Abstract
Colorectal cancer (CRC) is a highly malignant tumor associated with poor prognosis, yet the molecular mechanisms are not fully understood. In this study, we showed that LYAR, a nucleolar protein, is expressed at a higher level in CRC tissue than in adjacent normal tissue and that LYAR expression is closely associated with distant CRC metastasis. LYAR not only significantly promotes the migration and invasion of CRC cells in vitro, but knockdown (KD) of LYAR in CRC cells also inhibits xenograft tumor metastasis in vivo. Microarray analysis of LYAR KD cells combined with a chromatin immunoprecipitation (ChIP) assay, gene reporter assay, and rescue experiment indicated that FSCN1 (encoding fascin actin-bundling protein 1 (Fascin-1)) serves as a novel key regulator of LYAR-promoted migration and invasion of CRC cells. Knockdown of FSCN1 significantly inhibits subcutaneous tumorigenesis of CRC cells and leads to the downregulation of FASN and SCD, genes encoding key enzymes in fatty acid synthesis. In summary, this study reveals a novel mechanism by which LYAR promotes tumor cell migration and invasion by upregulating FSCN1 expression and affecting fatty acid metabolism in CRC.
1. Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed cancer, with 1.85 million new cases in 2018, and is the second most common cause of cancer-related mortality worldwide [IARC World Cancer Report 2020] [1]. With advances such as prognostic techniques, neoadjuvant chemoradiotherapy, cytotoxic therapy, and surgical therapy, great progress in treatment has been made. The five-year survival rate for CRC, however, remains poor (~64%), and in metastatic cases, it is even worse (14%) [2, 3]. Twenty-five percent of patients with CRC already have metastases at the time of diagnosis, and 50–60% of the remainder will develop metastases later in the course of the disease [4, 5]. The formation of metastasis is a multistep process, in which tumor cells leave the primary tumor site, invade and penetrate the surrounding extracellular matrix and endothelium, enter the blood and lymph vessels, survive migration, and finally attach at a distant site, where tumor cells begin to proliferate, induce angiogenesis, evade apoptotic death, and form a new tumor [6, 7]. These distant settlements of tumor cells, metastases, are the cause of 90% of human cancer deaths [6, 8–10]. Tumor recurrence and metastasis are the main causes of death in CRC. Approximately 30–50% of patients with CRC after curative surgery will relapse [11]. However, the molecular mechanism involved in the cascade of events during invasion and metastasis of CRC is still not fully understood.
LYAR was first identified in 1993 as a novel nucleolar oncoprotein, which consists of a zinc finger motif and three nuclear localization signals [12]. However, research was stagnant until a second LYAR article was published in 2009 [13]. Subsequently, there have been very few articles related to the function of LYAR [14–18], especially in the field of tumor research. In 2014, Ju et al. demonstrated that LYAR is a transcription factor with a DNA-binding motif (GGTTAT/G) that inhibits human fetal globin gene expression [19]. We further reported that LYAR promotes tumor cell migration and invasion by directly binding to the LYAR-binding motif (CTAACC; reverse complement GGTTAG) in the LGALS1 promoter to activate its expression in CRC cells [20]. LGALS1 encodes galectin-1, which is a member of the lectin family. Galectin-1 is a homodimer of 14 kD subunits and is characterized by its affinity for glycans containing β-galactosides. Galectin-1 has been detected in various malignancies such as colorectal, hepatocellular, prostate, ovarian, and breast cancers. Galectin-1 activation occurs via autocrine or paracrine sugar-dependent interactions with β-galactoside-containing glycoconjugates and participates in various key processes of carcinogenesis such as metastasis, angiogenesis, and immunosuppression [21].
The FSCN1 gene is located on chromosome 7p22 and encodes Fascin-1, a cytoskeleton protein with a relative molecular weight of 55kD [22]. Fascin-1 can promote the formation of filamentous pseudopodia, lamellar pseudopodia, and microspines of cell membrane after cross-linking with F-actin. This enhances the movement, metastasis, and invasion of tumor cells [23]. A mass spectrometry-based proteomic analysis predicted that FSCN1 may play an important role in metabolism, suggesting a new role of FSCN1 in tumors [24].
In the present study, we showed that LYAR, a key regulator of CRC, regulates a novel LYAR target, FSCN1, to promote the migration and invasion of CRC cells, which in turn positively regulates fatty acid metabolism. Our data demonstrated the presence of a LYAR/FSCN1/fatty acid metabolism axis which promoted the growth of CRC tumors in vivo. These findings suggest that LYAR could be used as a prognostic and therapeutic candidate target for the prevention and treatment of CRC.
2. Materials and Methods
2.1. Tissue Microarrays and Immunohistochemistry
CRC tissue arrays were purchased from Shanghai Outdo Biotech Co., Ltd., Shanghai, China. The immunohistochemistry experiment and analysis were carried out according to the procedure described in our previous literature [20]. The LYAR primary antibody (orb215217) and the anti-rabbit secondary antibody were purchased, respectively, from Biorbyt and Sigma-Aldrich. All researches involving human CRC tissues were approved by the Ethics Committee of Bengbu Medical College and were in accordance with the principles of the Declaration of Helsinki.
2.2. Cell Lines and Cell Culture
The HCT8, HCT15, and HCT116 human colon cancer cell lines were purchased from the Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences. HCT8 and HCT15 was maintained on gelatinized 10 cm-plate in RPMI-1640 Medium (Gibco) supplemented with 10% FBS (fetal bovine serum) (Gibco), 100 U penicillin/100 mg streptomycin (Gibco) at 37°C, and 5% CO2. HCT116 cells were cultured in McCoy's 5a Medium (Gibco).
2.3. Silencing of LYAR and FSCN1 by Transient Transfection siRNA
The siRNA sequences against human LYAR, FSCN1 and nonsilencing were designed and chemically synthesized by Genepharma (Shanghai, China). HCT15 cells were transfected with 100-200 pmol siRNA using 5-10 μL INTERFERin® transfection reagent (Polyplus) in a 6-well plate. After 24-48 hours, cells were collected to perform cell proliferation, cell cycle, apoptosis, colony formation, adhesion, migration, and invasion assays. The siRNAs sequences were as follows:
Human LYAR-siRNA1: 5′-GGGAGGUGAAGAAGAAUAA-3′
Human LYAR-siRNA2: 5′-GCACUCGGAAGUUGAAACA-3′
Human FSCN1-siRNA1: 5′-GCGCCUACAACAUCAAAGA-3′
Human FSCN1-siRNA2: 5′-GCCCAUGAUAGUAGCUUCA-3′
2.4. Microarray Analysis and RNA-Seq
Approximately 1 × 107 cells were collected and lysed with TRIzol® Reagent (Life Technologies) in the RNase-free Eppendorf tube. We then submitted the samples to KangChen-Biotech Corporation (Shanghai, China) for microarray analysis using the Human 12 × 135K Gene Expression Array (Roche NimbleGen). The experiment and data analysis for microarray analysis, including RNA isolation, microarray experiment, data processing, statistical analysis, and gene ontology analysis, were done by KangChen-Biotech Corporation.
FSCN1 siRNA and NC (negative control) were transfected into cells and cultured for 24-48 hours. Then, total RNA was isolated by the RNA pure Tissue and Cell Kit and the RNA was sequencing by the 150 bp paired method. Hisat2, samtools, and htseq-count packages were used to align 150 bp paired-end reads with hg19 (UCSC). The expression of genes and transcripts was quantified by using cufflinks tools.
