Abstract
Over a 2½-month period in 1999, 37 ceftazidime-resistant nonrepetitive enterobacterial isolates were collected from 37 patients in a Bangkok hospital, Thailand. Eighty-one percent of these strains expressed a clavulanic acid-inhibited extended-cephalosporin resistance profile. An identical extended-spectrum β-lactamase (ESBL), VEB-1, was found in 16 unrelated enterobacterial isolates (Escherichia coli, n = 10; Enterobacter cloacae, n = 2; Enterobacter sakazakii, n = 1; and Klebsiella pneumoniae, n = 3) and in two clonally related E. cloacae isolates. The blaVEB-1 gene was located on mostly self-conjugative plasmids (ca. 24 to 200 kb) that conferred additional non-β-lactam antibiotic resistance patterns. Additionally, the blaVEB-1 gene cassette was part of class 1 integrons varying in size and structure. The blaVEB-1-containing integrons were mostly associated with blaOXA-10-like and arr-2-like gene cassettes, the latter conferring resistance to rifampin. These data indicated the spread of blaVEB-1 in Bangkok due to frequent transfer of different plasmids and class 1 integrons and rarely to clonally related strains. Plasmid- and integron-mediated resistance to rifampin was also found in enterobacterial isolates.
Plasmid-mediated extended-spectrum β-lactamases (ESBLs) were first identified in a Klebsiella pneumoniae isolate in Germany in 1983 (15). Since then, the infections caused by ESBL-producing members of the family Enterobacteriaceae have rapidly increased (20). These enzymes confer variable degrees of protection against expanded-spectrum cephalosporins such as cefotaxime, ceftazidime, and the monobactam aztreonam. Their activity is inhibited by clavulanic acid in vitro (13, 15). Most of the ESBLs that are disseminated worldwide are derivatives of narrow-spectrum TEM- and SHV-type β-lactamases, with one or more amino acid substitutions surrounding their active site, thus explaining the extension of their hydrolytic profile (20). In addition to these ESBLs, non-TEM, non-SHV derivatives with weak structural relationships have been detected with specific geographical distributions, such as CTX-M derivatives in Europe and South America, TOHO-1 and TOHO-2 in Japan, PER-1 in Turkey, and PER-2 in South America (2, 3, 7, 8, 11, 19, 25, 40, 41).
Strains expressing the ESBL VEB-1 (also named CEF-1) have rarely been identified, i.e., one Escherichia coli and one K. pneumoniae isolate from the same Vietnamese patient and two Pseudomonas aeruginosa isolates from two patients hospitalized in Thailand (23, 30, 38). Genetic analysis of blaVEB-1 revealed either its chromosome or its plasmid location and always its integration within class 1 integrons of variable structure. Integrons contain a site-specific recombination system able to capture and express genes as gene cassettes (4, 9). The essential components of class 1 integrons are the 5′ conserved segment (5′-CS) that includes an integrase gene, intI, which encodes a site-specific recombinase, an adjacent site, attI, that is recognized by the integrase and acts as a receptor for gene cassettes, and a common promoter region(s), Pant (P1) and/or P2, from which integrated gene cassettes are expressed (5, 32). The 3′ conserved segment (3′-CS), located downstream of the integrated gene cassettes, usually contains a combination of the three genes qacE1 (antiseptic resistance), sulI (resistance to sulfonamides), and an open reading frame (orf5) of unknown function (27). Each gene cassette is associated with a site-specific recombination site designated the 59-base element (59-be) and located downstream of the gene. Among the cassette-integrated β-lactamase genes, most of them encode β-lactamases of Ambler class D (oxacillin-hydrolyzing β-lactamases) and only rarely of class B (such as IMP-1, VIM-1, and VIM-2 carbapenem-hydrolyzing β-lactamases) or of class A (carbenicillin-hydrolyzing β-lactamases) (10, 22, 31, 32). The blaVEB-1 and blaGES-1 gene cassettes are the only class A ESBL gene cassettes so far known as part of class 1 integrons (29, 30).
The aim of the present study was to evaluate the prevalence of nosocomial Enterobacteriaceae isolates that produced VEB-1 among nonrepetitive ceftazidime-resistant Enterobacteriaceae isolates over a 2½-month period in 1999 from patients hospitalized in a hospital in Bangkok, Thailand. To determine whether the enterobacterial isolates were epidemiologically related, they were compared for their β-lactamase content, their plasmid profile, and their genotype using arbitrary primer PCR analyses. Conjugation experiments (and/or electrotransformation) were performed to analyze the self-conjugative property of the plasmids. Finally, class 1 integrons were searched and analyzed in the VEB-1-positive isolates. This is, to our knowledge, the first work designed to study the distribution of an Ambler class A ESBL in this part of the world and the first study on the molecular epidemiology of an integron-located class A ESBL gene.
MATERIALS AND METHODS
Bacterial isolates.
Thirty-seven nonrepetitive ceftazidime-resistant entero-bacterial isolates were consecutively collected in the bacteriology laboratory in the Department for Microbiology at Siriraj Hospital, Bangkok (the biggest hospital facility in Thailand) from June to August 1999. The total number of enterobacterial isolates in the studied period was as follows: E. coli (n = 266), K. pneumoniae (n = 62), and Enterobacter sp. (n = 46). Isolates were identified by using the API20E system (bioMérieux SA, Marcy-l'Etoile, France). Electrocompetent E. coli DH10B (GIBCO BRL, Life Technologies, Cergy Pontoise, France) was used as a recipient strain in transformation experiments. Nalidixic acid-resistant E. coli JM109 was used as host in conjugation experiments (30). E. coli NCTC 50192, harboring 154-, 66-, 38-, and 7-kb plasmids, was used as a plasmid-containing reference strain (30). E. coli DH10B harboring recombinant plasmid pRLT1 that carries the blaVEB-1 gene was used as a VEB-1-producing reference strain, as described previously (30).
