Abstract
Virus-like particles (VLPs) are nano-scale particles that are morphologically similar to a live virus but which lack a genetic component. Since the pandemic spread of COVID-19, much focus has been placed on coronavirus (CoV)-related VLPs. CoVs contain four structural proteins, though the minimum requirement for VLP formation differs among virus species. CoV VLPs are commonly produced in mammalian and insect cell systems, sometimes in the form of chimeric VLPs that enable surface display of CoV epitopes. VLPs are an ideal model for virological research and have been applied as vaccines and diagnostic reagents to aid in clinical disease control. This review summarizes and updates the research progress on the characteristics of VLPs from different known CoVs, mainly focusing on assembly, in vitro expression systems for VLP generation, VLP chimerism, protein-based nanoparticles and their applications in basic research and clinical settings, which may aid in development of novel VLP vaccines against emerging coronavirus diseases such as SARS-CoV-2.
Keywords: Virus-like particles (VLPs), Coronavirus (CoV), Vaccine, Expression system
1. Introduction
Coronavirus disease 2019 (COVID-19), a novel highly infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported in Wuhan, China in December 2019 [1]. Since then, it spread rapidly into a worldwide pandemic. According to the WHO, as of October 15, 2021, there have been more than 2.3 billion confirmed COVID-19 cases around the world with over 4.8 million deaths, with a huge negative impact on public health, social and economic systems. In the past two decades, two other highly pathogenic CoVs – SARS-CoV and Middle East respiratory syndrome (MERS-CoV) [2], [3] – emerged from bats, and spilled over to humans through respective intermediate hosts, civet cats and camels [4], [5]. CoVs are enveloped viruses with a large single-stranded positive-sense RNA genome that mainly cause respiratory or enteric disease in infected hosts, although neurological or liver disease is found in some cases [6]. Genetically and serologically they can be classified into four genera: alpha-, beta-, gamma- and deltacoronavirus [7]. Whereas alpha- and betacoronavirus mainly infect mammals, gammacoronavirus tends to infect poultry and marine mammals, and deltacoronavirus can infect both poultry and mammals [8], [9]. Based on comparative genomic and phylogenetic analysis, bat CoVs are the source of alpha- and betacoronavirus and avian CoVs are the source of gamma- and deltacoronavirus [8].
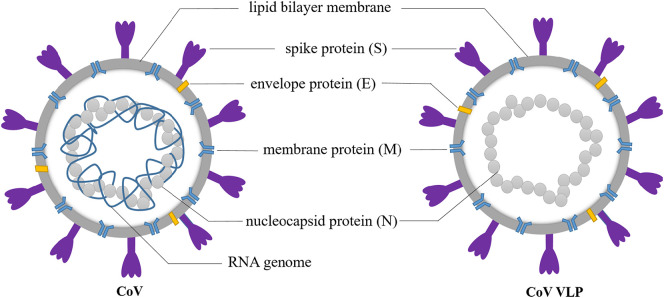
The large genome (25–32 kb) of CoVs has led to the development of a unique replication mechanism, which results in a high frequency of recombination [10], [11]. In addition, there is precedent for cross-species transmission of CoV, and thus a constant threat of spillover from bat or avian CoVs into humans [12], [13], [14]. For this reason, it is essential to study all aspects of CoVs in great detail, which will lead to novel vaccines, diagnostics and therapeutic technology. Virus-like particles (VLPs) are a nano-scale particle composed of viral structural proteins. They have a similar morphology and spatial structure to natural virions, but are not infectious or replicative due to lack of genetic material (Fig. 1 ), making them an ideal model for certain types of virological research. VLPs can be efficiently recognized by the immune system and induce specific cellular, humoral, and mucosal immune responses [15]. Chimeric VLPs that contain foreign epitopes or proteins have been developed, thus broadening the prospects for VLP-based vaccines. In addition, VLPs can be used as gene- or drug-delivery systems due to their ability to encapsulate nucleic acids or carry other molecules [16]. This review focuses on CoV-related VLPs and their potential use in basic research and clinical settings.
Fig. 1.
A schematic diagram of coronavirus (CoV) and CoV VLPs that are assembled from structural proteins. VLPs have a similar morphology and spatial structure to natural virions, but are not infectious or replicative due to a lack of genetic material.
2. Proteins involved in CoV VLP assembly
CoV particles contain four main structural proteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N) (Fig. 1) [17]. The S protein is a class I fusion protein that plays a critical role in virus entry by mediating receptor binding and membrane fusion [18], [19]. The S protein has access to the endoplasmic reticulum (ER) in eukaryotic cells due to its N-terminal signal peptide, and it is heavily N-glycosylated [20]. Spike proteins assemble into homotrimers on the surface of virions, which gives CoVs a crown-like appearance [21]. The identity and distribution of the specific cellular receptor recognized by the S protein are the key factors that determine host species susceptibility and tissue tropism of the virus [22]. Since the S protein contains epitopes that induce strongly neutralizing antibodies, it is the most common target for CoV vaccines. The M protein is the most abundant component of virions and plays an important role in the assembly and release of progeny virus [23]. The M protein has three transmembrane domains that facilitate dimer formation and create a scaffold for the viral envelope [24], [25], and it also interacts with the N protein to package the RNA genome within the virus particle [23]. Studies have shown that M protein can also stimulate the immune system to produce neutralizing antibodies [26]. The E protein is a transmembrane protein with ion channel activity that is found at low levels in virus particles [27], where it facilitates virion assembly and release [28] and also functions on inflammatory activation [29], [30]. The N protein is the only known protein constituent of the viral nucleocapsid [31], and both N- and C-terminal domains are able to bind viral genomic RNA [32]. The C-terminal RNA binding domain also specifically binds to the genomic packaging signal [33].
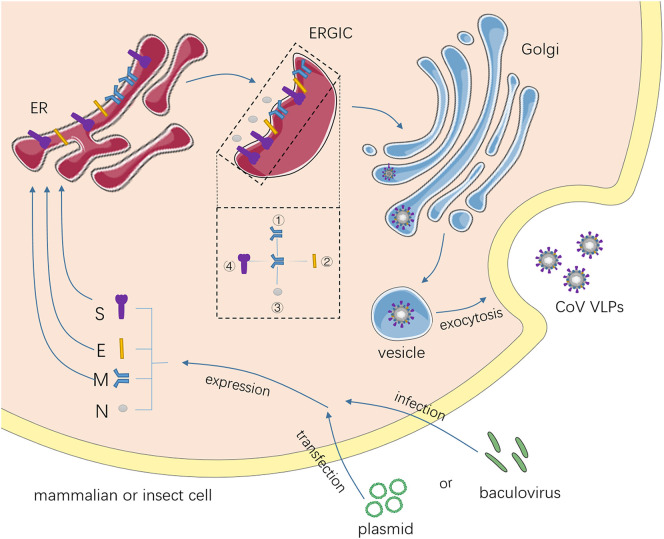
The process of virion assembly involves protein-protein (Fig. 2 ) and protein-RNA interactions [34]. The N protein is translated on free polysomes and interacts with genomic RNA to form helical nucleocapsids, whereas S, E and M are translated on membrane-bound polysomes, inserted into the ER, and transported into ER–Golgi intermediate compartments (ERGIC) [35]. There, M protein directs the assembly process by interacting with: (1) itself, to build the overall framework of the viral envelope; (2) the E protein, to induce membrane curvature and budding; and (3) nucleocapsids, to package the viral genome into the virion [36], [37]. S protein is incorporated into virions via interaction with M, but is not required for assembly. Following assembly, virions are transported to the cell surface in vesicles and exocytotically released [17].
Fig. 2.
The process of coronavirus (CoV) VLP assembly in eukaryotic cells. The structural genes of CoV are introduced into mammalian cell systems or baculovirus-insect cell systems. N proteins are translated on free polysomes. S, E and M proteins are translated on membrane-bound polysomes, inserted into the endoplasmic reticulum (ER), and transported into the ER–Golgi intermediate compartment (ERGIC). M protein directs the assembly process by interacting with every constituent of the VLP. The number in the dashed box indicates the order of interaction of each protein with M protein in the ERGIC. VLPs are transported to the cell surface in vesicles and exocytotically released.
For many CoVs, including mouse hepatitis virus (MHV) [38], transmissible gastroenteritis virus (TGEV) [39], bovine CoV (BCoV) [39] and infectious bronchitis virus (IBV) [40], the coexpression of M + E is the minimum requirement to form a VLP. But for SARS-CoV, studies have shown that either M + E [41], [42], [43] or M + N [44], [45] is sufficient for VLP formation.
3. VLP expression systems and resultant vaccination efficacy
VLP production generally proceeds via the following steps: construction and cloning of viral structural genes; expression in a recombinant heterologous system; separation and purification; and finally, characterization and verification. VLPs can be produced in prokaryotic (mainly bacterial) or eukaryotic (mammalian, yeast, plant, or baculovirus-insect cell) expression systems, and in some cases cell-free expression systems. VLPs can be divided into two categories determined by viral structure: non-enveloped and enveloped VLPs. Most non-enveloped VLPs can self-assemble by viral capsid proteins and contain no host components. For example, the capsid proteins of hepatitis E virus (HEV) [46], human papilloma virus (HPV) [47] and rotavirus (RV) [48] can form non-enveloped VLPs. On the other hand, enveloped VLPs are composed of capsid proteins and host cell membrane, and include those derived from influenza virus [49], hepatitis C virus (HCV) [50], CoV [42], etc. Some expression systems are more suitable for generation of different VLPs based on their variable characteristics (Table 1 ). For CoV-like particle production, mammalian and baculovirus-insect cell systems are the most common (Fig. 2), though plant systems have been reported to generate VLP-based vaccines against SARS-CoV-2.