We downloaded expression profiles of human colorectal cancer of TCGA and normal colon data of GTEx from UCSC Xena browser (https://xenabrowser.net/datapages/).
2.5. Plasmid Construction and Viral Infection
To build stable knockdown cells, the small hairpin RNAs (LYAR-shRNAs, FSCN1-shRNAs, and Control-shRNA) were designed correspondingly according to the sense and antisense sequences of the two LYAR-specific siRNAs and FSCN1 siRNAs (see above) and a nonspecific siRNA and synthesized by Invitrogen (Shanghai, China). These chemically synthesized oligonucleotides were annealed to generate double-stranded oligonucleotides using the touchdown program in the PCR instrument and inserted into the Xho I/Hpa I sites in the pLentiLox 3.7 vectors, which was subsequently confirmed by sequencing.
To overexpress FSCN1, the human FSCN1 coding sequence (CDS) was prepared by reverse transcription-polymerase chain reaction (RT-PCR) from normal colorectal tissues and cloned into the pLVX-IRES-mCherry Vector at Xba I and BamH I sites for overexpressing FSCN1 in LYAR-KD cells. Lentiviruses were packaged and produced in 293T cells. The supernatant of virus production was collected and filter-sterilized to infect the corresponding cells. The stably transfected cells were sorted and collected by flow cytometry using mCherry fluorescence.
2.6. Cell Migration and Invasion Assays
Equal amounts of cells (1 × 105 in 200 μL medium without FBS) were seeded on a fibronectin-coated polycarbonate membrane insert in a Transwell apparatus (Corning Costar), and medium supplemented with 20% FBS was added to the lower chamber. After incubation for 48 h at 37°C, the insert was washed with phosphate-buffered saline, and cells on the upper surface of the insert were removed by wiping with a wet cotton swab. Cells migrating to the lower membrane surface were fixed by an equal volume mixture of methanol and acetone and stained with 0.4% crystal violet and counted under a microscope (Nikon). The invasion assay was executed as described in the migration assay using upper chamber precoated with 50 μL matrigel solution that original matrigel (BD Biosciences) was diluted with FBS-free medium according to the ratio of 1 : 10.
2.7. Quantitative RT-PCR (RT-qPCR)
Total RNA was isolated from cells with TRIzol® Reagent (Life Technologies). cDNA was synthesized using HiScript® II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme). The RT-qPCR primers were designed by Primer Premier 6.0 software. RT-qPCR was carried out in a Rotorgene 6000 (Corbett Research) using the FastStart Universal SYBR Green Master Mix (Roche) in a final volume of 20 μL. The relative quantification was executed for the following genes using GAPDH as an internal reference in CRC cells. Each reaction was performed in triplicate and repeated at least two times.
The primers for human GAPDH were as follows:
Forward 5′-ACCATCTTCCAGGAGCGAGA-3′
Reverse 5′-GTTCACACCCATGACGAACATG-3′
The primers for human LYAR were as follows:
Forward 5′-GGAGGCACTCGGAAGTTGAAA-3′
Reverse 5′-GTTCCTCTTCGGATCTGTGATG-3′
The primers for human FSCN1 were as follows:
Forward 5′-CTGCTACTTTGACATCGAGTGG-3′
Reverse 5′-GGGCGGTTGATGAGCTTCA-3′
The primers for human FASN were as follows:
Forward 5′-GCTGGAAGGAGGAAGAGGTT-3′
Reverse 5′-CTCGAGTGGTCCGTGAGTTT-3′
The primers for human SCD were as follows:
Forward 5′-CTCAGTTCCTACGCTTCGCAT-3′
Reverse 5′-GTCGAGGTCAGTGAACAGCA-3′
2.8. Western Blotting
Cell extracts were prepared using the lysis buffer (Beyotime) from the CRC cell lines. Cell lysates (20-50 μg) were loaded and separated by 10-15% sodium dodecyl sulphate-polyacrylamide gel (Bio-Rad) and transferred onto nitrocellulose membranes (Millipore). After 1 h blocking with 5% nonfat milk (or 3% BSA (bovine serum albumin)) blocking solution prepared with PBST (1 × PBS with 0.1% Tween-20), the membranes were incubated overnight at 4°C with the primary antibodies against LYAR (orb215217, Biorbyt), FSCN1 (ab220195, Abcam), FASN (10624-2-AP, Proteintech), SCD (2794S, CST), and GAPDH (M171-3, MBL) as the internal control and followed by incubation with the secondary antibody (Sigma-Aldrich) for 1 h at room temperature. The specific bands were visualized with Thermo SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher Scientific Inc.).
2.9. Luciferase Reporter Assay
The FSCN1 promoter region with a 496-bp sequence containing the specific DNA-binding motif for LYAR was amplified from the genomic DNA by PCR. Subsequently, the 496-bp DNA fragment product was subcloned into pGL3-Basic (Promega) to construct a luciferase reporter plasmid (pGL3-FSCN1-496bp-LYAR-wildetype), and the sequence was confirmed by double-direction sequencing (Invitrogen). Based on the pGL3-FSCN1-496bp-LYAR-wildetype plasmid, the pGL3-FSCN1-496bp-LYAR-mutant plasmid was constructed using the Muta-direct™ site-directed mutagenesis kit (SBS Genetech) for mutating the LYAR binding site. Luciferase reporter assays were performed as described previously [25].
2.10. Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed following standard ChIP procedures [26]. Chromatin fractions from HCT15 cells were immunoprecipitated with LYAR primary antibody (a kind gift from the State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, China). Normal rabbit immunoglobulin G (IgG, Beyotime) served as the controls. The ChIP samples were analyzed by quantitative real-time PCR using the FastStart Universal SYBR Green Master Mix (Roche) and specific primers (Table S2) spanning the FSCN1 promoter. A standard curve was prepared for each set of primers using serial titrations of the input DNA. The percentage of ChIP DNA was calculated relative to the input DNA from primer-specific standard curves using the Rotor-Gene 6000 Series Software 1.7. Each experiment was performed at least two independent times.
2.11. Rescue Experiments
The supernatant of virus production of pLVX-IRES-mCherry (empty vector) and pLVX-IRES-mCherry-FSCN1-OE (exogenous expression of FSCN1) were used, respectively, to infect the stable LYAR-KD and LYAR-Control (only infected by empty vector) cells. Those stably infected cells were sorted and collected using mCherry fluorescence by flow cytometry, followed by cell migration and invasion assays (see above).
2.12. Animal Experiments
Mice were purchased from and housed at Suzhou University. All animal studies were approved by the ethical regulations of Institutional Animal Care and Use Committee (IACUC) of Suzhou University. For the tumor metastasis assay, stable HCT15 clones (Control or LYAR-KD1) were injected into six-week-old female NOD/SCID mice. For each group, 10 mice were injected with 1 × 106 cells per animal via the tail vein. The mice were sacrificed 6 weeks after injection, and the lung and liver metastases were examined. Metastastic nodules in lung and liver tissues were fixed in Bouin's solution (Applygen), and the number of metastases was counted. The tumor samples were embedded in paraffin, cut into 5 μm sections, and stained with hematoxylin and eosin (H&E).
For the tumor xenograft assay, 8 six-week-old female nude mice were processed by subcutaneous implantation of 1 × 106 HCT15 cells expressing either control shRNA or FSCN1-shRNA. Mice were maintained for 30 days, and tumor volumes were measured at indicated time points. At the end of experiments, mice were euthanized and xenograft tumors were dissected for further analyses.