Susceptibility testing and screening for ESBL-producing strains.
The antibiotic susceptibility of Enterobacteriaceae clinical isolates and their E. coli transformants was first determined by the disk diffusion method on Mueller-Hinton (MH) agar plates with β-lactam and non-β-lactam antibiotic-containing disks (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France), according to the guidelines of the antibiogram committee of the French Society for Microbiology (30). Since in some cases transconjugants were obtained, plasmid DNAs of the transconjugants were electroporated into E. coli DH10B in order to make valid comparisons of the MICs for all transformants in an identical E. coli genetic background. The double-disk synergy test was performed with cefotaxime, ceftazidime, aztreonam, and amoxicillin-clavulanic acid disks on MH agar plates, and the results were interpreted as described previously (13).
MICs were determined for selected β-lactams by an agar dilution technique on MH agar with an inoculum of 104 CFU per spot, as described previously (28). MICs of some β-lactams were determined alone or in combination with a fixed concentration of either clavulanic acid (2 μg/ml) or tazobactam (4 μg/ml). MIC results were interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (24).
Conjugation, electroporation, and plasmid DNA content analysis.
Conjugation experiments were performed between clinical isolates and E. coli JM109 in solid and liquid media at 37°C as reported previously (28). Transconjugants were selected on Trypticase soy (TS) agar plates containing 100 μg of nalidixic acid per ml and 2 μg of ceftazidime per ml. Non-self-conjugative plasmid DNAs of the enterobacterial isolates and plasmid DNAs of the transconjugants were extracted as described previously (28). They were electroporated into E. coli DH10B, and recombinant strains were selected on ceftazidime-containing (2 μg/ml) TS agar plates. Plasmid DNAs of these transformants were analyzed as described previously (28).
IEF analysis.
Cultures of the E. coli electroporants were grown overnight at 37°C in 10 ml of TS broth containing 2 μg of ceftazidime per ml. Then, 1 ml of each overnight culture was further grown for 3 h at 37°C in 10 ml of TS broth with ceftazidime. Analytical isoelectric focusing (IEF) was performed using an ampholine polyacrylamide gel as described previously (28).
PCR amplification for detection of ESBL genes, analysis of class 1 integrons, and sequencing.
Under standard PCR conditions (33), a series of primers was designed for detection of Ambler class A β-lactamase genes. Detection was performed for genes encoding TEM (TEM-A and TEM-B [29, 35]), SHV (SHV-F and SHV-B [21]), PER-1/PER-2 (PER-A and PER-B [25, 29]), VEB-1 (VEB-1A and VEB-1B [23]), CTX-M (CTXM-2A and CTXM2B [8]), TOHO-1/TOHO-2 (TOHO-A and TOHO-B [11, 29]), and GES-1 (GES-1A and GES-1B [29]) (Table 1). For each reaction, 0.5 μg of whole-cell DNA of the ESBL-possessing Enterobacteriaceae isolates or 0.5 μg of plasmid DNA from E. coli DH10B electroporants was used. Primers for the detection of class 1 integrons were located in the 5′-CS and in the 3′-CS regions (primers 5′-CS and 3′-CS [16]). A combination of 5′-CS or 3′-CS and blaVEB-1 primers was also used for the determination of the genetic content of the class 1 integrons (primers 5′-CS and VEB-B, or 3′-CS and VEB-F [16, 23]). Additionally, since oxa-10 and arr-2 genes had been found to be associated with the blaVEB-1 gene (23, 38), their detections by PCR amplification were performed (primers OXA-10casF and -casB [Table 1] and ARR-2-F and ARR-2-B [37]). Their position relative to the blaVEB-1 gene was determined by PCR using primers for blaVEB-1 (primers VEB-1A and VEB-1B [23] and VEB-INV4F and VEB-INFV3B [Table 1]), for blaOXA-10 (primers OXA-10promB [Table 1] and OPR-1 and OPR-2 [42]), and for arr-2 (primers ARR-2-F and ARR-2-B).
TABLE 1.
Unpublished PCR primers used in this study
| Gene | Primera | Sequence 5′ to 3′ | Accession no.b | Position of primer |
|---|---|---|---|---|
| blaVEB-1 | VEB-INV4F | ACGAAGAACAAATGCACAAGG | AF010416 | 660–681 |
| blaVEB-1 | VEB-INV3B | GAACAGAATCAGTTCCTCCG | AF010416 | 610–591 |
| blaOXA-10 | OXA-10promB | CGTTAATTCGACCTCAGAGGC | AF205943 | 7491–7471 |
| blaOXA-10 | OXA-10casF | TTAGGCCTCGCCGAAGCG | AF205943 | 7331–7348 |
| blaOXA-10 | OXA-10casB | CTTTGTTTTAGCCACCAATGATG | AF205943 | 8297–8319 |
F, forward nucleotide sequence; B, backward nucleotide sequence.
Accession numbers are from the published sequences in the GenBank and EMBL databases.
For direct DNA sequencing, PCR products using external primers for the blaVEB-1 gene cassette (primers VEBcas-F and VEBcas-B [23]) were purified with PCR purification columns (Qiagen). Sequencing reactions were performed using the same blaVEB-1 specific primers and an automated sequencer (ABI 377; Applied Biosystems, Foster City, Calif.). The nucleotide and deduced protein sequences were analyzed with software available over the Internet (29).
RAPD fingerprinting.
Random amplified polymorphism DNA (RAPD) analysis was performed and interpreted as described by Williams et al. (43) with some modifications as reported previously (28). The RAPD primers were ERIC-2 and 628 (28) and AP1 and AP4 (1).
Hybridizations.
