Abstract
Although infections among vaccinated individuals lead to milder COVID-19 symptoms relative to those in unvaccinated subjects, the specificity and durability of antibody responses elicited by breakthrough cases remain unknown. Here, we demonstrate that breakthrough infections induce serum-binding and -neutralizing antibody responses that are markedly more potent, durable, and resilient to spike mutations observed in variants than those in subjects who received only 2 doses of vaccine. However, we show that breakthrough cases, subjects who were vaccinated after infection, and individuals vaccinated three times have serum-neutralizing activity of comparable magnitude and breadth, indicating that an increased number of exposures to SARS-CoV-2 antigen(s) enhance the quality of antibody responses. Neutralization of SARS-CoV was moderate, however, underscoring the importance of developing vaccines eliciting broad sarbecovirus immunity for pandemic preparedness.
Keywords: SARS-CoV-2, COVID-19, Delta variant, vaccine, antibodies, neutralization, breakthrough infection
Graphical abstract
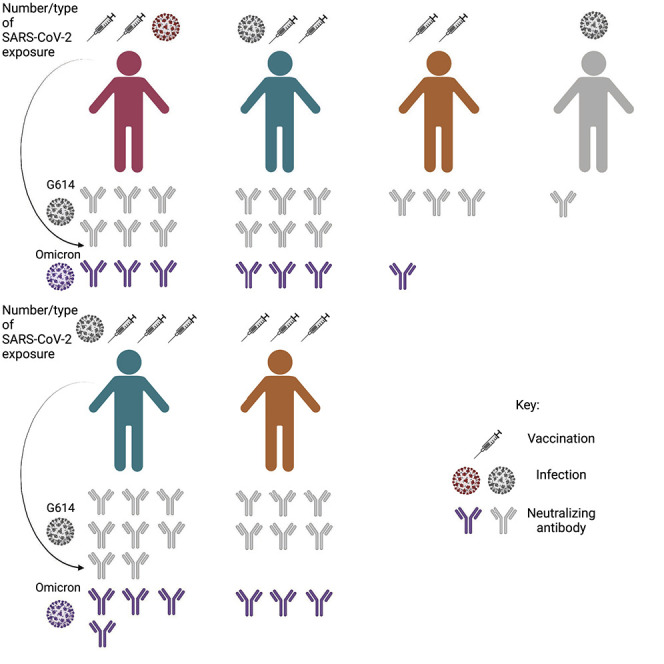
Individuals with breakthrough COVID-19 infections, previously infected/vaccinated individuals, and those vaccinated thrice have potent serum-binding and -neutralizing antibody responses against variants of concern, including Omicron. Neutralization of SARS-CoV, however, was moderate, thus urging the need for developing broad vaccines for pandemic preparedness.
Introduction
The SARS-CoV-2 Delta (B.1.617.2) variant of concern emerged at the end of 2020 and became dominant globally by mid-2021. Mutations in the spike (S) glycoprotein (Johnson et al., 2021; Walls et al., 2020a; Wrapp et al., 2020) and in the nucleoprotein (N) have been suggested to account for its enhanced transmissibility, replication kinetics, and viral loads in oropharyngeal and nose-throat swabs of infected individuals relative to the ancestral Wuhan-Hu-1 virus and other variants (Li et al., 2021; Liu et al., 2021b; Mlcochova et al., 2021; Saito et al., 2021; Syed et al., 2021). Moreover, multiple S mutations in the N-terminal domain and receptor-binding domain have been shown to promote immune evasion (McCallum et al., 2021a, 2021b; Mlcochova et al., 2021; Suryadevara et al., 2021; Ying et al., 2021). These characteristics combined with the waning of serum-neutralizing antibody titers over time in vaccinated individuals have resulted in breakthrough infections that are usually associated with much milder symptoms than infections of unvaccinated individuals (Levine-Tiefenbrun et al., 2021; Mlcochova et al., 2021).
Understanding the magnitude and breadth of immune responses following breakthrough infections is key to guiding vaccination policies and pandemic preparedness efforts (Collier et al., 2021). Serum-neutralizing antibody titers represent the current best correlate of protection against SARS-CoV-2 in animal challenge studies (Arunachalam et al., 2021; Case et al., 2020a; Corbett et al., 2021; Hassan et al., 2021; Khoury et al., 2021; McMahan et al., 2021; Winkler et al., 2020), and multiple clinical trials have shown the benefits of therapeutic administration of monoclonal antibodies in humans (Corti et al., 2021). Furthermore, serum-neutralizing antibodies are used in ongoing comparative clinical trials as key success metrics for the next generation of vaccines (e.g., NCT05007951 and NCT04864561 comparing GBP510 and VLA2001 with AZD1222, respectively). To understand whether the sequence of infection and/or vaccination as well as repeated exposures alters the durability, magnitude, and breadth of antibody responses, we followed and compared serum antibodies in individuals who were vaccinated, previously infected and then vaccinated, or vaccinated and then infected predominantly with the SARS-CoV-2 Delta variant.
