Kozak RA, Aftanas P, Raphenya AR, Smith KM, Maguire F, Maan H, Alruwaili M, Banerjee A, Mbareche H, Alcock BP, Knox NC, Mossman K, Wang B, Hiscox JA, McArthur AG, Mubareka S. 2020. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses 12:895. doi: 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2021. Guidance for reporting SARS-CoV-2 sequencing results. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/reporting-sequencing-guidance.html. Accessed 15 August 2021. [Google Scholar]
- 11.Bhoyar RC, Jain A, Sehgal P, Divakar MK, Sharma D, Imran M, Jolly B, Ranjan G, Rophina M, Sharma S, Siwach S, Pandhare K, Sahoo S, Sahoo M, Nayak A, Mohanty JN, Das J, Bhandari S, Mathur SK, Kumar A, Sahlot R, Rojarani P, Lakshmi JV, Surekha A, Sekhar PC, Mahajan S, Masih S, Singh P, Kumar V, Jose B, Mahajan V, Gupta V, Gupta R, Arumugam P, Singh A, Nandy A, P VR, Jha RM, Kumari A, Gandotra S, Rao V, Faruq M, Kumar S, Reshma GB, Varma GN, Roy SS, Sengupta A, Chattopadhyay S, Singhal K, Pradhan S, et al. 2021. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS One 16:e0247115. doi: 10.1371/journal.pone.0247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alessandrini F, Caucci S, Onofri V, Melchionda F, Tagliabracci A, Bagnarelli P, Di Sante L, Turchi C, Menzo S. 2020. Evaluation of the Ion AmpliSeq SARS-CoV-2 research panel by massive parallel sequencing. Genes (Basel) 11:929. doi: 10.3390/genes11080929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hourdel V, Kwasiborski A, Balière C, Matheus S, Batéjat CF, Manuguerra J-C, Vanhomwegen J, Caro V. 2020. Rapid genomic characterization of SARS-CoV-2 by direct amplicon-based sequencing through comparison of MinION and Illumina iSeq100TM system. Front Microbiol 11:571328. doi: 10.3389/fmicb.2020.571328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. 2021. Fact sheet for health care providers: Emergency Use Authorization (EUA) of bamlanivimab and etesevimab. Food and Drug Administration, Washington, DC. https://www.fda.gov/media/145802/download. Accessed 15 August 2021. [Google Scholar]
- 15.Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, López-Camacho C, Slon-Campos J, Walter TS, Skelly D, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert SC, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Paterson NG, Williams MA, Hall DR, Hulswit RJG, Bowden TA, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunleavy K. 27 May 2021. Eli Lilly’s COVID-19 antibody halted in 6 more states as Regeneron’s competing option sails along. FiercePharma https://www.fiercepharma.com/pharma/fda-halts-use-lilly-covid-19-antibody-treatment-6-states-where-variants-are-prevalent.
- 17.Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, Curran MD, Parmar S, Caller LG, Caddy SL, Khokhar FA, Yakovleva A, Hall G, Feltwell T, Forrest S, Sridhar S, Weekes MP, Baker S, Brown N, Moore E, Popay A, Roddick I, Reacher M, Gouliouris T, Peacock SJ, Dougan G, Török ME, Goodfellow I. 2020. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis 20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Løvestad AH, Jørgensen SB, Handal N, Ambur OH, Aamot HV. 2021. Investigation of intra-hospital SARS-CoV-2 transmission using nanopore whole genome sequencing. J Hosp Infect 111:107–116. doi: 10.1016/j.jhin.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sikkema RS, Pas SD, Nieuwenhuijse DF, O'Toole Á, Verweij J, van der Linden A, Chestakova I, Schapendonk C, Pronk M, Lexmond P, Bestebroer T, Overmars RJ, van Nieuwkoop S, van den Bijllaardt W, Bentvelsen RG, van Rijen MML, Buiting AGM, van Oudheusden AJG, Diederen BM, Bergmans AMC, van der Eijk A, Molenkamp R, Rambaut A, Timen A, Kluytmans JAJW, Oude Munnink BB, Kluytmans van den Bergh MFQ, Koopmans MPG. 2020. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect Dis 20:1273–1280. doi: 10.1016/S1473-3099(20)30527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryutov A, Gai X, Ostrow D, Maglinte DT, Flores J, Salas EJ, Glucoft M, Smit M, Dien Bard J. 19 April 2021. Utility of viral whole-genome sequencing for institutional infection surveillance during the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol doi: 10.1017/ice.2021.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klompas M, Baker MA, Rhee C, Tucker R, Fiumara K, Griesbach D, Bennett-Rizzo C, Salmasian H, Wang R, Wheeler N, Gallagher GR, Lang AS, Fink T, Baez S, Smole S, Madoff L, Goralnick E, Resnick A, Pearson M, Britton K, Sinclair J, Morris CA. 2021. A SARS-CoV-2 cluster in an acute care hospital. Ann Intern Med 174:794–802. doi: 10.7326/M20-7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan ER, Jones LD, Redmond SN, Navas ME, Kachaluba NM, Zabarsky TF, Bhullar D, Cadnum JL, Zimmerman PA, Donskey CJ. 3 May 2021. Use of whole-genome sequencing to investigate a cluster of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in emergency department personnel. Infect Control Hosp Epidemiol doi: 10.1017/ice.2021.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellingford JM, George R, McDermott JH, Ahmad S, Edgerley JJ, Gokhale D, Newman WG, Ball S, Machin N, Black GC. 2021. Genomic and healthcare dynamics of nosocomial SARS-CoV-2 transmission. Elife 10:e65453. doi: 10.7554/eLife.65453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Public Health Ontario. 2021. Interim guidance for infection prevention and control of SARS-CoV-2 variants of concern for health care settings. 1st revision. https://www.publichealthontario.ca/-/media/documents/ncov/voc/2021/02/pidac-interim-guidance-sars-cov-2-variants.pdf?la=en. Accessed 26 July 2021.
- 25.Alberta Health Services. 2021. IPC cohorting recommendations for COVID-19 in acute care. Alberta Health Services, Edmonton, Alberta, Canada. https://www.albertahealthservices.ca/assets/healthinfo/ipc/hi-ipc-covid-desgnd-unit.pdf. Accessed 15 August 2021. [Google Scholar]
- 26.Novazzi F, Baj A, Genoni A, Spezia PG, Colombo A, Cassani G, Zago C, Pasciuta R, Della Gasperina D, Ageno W, Severgnini P, Dentali F, Focosi D, Maggi F. 2021. SARS-CoV-2 B.1.1.7 reinfection after previous COVID-19 in two immunocompetent Italian patients. J Med Virol 93:5648–5649. doi: 10.1002/jmv.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matic N, Lowe CF, Ritchie G, Stefanovic A, Lawson T, Jang W, Young M, Dong W, Brumme ZL, Brumme CJ, Leung V, Romney MG. 2021. Rapid detection of SARS-CoV-2 variants of concern, including B.1.1.28/P.1, British Columbia, Canada. Emerg Infect Dis 27:1673–1676. doi: 10.3201/eid2706.210532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Food and Drug Administration. 2021. Coronavirus (COVID-19) update: FDA revokes Emergency Use Authorization for monoclonal antibody bamlanivimab. Food and Drug Administration, Washington, DC. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab. Accessed 15 August 2021. [Google Scholar]
- 29.Sharp PM. 2002. Origins of human virus diversity. Cell 108:305–312. doi: 10.1016/s0092-8674(02)00639-6. [DOI] [PubMed] [Google Scholar]
- 30.Joung J, Ladha A, Saito M, Kim N-G, Woolley AE, Segel M, Barretto RPJ, Ranu A, Macrae RK, Faure G, Ioannidi EI, Krajeski RN, Bruneau R, Huang M-LW, Yu XG, Li JZ, Walker BD, Hung DT, Greninger AL, Jerome KR, Gootenberg JS, Abudayyeh OO, Zhang F. 2020. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med 383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Stryke D, Bouquet J, Somasekar S, Linnen JM, Dodd R, Mulembakani P, Schneider BS, Muyembe-Tamfum J-J, Stramer SL, Chiu CY. 2015. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]