DNA-DNA hybridizations were performed as described by Sambrook et al. with a Southern transfer of an agarose gel containing plasmid DNA from E. coli DH10B electroporants as a template (33). The probe consisted of a 650-bp PCR fragment generated from recombinant plasmid pRLT-1 and internal to blaVEB-1 (30). Labeling of the probe and signal detection were carried out using a nonradioactive labeling and detection kit according to the manufacturer's instructions (Amersham Pharmacia Biotech).
RESULTS
Epidemiology and preliminary PCR detection of β-lactamase genes.
A total of 37 nonrepetitive ceftazidime-resistant enterobacterial isolates were collected from hospitalized patients in a 2,000-bed hospital in Bangkok from June to August 1999. Ceftazidime resistance accounted for 4.5% (12 of 266) of E. coli, 41% (19 of 46) of Enterobacter sp., and 9.6% (6 of 62) of K. pneumoniae isolates. Preliminary antibiotic susceptibility testing by disk diffusion showed synergy between ceftazidime- and clavulanate-containing disks for 30 of these 37 isolates. The seven isolates that did not show synergy image were Enterobacter cloacae. They likely had a derepressed expression of their cephalosporinase since the diameter around the ceftazidime disk increased when antibiograms were performed on oxacillin-containing MH plates (200 μg/ml) (data not shown).
Most of the synergy-positive isolates were not K. pneumoniae but Enterobacter sp. and E. coli (24 of 30 isolates). PCR experiments using primers specific for blaVEB-1, blaTEM, blaSHV, blaGES-1, blaPER-1, and blaCTX-M-1 gave blaVEB-1-positive results for 18 of these 30 strains (Table 2). Among the 12 blaVEB-1-negative strains, all E. cloacae isolates (n = 6) were blaSHV positive and 4 of these strains were also blaTEM positive, the Enterobacter sakazakii isolates were blaSHV positive and one of them was additionally blaTEM positive, the E. coli isolate was blaTEM and blaSHV positive, and the naturally blaSHV-positive K. pneumoniae strain was also blaTEM positive (data not shown). Thus, the 12 blaVEB-1-negative isolates that showed a positive synergy test possessed either a blaTEM- and/or a blaSHV-derivative gene that may explain their ESBL phenotype.
TABLE 2.
Clinical features of the ceftazidime-resistant blaVEB-1-positive enterobacterial isolatesa
| Isolate | Date of isolationb | Dates of hospitalizationb | Hospitalization unit | Source | Underlying disease | Treatment | Clinical outcome |
|---|---|---|---|---|---|---|---|
| E. coli 1 | 06-15-99 | 07-09-95 to 10-01-00 | Medicine | Urine | UTI | None | Improved |
| E. coli 2 | 06-16-99 | 05-21-99 to 06-25-99 | Medicine | Urine | Diabetes mellitus, septicemia with UTI | Imipenem | Dead |
| E. coli 3 | 07-09-99 | 05-09-99 to 08-10-99 | Medicine | Urine | Septicemia with UTI | Sulbactam-cefoperazone | Cured |
| E. coli 4 | 07-18-99 | 06-16-99 to 07-24-99 | Medicine | Urine | Diabetes mellitus, pneumonia, UTI | Amoxicillin-clavulanic acid | Cured |
| E. coli 5 | 07-20-99 | 06-26-99 to 08-03-99 | Medicine | Urine | Psychiatric disorder, UTI | None | Cured |
| E. coli 6 | 07-23-99 | 07-22-99 to 07-26-99 | Pediatrics | Urine | Neuroblastoma, UTI | None | Cured |
| E. coli 7 | 08-03-99 | 08-01-99 to 08-07-99 | Surgery | Urine | Chronic aortic dissection, UTI | None | Cured |
| E. coli 8 | 08-07-99 | 05-26-99 to 09-17-99 | Medicine | Urine | Diabetes mellitus, UTI | Ceftazidime | Cured |
| E. coli 9 | 08-09-99 | 07-30-99 to 08-06-99 | Pediatrics | Urine | UTI | Ceftazidime | Cured |
| E. coli 10 | 08-26-99 | 07-08-99 to 09-15-99 | Pediatrics | Pus | Scalp abscess | Gentamicin | Cured |
| E. cloacae 11 | 06-26-99 | 06-21-99 to 06-28-99 | Surgery ICU | Pus | Septicemia, congenital diaphragmatic hernia | Imipenem | Dead |
| E. cloacae 12 | 07-13-99 | 06-28-99 to 08-19-99 | Surgery | Pus | Neonatal intestinal obstruction | Cefotaxime-netilmicin | Cured |
| E. cloacae 13 | 07-31-99 | 06-17-99 to 10-03-99 | Surgery | Urine | UTI | Unknown | Unknown |
| E. cloacae 14 | 08-03-99 | 07-24-99 to 11-07-99 | Medicine ICU | Urine | Pneumonia, UTI | Imipenem, netilmicin | Cured |
| E. sakazakii 15 | 08-16-99 | 08-02-99 to 10-07-99 | Medicine | Pus | Triple vessel disease, diabetes mellitus | Ceftazidime, cefoperazone-sulbactam | Cured |
| K. pneumoniae 16 | 08-03-99 | 07-08-99 to 09-15-99 | Neurology ICU | Urine | Cerebral tumor, pneumonia, UTI | Imipenem | Dead |
| K. pneumoniae 17 | 08-17-99 | 06-16-99 to 08-28-99 | Surgery | Bile | Infected cholangiocarcinoma | Unknown | Dead |
| K. pneumoniae 18 | 08-20-99 | 08-06-99 to 09-16-99 | Surgery | Urine | Coronary artery disease, UTI | Ofloxacin | Cured |
ICU, intensive care unit; UTI, urinary tract infection.
Dates are given as month-day-year.