Results
We compared serum-binding titers following infection, vaccination, or both in groups of ∼15 individuals enrolled in the hospitalized or ambulatory adults with respiratory viral infections (HAARVI) longitudinal cohort study at the University of Washington in Seattle (Table S1). The breakthrough group (13/16 confirmed Delta infection, see STAR Methods) was composed of n = 1 Jansen Ad26.COV2.S, n = 1 Covishield (Oxford-Astrazeneca), n = 5 Moderna mRNA-1273, and n = 9 Pfizer Comirnaty vaccines. For the infected then vaccinated (infected/vaccinated) cohort, all samples were obtained prior to September 2020, indicating that these infections were likely with Washington-1-like isolates (according to outbreaks.info) and comprised n = 1 Jansen Ad26.COV2.S, n = 3 Moderna mRNA-1273, and n = 11 Pfizer Comirnaty vaccines. The vaccinated-only group was made up of n = 3 Moderna mRNA-1273 and n = 12 Pfizer Comirnaty vaccines (Table S1). Benchmarking of these samples was carried out with human convalescent plasma (HCP), which was collected prior to October 2020 in Washington State, indicating that these infections were likely with Washington-1-like isolates (Table S1). Eight infected/vaccinated individuals and eight vaccinated-only individuals received a matched third vaccine dose. These samples were compared with plasma from SARS-CoV-2 naive individuals whose blood was drawn prior to vaccination (Table S1), as confirmed by the lack of SARS-CoV-2 nucleocapsid (N) reactivity using the Roche anti-N immuno assay (only convalescent samples were positive) (Table S1).
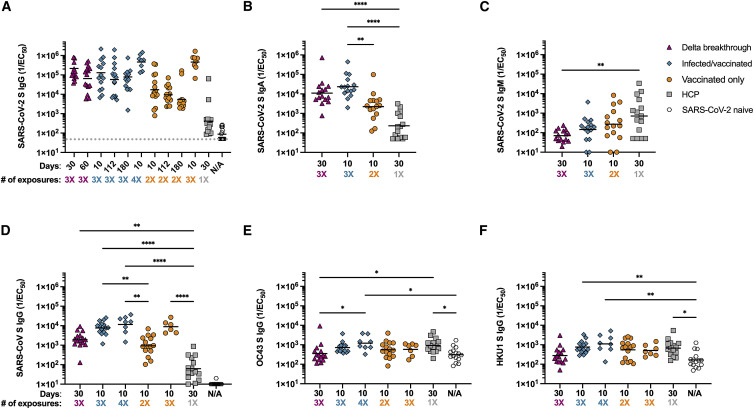
Serum-immunoglobulin (Ig) G-, IgA-, and IgM-binding titers were evaluated using ELISAs with the SARS-CoV-2 Hexapro S antigen (Hsieh et al., 2020). The cohorts were followed longitudinally for up to 180 days after the initial blood was drawn to evaluate differences in durability of antibody responses. Responses were highest among individuals who were exposed to SARS-CoV-2 S three or four times through vaccination or a combination of infection and vaccination. The magnitude of IgG responses for vaccinated individuals who experienced a breakthrough infection was greatest 30 days post positive PCR test and reduced ∼3-fold by day 60 (Figure 1 A; Tables 1 and S2; Data S1). Infected/vaccinated individuals had binding geometric mean titers (GMTs) which peaked 10 days after receiving a second vaccine dose, dropped ∼2-fold at day 112, and remained stable at day 180 (Figure 1A; Tables 1 and S2; Data S1). Peak binding titers were ∼1.6-fold lower than the breakthrough samples at 30 days post infection. Following a third vaccination, the infected/vaccinated individuals experienced a 3.6-fold rise in binding GMTs relative to day 10 post second shot (Figure 1A; Tables 1 and S2; Data S1). Vaccinated-only individuals, who received 2 doses, had binding GMTs 12- and ∼7.5-fold lower than breakthrough and infected/vaccinated individuals at peak time points (30 days post positive PCR or 10 days post second vaccination, respectively). Binding titers were reduced 1.7- and 3-fold at 112 and 180 days after the second vaccination, respectively. Following administration of a third vaccine dose, we observed a 27-fold binding GMT increase for vaccinated-only individuals relative to 10 days post second vaccination (Figure 1A; Tables 1 and S2; Data S1). Peak binding GMT for infected-only individuals (30 days post positive PCR) was well below all other groups, suggesting that infection alone or a single exposure do not induce robust antibody responses as compared with multiple vaccinations or a mixture of vaccination and infection (Bowen et al., 2021). SARS-CoV-2 naive individuals had very low responses overall (at the limit of detection of our assay) (Figure 1A; Tables 1 and S2; Data S1). These data suggest that repeated exposures through vaccination, infection or a combination of both induce potent polyclonal serum antibody-binding titers. Furthermore, we observed a greater durability of binding GMTs over 180 days for the infected/vaccinated relative to the vaccinated-only (2 doses) cohorts, which could result from increased number of exposures, different exposure spacing (Parry et al., 2021), or due to actual infection versus vaccination only.