Table 1.
Comparison of different VLP expression systems for viruses.
| Mammalian | Insect | Plant | Yeast | Bacteria | Cell-free | |
|---|---|---|---|---|---|---|
| VLP type | Non-enveloped and enveloped | Non-enveloped and enveloped | Non-enveloped and enveloped | Mostly Non-enveloped | Non-enveloped | Non-enveloped |
| Cost | ++++ | +++ | ++ | + | + | ++++ |
| PTMs | ++++ | +++ | ++ | + | None | + |
| Contamination | Mammalian virus | Baculovirus and nodavirus | None | None | Endotoxins | None |
Note: greater number of “+” = a higher level; PTM, post-translational modification.
3.1. Mammalian cell systems
Mammalian cells possess a capacity for proper protein folding, assembly and complex post-translational modification (PTM) including glycosylation, phosphorylation, acylation and disulfide bond formation, among others. Thanks to this advantage, mammalian cells have become the major system for production of recombinant proteins for clinical applications [51]. VLPs produced from mammalian cells maintain complete structure and function, and have greater immunogenicity compared to yeast-derived VLPs [52]. However, mammalian cell systems are limited by their high production costs, low expression efficiency and potential safety concerns due to contamination by other viruses [53], [54].
Xu et al. described the efficient creation of SARS-CoV-2 VLPs by plasmid-driven transfection of viral structural proteins in mammalian cells. They demonstrated that M + E are minimally required for efficient assembly and egress of SARS-CoV-2 VLPs and that the presence of E may potentiate the release efficiency of both M and S [55]. Moreover, the corona-like structure of SARS-CoV-2 VLPs derived from Vero E6 cells was more stable and unified relative to those from HEK-293 T cells under transmission electron microscopy (TEM) [55]. VLPs of MHV and feline infectious peritonitis virus (FIPV) were generated in recombinant vaccinia-T7-polymerase (vTF7-3)-infected OST7-1 cells that were co-transfected with DNA constructs separately encoding E and M of MHV-A59 and FIPV [38]. Bos et al. produced MHV VLPs by co-expression of MHV-A59 S, E, M and N in vTF7-3-infected mouse fibroblasts [56]. These MHV VLPs can package defective interfering (DI) RNA and become infectious [56]. Co-infection by recombinant vaccinia viruses encoding IBV E and M in BHK-21 cells resulted in the production of IBV VLPs [57]. However, particle release was extremely inefficient, indicating that other IBV components are required for efficient VLP formation [57]. Several studies have shown that SARS-CoV VLPs can be successfully produced by co-expression of N + M in 293 T cells without exogenous viruses, or E + M in vTF7-3-infected Vero E6 cells [43], [44]. The discrepancies among the reported roles of the SARS-CoV structural proteins in particle formation might be attributable to the use of different experimental expression systems [58]. Siu et al. demonstrated that E, N and M are required for the efficient production and release of SARS-CoV VLPs by transfected Vero E6 cells [59].
3.2. Baculovirus-insect cell systems
The baculovirus-insect cell expression system has been widely used in recent years to express foreign proteins because baculovirus is harmless to vertebrates due to its strict tropism for insect cells, and because its large genome can accommodate relatively large foreign genes. Proteins in this system are mainly expressed in soluble form with high efficiency and undergo almost the same PTM as in mammalian cells, which contributes to their strong immunogenicity [60], [61]. However, a major challenge faced by this system is the separation of purified target VLPs from co-produced baculovirus and contaminating nodavirus particles [62], [63].
A few different studies have prepared CoV-like particles using a baculovirus-insect cell expression system, with correct VLP morphology confirmed by electron microscopy. MERS-CoV and porcine epidemic diarrhea virus (PEDV) VLPs were successfully generated in Sf9 cells after infection with a recombinant baculovirus co-expressing the S, E and M genes of the MERS-CoV [64] and PEDV [65]. The shape and size of the VLPs were similar to those of the naive virus under TEM. Recently, Kim et al. found that the M protein alone is sufficient for the formation of PEDV VLPs in a HEK-293T cell expression system [66]. S monomers expressed in insect cells were slightly larger than those expressed in mammalian cells, suggesting incomplete glycosylation of these VLPs [64]. Inoculation of rhesus macaques with MERS-CoV VLPs plus alum adjuvant elicited neutralizing antibody titers up to 1:40 after the fourth immunization, and elicited specific IgG antibodies against the receptor binding domain (RBD) with endpoint titers reaching 1:1280. Besides, the detection of IFN-γ suggested that these MERS-CoV VLPs also induced a Th1-mediated response, raising hopes for a potential vaccine candidate [64]. PEDV VLPs induced a higher level of specific neutralizing antibody compared with inactivated PEDV, with titers reaching 563 and 282 after the second booster vaccination in mice. Furthermore, enhanced IL-4 levels demonstrated that a Th2 cell-mediated response was elicited by the PEDV VLPs [65]. In a slightly different strategy, IBV VLPs were generated through simultaneous co-infection of Sf9 cells with three recombinant baculoviruses, each expressing M, E or a recombinant S protein containing the S1 subunit fused to the transmembrane domain and the cytoplasmic tail (TM/CT) of S2 subunit [67]. A recombinant S was used because the interactions between the TM/CT domain of S and the CT domain of M are of critical importance for incorporation of S protein into VLPs. These VLPs induced IBV-specific neutralizing antibodies in chickens that were comparable to the inactivated IBV strains. SARS-CoV VLPs have been successfully generated in Sf21 cells by expression of S, E and M proteins using different combinations of recombinant baculovirus [42], [68]. Observations of immunogenicity in mice showed that SARS CoV VLPs induced high levels of neutralizing antibody and a Th1-dominated immune response. Sera from VLP-immunized mice reached 90% neutralization capacity at dilutions of 1:8 [68].
3.3. Plant systems
Plant expression systems have several advantages, including high expression levels, low cost, absence of mammalian pathogen contaminants, and the ability to produce proteins with correct folding and PTM. Proteins can be produced in plants by different strategies, including stable integration of a transgene into the nuclear or chloroplast genome, or transient transformation by plant viral vectors [69]. The most common gene transfer methods in plants utilize Agrobacterium-mediated transformation or particle bombardment [70].
The S protein of TGEV [71] was expressed in Arabidoposis plants after transformation with S cDNAs under the control of the cauliflower mosaic virus 35S (CaMV 35S) promoter. S proteins produced from plants elicited neutralizing antibodies in immunized mice [71]. Medicago Inc. has developed a tobacco plant system to produce VLP-based vaccine candidates, and has manufactured 10 million doses of a monovalent influenza vaccine within 1 month for the Defense Advanced Research Projects Agency (DARPA). A few months after the COVID-19 outbreak, Medicago announced production of VLP-based vaccines against SARS-CoV-2 using their proprietary plant–based platform.
3.4. Yeast systems
Yeast are another important eukaryotic expression system for production of recombinant proteins, and they have also been applied to the preparation of VLPs. Two licensed VLP-based vaccines developed by Merck & Co Inc., Recombivax HB® against hepatitis B virus (HBV) and Gardasil® against HPV, have been generated using this system [69]. Pichia pastoris, Saccharomyces cerevisiae and Hansenula yeast are commonly used, since they are easy to expand and cultivate, cost-effective, and can provide a certain degree of PTM. However, the VLP yield in yeast is lower than in bacteria, and their PTM functions are limited, especially in protein glycosylation [72]. Therefore, yeast systems are mostly used for the preparation of non-enveloped VLPs.
3.5. Bacterial systems
Bacteria are the most widely used expression system for the production of recombinant proteins, but they are not always desirable for VLP production due to key limitations such as a lack of PTM, protein solubility problems and contamination by endotoxins [72], [73]. They are commonly used to produce non-enveloped VLPs, whose components can self-assemble within the bacterial host. E. coli is the most common bacterial type for VLP production, and some approaches such as low-temperature cultivation and the use of different fusion protein systems can enhance expression levels and solubility [16], [74].
3.6. Cell-free systems
In some cases, the capsid proteins of non-enveloped, single-capsid VLPs can be assembled in a cell-free environment to form homogenous VLPs [75], [76], [77], or synthesized and assembled in an entirely cell-free environment [78], [79]. In one such system based on purified components, VLP precursors or capsomeres are expressed in E. coli, purified from host contaminants, and assembled into VLPs in a cell-free in vitro environment. A different system based on cell-free protein synthesis (CFPS) uses lysates from microbial, animal or plant cells as a source of energy and catalytic components necessary for target protein synthesis. The most commonly used cell-free extracts are from E. coli, wheat germ, rabbit reticulocytes and insect cells [80], [81], [82]. Cell-free systems have many advantages including time-saving, high yield of proteins and limited cellular contaminants [83]. In addition, CFPS system can expand the possibilities of protein engineering. For example, besides the 20 naturally occurring amino acids, unnatural amino acids, such as furanomycin, can be introduced into a protein structure for specifically designed purposes [84], [85], [86].
4. Chimeric CoV-related VLPs and their prospects for vaccines
VLPs can also be used as a platform to display foreign epitopes or proteins after genetic or chemical fusion to VLP proteins, resulting in so-called chimeric VLPs (cVLPs) that can be used to simultaneously stimulate immunity against multiple antigens (Fig. 3 ). Immunogenicity of some epitopes can also be improved by including them within such cVLPs. Here, we will focus on cVLPs for expression of CoV proteins (summarized in Table 2 ).