The study was then focused on the 18 blaVEB-1 PCR-positive isolates. Random PCR showed that these isolates were not clonally related within a given enterobacterial species except E. cloacae 11 and 12 from two patients hospitalized in the same surgery department at different times (Table 2).
MICs of β-lactam antibiotics and IEF analysis.
In all cases, the MICs of ceftazidime and aztreonam were higher than those of cefotaxime, which is consistent with the presence of VEB-1 β-lactamase (Table 3) (30). The addition of clavulanic acid consistently decreased the MICs of ceftazidime, cefotaxime, and aztreonam (Table 3). The MICs of β-lactams for transformants were very similar to those found for clinical isolates (Table 3). IEF analysis showed that the transformants expressed a β-lactamase pI value of 7.4 that corresponded to VEB-1 in all cases (Table 4). An additional β-lactamase with a pI value of 6.3 was found in all cases except for three transformants (E. cloacae strains 11, 12, and 14) (Table 4). In one case (transformant E. cloacae 14) a β-lactamase with a pI value of 6.5 instead of 6.3 was found. Additionally, a β-lactamase with a pI value of 5.4 that may correspond to a TEM derivative was found in four transformants (E. coli 1, E. cloacae 13, E. cloacae 15, and K. pneumoniae 18), all of them being blaTEM PCR positive (Table 4).
TABLE 3.
MICs of β-lactams for blaVEB-1-positive clinical isolates, their transformants in E. coli DH10B, and the E. coli DH10B reference strain
| Isolate | MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMX | PIP | TZB | CEF | FOX | CAZ | CAZ-CLA | CTX | CTX-CLA | AZT | |
| E. coli DH10B | 2 | 2 | 1 | 4 | 8 | 0.12 | 0.12 | 0.06 | 0.06 | 0.12 |
| E. coli 1 | >512 | 256 | 4 | 128 | 8 | 128 | 0.25 | 8 | 0.12 | 128 |
| Transformant | >512 | 128 | 8 | 64 | 4 | 128 | 0.25 | 4 | 0.06 | 64 |
| E. coli 2 | >512 | 64 | 4 | 256 | 32 | 128 | 0.25 | 16 | 0.25 | 256 |
| Transformant | 512 | 32 | 4 | 64 | 8 | 256 | 0.25 | 2 | 0.06 | 64 |
| E. coli 3 | >512 | 128 | 4 | 256 | 8 | 128 | 0.25 | 8 | 0.06 | 128 |
| Transformant | 512 | 16 | 4 | 32 | 8 | 128 | 0.25 | 2 | 0.06 | 64 |
| E. coli 4 | >512 | 64 | 2 | 64 | 4 | 128 | 0.12 | 2 | 0.06 | 64 |
| Transformant | 512 | 16 | 4 | 64 | 8 | 256 | 0.25 | 4 | 0.06 | 64 |
| E. coli 5 | >512 | 64 | 4 | 64 | 4 | 128 | 0.12 | 2 | 0.06 | 64 |
| Transformant | 512 | 16 | 4 | 32 | 4 | 128 | 0.25 | 2 | 0.06 | 32 |
| E. coli 6 | >512 | 64 | 2 | 128 | 4 | 64 | 0.25 | 8 | 0.06 | 64 |
| Transformant | 512 | 16 | 2 | 32 | 4 | 128 | 0.25 | 2 | 0.06 | 64 |
| E. coli 7 | >512 | >512 | 2 | 128 | 4 | 128 | 0.12 | 8 | 0.06 | 64 |
| Transformant | 512 | 16 | 4 | 32 | 4 | 128 | 0.12 | 4 | 0.12 | 64 |
| E. coli 8 | >512 | 256 | 4 | 64 | 4 | 128 | 0.5 | 4 | 0.06 | 32 |
| Transformant | 256 | 16 | 2 | 32 | 4 | 128 | 0.25 | 2 | 0.06 | 64 |
| E. coli 9 | >512 | 32 | 2 | 32 | 2 | 64 | 0.06 | 4 | 0.06 | 32 |
| Transformant | 512 | 16 | 2 | 32 | 4 | 128 | 0.25 | 4 | 0.12 | 64 |
| E. coli 10 | >512 | >512 | 32 | 128 | 4 | 128 | 0.25 | 4 | 0.06 | 64 |
| Transformant | 256 | 16 | 2 | 32 | 4 | 128 | 0.25 | 4 | 0.06 | 32 |
| E. cloacae 11 | >512 | 64 | 32 | >512 | 512 | 256 | 32 | 64 | 32 | 64 |
| Transformant | 256 | 16 | 4 | 128 | 4 | 64 | 0.25 | 1 | 0.06 | 16 |
| E. cloacae 12 | >512 | 64 | 64 | >512 | >512 | 128 | 32 | 64 | 64 | 64 |
| Transformant | 256 | 8 | 2 | 128 | 4 | 64 | 0.25 | 1 | 0.06 | 16 |
| E. cloacae 13 | >512 | >512 | 2 | >512 | 64 | 64 | 0.5 | 16 | 0.5 | 256 |
| Transformant | >512 | 128 | 2 | 256 | 4 | 64 | 0.25 | 1 | 0.06 | 128 |
| E. cloacae 14 | >512 | >512 | 256 | >512 | 256 | 512 | 128 | 256 | 256 | 128 |
| Transformant | >512 | 256 | 2 | 512 | 8 | >512 | 0.5 | 64 | 0.06 | >512 |
| E. sakazakii 15 | >512 | 512 | 8 | >512 | 64 | 512 | 8 | 16 | 16 | 128 |
| Transformant | >512 | 512 | 4 | 64 | 16 | 256 | 0.25 | 4 | 0.06 | 64 |
| K. pneumoniae 16 | >512 | 64 | 4 | 256 | 8 | 256 | 0.12 | 8 | 0.06 | 128 |
| Transformant | 512 | 16 | 4 | 32 | 8 | 18 | 0.25 | 4 | 0.06 | 64 |
| K. pneumoniae 17 | >512 | 128 | 8 | 128 | 8 | 512 | 0.5 | 8 | 0.12 | 256 |
| Transformant | >512 | 32 | 2 | 64 | 8 | 128 | 0.5 | 4 | 0.06 | 64 |
| K. pneumoniae 18 | >512 | 64 | 4 | 256 | 4 | 256 | 0.12 | 16 | 0.06 | 128 |
| Transformant | >512 | 64 | 2 | 128 | 8 | 128 | 0.25 | 4 | 0.06 | 64 |
Abbreviations: AMX, amoxicillin; PIP, piperacillin; TZB, piperacillin and tazobactam at a fixed concentration of 4 μg/ml; CEF, cephalothin; FOX, cefoxitin; CAZ, ceftazidime; CLA, clavulanic acid at a fixed concentration of 2 μg/ml; CTX, cefotaxime; AZT, aztreonam.