Figure 1.
Repeated exposures to SARS-CoV-2 antigens through vaccination or infection enhance S-specific serum-IgG- and IgA-binding titers
(A) Serum-IgG-binding titers at 30 or 60 days post infection or 10, 112, or 180 days post second or third vaccine dose were evaluated for longitudinal samples by ELISA using prefusion-stabilized SARS-CoV-2 S Hexapro as the antigen. Serum samples were obtained from individuals who had a breakthrough infection (n = 15, magenta triangle), were previously infected in 2020 and then vaccinated (n = 15, teal diamond), had only been vaccinated (n = 15, orange circle), were infected in 2020 in Washington state (n = 15, gray square), or were SARS-CoV-2 naive (samples taken prior to vaccination, n = 15, open hexagon). Demographics of each individual are shown in Table S1. Statistical significance was determined by Kruskal-Wallis and Dunn’s multiple comparisons test and is shown in Table S2.
(B) Serum-IgA-binding titers at 30 days post infection or 10 days post second vaccine dose were evaluated by ELISA using prefusion-stabilized SARS-CoV-2 S Hexapro as the antigen.
(C) Serum-IgM-binding titers at 30 days post infection or 10 days post second vaccine dose were evaluated by ELISA using prefusion-stabilized SARS-CoV-2 S Hexapro as the antigen.
(D–F) Serum-IgG-binding titers were evaluated by ELISA at 30 days post infection, 10 days post second or third vaccine dose or prior to SARS-CoV-2 exposure (SARS-CoV-2 naive) using prefusion-stabilized (D) SARS-CoV 2P S as the antigen, OC43 S (E) or HKU1 2P S (F) as the antigen. # of exposures: number of SARS-CoV-2 S exposures (infection or vaccination). Shown are representative GMTs from at least two independent experiments. Statistical significance was determined by Kruskal-Wallis and Dunn’s multiple comparisons test and is shown only when significant. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001. LOD is shown as a gray horizontal dotted line when above the x axis. Raw fits are shown in Data S1.
Table 1.
ELISA GMTs shown in Figure 1
| 30 days breakthrough | 60 days breakthrough | 10 days infected/vaccinated | 112 days infected/vaccinated | 180 days infected/vaccinated | 10 days post boost infected/vaccinated | 10 days vaccinated only | 112 days vaccinated only | 180 days vaccinated only | 10 days post boost vaccinated only | HCP | SARS-CoV-2 naive | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 IgG GMT | 2.1 × 105 | 6.5 × 104 | 1.3 × 105 | 5.8 × 104 | 7.9 × 104 | 4.7 × 105 | 1.7 × 104 | 9.5 × 103 | 5.4 × 103 | 4.6 × 105 | 4.2 × 102 | 8.8 × 101 |
| SARS-CoV-2 IgA GMT | 1.1 × 104 | 2.3 × 104 | 2.2 × 103 | 2.3 × 102 | ||||||||
| SARS-CoV-2 IgM GMT | 7.0 × 101 | 1.5 × 102 | 2.7 × 102 | 7.1 × 102 | ||||||||
| SARS-CoV IgG GMT | 1.9 × 103 | 7.7 × 103 | 9.3 × 102 | 8.9 × 103 | 1.2 × 104 | 6.3 × 101 | 1.0 × 101 | |||||
| OC43 IgG GMT | 3.6 × 102 | 7.2 × 102 | 5.6 × 102 | 5.8 × 102 | 1.2 × 103 | 9.2 × 102 | 3.1 × 102 | |||||
| HKU1 IgG GMT | 2.8 × 102 | 7.6 × 102 | 5.5 × 102 | 5.1 × 102 | 5.1 × 102 | 6.7 × 102 | 1.7 × 102 |
Peak S-specific IgA-binding GMTs for breakthrough cases, infected/vaccinated individuals, vaccinated-only individuals, and for HCP followed a trend roughly similar to IgG-binding titers (Figure 1B; Data S1). Conversely, IgM-binding titers were approximately an order of magnitude higher for HCP relative to other cohorts (Figure 1C; Data S1), presumably as a result of extensive immunoglobulin class switching and affinity maturation occurring in the latter cases, mirroring findings made with memory B cells in vaccinated individuals (Cho et al., 2021). Collectively, these data indicate that breakthrough infections, vaccination following infection, or vaccination only trigger similarly robust anamnestic responses.