Fig. 3.
A schematic diagram of existing clinical applications of coronavirus (CoV)-related VLPs. CoV VLPs and chimeric VLPs (which both carry CoV antigens) are promising vaccine candidates due to their ability to stimulate strong humoral and cellular immunity. VLPs that package target regions of CoV RNAs can serve as positive controls for all steps of clinical testing, from RNA extraction to PCR amplification.
Table 2.
The examples of chimeric VLPs carrying CoV proteins.
| VLP platform | Basic composition | Foreign protein | Ref. |
|---|---|---|---|
| MHV | E, M and N | S of SARS-CoV | [87] |
| SARS-CoV | E and M | S of SARS-like CoV | [41] |
| Influenza virus | M1 and CT and TM domain of HA | S of SARS-CoV | [95] |
| Influenza virus | M1 and CT and TM domain of NA | S1 of IBV | [96] |
| CPV | VP2 | RBD of MERS-CoV | [98] |
| HBV | HBcAg | B-cell epitopes of PEDV | [104], [105] |
| HBV | HBcAg | Immunogenic epitopes of SARS-CoV-2 | [106] |
| NDV | M, NP and CT and TM domain of F | S of SARS-CoV-2 | [110] |
| MLV | Gag | S of SARS-CoV-2 | [113] |
Abbreviations: MHV, mouse hepatitis virus; SARS-CoV, severe acute respiratory syndrome coronavirus; CPV, canine parvovirus; HBV, hepatitis B virus; NDV, Newcastle disease virus; MLV, murine leukemia virus; IBV, infectious bronchitis virus; MERS-CoV, Middle East respiratory syndrome coronavirus; PEDV, porcine epidemic diarrhea virus; RBD, receptor binding domain; CT, carboxyl terminus; TM, transmembrane; HBcAg, hepatitis B core antigen; NP, nucleocapsid; HA, hemagglutinin; NA, neuraminidase; M1, matrix 1; S, spike; E, envelope; N, nucleocapsid; M, membrane.
Co-expression of structural proteins from different CoVs origins results in self-assembly of cVLPs. Since the efficiency of SARS-CoV VLP production in mammalian cells is low, Lokugamage et al. developed a cVLP carrying the MHV E, N and M proteins with the SARS-CoV S protein by cotransfection with four respective plasmids in 293 T or CHO cells [87]. The efficiency of the cVLP production was about 8 times better than that of VLPs made from SARS-CoV proteins alone [87]. Mice inoculated with cVLPs were protected from SARS-CoV challenge and produced high levels of specific neutralizing antibodies [87]. Bai et al. utilized the baculovirus-insect cells system to generate a cVLP containing S protein of a SARS-like (SL)-CoV from bats and E and M of SARS-CoV [41]. In the absence of a cell culture model for SL-CoV, this cVLP can be used to study the interaction between SL-CoV and host cells.
Since influenza virus is a huge threat to public health, flu VLP development has been a hot research topic in recent years. Influenza VLPs have been generated by co-expressing its structural proteins: matrix 1 (M1) + hemagglutinin (HA) [88], [89], HA + neuraminidase (NA) [90], HA + NA + M1 [91], [92], [93], or HA + NA + M1 + matrix 2 (M2) [94]. Influenza VLPs have been shown to induce protective immune responses and can serve as a platform for the expression of foreign proteins. Liu et al. produced a cVLP containing a chimeric SARS-CoV S protein and the avian influenza M1 protein in a recombinant baculovirus system [95]. The chimeric SARS-CoV S protein contained an influenza HA transmembrane (TM) and carboxyl terminus (CT) to maximize the interaction between the SARS S and influenza M1. These chimeric SARS-CoV VLPs with a corona-like spike structure on the outer rim have a similar size and morphology to wild-type SARS CoV and protected 100% of the immunized mice from challenge of SARS-CoV in the absence of adjuvant. Lv et al. generated a cVLP containing the influenza M1 protein and an IBV S1 protein fused to the CT and TM domain of an avian influenza NA protein [96], with a morphology similar to standard influenza VLPs. The cVLPs elicited higher S1-specific antibody responses in mice and chickens, higher neutralization antibody levels in chickens and higher IL-4 production in mice than inactivated IBV viruses [96].
VP2, the main structural protein of canine parvovirus (CPV), can be assembled into VLPs using a baculovirus expression system [97]. CPV-based cVLPs can be produced by fusing a foreign epitope to the N-terminus of VP2 protein; Wang et al. fused the MERS-CoV RBD to produce a novel cVLP displaying MERS-CoV RBD on the surface [98]. This cVLP retained certain characteristics of parvovirus, such as agglutination of porcine erythrocytes and CPV virion morphology, while immunization of mice induced RBD-specific humoral and cellular immune responses [98].
Expression of the nucleocapsid of HBV, also called HBV core antigen (HBcAg), in Escherichia coli self-assembles into core particles [99]. HBcAg is highly immunogenic and is known to induce a significant T cell response and high antibody titers [100], [101]. Foreign epitopes are often inserted at a superficial loop around amino acid residues 74–83 of HBcAg, considered to be the major immunodominant region (MIR) [102]. Insertion of foreign epitopes in the MIR has been shown to induce a stronger immune response than insertion at the N- or C-terminus [103]. There have been four small, linear and fairly conserved B-cell epitopes (named by S1, S2, S3, M respectively) identified in PEDV. Gillam et al. reported that insertion of these four epitopes into the MIR of HBcAg individually and collectively resulted in successful production of cVLPs in an E. coli expression system [104]. Each cVLP, when delivered by IP injection, was able to elicit a strong immunogenic response specific to the inserted PEDV epitopes in mice. Only the HBcAg + S1 and the HBcAg + S1S2S3M VLPs could induce an immune response capable of PEDV neutralization. Futhermore, Gillam et al. tried to make some design changes to the HBcAg-based cVLPs to increase the overall immune response to the inserted PEDV epitopes. New cVLPs carrying substitutions at two residues, D29C and R127C, in the HBcAg protein were able to elicit a strong antibody response to the S1 epitope that had increased ability to neutralize virus [105]. Additionally, placement of foreign epitopes within the HBcAg vector protein influences the immune reaction [105]. Ghorbani et al. predicted the immunogenic epitopes of SARS-CoV-2 and designed an HBcAg-based VLP with insertion in the MIR [106]. One shortcoming of using the HBcAg-based cVLPs as vaccines is that any pre-existing circulating anti-HBcAg antibodies in the host would greatly reduce their efficacy.
Newcastle disease virus (NDV) is an avian pathogen that causes severe economic losses in the poultry industry worldwide. NDV has six structural proteins including nucleocapsid (NP), phosphoprotein (P), matrix (M), fusion (F), haemagglutinin–neuraminidase (HN), and the large polymerase (L) [107]. The NDV M protein is the main driving force for budding of progeny virions, and its expression alone is sufficient for effective formation and release of VLPs [108], [109]. In 293T cells, Yang et al. co-expressed NDV M and NP proteins with a SARS-CoV-2S protein whose TM and CT domains were replaced with those from the NDV F protein, generating a cVLP that displayed the prefusion-stabilized SARS-CoV-2 spike ectodomain [110]. Immunoassays in mice showed this cVLP could be a promising COVID-19 vaccine candidate [110].
VLPs have also been created from expression of the major retrovirus capsid, gag, in a number of systems including bacterial, insect and mammalian cells [111]. During the budding process, gag VLPs can be incorporated with glycoproteins present in the plasma membrane of the producer cells [112]. This phenomenon in which viral envelope proteins are replaced by another envelope glycoprotein is called pseudotyping. Roy et al. produced Moloney murine leukemia virus (MLV)-based VLPs carrying pseudotyped SARS-CoV-2 spike protein [113]. Stable expression of S protein in 293 cells expressing MLV Gag-pol led to a more efficient production of VLPs compared to transient expression [113].
5. VLPs as tools for basic CoV research
VLP systems are valuable tools for the study of protein function, viral assembly, protein-protein and protein-RNA interaction. For example, an MHV VLP system containing reporter DI RNAs was used to prove that infectivity of VLPs was dependent on S protein. Several mutant S proteins that were shown to be uncleaved were inserted into MHV VLPs, proving that S protein cleavage is not required for infectivity of MHV [114]. The observation that only M and E are required for formation of MHV VLPs indicated the novel principle that the assembly of CoV envelopes is nucleocapsid-independent, which is a novel budding principle compared to the two other well-studied models, type D retroviruses and alphaviruses, whose nucleocapsids play a central role [38]. Some have reported the release of MHV and IBV E proteins in lipid vesicles when expressed alone, suggesting that E protein likely plays an important role in CoV envelope formation and budding [57], [115]. For SARS-CoV, M proteins are capable of self-assembly and release in the form of vesicles when expressed alone, with lower densities than VLPs formed from M + N [116]. Site-directed mutagenesis results showed exactly which amino acid residues are important for M self-assembly, dispersed along the C-terminal endodomain and the N-terminal region, including the transmembrane domains [34]. This finding is consistent with previous deletion analysis results that multiple SARS-CoV M regions are involved in M self-assembly [116]. To identify important M amino acid residues for VLP assembly, genetic analysis and the ability of mutant M proteins to participate in VLP assembly were investigated in an MHV model. A conserved, 12-amino-acid domain in the M protein tail was reported to be functionally important for it to participate in virus assembly [117]. The study, performed in SARS-CoV, showed that cytoplasmic M tail dileucine residues are involved in the packaging of N into VLPs [34]. Two chimeric MHV-FIPV S proteins, consisting of the ectodomain of one virus and the transmembrane and endodomain of the other, were found to only assemble into viral particles of the species from which their C-terminal domain originated [118]. This indicates that assembly of spikes into the CoV envelope is governed by the S protein's C-terminal domain [118]. The result that the cytoplasmic tails of IBV E and M proteins mediate their interaction was confirmed by examining the ability of mutant and chimeric E and M proteins to form VLPs [40]. A cVLP assembly system in which the HIV-1 nucleocapsid (NC) domain was replaced by a SARS-CoV N-coding sequence was employed to test SARS-CoV N protein's self-interaction capacity, finding that the C-terminal half directed VLP assembly and release [119].