TABLE 4.
Genotypic characterization of enterobacterial isolates and their transformants, β-lactamase isoelectric points and plasmid DNA sizes of transformants, and phenotypic characterization of non-β-lactam antibiotic resistance markers
| Isolate | PCRa
|
PCR (kb)a
|
Isoelectric point(s) | Plasmid size (kb) | Associated non-β-lactam antibiotic resistance markersb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| blaTEM | blaSHV | blaOXA-10 | arr-2 | 5′ CS-blaVEB-1 | blaVEB-1-3′ CS | blaVEB-1-arr-2 | blaVEB-1-blaOXA-10 | ||||
| E. coli 1 | + | − | + | + | Sp Ka Tb Ss Sx Cl Na Pe Ra | ||||||
| Transformant | + | − | + | + | 0.1 | 6.3; 0.8 | 0.7 | 3 | 7.4; 6.3; 5.4 | 210 | Sp Ka Ss Cl Ra |
| E. coli 2 | + | − | + | + | Sp Ka Tb Ss Sx Cl Te Na Pe Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 4.5 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Cl Ra |
| E. coli 3 | − | + | + | + | Sp Ka Tb Ss Sx Cl Te Na Pe Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 8.2; 2.3 | 1.8 | 5 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Cl Ra |
| E. coli 4 | + | − | + | + | Sp Ka Tb Ss Sx Cl Na Pe Ra | ||||||
| Transformant | − | − | + | + | 4.2 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Cl Ra |
| E. coli 5 | + | − | + | + | Sp Ka Tb Ss Sx Cl Te Na Pe Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 210 | Sp Ka Tb Ss Sx Cl Te Ra |
| E. coli 6 | + | − | + | + | Sp Ka Tb Ss Sx Cl Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Cl Ra |
| E. coli 7 | + | − | + | + | Sp Ka Tb Ss SX Cl Te Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Cl Ra |
| E. coli 8 | + | − | + | + | Sp Ka Tb Ss Sx Cl Te Na Pe Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 8.6 | − | 5 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Te |
| E. coli 9 | − | − | + | + | Sp Ka Tb Ss Cl Te Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Cl Ra |
| E. coli 10 | + | − | + | + | Sp Ka Tb Ss Sx Cl Te Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 210 | Sp Cl Ra |
| E. cloacae 11 | − | − | − | + | Ka Tb Ra | ||||||
| Transformant | − | − | − | + | 1.3 | 3.9 | − | − | 7.4 | 150 | Ka |
| E. cloacae 12 | − | − | − | + | Ka Tb Ra | ||||||
| Transformant | − | − | − | + | 1.3 | 3.9 | − | − | 7.4 | 150 | Ka |
| E. cloacae 13 | + | + | + | + | Sp Ka Tb Ss Sx Cl Te Ra | ||||||
| Transformant | + | − | − | + | 0.1 | 0.8 | − | 3 | 7.4; 6.3; 5.4 | 100 | Ka Tb Ss Sx Ra |
| E. cloacae 14 | + | − | − | + | Sp Ka Tb Ss Sx Cl Ra | ||||||
| Transformant | − | − | − | + | 0.1 | 0.8 | − | − | 7.4; 6.5 | 24 | Ss |
| E. sakazakii 15 | + | − | + | + | Sp Ka Tb Ss Sx Cl Te Na Pe Ra | ||||||
| Transformant | + | − | + | + | 1.7 | 4.8 | 0.7 | 3 | 7.4; 6.3; 5.4 | 150 | Ka Tb Te Ra |
| K. pneumoniae 16 | − | + | + | + | Sp Ka Tb Ss Sx Ra | ||||||
| Transformant | − | − | + | + | 2.3 | 6.5 | 0.7 | 3 | 7.4; 6.3 | 150 | Sp Ka Tb Ss Ra |
| K. pneumoniae 17 | − | + | + | + | Sp Ka Tb Ss Sx Cl Te Na Pe Ra | ||||||
| Transformant | − | − | + | + | 1.4 | 4.8 | 0.7 | 3 | 7.4; 6.3 | 210 | Sp Ka Tb Ss Sx Cl Te Ra |
| K. pneumoniae 18 | + | + | + | + | Sp Ka Tb Ss Sx Cl Te Ra | ||||||
| Transformant | + | − | + | + | 0.1 | 6.3; 0.8 | 0.7 | 3 | 7.4; 6.3; 5.4 | 210 | Sp Ka Ss Cl Ra |
Minus (−) indicates that no PCR product was obtained.
Antibiotic resistance markers: Sp, spectinomycin; Ka, kanamycin; Tb, tobramycin; Ss, sulfonamides; Sx, trimethroprim-sulfamethoxazole; Cl, chloramphenicol; Te, tetracycline; Na, nalidixic acid; Pe, pefloxacin; Ra, rifampin.