2.13. Statistical Analysis
All statistical analyses were performed using SPSS 22.0 software (SPSS Inc. Chicago, IL, USA). The experimental results were statistically evaluated using Student'st-test for comparisons between two groups or ANOVA for comparisons between more than two groups. Kaplan-Meier survival curves were generated to determine the relationship between LYAR levels and the overall survival of CRC patients, and the differences between the curves were calculated using the log-rank test. Multiple Cox proportional hazards regression was carried out to identify the independent factors with a significant impact on patient survival. p values < 0.05 were considered statistically significant.
3. Results
3.1. LYAR Is Highly Expressed in Human CRC Tissue and Promotes CRC Metastasis
Based on data from the Oncomine database (https://www.oncomine.org/) and a previous study [20], we found that LYAR was highly expressed and promoted cell mobility in CRC cells. However, the relationship between the high expression level of LYAR and the survival rate of CRC patients is unclear. To answer this question, we performed immunohistochemistry (IHC) analysis of LYAR on tissue arrays of 166 paraffin-embedded adjacent sections of normal colorectal tissues and CRC tissues. Tissues that displayed moderate or strong immunostaining were classified as having high LYAR expression and those that displayed negative or weak immunostaining as having low LYAR expression. There was strong LYAR staining in only 4.8% (8/166) of the adjacent normal epithelial tissues, whereas ~47.0% (78/166) of total CRC tissues had high expression levels of LYAR (Figure 1(a), Table 1, and Table S1). Furthermore, high LYAR expression was significantly correlated with poor prognosis (Figure 1(b)), which was consistent with the survival curve analysis of CRC data from The Cancer Genome Atlas (TCGA) database (Figure S1). Notably, LYAR expression was positively correlated with a higher metastasis status (Figure 1(c) and Table S1). Taken together, these results indicate that LYAR is highly expressed in CRC tissues, particularly metastatic tissues. This suggests that LYAR may be involved in metastasis of CRC cells, with the potential to be a novel prognostic biomarker for CRC.
Figure 1.
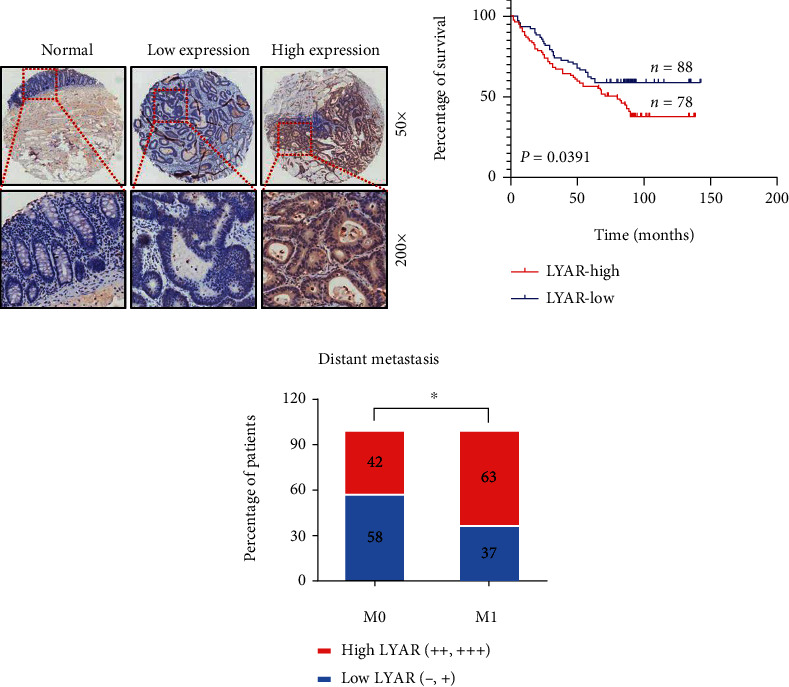
LYAR is highly expressed in human CRC tissues. (a) Immunohistochemical staining showing the LYAR protein (brown) in adjacent normal colorectal tissue and carcinoma tissue from CRC patients. Representative micrographs are shown at the original magnification (50x and 200x). Low and high LYAR expressions were defined based on immunostaining scores [20]. (b) The prognostic curve of LYAR. (c) The percentage of patients with different metastasis statuses (M0: no regional or distant metastasis; M1: regional or distant metastasis). ∗p < 0.05.
Table 1.
LYAR is highly expressed in human CRC tissues.
| Total | Positive (LYAR high expression) | Positive rate | χ 2 | p | |
|---|---|---|---|---|---|
| Adjacent | 166 | 8 | 4.8% | 81.806 | <0.001 |
| CRC | 166 | 78 | 47.0% |
LYAR has zinc finger DNA-binding motifs and binds to DNA at the GGTTAT/G consensus sequence [12, 19]. To perform a systematic analysis and to take our previous research a step forward, we verified that LYAR promoted migration and invasion of HCT15 cells in vitro (Figures 2(a)–2(c) and Figure S2-4). More importantly, we found that LYAR enhanced the lung and liver metastasis of CRC cells through a mouse tail vein assay in vivo. (Figures 2(d)–2(f)).
Figure 2.
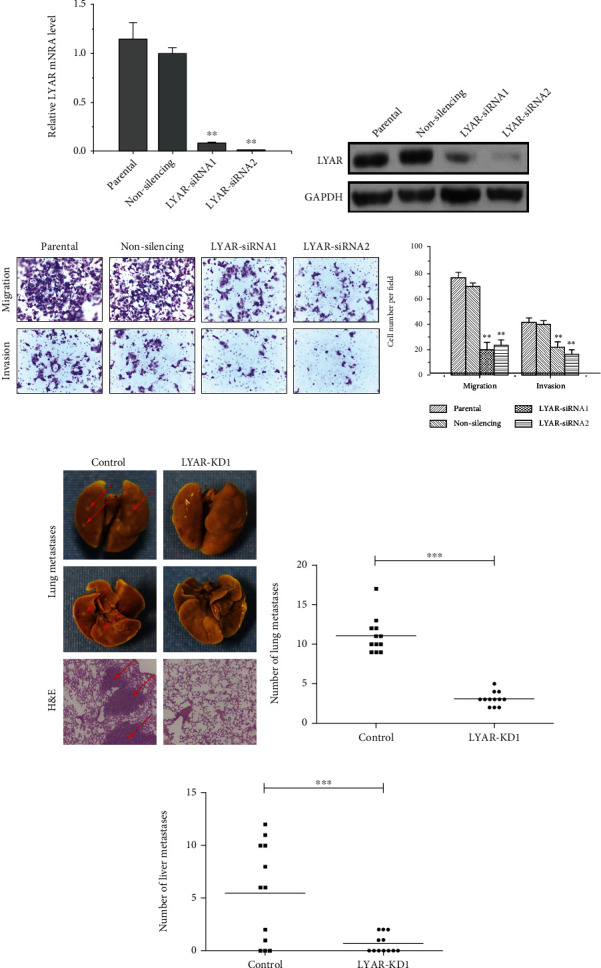
LYAR promotes metastasis of CRC cells. (a) Quantitative RT-PCR analysis of LYAR expression normalized to GAPDH expression in LYAR knockdown HCT15 cells. Results shown are the mean ± standard deviation (n = 3). ∗∗p < 0.01 compared with the nonspecific siRNA control. (b) Western blot assay showing LYAR protein expression in LYAR knockdown HCT15 cells. GAPDH served as the loading control. (c) At left, representative photos of haptotactic migration assay and matrigel chemoinvasion assay using LYAR knockdown HCT15 cells. Original magnification, 200x. At right, the percentage of migrated and invaded LYAR knockdown HCT15 cells compared to the control. ∗∗p < 0.01. (d) Representative images of metastastic nodules (indicated with arrows) in lung tissues after Bouin's fixation (upper four panels) and hematoxylin and eosin (H&E) staining of tissue sections (lower two panels). (e) Visible lung metastasis counts. ∗∗∗p < 0.001. (f) Visible liver metastasis counts. ∗∗p < 0.01.