Palmitoylation is a common PTM in which a saturated fatty acid is linked to a cysteine residue by a thioester bond [120]. In viral proteins, this modification has been shown to affect assembly or replication. CoV S proteins are palmitoylated at a cysteine-rich motif in the cytoplasmic domain, adjacent to the transmembrane region [121]. It has been shown that palmitoylation of the MHV S protein is necessary for its interaction with M protein, which is thought to mediate MHV S incorporation into virions [121]. Accordingly, a SARS-CoV VLP system was utilized to determine the functional role of S protein palmitoylation. Results showed that SARS-CoV S mutants lacking two palmitoylated cysteine residues failed to be incorporated into VLPs but could co-localize with the M protein, indicating that M–S co-localization occurs independently of S incorporation in SARS-CoV assembly [122], which is in contrast to studies of the MHV S protein. A similar VLP assay was performed on TGEV, in which a mutant S protein with all cysteine residues substituted by alanines failed to be incorporated into VLPs [120]. However, the S-M interaction was not impaired by lack of palmitoylation; thus, palmitoylation of the S protein of TGEV is dispensable for S–M interaction, but required for the production of infectious virions [120].
VLP systems have been applied to identify the RNA packaging signal (PS) of CoV. The putative packaging signal of CoV genomic RNA was inserted into the 3′ noncoding region of the green fluorescent protein (GFP) gene, allowing GFP to act as a tracker for the packaging of RNA into VLPs. For MERS-CoV and SARS-CoV, assembly of the RNA PS into VLPs is dependent on the viral N protein [43], [123]. However, for MHV, the RNA PS can be packaged into VLPs in the absence of N protein because of the selective interaction of M protein with PS-containing RNA [124], [125]. Furthermore, a functional SARS-CoV RNA packaging signal failed to assemble into the MERS VLPs, indicating that there is virus-specific assembly of the RNA genome [123].
6. Clinical applications of VLP for CoV diseases
6.1. Current state of VLP-based vaccines
At present, vaccination is still the most effective means to prevent and control infectious diseases, and live-attenuated and inactivated virus vaccines are the most widely used. Live-attenuated vaccines mimic natural infection and thus induce strong cellular and humoral immune responses [126], [127]. They are typically highly efficacious, but carry safety concerns of reversion to virulence and require strict refrigeration environment throughout storage and distribution [127]. Inactivated virus vaccines lose their pathogenicity and retain partial immunogenicity. They tend to stimulate a relatively low level of immune response and do not generate a full cellular immune response [127]. They often contain adjuvants and multiple doses are needed to enhance the immune effect [127], [128]. Besides these traditional vaccine types, protein subunit, nucleic acid (DNA or mRNA) and viral-vectored vaccines are other strategies for vaccine development. Protein subunit vaccines are safer than live-attenuated and inactivated vaccines since there is no live particle involved, but its immunogenicity is weak [129]. DNA and mRNA vaccines have received increased attention owing to their safety, efficacy, and ease in mass manufacturing [129], [130]. However, the stability of RNA remains a problem [131]. Viral-vectored vaccines can induce strong humoral and cellular immune responses but possess the problems of pre-existing anti-vector immunity and a risk of mutagenesis [132], [133]. As a novel type of vaccine, VLPs have better immunogenicity and safety. VLPs can be taken up efficiently by antigen-presenting cells (APCs) due to their appropriate size, triggering the adaptive immune system, and can stimulate strong humoral and cellular immunity against multiple antigens and specific structures (Fig. 3) [16], [106], [134], [135]. VLPs are free of viral nucleic acid and thus are not capable of infection and replication. Since only a short time is required for the process of VLP design and expression, VLP vaccines can quickly respond to epidemics of viral disease [15]. However, VLP technology has been limited by low expression yield and the need for purification from the various protein expression systems [136]. The high cost of production and purification presently restricts their widespread use [137]. Therefore, for economic reasons, the development of VLP-based vaccines is usually only suitable for widespread viruses that cause great damage. In the future, it will be necessary to reduce costs by improving production procedures, so as to expand the range of feasibility for VLP-based vaccines.
Over the past few decades, there have been different VLPs studied to fight against important pathogens such as ebolavirus (EBOV), Chikungunya, Dengue, influenza A, respiratory syncytial virus, and Zika [138]. Several VLP-based vaccines including Recombivax HB and Engerix-B for HBV, Gardasil, Cervarix, and Gardasil-9 for HPV, Hecolin for HEV, as well as in the veterinary field, Ingelvac CircoFLEX for PCV2 are licensed and commercialized [16], [83]. Before the outbreak of COVID-19, there were no vaccines approved for a human CoV, let alone a VLP-based option. When SARS and MERS first emerged, there was a rush to develop vaccines against SARS-CoV and MERS-CoV. However, since both epidemics were quickly and effectively controlled, the development of vaccines by researchers was suspended, and vaccines against animal CoVs mostly remain in preclinical trials. The disaster triggered by the rapid spread of SARS-CoV-2 around the world spurred an unprecedented global race towards a vaccine [139]. According to the WHO, as of October 15, 2021, 194 vaccines against SARS-CoV-2 are in pre-clinical development and 126 are in clinical development, among which 5 VLP candidates are in clinical trials (Table 3 ) and 18 VLP candidates are in preclinical evaluation. CoVLP is a plant-derived COVID-19 vaccine developed by Medicago Inc. In a phase 2 clinical trial, Medicago's vaccine exhibited an acceptable safety profile, and adverse events were primarily mild and moderate, and the duration was short. In terms of immunogenicity, the vaccine candidate induced a significant humoral immune response in both age cohorts (adults aged 18–64 and older adults aged 65+) after two doses, with neutralizing antibody titers that were about 10 times higher than those in a panel of sera from patients recovering from COVID-19 (https://medicago.com/en/press-release/1751/). VBI-2902a (VBI Vaccine Inc.) is a monovalent enveloped VLP vaccine expressing the native SARS-CoV-2 spike protein. In the phase 1 portion of an ongoing phase 1/2 study in 61 healthy adults (age 18–54 years), after two doses, VBI-2902a induced neutralization titers in 100% of participants, with 4.3× higher geometric mean titer (GMT) than that of the convalescent serum panel, and peak antibody binding GMT was 1:4047 (https://www.vbivaccines.com/press-releases/covid-19-vaccine-phase-1-data/). Undoubtedly, VLPs are acquiring prominence as a vaccine platform for CoVID-19.
Table 3.
VLP vaccine candidates against SARS-CoV-2 in clinical development.
| Name of vaccine | Description | Phase | Identifier | Developers |
|---|---|---|---|---|
| RBD SARS-CoV-2 HBsAg VLP | HBsAg VLPs displaying the SARS-CoV-2 RBD | Phase 1/2 | ACTRN12620000817943 | Serum Institute of India &Accelagen Pty&SpyBiotech |
| CoVLP | CoV-like particles derived from plant - nicotiana benthamiana | Phase 2/3 | NCT04636697 | Medicago |
| VBI-2902a | An enveloped VLP of SARS-CoV-2 spike glycoprotein and aluminum phosphate adjuvant. | Phase 1/2 | NCT04773665 | VBI Vaccines Inc. |
| No name | SARS-CoV-2 VLP vaccine | Phase 2 | NCT04818281 | The Scientific and Technological Research Council of Turkey |
| ABNCoV2 | Employs a novel plug-&-play capsid cVLP platform | Phase 1 | NCT04839146 | Radbound University |
Abbreviations: HBsAg, hepatitis B surface antigen; RBD, receptor binding domain.
6.2. Protein-based nanoparticles – one of the most appealing platforms for COVID-19 vaccine design
Protein-based nanoparticles are a popular tool in vaccine design, as they allow the repetitive surface presentation of antigens in their native conformations [140]. Protein-based nanoparticle vaccines (NPVs) can be roughly divided into three categories according to their scaffold protein: VLP proteins, non-viral naturally oligomeric proteins and engineered proteins. Among them, VLP proteins like hepatitis B surface antigen (HBsAg) from HBV were early platforms utilized in vaccines [141]. In addition to VLPs, many naturally oligomeric proteins such as ferritin, lumazine synthase, and C4b-binding protein (C4bp) orthologs have been used as platforms [140], [142]. Recently, a new development in this area is computational protein design, through which proteins are engineered that can assemble into large, highly oligomeric complexes. This design offers greater control over antigen stoichiometry, spacing, and particle size [142]. Similar to VLPs, NPVs are likely not widely used due to the high cost of production and purification in the present stage.