Transfer of resistance and plasmid analysis.
The transmissibility of the ceftazidime resistance marker was tested by conjugation. The transfer of resistance was obtained for 8 of 18 strains at a high frequency (10−3 to 10−4). For the remaining 10 strains, transfer of the ceftazidime resistance marker was obtained by electroporation using plasmid extracts from the clinical isolates. Plasmid DNAs of the transconjugants were extracted and retransformed into E. coli DH10B in order to make a valid comparison between transconjugants and transformants. All transformants were checked to be blaVEB-1 positive by hybridization (data not shown). The plasmid sizes of the transformants ranged from ca. 24 to 210 kb (Table 4). A correlation was established between plasmids of similar size and non-β-lactam antibiotic resistance markers of the transformants (Table 4). This was the case for transformants of E. coli strains 2, 3, 4, 6, 7, and 9 on the one hand and transformants of E. coli 1 and K. pneumoniae 18 on the other hand. It is likely that, in some cases, plasmid transfer may explain interspecies transfer of blaVEB-1. Additionally, E. cloacae 11 and 12 were clonally related and harbored identically sized plasmid conferring identical non-β-lactam antibiotic resistance phenotypes in their transformants (Table 4).
Identification of blaVEB-1 and integron structure determination.
From each of the 18 blaVEB-1-positive strains, cassette-located external primers for the blaVEB-1 gene were used to PCR amplify and sequence the entire blaVEB-1 genes. An identical blaVEB-1 gene was identified in all cases, showing the widespread distribution of this ESBL gene and underlining the genetic stability of this coding sequence and its cassette (data not shown). Since the blaVEB-1 gene cassette had been reported as integron located, PCR amplifications were performed to identify its precise location. Using 5′-CS, 3′-CS, and blaVEB-1 primers, PCR fragments of various sizes were obtained (Table 4), indicating that blaVEB-1 was part of class 1 integrons. Taking into account the size of the amplified fragments, it could be deduced that the surrounding sequences upstream of blaVEB-1 varied from 0.1 to 4.2 kb. Since the MICs of β-lactams were similar for each of the transformants (except E. cloacae 14 [Table 4]), the expression of VEB-1 was not related to the position of the blaVEB-1 cassette relative to that of the 5′-CS where the integron promoter sequences are located.
Using primers located in the 3′-CS and in the blaVEB-1 gene, amplimers were obtained ranging from 0.8 to 8.2 kb. Thus, the sizes of the blaVEB-1-containing integrons were on the average around 10 kb, a size that fits with that previously described for other blaVEB-1-containing integrons (23, 29, 38). Interestingly, in some cases (transformants E. coli 1 and 3 and K. pneumoniae 18), two PCR fragments were obtained using primers for blaVEB-1 and 3′-CS. This result may indicate the presence of two different blaVEB-1-containing integrons within the same plasmid or spontaneous deletion of the gene cassette within integrons.
Since blaOXA-10 is associated with blaVEB-1 in P. aeruginosa JES isolates (23), PCR experiments using blaOXA-10 primers were performed. In all cases but three (E. cloacae 11, 12, and 14 and their transformants), a blaOXA-10-like gene was identified (Table 4). A correlation was established between expression of a β-lactamase with a pI value of 6.3 and blaOXA-10-like positive PCR results (Table 3). These blaOXA-10-like genes likely coded for the narrow-spectrum clavulanic acid-resistant β-lactamase OXA-10 because (i) the MICs of ceftazidime, cefotaxime, and aztreonam for the transformants were decreased by the addition of clavulanic acid (almost at the level for wild-type E. coli DH10B) and (ii) the pI value of 6.3 corresponded to OXA-10. In the case of transformant E. cloacae 14, the β-lactamase with a pI value of 6.5 did not correspond to any of the class A ESBL genes tested nor to oxa-1, oxa-2, and pse-1 derivatives (data not shown).
PCR detection of the arr-2 gene that conferred resistance to rifampin was found for all clinical isolates and transformants, thus indicating a plasmid location for this gene. Expression of rifampin resistance was found for all transformants except E. coli 8 and E. cloacae 11, 12 and 14. These transformants and transformant E. cloacae 13 gave a weakened PCR signal compared to the other transformants (data not shown).
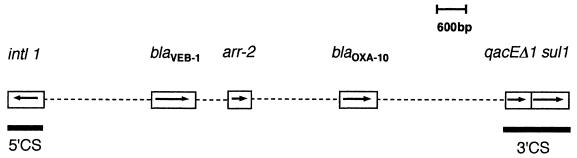
By performing PCR experiments with primers for blaVEB-1, blaOXA-10, and arr-2 genes and comparing the results with those obtained with primers for blaVEB-1, 5′-CS, and 3′-CS, the structures of integrons were deduced. The most common structure of the integrons was the downstream location of the arr-2 gene from blaVEB-1 followed by a blaOXA-10-like gene (transformants E. coli 1, 2, 3, 4, 5, 6, 7, 9, and 10, E. cloacae 15, and K. pneumoniae 16, 17, and 18) (Table 4 and Fig. 1). In one case (transformant E. cloacae 13), a blaOXA-10-like gene was located outside the 3′-CS sequence (Table 4). For transformants E. coli 8 and E. cloacae 11, 12, and 13, which gave a weak PCR signal for the arr-2 gene, no blaVEB-1–arr-2 fragment was amplified by PCR. In these cases, an arr2-like gene may be involved in conferring either no or a low level of resistance to rifampin (Table 4). Although the sul1 gene is associated with class 1 integrons, expression of resistance to sulfonamides was not found in several transformants (E. coli 10, E. cloacae 11 and 12, and E. sakazakii 15 [Table 4]). In these cases, the sul1 gene is either lacking or not expressed, as reported previously (4, 5, 32).