3.2. FSCN1 Is a Novel Target of LYAR
Heterogeneity is a distinctive feature of solid tumors. Therefore, we speculated that there is more than one molecular mechanism by which LYAR promotes CRC migration and invasion, in addition to upregulating LGALS1. In order to comprehensively identify regulatory roles of LYAR in metastatic CRC, a whole-genome microarray analysis of gene expression was performed in LYAR knockdown (KD) and control HCT15 cells (Figure 3(a)). Gene Set Enrichment Analysis (GSEA) showed that differentially expressed genes (DEGs) in the LYAR-KD line were significantly enriched in cholesterol homeostasis, a lipid metabolism-related pathway (Figure 3(b)). We then screened for genes associated with metastasis and metabolism among the DEGs. The upstream promoter region (2000 bp upstream of the transcription start site) of each candidate gene was searched for the GGTTAT/G motif, which is the consensus DNA-binding site of LYAR. Candidate genes with this motif were selected for further analysis in HCT15 cells. Quantitative RT-PCR (RT-qPCR) revealed that the expression of FSCN1, one of the candidate genes, was consistently lower in LYAR-KD cells compared to the control (Figure 3(c)). Downstream involvement of FSCN1 in LYAR-regulated CRC would be a novel finding; we therefore conducted further experiments to verify this relationship between LYAR and FSCN1. We detected FSCN1 expression after LYAR knockdown by siRNA in all three cell lines and found that FSCN1 was only downregulated in HCT15 (Figure 3(d) and Figure S5). We therefore performed the subsequent experiments only in HCT15 cells. Together, these results indicated that FSCN1 is a potential target of LYAR in CRC cells.
Figure 3.
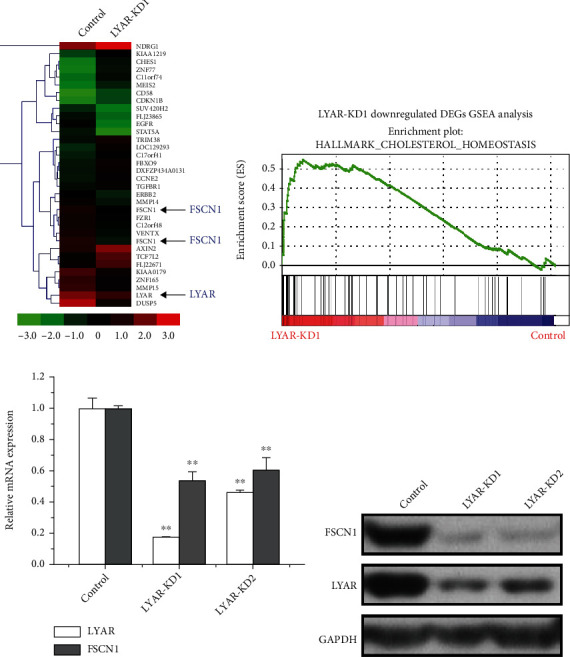
FSCN1 is a novel target of LYAR. (a) Heatmap representation of gene expression for LYAR and FSCN1 (indicated with arrows) and selected other genes in the LYAR knockdown or control HCT15 cells. Color represents expression values, with green indicating low, black indicating medium, and red indicating high expression. (b) Gene Set Enrichment Analysis (GSEA) plots of RNA-Seq in LYAR knockdown HCT15 cells. (c) Quantitative real-time PCR analysis of LYAR and FSCN1 from LYAR knockdown or control HCT15 cells. Results shown are the mean ± standard deviation (n = 3). ∗∗p < 0.01. (d) Western blot assay showing LYAR and FSCN1 protein expression in LYAR knockdown HCT15 cells. GAPDH served as the loading control. KD1 and KD2 refer to stable LYAR knockdown lines LYAR-siRNA1 and LYAR-siRNA2, respectively.
3.3. LYAR Binds to the FSCN1 Promoter in CRC Cells
We identified a consensus DNA-binding motif of LYAR (GGTTAG) at -237 bp upstream of the FSCN1 gene transcriptional start site (Figure 4(a) and Table S2). Chromatin immunoprecipitation (ChIP) analysis spanning the promoter of the FSCN1 gene was performed to determine if LYAR binds to the FSCN1 promoter and regulates its expression. LYAR indeed bound to the FSCN1 promoter region between -337 and -183 bp in HCT15 cells (Figure 4(b)). When LYAR was knocked down in HCT15 cells with shRNAs, LYAR enrichment on the FSCN1 promoter was significantly reduced (Figure 4(c)). Furthermore, a luciferase reporter assay showed that the extent of LYAR binding to the FSCN1promoter corresponded to changes in LYAR expression in HCT15 cells transfected with a wild-type FSCN1 promoter-driven luciferase construct (Figures 4(d) and 4(e)). Mutating the LYAR binding motif from GGTTAG to GACTAG abolished corresponding FSCN1 promoter activity changes (Figures 4(d) and 4(e)). In summary, these results demonstrated that LYAR directly regulates FSCN1 gene expression and that the LYAR-binding motif (GGTTAG) in the promoter of FSCN1 plays a critical role in LYAR-mediated transcriptional activation of FSCN1.
Figure 4.
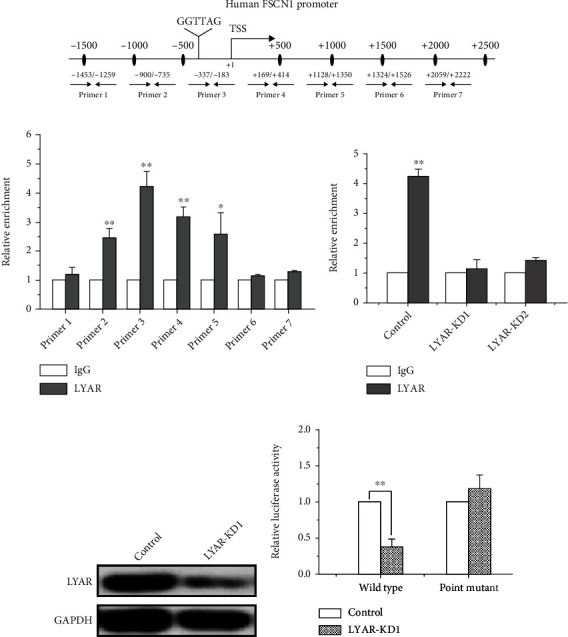
LYAR binds to the FSCN1 promoter and directly activates the expression of FSCN1. (a) A schematic diagram showing seven primer pairs spanning the FSCN1 promoter that were designed for ChIP. (b) ChIP analysis of LYAR on the FSCN1 promoter in HCT15 cells. Normal rabbit IgG served as the control. Results are shown as the mean ± standard deviation (n = 3). ∗∗p < 0.01 and ∗p < 0.05 compared to the IgG control. (c) ChIP analysis of LYAR on the FSCN1 promoter in LYAR knockdown and control HCT15 cells. Normal rabbit IgG served as the control. Results are shown as the mean ± standard deviation (n = 3). ∗∗p < 0.01 compared to the IgG control. (d) Western blot assay showing LYAR protein expression in LYAR knockdown HCT15 cells. GAPDH served as the loading control. (e) Luciferase reporter analyses of wild-type FSCN1 promoter (“Wild type”) and mutant FSCN1 promoter (“Point mutant”) in LYAR knockdown HCT15 cells. Results are shown as the mean ± standard deviation (n = 3). ∗∗p < 0.01 compared with the scrambled shRNA control.