SpyBiotech and Serum Institute of India developed a vaccine candidate using SpyCatcher/SpyTag technology to display the SARS-CoV-2 RBD on the surface of HBsAg VLPs. SpyCatcher/SpyTag is a novel antigen-displaying technology distinct from two older methods, chemical conjugation and genetic fusion. SpyCatcher is an engineered protein designed to spontaneously form an irreversible isopeptide bond to its peptide partner (SpyTag) upon mixing [143]. When used for antigen attachment, SpyCatcher is fused to a nanoparticle scaffold for further coupling with SpyTag-fused antigens [143], [144], [145]. Tan et al. utilized the SpyCatcher/SpyTag technology to assemble the SARS-CoV-2 RBD on a protein nanoparticle platform, SpyCatcher003-mi3 VLPs [146]. A nanoparticle scaffold was computationally engineered using an aldolase from thermophilic bacteria that spontaneously assembles into a hollow 36-nm dodecahedral cage. The RBD-SpyVLP has been shown to be highly immunogenic in mice and pigs, inducing robust SARS-CoV-2-neutralizing antibody responses that are superior to convalescent human sera [146], [147].
Ferritin is a kind of iron storage protein that is found widely in organisms. Ferritin self-assembles into nanoparticles with strong thermal and chemical stability [141]. Saunders et al. designed a VLP-like vaccine that conjugated the 24-mer SARS-CoV-2 RBD to a ferritin scaffold using a sortase A reaction [140]. Vaccination of macaques with these nanoparticles elicited a cross-neutralizing antibody responses against bat CoVs, SARS-CoV and SARS-CoV-2 and resulted in a 50% inhibitory reciprocal serum dilution (ID50) neutralization GMT of 47,216 for SARS-CoV-2, as well as in protection against SARS-CoV-2 in the upper and lower respiratory tracts [140].
Most recently, Walls et al. designed a vaccine that displays 60 copies of the SARS-CoV-2 RBD on the surface of a computationally designed nanoparticle, I53-50, which is a 120-subunit complex with icosahedral symmetry constructed from trimeric (I53-50A) and pentameric (I53-50B) components [148]. These nanoparticle vaccines induced protective antibody responses in mice, with neutralizing titers ten-fold higher than soluble prefusion-stabilized S despite the ~five-fold lower dose. Antibodies recognize multiple distinct RBD epitopes, suggesting that they may not be easily prone to escape mutations [148].
6.3. Progress in development of VLP-based diagnostic reagents
Rapid, large-scale and accurate diagnostic testing is increasingly essential for early detection of infections to control the spread of epidemics from the source. Nucleic acid testing, especially real-time reverse transcription polymerase chain reaction (RT-qPCR), is considered the gold standard for detection of respiratory viruses [149], [150], and various RT-qPCR primer/probe sets targeting distinct regions of the viral genome have been reported for the detection of SARS-CoV-2. To ensure the accuracy of RT-qPCR test, positive controls must be run in parallel with clinical samples [151], and the most commonly used are synthetic RNA transcripts or plasmids. RNA is unstable and easily degraded, requiring cold chain transportation and storage. Moreover, neither plasmid DNA or RNA can serve as an appropriate control for extraction from clinical samples.
In order to solve these problems, VLPs that package RNA fragments containing the appropriate nucleic acids for binding of RT-qPCR primers and probes have been developed to serve as positive controls (Fig. 3). This approach was previously applied in the detection of foot-and-mouth disease virus (FMDV), EBOV, influenza virus and SARS-CoV [152], [153], [154]. In response to COVID-19, VLPs derived from Qβ bacteriophage [151], MS2 [155], plant virus cowpea chlorotic mottle virus (CCMV) [151] and cowpea mosaic virus (CPMV) [135] were explored to encapsidate target RNA and used as positive controls for SARS-CoV-2 detection. These VLP-based positive controls are highly stable, able to be mass-produced, and can serve as a standard to monitor the full process of nucleic acid testing, from RNA extraction to PCR amplification.
7. Conclusions
In view of the great threat of CoVs to human and animal health, VLPs incorporating their proteins merit further study. The minimal structural protein requirement for CoV VLP formation is still controversial, though this is related to differences between expression and assembly systems used. Because VLPs have the advantage of a similar structure to natural virions, lack of possible infectivity, excellent immunogenicity, good structural stability and the unique ability to display foreign molecules, they have broad potential application in biomedical research and clinical care. VLPs are a safe and accessible alternative to the use of live virus in basic CoV research. Licensed VLP vaccines, such as Recombivax HB and Engerix-B for HBV, Gardasil, Cervarix, and Gardasil-9 for HPV demonstrate that VLPs are an effective platform for vaccine development. However, at present there is no approved VLP-based vaccine product for human CoV, as the safety and efficacy of VLP-based vaccines remain to be verified and improved for use in animal and human populations. With the development of nanotechnology and protein engineering, VLPs are emerging to be a robust and flexible vaccine platform for the targeting of different diseases. Computational design provides accurate prediction and control for the composition and assembly of VLPs, such as the I53–50 nanoparticle vaccines for SARS-CoV-2 [148], thus broadening the range of antigens that can be displayed. Moreover, new bioprocessing technologies like in vitro assembly and cell-free expression provide new avenues for improvement of the vaccine production process [156]. VLPs have already been used in the development of vaccines and diagnostic reagents for CoV diseases due to their ability to carry antigens and encapsulate desired nucleic acids. In addition, the successful packaging of synthetic drugs inside VLPs has opened up a prospective role in drug delivery [157], and the surface of the VLP can be changed so that it targets specific tissues or organs [83]. In summary, the application of VLPs in the field of CoVs has only just begun, and will play a great role in the prevention and treatment of future epidemics.
CRediT authorship contribution statement
Wan Lu: Conceptualization, Writing – original draft, Validation. Zhuangzhuang Zhao: Writing – review & editing, Validation. Yao-Wei Huang: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Validation. Bin Wang: Conceptualization, Supervision, Writing – review & editing, Validation.
Acknowledgements
This work was supported by the Laboratory of Lingnan Modern Agriculture Project (NG2022001), the National Natural Science Foundation of China (32041003), and the Zhejiang Provincial Natural Science Foundation (LZ22C180002). We thank the professional editing service of NB Revisions for technical preparation of the text prior to submission.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., I. China Novel Coronavirus. T. Research A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Reusken C.B., Raj V.S., Koopmans M.P., Haagmans B.L. Cross host transmission in the emergence of MERS coronavirus. Curr. Opin. Virol. 2016;16:55–62. doi: 10.1016/j.coviro.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Artika I.M., Dewantari A.K., Wiyatno A. Molecular biology of coronaviruses: current knowledge. Heliyon. 2020;6(8) doi: 10.1016/j.heliyon.2020.e04743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams M.J., Carstens E.B. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2012) Arch. Virol. 2012;157(7):1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y.L., Yu J.Q., Huang Y.W. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): an update three years after its discovery. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai M.M.C. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 1996;7(6):381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian P.F., Jin Y.L., Xing G., Qv L.L., Huang Y.W., Zhou J.Y. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg. Infect. Dis. 2014;20(10):1735–1738. doi: 10.3201/eid2010.140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y.L., Qin P., Wang B., Liu Y., Xu G.H., Peng L., Zhou J., Zhu S.J., Huang Y.W. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J. Virol. 2019;93(24) doi: 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Wang B., Liang Q.Z., Shi F.S., Ji C.M., Yang X.L., Yang Y.L., Qin P., Chen R., Huang Y.W. Roles of two major domains of the porcine deltacoronavirus S1 subunit in receptor binding and neutralization. J. Virol. 2021;95:e01118–e01121. doi: 10.1128/JVI.01118-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opriessnig T., Huang Y.W. Update on possible animal sources for COVID-19 in humans. Xenotransplantation. 2020;27(3) doi: 10.1111/xen.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan D., Wei Y.Q., Guo H.C., Sun S.Q. The application of virus-like particles as vaccines and biological vehicles. Appl. Microbiol. Biotechnol. 2015;99(24):10415–10432. doi: 10.1007/s00253-015-7000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirbaghaee Z., Bolhassani A. Different applications of virus-like particles in biology and medicine: vaccination and delivery systems. Biopolymers. 2016;105(3):113–132. doi: 10.1002/bip.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64(11):5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Haan C.A.M., Vennema H., Rottier P.J.M. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane-protein, E1-glycoprotein, from a coronavirus. Nature. 1984;308(5961):751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z.B., Chen J.F., Shi H.Y., Chen X.J., Shi D., Feng L., Yang B. Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus. Virol. J. 2012;9 doi: 10.1186/1743-422X-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdia-Baguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortego J., Ceriani J.E., Patino C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368(2):296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X.G., Zhang H.L., Zhang Q., Dong J., Liang Y.B., Huang Y., Liu H.J., Tong D.W. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10 doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieto-Torres J.L., DeDiego M.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo L.L., Masters P.S. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 2013;87(9):5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83(14):7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molenkamp R., Spaan W.J.M. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tseng Y.T., Chang C.H., Wang S.M., Huang K.J., Wang C.T. Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krijnse-Locker J., Ericsson M., Rottier P.J., Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 1994;124(1–2):55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raamsman M.J., Locker J.K., de Hooge A., de Vries A.A., Griffiths G., Vennema H., Rottier P.J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000;74(5):2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baudoux P., Carrat C., Besnardeau L., Charley B., Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998;72(11):8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312(1):25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai B., Hu Q., Hu H., Zhou P., Shi Z., Meng J., Lu B., Huang Y., Mao P., Wang H. Virus-like particles of SARS-like coronavirus formed by membrane proteins from different origins demonstrate stimulating activity in human dendritic cells. PLoS One. 2008;3(7) doi: 10.1371/journal.pone.0002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318(4):833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79(22):13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y., Yang Z.Y., Kong W.P., Nabel G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 2004;78(22):12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatakeyama S., Matsuoka Y., Ueshiba H., Komatsu N., Itoh K., Shichijo S., Kanai T., Fukushi M., Ishida I., Kirikae T., Sasazuki T., Miyoshi-Akiyama T. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology. 2008;380(1):99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T.C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M.A., Uchida T., Takeda N., Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 1997;71(10):7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasagawa T., Pushko P., Steers G., Gschmeissner S.E., Hajibagheri M.A., Finch J., Crawford L., Tommasino M. Synthesis and assembly of virus-like particles of human papillomaviruses type 6 and type 16 in fission yeast Schizosaccharomyces pombe. Virology. 1995;206(1):126–135. doi: 10.1016/s0042-6822(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 48.Labbe M., Charpilienne A., Crawford S.E., Estes M.K., Cohen J. Expression of rotavirus VP2 produces empty corelike particles. J. Virol. 1991;65(6):2946–2952. doi: 10.1128/jvi.65.6.2946-2952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen B.J., Leser G.P., Morita E., Lamb R.A. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 2007;81(13):7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumert T.F., Ito S., Wong D.T., Liang T.J. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 1998;72(5):3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wurm F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22(11):1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 52.Diminsky D., Schirmbeck R., Reimann J., Barenholz Y. Comparison between hepatitis B surface antigen (HBsAg) particles derived from mammalian cells (CHO) and yeast cells (Hansenula polymorpha): composition, structure and immunogenicity. Vaccine. 1997;15(6–7):637–647. doi: 10.1016/s0264-410x(96)00239-3. [DOI] [PubMed] [Google Scholar]
- 53.Grillberger L., Kreil T.R., Nasr S., Reiter M. Emerging trends in plasma-free manufacturing of recombinant protein therapeutics expressed in mammalian cells. Biotechnol. J. 2009;4(2):186–201. doi: 10.1002/biot.200800241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuenmayor J., Godia F., Cervera L. Production of virus-like particles for vaccines. New Biotechnol. 2017;39(Pt B):174–180. doi: 10.1016/j.nbt.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu R., Shi M., Li J., Song P., Li N. Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol. 2020;8:862. doi: 10.3389/fbioe.2020.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218(1):52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74(9):4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakauchi M., Kariwa H., Kon Y., Yoshii K., Maeda A., Takashima I. Analysis of severe acute respiratory syndrome coronavirus structural proteins in virus-like particle assembly. Microbiol. Immunol. 2008;52(12):625–630. doi: 10.1111/j.1348-0421.2008.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82(22):11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy P., Noad R. Virus-like particles as a vaccine delivery system: myths and facts. Pharm. Biotechnol. 2009;655:145–158. doi: 10.1007/978-1-4419-1132-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ong H.K., Tan W.S., Ho K.L. Virus like particles as a platform for cancer vaccine development. PeerJ. 2017;5 doi: 10.7717/peerj.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hervas-Stubbs S., Rueda P., Lopez L., Leclerc C. Insect baculoviruses strongly potentiate adaptive immune responses by inducing type I IFN. J. Immunol. 2007;178(4):2361–2369. doi: 10.4049/jimmunol.178.4.2361. [DOI] [PubMed] [Google Scholar]
- 63.Li T.C., Scotti P.D., Miyamura T., Takeda N. Latent infection of a new alphanodavirus in an insect cell line. J. Virol. 2007;81(20):10890–10896. doi: 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H., Feng N., Chi H., Qiu B., Li N., Wang T., Gao Y., Yang S., Xia X. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017;8(8):12686–12694. doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C., Yan F., Zheng X., Wang H., Jin H., Wang C., Zhao Y., Feng N., Wang T., Gao Y., Yang S., Xia X. Porcine epidemic diarrhea virus virus-like particles produced in insect cells induce specific immune responses in mice. Virus Genes. 2017;53(4):548–554. doi: 10.1007/s11262-017-1450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J., Yoon J., Park J.E. Construction of porcine epidemic diarrhea virus-like particles and its immunogenicity in mice. Vaccines (Basel) 2021;9(4) doi: 10.3390/vaccines9040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu P.W., Wu X., Wang H.N., Ma B.C., Ding M.D., Yang X. Assembly and immunogenicity of baculovirus-derived infectious bronchitis virus-like particles carrying membrane, envelope and the recombinant spike proteins. Biotechnol. Lett. 2016;38(2):299–304. doi: 10.1007/s10529-015-1973-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu X., Chen Y., Bai B., Hu H., Tao L., Yang J., Chen J., Chen Z., Hu Z., Wang H. Immune responses against severe acute respiratory syndrome coronavirus induced by virus-like particles in mice. Immunology. 2007;122(4):496–502. doi: 10.1111/j.1365-2567.2007.02676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kushnir N., Streatfield S.J., Yusibov V. Virus-like particles as a highly efficient vaccine platform: diversity of targets and production systems and advances in clinical development. Vaccine. 2012;31(1):58–83. doi: 10.1016/j.vaccine.2012.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yusibov V., Rabindran S. Recent progress in the development of plant derived vaccines. Expert Rev. Vaccines. 2008;7(8):1173–1183. doi: 10.1586/14760584.7.8.1173. [DOI] [PubMed] [Google Scholar]
- 71.Gomez N., Carrillo C., Salinas J., Parra F., Borca M.V., Escribano J.M. Expression of immunogenic glycoprotein S polypeptides from transmissible gastroenteritis coronavirus in transgenic plants. Virology. 1998;249(2):352–358. doi: 10.1006/viro.1998.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeltins A. Construction and characterization of virus-like particles: a review. Mol. Biotechnol. 2013;53(1):92–107. doi: 10.1007/s12033-012-9598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohsen M.O., Zha L., Cabral-Miranda G., Bachmann M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin. Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 74.Huang X., Wang X., Zhang J., Xia N., Zhao Q. Escherichia coli-derived virus-like particles in vaccine development. NPJ Vaccines. 2017;2:3. doi: 10.1038/s41541-017-0006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X.S., Casini G., Harrison S.C., Garcea R.L. Papillomavirus capsid protein expression in Escherichia coli: purification and assembly of HPV11 and HPV16 L1. J. Mol. Biol. 2001;307(1):173–182. doi: 10.1006/jmbi.2000.4464. [DOI] [PubMed] [Google Scholar]
- 76.Hwang B.J., Jang Y., Kwon S.B., Yu J.E., Lim J., Roh Y.H., Seong B.L. RNA-assisted self-assembly of monomeric antigens into virus-like particles as a recombinant vaccine platform. Biomaterials. 2021;269 doi: 10.1016/j.biomaterials.2021.120650. [DOI] [PubMed] [Google Scholar]
- 77.Salunke D.M., Caspar D.L.D., Garcea R.L. Self-assembly of purified polyomavirus capsid protein-Vp1. Cell. 1986;46(6):895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 78.Lua L.H.L., Connors N.K., Sainsbury F., Chuan Y.P., Wibowo N., Middelberg A.P.J. Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 2014;111(3):425–440. doi: 10.1002/bit.25159. [DOI] [PubMed] [Google Scholar]
- 79.Spice A.J., Aw R., Bracewell D.G., Polizzi K.M. Synthesis and assembly of hepatitis B virus-like particles in a Pichia pastoris cell-free system. Front. Bioeng. Biotechnol. 2020;8:72. doi: 10.3389/fbioe.2020.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 2012;30(5):1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pelham H.R., Jackson R.J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 1976;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- 82.Swartz J. Developing cell-free biology for industrial applications. J. Ind. Microbiol. Biotechnol. 2006;33(7):476–485. doi: 10.1007/s10295-006-0127-y. [DOI] [PubMed] [Google Scholar]