To assess how vaccination and infection affect the breadth of binding antibody responses, we performed ELISAs using prefusion diproline-stabilized (2P) SARS-CoV S as antigen (Kirchdoerfer et al., 2018; Pallesen et al., 2017). We determined binding GMTs for breakthrough cases, infected/vaccinated individuals, and vaccinated-only subjects at 30 days post positive PCR test or 10 days post vaccination. Infected/vaccinated individuals had higher SARS-CoV S-binding GMTs compared with breakthrough and vaccinated-only subjects (Figure 1D; Data S1). Upon receiving a third immunization, SARS-CoV S-specific responses increased 1.5- and 9.5-fold for infected/vaccinated and vaccinated-only subjects, respectively. We observed the lowest SARS-CoV S-binding responses for HCP and for SARS-CoV-2 naive individuals (limit of detection). These results suggest that antibody cross-reactivity within the sarbecovirus subgenus improves with repeated exposures to SARS-CoV-2 S (Figure 1D; Data S1). We also investigated antibody cross-reactivity beyond the sarbecovirus subgenus using the prefusion-stabilized S trimers of the common-cold causing embecoviruses OC43 (Tortorici et al., 2019) and HKU1 as ELISA antigens. We determined that GMTs were not markedly enhanced with repeated SARS-CoV-2 S exposures relative to SARS-CoV-2 naive individuals (Figures 1E and 1F; Data S1). These findings show that exposure to SARS-CoV-2 S does not enhance emebecovirus cross-reactive antibody responses substantially in the cohorts examined, even compared with SARS-CoV-2 naive individuals who might have been previously exposed to seasonal coronaviruses.
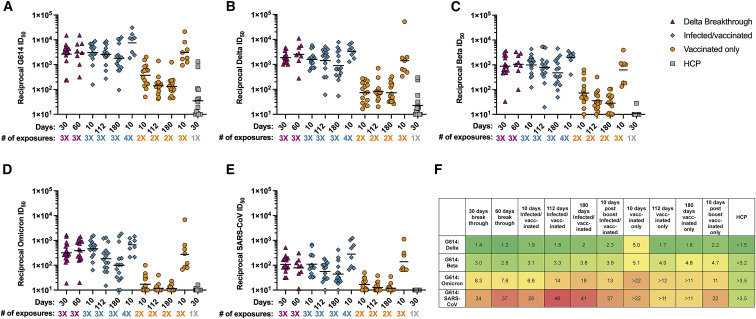
To understand the magnitude and durability of neutralizing antibody responses among the different groups, we determined serum-neutralizing activity for all samples obtained longitudinally using vesicular stomatitis virus (VSV) pseudotyped with SARS-CoV-2 G614 S using VeroE6-TMPRSS2 target cells (Crawford et al., 2020; Kaname et al., 2010; Lempp et al., 2021). Breakthrough infections resulted in potent neutralizing activity for up to 60 days following positive PCR test (Figure 2 A; Tables 2 and S2; Data S1). Infected/vaccinated individuals had similarly potent neutralizing activity and a 2-fold neutralizing activity decay after 180 days (Figure 2A; Tables 2 and S2; Data S1). 10 days following a third vaccination, neutralizing GMT for infected/vaccinated individuals increased 2.5-fold relative to 10 days following their second dose (Figure 2A; Tables 2 and S2; Data S1). Vaccinated-only (2 doses) individuals had serum-neutralizing activity ∼10-fold lower than breakthrough or infected/vaccinated individuals at peak time points, which decayed 3-fold after 180 days (Figure 2A; Tables 2 and S2; Data S1). Following a third vaccination, neutralizing GMT for vaccinated-only individuals rose 8.4-fold compared with 10 days after receiving the second dose, reaching levels comparable with breakthrough cases 30 days after infection and infected/vaccinated subjects 10 days post second vaccine dose (Figure 2A; Tables 2 and S2; Data S1). HCP had weak neutralizing potency, in line with their low binding titers (Figure 2A; Table 2 and S2; Data S1). These data suggest that the number of exposures to S is more important than the type of exposure (infection versus vaccination) to determine the magnitude of serum-neutralizing activity. These findings are in agreement with studies of individuals infected and subsequently vaccinated with mRNA vaccines or Jansen Ad26.COV2.S (Keeton et al., 2021; Krammer et al., 2021; Saadat et al., 2021; Stamatatos et al., 2021; Tan et al., 2021a). However, the durability of neutralizing antibody responses for breakthrough and infected/vaccinated cases sets these cohorts apart from vaccinated-only individuals who experienced a decay of ∼65% over 6 months after the second vaccine dose. It remains to be determined if waning of binding and neutralizing GMTs will occur with similar kinetics after the third immunization of vaccinated-only subjects as it did after 2 doses or if they will mirror the durable antibody responses observed in breakthrough and infected/vaccinated cohorts. Hence, multiple types of exposure or increased number of exposures to SARS-CoV-2 antigen(s) enhance the magnitude and durability of serum-neutralizing activity possibly due to sustained antibody production by long lived plasma cells, increased affinity maturation yielding antibodies with greater neutralization potency, or a combination of both (Cho et al., 2021; Turner et al., 2021a, 2021b).