3.4. Ectopic Expression of FSCN1 Restores Migration and Invasion Potential of LYAR-KD Cells
FSCN1 has been reported to increase CRC migration and invasion in cell cultures and cause cell dissemination and metastasis in vivo [27]. In this study, we found that knockdown of LYAR by shRNAs in HCT15 cells led to downregulation of FSCN1 (Figures 3(c) and 3(d)) and that the cell migration and invasion potential were correspondingly decreased (Figures 2(c) and S4). To investigate the role of FSCN1 in LYAR-promoted CRC cell migration and invasion, rescue experiments were performed in LYAR-KD cells in which FSCN1 was overexpressed by a stably transfected FSCN1-coding sequence. Overexpression of FSCN1 and knockdown of LYAR were confirmed by Western blot in HCT15 cells (Figure 5(a)). Compared with the control LYAR-KD cells (LYAR-KD-Vector), the migration and invasion potential of the HCT15 cells with FSCN1 overexpression was significantly increased (Figures 5(b) and 5(c)), indicating that exogenous expression of FSCN1 partially restored cell migration and invasion potential of LYAR-KD cells. To test this effect in vivo, stable FSCN-KD or control HCT15 cells were subcutaneously adoptively transferred to nude mice. Xenografts in the FSCN1-KD group were significantly inhibited compared to those in the control group (Figures 5(d)–5(f)). In summary, these results indicated that FSCN1 plays a key role in mediating LYAR-promoted CRC cell migration and invasion.
Figure 5.
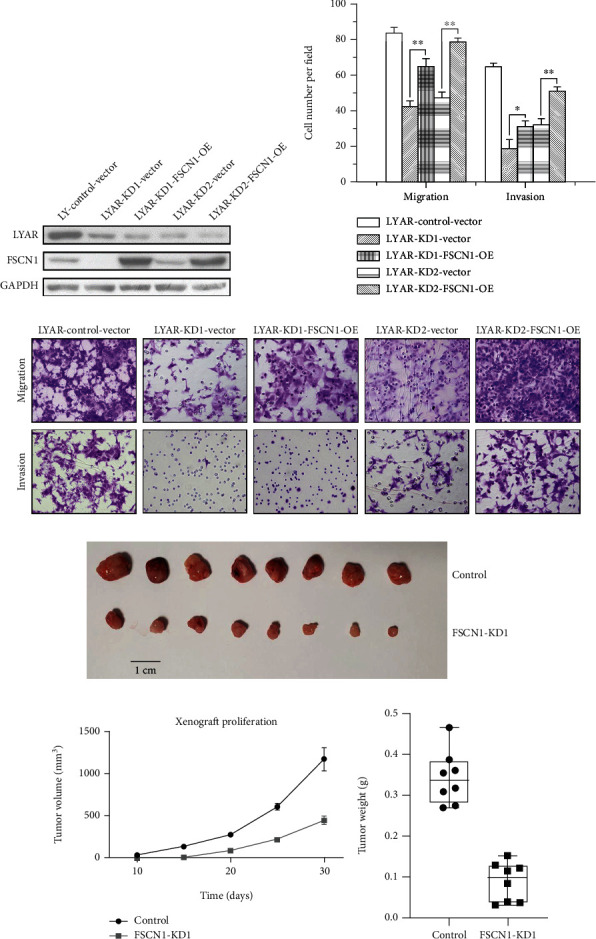
FSCN1 overexpression restores the metastatic potential of LYAR knockdown cells. (a) Western blot assay showing LYAR and FSCN1 protein expression in LYAR knockdown (KD) HCT15 cells with and without FSCN1 overexpression (OE). GAPDH served as the loading control. (b) Representative photos of haptotactic migration assay and matrigel chemoinvasion assay using LYAR knockdown HCT15 cells with and without FSCN1 overexpression. Original magnification, 200x. (c) Results of the migration and invasion assays shown in (b). Data are shown as the mean ± standard deviation (n = 3). ∗∗p < 0.01 and ∗p < 0.05 compared with the empty vector control. (d) Xenografted tumors of negative control (NC) cells and stable FSCN1-KD HCT15 cells (n = 8). (e) Tumor volumes over time. (f) Xenografted tumors were resected and weighed at the end of the experiment. Results are shown as the mean ± standard deviation (n = 8). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. OE: stable FSCN1 overexpression in LYAR knockdown (KD) HCT15 cells.
3.5. Reduction of FSCN1 Expression in CRC Cells Inhibits the Expression of Some Key Enzymes in Fatty Acid Metabolism
To clarify the mechanism of FSCN1 in promoting tumor invasion and metastasis, RNA-Seq was used to search for potential FSCN1 targets and signaling pathways. In the FSCN1-KD line, metabolic pathways, including lipid metabolism, were enriched (Figure 6(a)). Because our previous analysis showed that LYAR knockdown caused a reduction in cholesterol homeostasis, which is also a lipid metabolism pathway (Figure 3(b)), we further investigated the influence of FSCN1 inhibition on key enzymes in lipid metabolism. Among the lipid metabolism-related genes, the expression of fatty acid synthase (FASN) and stearoyl-CoA desaturase (SCD) were consistently decreased in FSCN1-KD HCT15 cells compared to the control at both the mRNA (Figure 6(b)) and protein (Figure 6(c)) levels. It was previously reported that de novo synthesis of palmitic acid from acetyl-CoA is mainly catalyzed by FASN; moreover, saturated fatty acids (SFAs) are converted into monounsaturated fatty acids (MUFAs) by SCD, and their chains are elongated by elongases [28]. These data suggest that FSCN1 positively regulates fatty acid metabolism.
Figure 6.
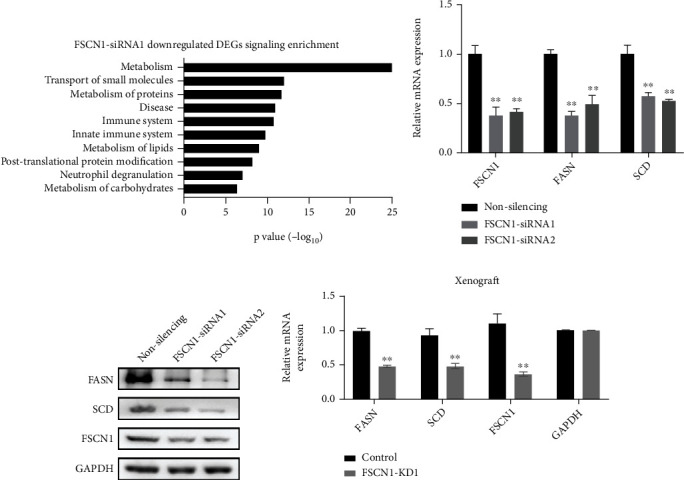
Reduction of FSCN1 expression in CRC cells inhibits the expression of some key enzymes in fatty acid metabolism. (a) Downregulated genes (fold change > 2) were enriched in several pathways (based on the Reactome pathway database). (b) Quantitative real-time PCR analysis of FSCN1, FASN, and SCD from FSCN1 knockdown or control HCT15 cells. Results are shown as the mean ± standard deviation (n = 3). ∗∗p < 0.01 compared with the control. (c) Western blot assay showing FSCN1, FASN, and SCD protein expression in FSCN1 knockdown HCT15 cells. GAPDH served as the loading control. (d) Quantitative real-time PCR analysis of FASN, SCD, and FSCN1 in FSCN1 knockdown or control HCT15 cells from xenograft. ∗∗p < 0.01 compared with the control.