FIG. 1.
Schematic representation of the most common class 1 integron that carries the blaVEB-1 gene cassette as found in transformants of E. coli 5, 6, 7, 9, and 10 and K. pneumoniae 16. The coding regions are represented as boxes, with arrows indicating their transcription direction. Dashed lines represent undetermined nucleotide sequences but are proportional to the gene distance.
Different groups of Enterobacteriaceae with integrons of identical structure were found: (i) transformants E. coli 1 and K. pneumoniae 18 and (ii) E. coli 5, 6, 7, 9, and 10 and K. pneumoniae 16 (Fig. 1). The presence of a similar-sized plasmid conferring identical non-β-lactam antibiotic-resistance markers in the transformants and similar integrons was found for E. coli 1 and K. pneumoniae 18 on the one hand and for E. coli 6, 7, and 9 on the other hand (Table 4).
Thus, the spread of the blaVEB-1 gene among these nosocomial isolates may be explained rarely by the spread of clonally related strains and often by similar plasmids and integrons.
DISCUSSION
Taking into account the total number of enterobacterial isolates in the studied period, the prevalence of ESBL-producing organisms was 4.5% for E. coli, 9.6% for K. pneumoniae, and 26% for Enterobacter sp. These values ranged within those reported for isolates of North American and European hospitals (12, 18, 20, 36, 46). In a multicenter study performed in Thailand, the prevalence of ceftazidime-resistant E. coli strains ranged from 0 to 62.5% (37). In this same study, the prevalence of ceftazidime-resistant K. pneumoniae was 45% on average, and that is higher than the value reported here. In Korea, the prevalence of ESBL-producing E. coli was 4.8% with mostly TEM- and SHV-type ESBLs and was 22% in K. pneumoniae isolates (14). In Taiwan, the prevalence of ESBL-producing K. pneumoniae was quite high (30%), mostly involving TEM-type ESBLs (17, 26, 45). On the contrary, in Japan ESBL-producing organisms are rarely encountered (<0.001%) and the ESBLs are mostly of the TOHO-2 type and rarely of the TEM and SHV type (44). In China, ESBLs have been reported but their prevalence is unknown (34).
In Bangkok, as reported previously in other countries (39), Enterobacter sp. but not K. pneumoniae may represent the main reservoir of ESBL-producing enteric isolates. This may result from either an outbreak or a situation in which such organisms are endemic. The blaVEB-1 gene seemed to be highly prevalent in these ceftazidime-resistant Thai isolates since it accounted for 60% of the ESBL-possessing isolates. However, frequent identifications of blaTEM and blaSHV derivatives may correspond also to additional ESBL genes in the same clinical isolates. The epidemiological analysis at the strain and plasmid levels indicated that blaVEB-1 had spread among various enterobacterial species. Its dissemination was not due to a single strain or a single plasmid type. Most of the blaVEB-1-positive isolates harbored a self-conjugative plasmid of large size, as found for E. coli MG-1 from Vietnam (30).
The present work established the dissemination of an Ambler class A ESBL gene via various structures of class 1 integrons. A dissemination of an unusual Ambler class A ESBL gene has been reported for blaPER-1 in Turkey (40, 41). However, in this case the plasmid location but not the integron location of blaPER-1 is known. In the case of the Ambler class B IMP-1 β-lactamase, its integron-located gene has been reported extensively in Japan (10). However, as opposed to blaIMP-1 (10), the blaVEB-1 location on a class 1 integron is always followed by its expression.
Structure analysis of the blaVEB-1-containing integrons showed their variability compared to the three other known blaVEB-1-containing integrons, which are from E. coli MG-1 (from Vietnam) and P. aeruginosa JES-1 and Thl-1 (from Thailand) (23, 30, 38). Various structures of class 1 integrons have been detected in Vibrio cholerae isolates from Thailand carrying a carbenicillinase gene not related to blaVEB-1 (6). Spread of another class A β-lactamase (blaPSE-1) has also been reported to be related to integron location in Salmonella enterica serotype Typhimurium DT104 (28).
In the present study, most of the integrons containing the blaVEB-1 gene cassette possessed arr-2 and blaOXA-10 gene cassettes. This result raises the question of how these genes have evolved and if they have been transferred among soil organisms such as mycobacteria, Enterobacteriaceae, and Pseudomonas species. Indeed, arr-2 shares a structural relationship with arr-1 from Mycobacterium smegmatis (38) and blaOXA-10, and their extended-spectrum derivative genes are mostly found in P. aeruginosa isolates (22). Once located on integrons, these resistance genes may have been transferred in block through a transposition-related mechanism. It would be interesting to test if blaVEB-1-carrying integrons are located on transposons such as Tn21 derivatives, as described for some class 1 integrons (4).
Our results also raise the question of whether antibiotic selective pressure in hospitals in Thailand may have led to dissemination of this integron-located gene. β-Lactam as well as aminoglycoside, sulfonamide, and disinfectant resistance genes are associated with class 1 integrons that may enhance the in vivo dissemination of these integrons. Restricted clinical use of broad-spectrum antibiotics and rigorous hygiene measures are the most important means to prevent the spread of this ESBL gene as much as possible. Since blaVEB-1 has been detected also in two P. aeruginosa isolates from Thailand, a further study would evaluate its spread among other gram-negative rods.
The important prevalence of blaVEB-1 among enterobacterial isolates may lead to heavy use of carbapenems in this Bangkok hospital, which in turn may favor the selection of, at least, carbapenem-resistant P. aeruginosa isolates.
ACKNOWLEDGMENTS
We thank Venus Tolun for help in preliminary experiments.