Figure 2.
Breakthrough, infected/vaccinated, and triple vaccinated individuals have exceptionally high serum-neutralizing activity
Serum samples were obtained from individuals who had a breakthrough infection (n = 15, magenta triangle), who were previously infected in 2020 and then vaccinated (n = 15, teal diamond, infected/vaccinated), who had been vaccinated only (n = 15, orange circle), or who were infected only in 2020 in Washington state (n = 15, gray square, HCP). All neutralization assays were performed using VeroE6-TMPRSS2 as target cells at least in duplicate. Shown are representative GMTs from at least two independent experiments. Demographics of each individuals are shown in Table S1. Statistical significance was determined by the Kruskal-Wallis and Dunn’s multiple comparison test and are shown in Table S2. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001. Normalized curves and fits are shown in Data S1.
(A) SARS-CoV-2 G614 S VSV pseudotype neutralization.
(B) SARS-CoV-2 Delta S VSV pseudotype neutralization.
(C) SARS-CoV-2 Beta S VSV pseudotype neutralization.
(D) SARS-CoV-2 Omicron S VSV pseudotype neutralization.
(E) SARS-CoV S VSV pseudotype neutralization. # of exposures: number of SARS-CoV-2 S exposures (infection or vaccination).
(F) Fold change of 614G:Delta/Beta/Omicron/SARS-CoV colored green (small fold change) to red (large fold change). A greater sign is used when the GMTs at the limit of detection may cause aberrant results.
Table 2.
Neutralization GMTs shown in Figure 2
| 30 days breakthrough | 60 days breakthrough | 10 days infected/vaccinated | 112 days infected/vaccinated | 180 days infected/vaccinated | 10 days post boost infected/vaccinated | 10 days vaccinated only | 112 days vaccinated only | 180 days vaccinated only | 10 days post boost vaccinated only | HCP | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 614G | 2.6 × 103 | 2.9 × 103 | 3.1 × 103 | 2.6 × 103 | 1.8 × 103 | 7.6 × 103 | 3.7 × 102 | 1.4 × 102 | 1.3 × 102 | 3.1 × 103 | 3.5 × 101 |
| SARS-CoV-2 Delta | 1.9 × 103 | 2.5 × 103 | 1.6 × 103 | 1.4 × 103 | 8.9 × 102 | 3.3 × 103 | 7.5 × 101 | 8.2 × 101 | 7.4 × 101 | 1.4 × 103 | 2.3 × 101 |
| SARS-CoV-2 Beta | 8.8 × 102 | 1.1 × 103 | 9.9 × 102 | 7.9 × 102 | 4.8 × 102 | 1.9 × 103 | 7.3 × 101 | 3.5 × 101 | 2.8 × 101 | 6.6 × 102 | 1.1 × 101 |
| SARS-CoV-2 Omi- cron | 3.2 × 102 | 3.9 × 102 | 4.6 × 102 | 1.8 × 102 | 9.9 × 101 | 6.0 × 102 | 1.7 × 101 | 1.2 × 101 | 1.2 × 101 | 2.7 × 102 | 1.0 × 101 |
| SARS-CoV | 1.1 × 102 | 7.9 × 101 | 1.1 × 102 | 5.6 × 101 | 4.4 × 101 | 2.8 × 102 | 1.7 × 101 | 1.3 × 101 | 1.2 × 101 | 1.4 × 102 | 1.0 × 101 |
We also observed durable neutralizing activity for the breakthrough serum samples against both Delta S and Beta S VSV pseudoviruses (Figures 2B and 2C; Tables 2 and S2; Data S1). Neutralizing GMTs increased against Delta and Beta from 30 to 60 days post positive PCR test. The dampening in neutralization potency relative to G614 S VSV for both Delta S (1.2- to 1.4-fold GMT reduction) and Beta S (2.8- to 3.0-fold GMT reduction) was smaller than previous findings with vaccinated-only or convalescent cohorts, possibly due to the heterologous S exposure (Cele et al., 2021; Madhi et al., 2021; McCallum et al., 2021a; Planas et al., 2021a; Walls et al., 2021; Wibmer et al., 2021). For infected/vaccinated subjects, we determined that neutralizing GMTs against Delta were reduced 1.8- to 2.0-fold relative to G614 S VSV (Figures 2A, 2B, and 2F; Tables 2 and S2; Data S1). After receiving a third vaccine dose, these individuals experienced a 2.0-fold increase in neutralizing titers against Delta S VSV, corresponding to a 2.3-fold GMT reduction relative to G614 S VSV (Figures 2A, 2B, and 2F; Tables 2 and S2; Data S1). Therefore, although the magnitude of neutralizing GMT increased, the breadth of broadly neutralizing antibody responses remained similar against Delta S VSV (Figures 2A and 2B). Beta S VSV neutralizing GMTs for infected/vaccinated individuals were reduced 