In xenograft samples, FASN and SCD were also downregulated in the FSCN1-KD line (Figure 6(d)). Intriguingly, the analysis of TCGA CRC data revealed that the expression levels of LYAR, FSCN1, FASN, and SCD in CRC tissues were all higher than those in normal tissues (Figure S6). This suggests that LYAR may promote CRC progression by upregulating FSCN1 expression and subsequent fatty acid metabolism, which is logically consistent with the results of our in vitro experiments (Figures 6(b)–6(d)). In conclusion, our data demonstrated the presence of a LYAR/FSCN1/fatty acid metabolism axis which promoted the growth of CRC tumors in vivo.
4. Discussion
The formation of metastasis is a complex, multistep process, and the molecular mechanisms of CRC metastasis are still not fully understood. In recent years, some studies have found that LYAR plays an important role in different biological processes, such as RNA synthesis and the development of erythroid cells [14–18, 29, 30]. LYAR may be involved in the MYCN signaling pathway in the development of medulloblastoma [31] and induces neuroblastoma cell proliferation and survival [32], and LYAR together with other five genes (PDIA3, NOP14, NCALD, MTSS1, and CYP1B1) can be used as potential prognostic biomarkers for curative and postoperative supportive therapies for ovarian cancer [33]. However, aside from these, there have been no other reports on the function of LYAR in cancer.
We previously found that LYAR was expressed at a higher level in metastatic CRC tissues and that it promotes tumor cell migration and invasion by upregulating LGALS1 expression in CRC cell lines [20]. However, that study lacked in vivo experiments; the potential of LYAR to promote metastasis could not be verified, and retrospective analysis could not be conducted on the survival of clinical CRC patients to hypothesize whether LYAR was an independent prognostic factor. In addition, the heterogeneity of solid tumors makes it unlikely that upregulating the expression of LGALS1 is the only molecular mechanism by which LYAR promotes CRC migration and invasion among all colorectal cancers with high LYAR expression.
In this study, we used large-scale immunohistochemical tissue microarrays and found that LYAR is highly expressed in CRC tissues. This high expression is positively correlated with the metastatic rate of CRC and is significantly correlated with poor CRC prognosis. Moreover, we performed a genome-wide expression profile analysis to further study heterogeneity in molecular mechanisms through which LYAR participates in the progression of colorectal cancer, demonstrating that LYAR plays a role in the promotion of CRC invasion and metastasis by upregulating FSCN1 expression. However, we detected the expression of FSCN1 after LYAR siRNA knockdown in three different cell lines and found that FSCN1 was only downregulated in HCT15. This suggested that LYAR regulates FSCN1 expression in only a subset of colorectal cancers. This phenomenon may be due to differences in race, gender, and age of the patients from whose tissue different colorectal cancer cell lines are derived; these can lead to differences in molecular features between cell lines. This result again illustrates that solid tumors exhibit significant heterogeneity.
FSCN1 has recently been shown to promote cancer cell migration and invasion through its role in formation of cellular protrusions such as filopodia and invadopodia [34]. Forced expression of FSCN1 in CRC cells increased their migration and invasion in vitro and caused cell dissemination and metastasis in vivo [27]. Another recent study showed that imipramine, a novel FSCN1 inhibitor, has been confirmed to have significant antitumor effects both in vivo and in vitro, which lays the foundation for molecular targeted therapy of serrated adenocarcinoma (SAC) and other FSCN1-overexpressing tumors [35]. In this study, we revealed that LYAR positively regulates FSCN1 expression. We also found that LYAR knockdown in CRC cells led to the reduction of FSCN1 levels and inhibited metastasis to the lungs and liver in NOD/SCID mice. This is consistent with the report that forced expression of FSCN1 in CRC cells caused cell dissemination and metastasis in vivo.
There is previous evidence in the literature of FSCN1 involvement in tumor metabolism. In lung cancer, FSCN1 promotes cancer growth and metastasis by enhancing glycolysis and PFKFB3 expression through YAP1 activation [36]. In the context of PIK3CA mutation or amplification, high expression of FSCN1 is associated with poor prognosis and radiotherapy response in cancer patients; mutant PIK3CA (E542K and E545K) can enhance glucose metabolism and cell proliferation in cancer cells [37, 38]. In the present study, separate from glucose metabolism, we demonstrated that FSCN1 was related to lipid metabolism in CRC. FSCN1 knockdown reduced the expression of FASN and SCD, which are key genes in de novo fatty acid synthesis, suggesting that FSCN1 promoted CRC by affecting lipid metabolism.
This regulatory action of FSCN1 may be because CRC is closely related to obesity and lipid metabolism [39, 40]. Fatty acids are indispensable for the biosynthesis of most lipids, such as membrane lipids and lipid signaling molecules, in addition to acting as substrates for energy production [41]. De novo synthesis of palmitic acid from acetyl-CoA (acetyl-coenzyme A) is mainly catalyzed by fatty acid synthase (FASN). SFAs are converted into MUFAs by stearoyl-CoA desaturase 1 (SCD), and their chains are elongated by elongases [28]. A high-fat diet increases palmitic acid levels, which in turn increase β2AR expressions in a Sp-1 dependent manner. Subsequently, the cAMP/PKA axis is activated and hormone sensitive lipase (HSL) is phosphorylated at S552 to increase energy production, which promotes CRC growth [42]. In addition, there may be another FSCN1-lipid metabolism axis independent of LYAR, which would require future studies to confirm.
In summary, we demonstrated that FSCN1 is a direct target of LYAR. A novel pathway for function of the transcription factor LYAR in CRC has also been revealed: LYAR promotes tumor migration and invasion by upregulating FSCN1 expression, which in turn positively regulates fatty acid metabolism. These findings suggest that LYAR could be used as a prognostic and therapeutic candidate target for the prevention and treatment of CRC.
Acknowledgments
This work was supported by the Natural Science Research Project of Anhui Educational Committee (KJ2019A0318), Funds for the Construction of Medical Technology Disciplines of Bengbu Medical College, Science Research Project of Bengbu Medical College (BYKF1804), and the Hefei National Laboratory for Physical Sciences at the Microscale (KF2020011). We are grateful to members of Dr. Wang Mingrong Laboratories (State Key Laboratory of Molecular Oncology, Cancer Hospital and Institute, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China) and Dr. Zhao Quan Laboratories (State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, China) for technical assistance and helpful discussions.
Contributor Information
Yupeng Wu, Email: wuyupeng@bbmc.edu.cn.
Xiaoyuan Song, Email: songxy5@ustc.edu.cn.
Sai Ma, Email: marseillems@outlook.com.