- 83.Nooraei S., Bahrulolum H., Hoseini Z.S., Katalani C., Hajizade A., Easton A.J., Ahmadian G. Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology. 2021;19(1):59. doi: 10.1186/s12951-021-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noren C.J., Anthonycahill S.J., Griffith M.C., Schultz P.G. A general-method for site-specific incorporation of unnatural amino-acids into proteins. Science. 1989;244(4901):182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 85.Kim D.M., Kigawa T., Choi C.Y., Yokoyama S. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur. J. Biochem. 1996;239(3):881–886. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- 86.Kohno T., Kohda D., Haruki M., Yokoyama S., Miyazawa T. Nonprotein amino acid furanomycin, unlike isoleucine in chemical structure, is charged to isoleucine tRNA by isoleucyl-tRNA synthetase and incorporated into protein. J. Biol. Chem. 1990;265(12):6931–6935. [PubMed] [Google Scholar]
- 87.Lokugamage K.G., Yoshikawa-Iwata N., Ito N., Watts D.M., Wyde P.R., Wang N., Newman P., Kent Tseng C.T., Peters C.J., Makino S. Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine. 2008;26(6):797–808. doi: 10.1016/j.vaccine.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quan F.S., Huang C., Compans R.W., Kang S.M. Virus-like particle vaccine induces protective immunity against homologous and heterologous strains of influenza virus. J. Virol. 2007;81(7):3514–3524. doi: 10.1128/JVI.02052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hossain M.J., Bourgeois M., Quan F.S., Lipatov A.S., Song J.M., Chen L.M., Compans R.W., York I., Kang S.M., Donis R.O. Virus-like particle vaccine containing hemagglutinin confers protection against 2009 H1N1 pandemic influenza. Clin. Vaccine Immunol. 2011;18(12):2010–2017. doi: 10.1128/CVI.05206-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li X., Ju H., Liu J., Yang D., Qi X., Yang X., Qiu Y., Zheng J., Ge F., Zhou J. Influenza virus-like particles harboring H9N2 HA and NA proteins induce a protective immune response in chicken. Influenza Other Respir. Viruses. 2017;11(6):518–524. doi: 10.1111/irv.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bright R.A., Carter D.M., Daniluk S., Toapanta F.R., Ahmad A., Gavrilov V., Massare M., Pushko P., Mytle N., Rowe T., Smith G., Ross T.M. Influenza virus-like particles elicit broader immune responses than whole virion inactivated influenza virus or recombinant hemagglutinin. Vaccine. 2007;25(19):3871–3878. doi: 10.1016/j.vaccine.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 92.Tao P., Luo M., Zhu D., Qu S., Yang Z., Gao M., Guo D., Pan Z. Virus-like particle vaccine comprised of the HA, NA, and M1 proteins of an avian isolated H5N1 influenza virus induces protective immunity against homologous and heterologous strains in mice. Viral Immunol. 2009;22(4):273–281. doi: 10.1089/vim.2009.0017. [DOI] [PubMed] [Google Scholar]
- 93.Pushko P., Tumpey T.M., Bu F., Knell J., Robinson R., Smith G. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine. 2005;23(50):5751–5759. doi: 10.1016/j.vaccine.2005.07.098. [DOI] [PubMed] [Google Scholar]
- 94.Latham T., Galarza J.M. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 2001;75(13):6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y.V., Massare M.J., Barnard D.L., Kort T., Nathan M., Wang L., Smith G. Chimeric severe acute respiratory syndrome coronavirus (SARS-CoV) S glycoprotein and influenza matrix 1 efficiently form virus-like particles (VLPs) that protect mice against challenge with SARS-CoV. Vaccine. 2011;29(38):6606–6613. doi: 10.1016/j.vaccine.2011.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lv L., Li X., Liu G., Li R., Liu Q., Shen H., Wang W., Xue C., Cao Y. Production and immunogenicity of chimeric virus-like particles containing the spike glycoprotein of infectious bronchitis virus. J. Vet. Sci. 2014;15(2):209–216. doi: 10.4142/jvs.2014.15.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng H., Hu G.Q., Wang H.L., Liang M., Liang H., Guo H., Zhao P., Yang Y.J., Zheng X.X., Zhang Z.F., Zhao Y.K., Gao Y.W., Yang S.T., Xia X.Z. Canine parvovirus VP2 protein expressed in silkworm pupae self-assembles into virus-like particles with high immunogenicity. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0079575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang C., Zheng X., Gai W., Wong G., Wang H., Jin H., Feng N., Zhao Y., Zhang W., Li N., Zhao G., Li J., Yan J., Gao Y., Hu G., Yang S., Xia X. Novel chimeric virus-like particles vaccine displaying MERS-CoV receptor-binding domain induce specific humoral and cellular immune response in mice. Antivir. Res. 2017;140:55–61. doi: 10.1016/j.antiviral.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cohen B.J., Richmond J.E. Electron microscopy of hepatitis B core antigen synthesized in E. coli. Nature. 1982;296(5858):677–679. doi: 10.1038/296677a0. [DOI] [PubMed] [Google Scholar]
- 100.Milich D.R., Mclachlan A., Moriarty A., Thornton G.B. Immune-response to hepatitis-B virus core antigen (Hbcag) - localization of T-cell recognition sites within Hbcag/Hbeag. J. Immunol. 1987;139(4):1223–1231. [PubMed] [Google Scholar]
- 101.Yi X., Yuan Y., Li N., Yi L., Wang C., Qi Y., Gong L., Liu G., Kong X. A mouse model with age-dependent immune response and immune-tolerance for HBV infection. Vaccine. 2018;36(6):794–801. doi: 10.1016/j.vaccine.2017.12.071. [DOI] [PubMed] [Google Scholar]
- 102.Borisova G., Borschukova Wanst O., Mezule G., Skrastina D., Petrovskis I., Dislers A., Pumpens P., Grens E. Spatial structure and insertion capacity of immunodominant region of hepatitis B core antigen. Intervirology. 1996;39(1–2):16–22. doi: 10.1159/000150470. [DOI] [PubMed] [Google Scholar]
- 103.Schodel F., Moriarty A.M., Peterson D.L., Zheng J., Hughes J.L., Will H., Leturcq D.J., Mcgee J.S., Milich D.R. The position of heterologous epitopes inserted in hepatitis-B virus core particles determines their immunogenicity. J. Virol. 1992;66(1):106–114. doi: 10.1128/jvi.66.1.106-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gillam F., Zhang J., Zhang C. Hepatitis B core antigen based novel vaccine against porcine epidemic diarrhea virus. J. Virol. Methods. 2018;253:61–69. doi: 10.1016/j.jviromet.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 105.Gillam F., Zhang C. Epitope selection and their placement for increased virus neutralization in a novel vaccination strategy for porcine epidemic diarrhea virus utilizing the hepatitis B virus core antigen. Vaccine. 2018;36(30):4507–4516. doi: 10.1016/j.vaccine.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ghorbani A., Zare F., Sazegari S., Afsharifar A., Eskandari M.H., Pormohammad A. Development of a novel platform of virus-like particle (VLP)-based vaccine against COVID-19 by exposing epitopes: an immunoinformatics approach. New Microbes New Infect. 2020;38 doi: 10.1016/j.nmni.2020.100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu X., Qian J., Qin L., Li J., Xue C., Ding J., Wang W., Ding W., Yin R., Jin N., Ding Z. Chimeric Newcastle disease virus-like particles containing DC-binding peptide-fused haemagglutinin protect chickens from virulent Newcastle disease virus and H9N2 avian influenza virus challenge. Virol. Sin. 2020;35(4):455–467. doi: 10.1007/s12250-020-00199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Takimoto T., Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106(2):133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 109.Pantua H.D., McGinnes L.W., Peeples M.E., Morrison T.G. Requirements for the assembly and release of Newcastle disease virus-like particles. J. Virol. 2006;80(22):11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang Y., Shi W., Abiona O.M., Nazzari A., Olia A.S., Ou L., Phung E., Stephens T., Tsybovsky Y., Verardi R., Wang S., Werner A., Yap C., Ambrozak D., Bylund T., Liu T., Nguyen R., Wang L., Zhang B., Zhou T., Chuang G.Y., Graham B.S., Mascola J.R., Corbett K.S., Kwong P.D. Newcastle disease virus-like particles displaying prefusion-stabilized SARS-CoV-2 spikes elicit potent neutralizing responses. Vaccines (Basel) 2021;9(2) doi: 10.3390/vaccines9020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adamson C.S., Davies A., Soneoka Y., Nermut M., Mitrophanous K., Jones I.M. A block in virus-like particle maturation following assembly of murine leukaemia virus in insect cells. Virology. 2003;314(2):488–496. doi: 10.1016/s0042-6822(03)00485-9. [DOI] [PubMed] [Google Scholar]
- 112.Chua A.J., Vituret C., Tan M.L., Gonzalez G., Boulanger P., Ng M.L., Hong S.S. A novel platform for virus-like particle-display of flaviviral envelope domain III: induction of dengue and West Nile virus neutralizing antibodies. Virol. J. 2013;10:129. doi: 10.1186/1743-422X-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roy S., Ghani K., de Campos-Lima P.O., Caruso M. A stable platform for the production of virus-like particles pseudotyped with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein. Virus Res. 2021;295 doi: 10.1016/j.virusres.2021.198305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bos E.C., Luytjes W., Spaan W.J. The function of the spike protein of mouse hepatitis virus strain A59 can be studied on virus-like particles: cleavage is not required for infectivity. J. Virol. 1997;71(12):9427–9433. doi: 10.1128/jvi.71.12.9427-9433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maeda J., Maeda A., Makino S. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology. 1999;263(2):265–272. doi: 10.1006/viro.1999.9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tseng Y.T., Wang S.M., Huang K.J., Lee A.I., Chiang C.C., Wang C.T. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J. Biol. Chem. 2010;285(17):12862–12872. doi: 10.1074/jbc.M109.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84(21):11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Godeke G.J., de Haan C.A.M., Rossen J.W.A., Vennema H., Rottier P.J.M. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 2000;74(3):1566–1571. doi: 10.1128/jvi.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang S.M., Chang Y.F., Chen Y.M., Wang C.T. Severe acute respiratory syndrome coronavirus nucleocapsid protein confers ability to efficiently produce virus-like particles when substituted for the human immunodeficiency virus nucleocapsid domain. J. Biomed. Sci. 2008;15(6):719–729. doi: 10.1007/s11373-008-9265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gelhaus S., Thaa B., Eschke K., Veit M., Schwegmann-Wessels C. Palmitoylation of the alphacoronavirus TGEV spike protein S is essential for incorporation into virus-like particles but dispensable for S-M interaction. Virology. 2014;464–465:397–405. doi: 10.1016/j.virol.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thorp E.B., Boscarino J.A., Logan H.L., Goletz J.T., Gallagher T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006;80(3):1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ujike M., Huang C., Shirato K., Matsuyama S., Makino S., Taguchi F. Two palmitylated cysteine residues of the severe acute respiratory syndrome coronavirus spike (S) protein are critical for S incorporation into virus-like particles, but not for M-S co-localization. J. Gen. Virol. 2012;93(Pt 4):823–828. doi: 10.1099/vir.0.038091-0. [DOI] [PubMed] [Google Scholar]