3.1- to 3.8-fold, compared with G614 S VSV, and were of overall similar magnitude to those of breakthrough cases. Following the third vaccination, these individuals had a 2.0-fold increase in neutralizing titers against Beta S VSV, yielding a 3.9-fold GMT reduction relative to G614 S VSV (Figures 2A, 2B, and 2F; Tables 2 and S2; Data S1). Prior to boosting, this cohort experienced a ∼2-fold reduction of neutralizing GMTs over 180 days against both Delta S and Beta S VSV pseudoviruses, mirroring neutralization of G614 S VSV (Figures 2A, 2C, and 2F; Tables 2 and S2; Data S1). The vaccinated-only cohort had neutralizing GMTs against Delta S VSV that were reduced 1.7- to 5-fold relative to G614 S VSV and of an overall magnitude ∼20-fold lower than breakthrough or infected/vaccinated subjects at similar timepoints (Figures 2B and 2F; Tables 2 and S2; Data S1). Triple vaccinated-only subjects displayed an 18.8-fold increase compared to individuals after receiving only two vaccine doses at the same time point, bringing their Delta neutralizing GMTs close to individuals who had three exposures through both infection and vaccination (Figures 2B and 2F; Tables 2 and S2; Data S1). Beta S VSV pseudovirus neutralizing GMTs for vaccinated-only individuals were 4- to 5.1-fold lower relative to G614 S VSV (Figures 2C and 2F; Tables 2 and S2; Data S1). Triple vaccinated-only subjects displayed a 9.0-fold neutralizing GMT increase compared with the same time point after receiving only two vaccine doses, consistent with the overall enhancement of neutralizing titers against all pseudoviruses tested.
The SARS-CoV-2 Omicron variant was first detected in November 2021 and was shown to evade antibody-mediated immunity to an unprecedented level due to 37 S amino acid mutations (Cameroni et al., 2021; Cao et al., 2021; Liu et al., 2021a; Planas et al., 2021b; VanBlargan et al., 2021). Therefore, we set out to evaluate serum-neutralizing activity of breakthrough, infected/vaccinated, or vaccinated individuals against Omicron. Breakthrough cases experienced a ∼7- to 8-fold decrease in neutralization potency compared with G614 VSV S (Figures 2D and 2F; Tables 2 and S2; Data S1). Infected/vaccinated individuals experienced a ∼6.6-fold decrease in neutralization capacity after 10 days, a 14-fold decrease after 112 days, and an 18-fold decrease after 180 days compared with G614 VSV S, showing greater waning than against G614 (Figures 2D and 2F; Tables 2 and S2; Data S1). Vaccinated-only subjects experienced ≥~11- to 22-fold reductions of neutralizing GMTs against Omicron versus G614 S VSV, which are likely underestimates due to the limit of detection of our assay (Figures 2D and 2F; Tables 2 and S2; Data S1). Following a third vaccination, we observed enhanced neutralizing activity against Omicron for both infected/vaccinated subjects (1.3-fold GMT rise compared with 10 days post second dose) and vaccinated-only individuals (16-fold GMT rise compared with 10 days post second dose) (Figures 2D and 2F; Tables 2 and S2; Data S1), which is in agreement with recent data (Garcia-Beltran et al., 2021). The latter group had neutralizing GMTs against Omicron S VSV after 3 doses which were comparable to GMTs against G614 S VSV after 2 doses. No Omicron neutralization was detected for HCP (Figures 2D and 2F; Tables 2 and S2; Data S1), which concurs with other studies and low neutralization against other variants (Cameroni et al., 2021). These data suggest that repeated exposures, even to a distinct antigen, improve the potency and resilience of serum-neutralizing activity to variants, including against Beta S and Omicron S VSV, in line with previous studies of infected/vaccinated individuals (Keeton et al., 2021; Stamatatos et al., 2021). Finally, we observed a subtle advantage for subjects who experienced both infections and vaccination (irrespective of the order) in terms of neutralization breadth against SARS-CoV-2 variants relative to triple vaccinated-only individuals.