Data Availability
The data generated or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
All research involving human CRC tissues was approved by the Ethics Committee of Bengbu Medical College and was in accordance with the principles of the Declaration of Helsinki. All animal studies were approved by the ethical regulations of Institutional Animal Care and Use Committee (IACUC) of Suzhou University.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
S. Ma, X.Y. Song, and Y.P. Wu conceived the idea; Y. Zhou performed the animal experiments; H.Y. Gao and Y.P. Wu performed development of methodology and acquisition of data; Q.Y. Cheng and S.K. Jian finished acquisition of data and interpretation of data; Q. Ding and W. Gu finished acquisition of data; Y.X. Yao and X.H. Tong conducted the statistical analysis; J. Ma, W.J. Wu, and Y.Y. Li conducted the bioinformatics analyses; Y.P. Wu, H.Y. Gao, Y.J. Wang, X.Y. Song, and S. Ma wrote the manuscript and all of the authors read and reviewed and approved the manuscript. Yupeng Wu, Yu Zhou, and Haiying Gao contributed equally to this work.
Supplementary Materials
Figure S1: Overall survival curve of those with colorectal cancer. Data were obtained from TCGA database and analyzed to generate Kaplan-Meier curves, which show a correlation between patients with tumors expressing low levels of LYAR and higher survival rates. Figure S2: LYAR expression in stable LYAR knockdown HCT15 cells. (a) RT-qPCR analysis of LYAR gene expression normalized to GAPDH gene expression in stable LYAR knockdown HCT15 cells. ∗∗p < 0.01 compared with the scrambled control. (b) Western blot assay showing LYAR protein expression in stable LYAR knockdown HCT15 cells. GAPDH served as the loading control. Figure S3: Detection of tumor cell phenotype in transient LYAR knockdown HCT15 cells. LYAR knockdown had no effect on cell proliferation (a), cell adhesion (b), cell cycle (c), apoptosis (d), or colony formation (e) of HCT15 cells. Parental refers to the control, nonsilencing refers to cells transfected with a nonspecific siRNA, and LYAR-siRNA1 and LYAR-siRNA2 refer to cells transfected with LYAR-specific siRNA1 and LYAR-specific siRNA2, respectively. Figure S4: Detection of migration and invasion in stable LYAR knockdown HCT15 cells. (a) Representative photos of haptotactic migration assay and matrigel chemoinvasion assay using stable LYAR knockdown HCT15 cells. Original magnification, 200x. (b) Results of migration and invasion assays. Data shown are the mean ± standard deviation (n = 3). ∗∗p < 0.01 compared to the scrambled control. Figure S5: Western blot assay showing FSCN1 expression after LYAR knockdown by siRNA. (a) LYAR and FSCN1 expression after LYAR knockdown in HCT8 cells. (b) LYAR and FSCN1 expression after LYAR knockdown in HCT116 cells. KD1 and KD2 refer to the stable LYAR knockdown lines LYAR-siRNA1 and LYAR-siRNA2, respectively. Figure S6: Expression of LYAR and downstream genes in colorectal cancer. The expression profiles of human colorectal cancer and normal colon tissue were obtained from TCGA and GTEx databases, respectively. The data show differential expression of LYAR (a), FSCN1 (b), FASN (c), and SCD (d) in colorectal cancer tissues compared to adjacent normal tissues. ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. Table S1: The relationship between LYAR protein expression and clinicopathological features in CRC patients. Table S2: ChIP primer sequences for the FSCN1 promoter.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians . 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kuipers E. J., Grady W. M., Lieberman D., et al. Colorectal cancer. Nature Reviews. Disease Primers . 2015;1(1):p. 15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA: a Cancer Journal for Clinicians . 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Lee R. M., Cardona K., Russell M. C. Historical perspective: two decades of progress in treating metastatic colorectal cancer. Journal of Surgical Oncology . 2019;119(5):549–563. doi: 10.1002/jso.25431. [DOI] [PubMed] [Google Scholar]
- 5.Tsitskari M., Filippiadis D., Kostantos C. The role of interventional oncology in the treatment of colorectal cancer liver metastases. Annals of Gastroenterology . 2019;32(2):147–155. doi: 10.20524/aog.2019.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan X. Cancer metastases: challenges and opportunities. Acta Pharmaceutica Sinica B . 2015;5(5):402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steeg P. S. Tumor metastasis: mechanistic insights and clinical challenges. Nature Medicine . 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell . 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.on behalf of the Cancer Research UK and Cancer Therapeutics CRC Australia Metastasis Working Group, Anderson R. L., Balasas T., et al. A framework for the development of effective anti-metastatic agents. Nature Reviews. Clinical Oncology . 2019;16(3):185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaffer C. L., Weinberg R. A. A perspective on cancer cell metastasis. Science . 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 11.Guraya S. Y. Pattern, stage, and time of recurrent colorectal cancer after curative surgery. Clinical Colorectal Cancer . 2019;18(2):e223–e228. doi: 10.1016/j.clcc.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Su L., Hershberger R. J., Weissman I. L. LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes & Development . 1993;7(5):735–748. doi: 10.1101/gad.7.5.735. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Wang B., Yang A., et al. Ly-1 antibody reactive clone is an important nucleolar protein for control of self-renewal and differentiation in embryonic stem cells. Stem Cells . 2009;27(6):1244–1254. doi: 10.1002/stem.55. [DOI] [PubMed] [Google Scholar]
- 14.Luna-Pelaez N., Garcia-Dominguez M. Lyar-mediated recruitment of Brd2 to the chromatin attenuates _Nanog_ downregulation following induction of differentiation. Journal of Molecular Biology . 2018;430(8):1084–1097. doi: 10.1016/j.jmb.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 15.Izumikawa K., Ishikawa H., Yoshikawa H., et al. LYAR potentiates rRNA synthesis by recruiting BRD2/4 and the MYST-type acetyltransferase KAT7 to rDNA. Nucleic Acids Research . 2019;47(19):10357–10372. doi: 10.1093/nar/gkz747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonezawa K., Sugihara Y., Oshima K., Matsuda T., Nadano D. Lyar, a cell growth-regulating zinc finger protein, was identified to be associated with cytoplasmic ribosomes in male germ and cancer cells. Molecular and Cellular Biochemistry . 2014;395(1-2):221–229. doi: 10.1007/s11010-014-2128-x. [DOI] [PubMed] [Google Scholar]
- 17.Datta D., Anbarasu K., Rajabather S., Priya R. S., Desai P., Mahalingam S. Nucleolar GTP-binding protein-1 (NGP-1) promotes G1 to S phase transition by activating cyclin-dependent kinase inhibitor p21Cip1/Waf1∗. The Journal of Biological Chemistry . 2015;290(35):21536–21552. doi: 10.1074/jbc.M115.637280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C., Liu X., Cheng T., et al. LYAR suppresses beta interferon induction by targeting phosphorylated interferon regulatory factor 3. Journal of virology . 2019;93(21) doi: 10.1128/JVI.00769-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju J., Wang Y., Liu R., et al. Human fetal globin gene expression is regulated by LYAR. Nucleic Acids Research . 2014;42(15):9740–9752. doi: 10.1093/nar/gku718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Liu M., Li Z., et al. LYAR promotes colorectal cancer cell mobility by activating galectin-1 expression. Oncotarget . 2015;6(32):32890–32901. doi: 10.18632/oncotarget.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Astorgues-Xerri L., Riveiro M. E., Tijeras-Raballand A., et al. Unraveling galectin-1 as a novel therapeutic target for cancer. Cancer Treatment Reviews . 2014;40(2):307–319. doi: 10.1016/j.ctrv.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Adams J. C. Roles of fascin in cell adhesion and motility. Current Opinion in Cell Biology . 2004;16(5):590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Jayo A., Parsons M. Fascin: a key regulator of cytoskeletal dynamics. The International Journal of Biochemistry & Cell Biology . 2010;42(10):1614–1617. doi: 10.1016/j.biocel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Liu H., Cui J., Zhang Y., et al. Mass spectrometry-based proteomic analysis of FSCN1-interacting proteins in laryngeal squamous cell carcinoma cells. IUBMB Life . 2019;71(11):1771–1784. doi: 10.1002/iub.2121. [DOI] [PubMed] [Google Scholar]