- 123.Hsin W.C., Chang C.H., Chang C.Y., Peng W.H., Chien C.L., Chang M.F., Chang S.C. Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J. Biomed. Sci. 2018;25(1):47. doi: 10.1186/s12929-018-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75(19):9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77(5):2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kandimalla R., Chakraborty P., Vallamkondu J., Chaudhary A., Samanta S., Reddy P.H., De Feo V., Dewanjee S. Counting on COVID-19 vaccine: insights into the current strategies, progress and future challenges. Biomedicines. 2021;9(11) doi: 10.3390/biomedicines9111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tlaxca J.L., Ellis S., Remmele R.L., Jr. Live attenuated and inactivated viral vaccine formulation and nasal delivery: potential and challenges. Adv. Drug Deliv. Rev. 2015;93:56–78. doi: 10.1016/j.addr.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 128.Baxter D. Active and passive immunity, vaccine types, excipients and licensing. Occup. Med. (Lond.) 2007;57(8):552–556. doi: 10.1093/occmed/kqm110. [DOI] [PubMed] [Google Scholar]
- 129.Malik J.A., Mulla A.H., Farooqi T., Pottoo F.H., Anwar S., Rengasamy K.R.R. Targets and strategies for vaccine development against SARS-CoV-2. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lundstrom K. The current status of COVID-19 vaccines. Front. Genome Ed. 2020;2 doi: 10.3389/fgeed.2020.579297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136(4):763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 132.Zak D.E., Andersen-Nissen E., Peterson E.R., Sato A., Hamilton M.K., Borgerding J., Krishnamurty A.T., Chang J.T., Adams D.J., Hensley T.R., Salter A.I., Morgan C.A., Duerr A.C., De Rosa S.C., Aderem A., McElrath M.J. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc. Natl. Acad. Sci. U. S. A. 2012;109(50):E3503–E3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Leitner W.W., Ying H., Restifo N.P. DNA and RNA-based vaccines: principles, progress and prospects. Vaccine. 1999;18(9–10):765–777. doi: 10.1016/s0264-410x(99)00271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grgacic E.V., Anderson D.A. Virus-like particles: passport to immune recognition. Methods. 2006;40(1):60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Capell T., Twyman R.M., Armario-Najera V., Ma J.K., Schillberg S., Christou P. Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. 2020;25(7):635–643. doi: 10.1016/j.tplants.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Woodrow K.A., Bennett K.M., Lo D.D. Mucosal vaccine design and delivery. Annu. Rev. Biomed. Eng. 2012;14:17–46. doi: 10.1146/annurev-bioeng-071811-150054. [DOI] [PubMed] [Google Scholar]
- 137.Ramqvist T., Andreasson K., Dalianis T. Vaccination, immune and gene therapy based on virus-like particles against viral infections and cancer. Expert. Opin. Biol. Ther. 2007;7(7):997–1007. doi: 10.1517/14712598.7.7.997. [DOI] [PubMed] [Google Scholar]
- 138.Queiroz A.M.V., Oliveira J.W.F., Moreno C.J., Guerin D.M.A., Silva M.S. VLP-based vaccines as a suitable technology to target trypanosomatid diseases. Vaccines (Basel) 2021;9(3) doi: 10.3390/vaccines9030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Karpinski T.M., Ozarowski M., Seremak-Mrozikiewicz A., Wolski H., Wlodkowic D. The 2020 race towards SARS-CoV-2 specific vaccines. Theranostics. 2021;11(4):1690–1702. doi: 10.7150/thno.53691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., Alam S.M., Sutherland L.L., Cai F., Sanzone A.M., Berry M., Manne K., Bock K.W., Minai M., Nagata B.M., Kapingidza A.B., Azoitei M., Tse L.V., Scobey T.D., Spreng R.L., Rountree R.W., DeMarco C.T., Denny T.N., Woods C.W., Petzold E.W., Tang J., Oguin T.H., 3rd, Sempowski G.D., Gagne M., Douek D.C., Tomai M.A., Fox C.B., Seder R., Wiehe K., Weissman D., Pardi N., Golding H., Khurana S., Acharya P., Andersen H., Lewis M.G., Moore I.N., Montefiori D.C., Baric R.S., Haynes B.F. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594(7864):553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lopez-Sagaseta J., Malito E., Rappuoli R., Bottomley M.J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct.Biotechnol. J. 2016;14:58–68. doi: 10.1016/j.csbj.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nguyen B., Tolia N.H. Protein-based antigen presentation platforms for nanoparticle vaccines. NPJ Vaccines. 2021;6(1):70. doi: 10.1038/s41541-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brune K.D., Leneghan D.B., Brian I.J., Ishizuka A.S., Bachmann M.F., Draper S.J., Biswas S., Howarth M. Plug-and-display: decoration of virus-like particles via isopeptide bonds for modular immunization. Sci. Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bruun T.U.J., Andersson A.C., Draper S.J., Howarth M. Engineering a rugged nanoscaffold to enhance plug-and-display vaccination. ACS Nano. 2018;12(9):8855–8866. doi: 10.1021/acsnano.8b02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rahikainen R., Rijal P., Tan T.K., Wu H.J., Andersson A.C., Barrett J.R., Bowden T.A., Draper S.J., Townsend A.R., Howarth M. Overcoming symmetry mismatch in vaccine nanoassembly through spontaneous amidation. Angew. Chem. Int. Ed. Engl. 2021;60(1):321–330. doi: 10.1002/anie.202009663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tan T.K., Rijal P., Rahikainen R., Keeble A.H., Schimanski L., Hussain S., Harvey R., Hayes J.W.P., Edwards J.C., McLean R.K., Martini V., Pedrera M., Thakur N., Conceicao C., Dietrich I., Shelton H., Ludi A., Wilsden G., Browning C., Zagrajek A.K., Bialy D., Bhat S., Stevenson-Leggett P., Hollinghurst P., Tully M., Moffat K., Chiu C., Waters R., Gray A., Azhar M., Mioulet V., Newman J., Asfor A.S., Burman A., Crossley S., Hammond J.A., Tchilian E., Charleston B., Bailey D., Tuthill T.J., Graham S.P., Duyvesteyn H.M.E., Malinauskas T., Huo J., Tree J.A., Buttigieg K.R., Owens R.J., Carroll M.W., Daniels R.S., McCauley J.W., Stuart D.I., Huang K.A., Howarth M., Townsend A.R. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021;12(1):542. doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hsia Y., Bale J.B., Gonen S., Shi D., Sheffler W., Fong K.K., Nattermann U., Xu C., Huang P.S., Ravichandran R., Yi S., Davis T.N., Gonen T., King N.P., Baker D. Design of a hyperstable 60-subunit protein dodecahedron. Nature. 2016;535(7610):136–139. doi: 10.1038/nature18010. [corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., Navarro M.J., Miranda M.C., Pettie D., Ravichandran R., Kraft J.C., Ogohara C., Palser A., Chalk S., Lee E.C., Guerriero K., Kepl E., Chow C.M., Sydeman C., Hodge E.A., Brown B., Fuller J.T., Dinnon K.H., 3rd, Gralinski L.E., Leist S.R., Gully K.L., Lewis T.B., Guttman M., Chu H.Y., Lee K.K., Fuller D.H., Baric R.S., Kellam P., Carter L., Pepper M., Sheahan T.P., Veesler D., King N.P. Elicitation of potent neutralizing antibody Responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183(5):1367–1382 e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert. Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jung Y., Park G.S., Moon J.H., Ku K., Beak S.H., Lee C.S., Kim S., Park E.C., Park D., Lee J.H., Byeon C.W., Lee J.J., Maeng J.S., Kim S.J., Kim S.I., Kim B.T., Lee M.J., Kim H.G. Comparative analysis of primer-probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2) ACS Infect. Dis. 2020;6(9):2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- 151.Chan S.K., Du P., Ignacio C., Mehta S., Newton I.G., Steinmetz N.F. Biomimetic virus-like particles as severe acute respiratory syndrome coronavirus 2 diagnostic tools. ACS Nano. 2021;15(1):1259–1272. doi: 10.1021/acsnano.0c08430. [DOI] [PubMed] [Google Scholar]
- 152.Lam P., Gulati N.M., Stewart P.L., Keri R.A., Steinmetz N.F. Bioengineering of tobacco mosaic virus to create a non-infectious positive control for ebola diagnostic assays. Sci. Rep. 2016;6:23803. doi: 10.1038/srep23803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yu X.F., Pan J.C., Ye R., Xiang H.Q., Kou Y., Huang Z.C. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2008;46(3):837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Madi M., Mioulet V., King D.P., Lomonossoff G.P., Montague N.P. Development of a non-infectious encapsidated positive control RNA for molecular assays to detect foot-and-mouth disease virus. J. Virol. Methods. 2015;220:27–34. doi: 10.1016/j.jviromet.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Crone M.A., Priestman M., Ciechonska M., Jensen K., Sharp D.J., Anand A., Randell P., Storch M., Freemont P.S. A role for biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 2020;11(1):4464. doi: 10.1038/s41467-020-18130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Charlton Hume H.K., Vidigal J., Carrondo M.J.T., Middelberg A.P.J., Roldao A., Lua L.H.L. Synthetic biology for bioengineering virus-like particle vaccines. Biotechnol. Bioeng. 2019;116(4):919–935. doi: 10.1002/bit.26890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zdanowicz M., Chroboczek J. Virus-like particles as drug delivery vectors. Acta Biochim. Pol. 2016;63(3):469–473. doi: 10.18388/abp.2016_1275. [DOI] [PubMed] [Google Scholar]