To assess the breadth of serum-neutralizing activity among these different cohorts against viruses belonging to a different sarbecovirus clade than SARS-CoV-2, we used a SARS-CoV S VSV pseudotype. Individuals with three hybrid exposures had neutralizing GMTs reductions of 24- to 37-fold (breakthrough) (Figures 2E and 2F; Tables 2 and S2; Data S1) and 28- to 46-fold (infected/vaccinated) (Figures 2E and 2F; Tables 2 and S2; Data S1) compared with G614 S VSV, which is approximately twice the magnitude of immune evasion observed with Omicron (Cameroni et al., 2021). After receiving a third vaccine dose, infected/vaccinated individuals had ∼2.5-fold enhanced serum-neutralizing responses against SARS-CoV S VSV relative to the same subjects at 10 days post second vaccination (Figures 2E and 2F; Tables 2 and S2; Data S1). Although it amounts to a 27-fold reduction compared with peak neutralizing G614 S VSV GMT, this neutralizing activity approaches that of vaccinated-only individuals against G614 S VSV after 2 doses (Figures 2A, 2E, and 2F). The vaccinated-only subjects had low neutralizing activity until receiving a third vaccine dose yielding GMT matching those of breakthrough cases or of infected/vaccinated individuals (after two vaccinations) (Figures 2E and 2F; Tables 2 and S2; Data S1), further suggesting that the number of exposures is more important than the type of exposure to determine the overall magnitude and breadth of serum-neutralizing activity. No SARS-CoV neutralization was detected for HCP, in line with the weak binding and neutralizing activity of this cohort even against SARS-CoV-2 (Figures 2E and 2F; Tables 2 and S2; Data S1).
Discussion
We show that breakthrough cases or infected/vaccinated subjects are endowed with greater potency, breadth, and durability of serum-neutralizing activity relative to individuals who received 2 doses of COVID-19 vaccine or those who were infected only by SARS-CoV-2 in 2020. Serum-neutralizing GMTs for the breakthrough cases and infected/vaccinated subjects were greater against the Delta and Beta S variants at all time points than those of vaccinated-only individuals (after 2 doses) against G614 S pseudovirus at peak titers. The latter levels of neutralization potency were associated with ∼95% protection from symptomatic COVID-19 disease in phase 3 clinical trials of the Pfizer Cominarty (Polack et al., 2020) and Moderna mRNA-1273 (Baden et al., 2021) vaccines and are reminiscent of observations made with infected/vaccinated samples (Stamatatos et al., 2021). The marked enhancement of serum-neutralizing activity for vaccinated-only subjects after receiving three doses, to levels comparable with breakthrough and infected/vaccinated cases, suggests that the number of exposures and/or delay between exposures to SARS-CoV-2 S, whether through vaccination or infection, correlates with the strength of neutralizing antibody responses, as well as resilience to variants. It remains to be determined whether the number of exposures also correlates with the durability of antibody responses for vaccinated-only individuals, as shown here with breakthrough cases up to 60 days and infected/vaccinated subjects for 180 days. The reduction of serum-neutralizing activity against SARS-CoV presented here following up to three SARS-CoV-2 S exposures, along with previous studies (Martinez et al., 2021; Stamatatos et al., 2021; Walls et al., 2021), suggests that combinations of COVID-19 disease and vaccination would leave the population more vulnerable to infection by a genetically divergent sarbecovirus due to the more extensive sequence differences between SARS-CoV-2 S and SARS-CoV S compared with any two SARS-CoV-2 S variants. However, four exposures to SARS-CoV-2 S, through infection followed by three vaccinations, elicit SARS-CoV serum-neutralizing titers with a magnitude that is equivalent to protective levels for SARS-CoV-2 based on clinical trial evaluation of COVID-19 vaccines (Baden et al., 2021; Polack et al., 2020). These findings suggest that repeated exposures may improve elicitation of broadly neutralizing sarbecovirus antibodies but not broadly reactive beta-coronavirus antibodies based on the comparable cross-reactive responses with OC43 and HKU1 S observed for all cohorts. Moreover, a recent study indicated that survivors of SARS-CoV infection who subsequently received a COVID-19 vaccine had broader sarbecovirus-neutralizing antibody responses than subjects only exposed to the SARS-CoV-2 virus or vaccine (Tan et al., 2021b). These data lend further support to the ongoing development of several vaccine candidates (Cohen et al., 2021; Martinez et al., 2021; Walls et al., 2021) designed to specifically elicit broad sarbecovirus immunity and could protect against SARS-CoV-2 variants as well as putative future zoonotic sarbecoviruses.