This work was financed by a grant from the Ministères de l'Education Nationale et de la Recherche (grant UPRES, JE-2227), Université Paris XI, France.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T V, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Mangold P, Amann S, Akalin E, Ang O, Bal C, Casellas J M. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob Agents Chemother. 1996;40:616–620. doi: 10.1128/aac.40.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett P M. Integrons and gene cassette: a genetic construction kit for bacteria. J Antimicrob Chemother. 1999;43:1–4. [PubMed] [Google Scholar]
- 5.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalsgaard A, Forslund A, Serichantalergs O, Sandvang D. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob Agents Chemother. 2000;44:1315–1321. doi: 10.1128/aac.44.5.1315-1321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gniadkowski M, Schneider I, Palucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 10.Hirakata Y, Izumikawa K, Yamaguchi T, Takemura H, Tanaka H, Yoshida R, Matsuda J, Nakano M, Tomono K, Maesaki S, Kaku M, Yamada Y, Kamihira S, Kohno S. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1998;42:2006–2011. doi: 10.1128/aac.42.8.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby G A, Han P. Detection of extended-spectrum β-lactamase in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Kwon Y, Pai H, Kim J-W, Cho D-T. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J Clin Microbiol. 1998;36:1446–1449. doi: 10.1128/jcm.36.5.1446-1449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knothe H, Shah P, Kremery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 16.Lévesque C, Piché L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu P Y, Tung J C, Ke S C, Chen S L. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a district hospital in Taiwan. J Clin Microbiol. 1998;36:2759–2762. doi: 10.1128/jcm.36.9.2759-2762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore D M, Yuan M. Antibiotic resistance and production of extended-spectrum β-lactamase amongst Klebsiella spp. from intensive care units in Europe. J Antimicrob Chemother. 1996;38:409–424. doi: 10.1093/jac/38.3.409. [DOI] [PubMed] [Google Scholar]
- 19.Ma L, Ishii Y, Ishiguro M, Matsuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding TOHO-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl.):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 21.Mercier J, Lévesque R C. Cloning of SHV-2, OHIO-1 and OXA-6 β-lactamase and cloning and sequencing of SHV-1 β-lactamase. Antimicrob Agents Chemother. 1990;34:1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naas T, Nordmann P. OXA-type β-lactamases. Curr Pharm Design. 1999;5:865–879. [PubMed] [Google Scholar]
- 23.Naas T, Poirel L, Karim A, Nordmann P. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1999;176:411–419. doi: 10.1111/j.1574-6968.1999.tb13691.x. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 25.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pai H, Lyu S, Lee J H, Kim J, Kwon Y, Kim J-W, Choe K W. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J Clin Microbiol. 1999;37:1758–1763. doi: 10.1128/jcm.37.6.1758-1763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen I T, Littlejohn T G, Radström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L, Guibert M, Bellais S, Naas T, Nordmann P. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob Agents Chemother. 1999;43:1098–2004. doi: 10.1128/aac.43.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical-sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Shen D, Biedenbach D J, Winokur P L, Pfaller M A, Jones R N. Phenotypic and genotypic characterization of Chinese strains of Escherichia coli producing extended-spectrum β-lactamases. Diagn Microbiol Infect Dis. 1999;34:159–164. doi: 10.1016/s0732-8893(99)00018-8. [DOI] [PubMed] [Google Scholar]
- 35.Siu L K, Ho P L, Yuen K Y, Wong S S, Chau P Y. Transferable hyperproduction of TEM-1 β-lactamase in Shigella flexneri due to a point mutation in the Pribnow box. Antimicrob Agents Chemother. 1997;41:468–470. doi: 10.1128/aac.41.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover F C, Mohammed M J, Gorton T S, Dembek Z F. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J Clin Microbiol. 1999;37:4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biedenbach D J, Johnson D M, Jones R N The Thailand Antimicrobial Resistance Study Group. In vitro evaluation of cefepime and other broad-spectrum β-lactams in eight medical centers in Thailand. Diagn Microbiol Infect Dis. 1999;35:325–331. doi: 10.1016/s0732-8893(99)00123-6. [DOI] [PubMed] [Google Scholar]
- 38.Tribuddharat C, Fennewald M. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:960–962. doi: 10.1128/aac.43.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol. 2000;38:542–546. doi: 10.1128/jcm.38.2.542-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vahaboglu H, Dodanli S, Eroglu C, Öztürk R, Soyletir G, Yildirim I, Avkan V. Characterization of multiple antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J Clin Microbiol. 1996;34:2942–2946. doi: 10.1128/jcm.34.12.2942-2946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vahaboglu H, Öztürk R, Aygun G, Coskunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamase among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vahaboglu H, Öztürk R, Akbal H, Saribas S, Tansel O, Coskunkan F. Practical approach for detection and identification of OXA-10-derived ceftazidime-hydrolyzing extended-spectrum β-lactamases. J Clin Microbiol. 1998;36:827–829. doi: 10.1128/jcm.36.3.827-829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams J G, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagi T, Kurokawa H, Shibata N, Shibayama K, Arakawa Y. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol Lett. 2000;184:53–56. doi: 10.1111/j.1574-6968.2000.tb08989.x. [DOI] [PubMed] [Google Scholar]
- 45.Yan J J, Wu S M, Tsai S H, Wu J J, Su I J. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in Southern Taiwan. Antimicrob Agents Chemother. 2000;44:1438–1442. doi: 10.1128/aac.44.6.1438-1442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan M, Aucken H, Hall L M, Pitt T L, Livermore D M. Epidemiological typing of Klebsiellae with extended-spectrum β-lactamases from European intensive care units. J Antimicrob Chemother. 1998;41:527–539. doi: 10.1093/jac/41.5.527. [DOI] [PubMed] [Google Scholar]