- 25.Xu Z., Gao X., He Y., et al. Synergistic effect of SRY and its direct target, WDR5, on Sox9 expression. PLoS One . 2012;7(4, article e34327) doi: 10.1371/journal.pone.0034327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Q., Rank G., Tan Y. T., et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature Structural & Molecular Biology . 2009;16(3):304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vignjevic D., Schoumacher M., Gavert N., et al. Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Research . 2007;67(14):6844–6853. doi: 10.1158/0008-5472.CAN-07-0929. [DOI] [PubMed] [Google Scholar]
- 28.Currie E., Schulze A., Zechner R., Walther T. C., Farese R. V., Jr. Cellular fatty acid metabolism and cancer. Cell Metabolism . 2013;18(2):153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G., Fulkerson C. M., Malek R., Ghassemifar S., Snyder P. W., Mendrysa S. M. Mutations in Lyar and p53 are synergistically lethal in female mice. Part A, Clinical and molecular teratology . 2012;94(9):729–737. doi: 10.1002/bdra.23048. [DOI] [PubMed] [Google Scholar]
- 30.Abetov D. A., Kiyan V. S., Zhylkibayev A. A., et al. Editors’ pick: native mammalian preribosomal complexes. The Journal of Biological Chemistry . 2019;294(28):10746–10757. doi: 10.1074/jbc.AC119.008378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swartling F. J., Grimmer M. R., Hackett C. S., et al. Pleiotropic role for MYCN in medulloblastoma. Genes & Development . 2010;24(10):1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y., Atmadibrata B., Yu D., et al. Upregulation of LYAR induces neuroblastoma cell proliferation and survival. Cell Death and Differentiation . 2017;24(9):1645–1654. doi: 10.1038/cdd.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isaksson H. S., Sorbe B., Nilsson T. K. Whole genome expression profiling of blood cells in ovarian cancer patients -prognostic impact of the CYP1B1, MTSS1, NCALD, and NOP14. Oncotarget . 2014;5(12):4040–4049. doi: 10.18632/oncotarget.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sui X., Zhu J., Tang H., et al. p53 controls colorectal cancer cell invasion by inhibiting the NF-κB-mediated activation of Fascin. Oncotarget . 2015;6(26):22869–22879. doi: 10.18632/oncotarget.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alburquerque-González B., Bernabé-García M., Montoro-García S., et al. New role of the antidepressant imipramine as a Fascin1 inhibitor in colorectal cancer cells. Experimental & Molecular Medicine . 2020;52(2):281–292. doi: 10.1038/s12276-020-0389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S., Li Y., Wang D., et al. Fascin promotes lung cancer growth and metastasis by enhancing glycolysis and PFKFB3 expression. Cancer Letters . 2021;518:230–242. doi: 10.1016/j.canlet.2021.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., Huang X. T., Wang M. Y., et al. FSCN1 promotes radiation resistance in patients with PIK3CA gene alteration. Frontiers in Oncology . 2021;11, article 653005 doi: 10.3389/fonc.2021.653005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W., He T., Liu S., et al. The PIK3CA E542K and E545K mutations promote glycolysis and proliferation via induction of the β-catenin/SIRT3 signaling pathway in cervical cancer. Journal of Hematology & Oncology . 2018;11(1):p. 139. doi: 10.1186/s13045-018-0674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keshk W. A., Zineldeen D. H., Wasfy R. E., el-Khadrawy O. H. Fatty acid synthase/oxidized low-density lipoprotein as metabolic oncogenes linking obesity to colon cancer via NF-kappa B in Egyptians. Medical Oncology . 2014;31(10):p. 192. doi: 10.1007/s12032-014-0192-4. [DOI] [PubMed] [Google Scholar]
- 40.Pu X., Chen D. Targeting adipokines in obesity-related tumors. Frontiers in Oncology . 2021;11, article 685923 doi: 10.3389/fonc.2021.685923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo X., Cheng C., Tan Z., et al. Emerging roles of lipid metabolism in cancer metastasis. Molecular Cancer . 2017;16(1):p. 76. doi: 10.1186/s12943-017-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatima S., Hu X., Huang C., et al. High-fat diet feeding and palmitic acid increase CRC growth in β2AR- dependent manner. Cell Death & Disease . 2019;10(10):p. 711. doi: 10.1038/s41419-019-1958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Overall survival curve of those with colorectal cancer. Data were obtained from TCGA database and analyzed to generate Kaplan-Meier curves, which show a correlation between patients with tumors expressing low levels of LYAR and higher survival rates. Figure S2: LYAR expression in stable LYAR knockdown HCT15 cells. (a) RT-qPCR analysis of LYAR gene expression normalized to GAPDH gene expression in stable LYAR knockdown HCT15 cells. ∗∗p < 0.01 compared with the scrambled control. (b) Western blot assay showing LYAR protein expression in stable LYAR knockdown HCT15 cells. GAPDH served as the loading control. Figure S3: Detection of tumor cell phenotype in transient LYAR knockdown HCT15 cells. LYAR knockdown had no effect on cell proliferation (a), cell adhesion (b), cell cycle (c), apoptosis (d), or colony formation (e) of HCT15 cells. Parental refers to the control, nonsilencing refers to cells transfected with a nonspecific siRNA, and LYAR-siRNA1 and LYAR-siRNA2 refer to cells transfected with LYAR-specific siRNA1 and LYAR-specific siRNA2, respectively. Figure S4: Detection of migration and invasion in stable LYAR knockdown HCT15 cells. (a) Representative photos of haptotactic migration assay and matrigel chemoinvasion assay using stable LYAR knockdown HCT15 cells. Original magnification, 200x. (b) Results of migration and invasion assays. Data shown are the mean ± standard deviation (n = 3). ∗∗p < 0.01 compared to the scrambled control. Figure S5: Western blot assay showing FSCN1 expression after LYAR knockdown by siRNA. (a) LYAR and FSCN1 expression after LYAR knockdown in HCT8 cells. (b) LYAR and FSCN1 expression after LYAR knockdown in HCT116 cells. KD1 and KD2 refer to the stable LYAR knockdown lines LYAR-siRNA1 and LYAR-siRNA2, respectively. Figure S6: Expression of LYAR and downstream genes in colorectal cancer. The expression profiles of human colorectal cancer and normal colon tissue were obtained from TCGA and GTEx databases, respectively. The data show differential expression of LYAR (a), FSCN1 (b), FASN (c), and SCD (d) in colorectal cancer tissues compared to adjacent normal tissues. ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. Table S1: The relationship between LYAR protein expression and clinicopathological features in CRC patients. Table S2: ChIP primer sequences for the FSCN1 promoter.
Data Availability Statement
The data generated or analyzed during the current study are available from the corresponding author on reasonable request.