Limitations of the study
Although this study uses pseudotyped virus assays rather than authentic viruses, good correlation was established between the two assays and with protection from challenge (Arunachalam et al., 2021; Case et al., 2020b). This study is based on cohorts of 15 individuals or fewer and may be limited by sample size. Finally, we emphasize that our study is focused on serum antibody responses and that other aspects of the immune system participate in controlling viral infections, including memory B cells and T cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgA-HRP | Southern BioTech | Cat# 2050-05, RRID AB_2687526 |
| Goat anti-human IgM-HRP | Southern BioTech | Cat# 2020-05, RRID AB_2795603 |
| Goat anti-human HRP | Invitrogen | Cat #A18817 Lot #65-180-071919, RRID AB_2535594 |
| 1-Hybridomamouse hybridoma | ATCC | CRL-2700 |
| Biological samples | ||
| SARS-CoV-2 Convalescent and vaccinated human sera and plasma | HAARVI study (Data S1) | N/A |
| VSV (G∗ΔG-luciferase) | Kaname et al., 2010 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TMB | SeraCare | Cat# 5120-0083 |
| VSV (G∗ΔG-luciferase) | Kaname et al., 2010 | N/A |
| Desthiobiotin | Fisher Scientific | Cat# 12763064 |
| Experimental models: Cell lines | ||
| Expi 293F | ThermoFisher | Cat #A14527 |
| VeroE6-TMPRSS2 | Gift from VirBiotech | Lempp et al., 2021 |
| HEK293T/17 Adherent | ATCC | Cat# CRL-11268 |
| Recombinant DNA | ||
| Hexapro S | Hsieh et al., 2020 | N/A |
| SARS-CoV S-2P trimer | GeneArt (Walls et al., 2020b) | N/A |
| OC43 S trimer | GeneArt (Tortorici et al., 2019) | N/A |
| HKU1 S2P trimer | GeneArt | N/A |
| SARS-CoV-2 S full-length D614G (YP 009724390.1) | Crawford et al., 2020 | Vector# BEI NR-52514 |
| SARS-CoV-2 Delta | GenScript (McCallum et al., 2021a) | N/A |
| SARS-CoV-2 Beta | GenScript (Cameroni et al., 2021) | Vector# BEI NR-52517 |
| SARS-CoV-2 Omicron | Cameroni et al., 2021 | Vector# BEI NR-52516 |
| SARS-CoV S full-length (YP 009825051.1) | GeneArt (Walls et al., 2020a) | N/A |
| Software and algorithms | ||
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Superdex 200 Increase SEC column | Cytiva | Cat# 28-9909-44 |
| Superose 6 Increase SEC column | Cytiva | Cat# 29091596 |
| ExpiFectamine 293 Transfection Kit | ThermoFisher | Cat# A14525 |
| HisTrapHP column | Cytiva | Cat# 17524701 |
| Biotin Blocking Solution | IBA | Cat# 2-0205-050 |
| StrepTrap HP column | Cytiva | Cat# 28907546 |
| Lipofectamine 2000 | Life Technologies | Cat# 11668019 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Veesler (dveesler@uw.edu).
Materials availability
Materials generated in this study will be made available on request after signing a materials transfer agreement with the University of Washington. This work is licensed under a Creative CommonsAttribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply tofigures/photos/artwork or other content included in the article that is credited to a third party;obtain authorization from the rights holder before using such material.
Experimental model and subject details
Cell lines
Expi293F (derived from 293 cells which are female) cells are derived from the HEK293F cell line (Life Technologies). Expi293F cells were grown in Expi293 Expression Medium (Life Technologies), cultured at 36.5°C with 8% CO2 and shaking at 130 rpm. HEK293T/17 is a female human embryonic kidney cell line (ATCC). The VeroE6-TMPRSS2 cell line is an African Green monkey Kidney cell line expressing TMPRSS2 (Lempp et al., 2021). All adherent cells were cultured at 37°C with 5% CO2 in flasks with DMEM + 10% FBS (Hyclone) + 1% penicillin-streptomycin. Cell lines were not tested for mycoplasma contamination nor authenticated.
Human subjects sera/plasma
Blood samples were collected from participants as part of the Hospitalized or Ambulatory Adults with Respiratory Viral Infections (HAARVI) study and was approved by the University of Washington Human Subjects Division Institutional Review Board (STUDY00000959). Baseline socio-demographic and clinical data for these individuals are summarized in Data S1. All samples were sera except for the convalescent and 60 day breakthrough samples which were ACD plasma. Naive samples were obtained from INGM, Ospedale Maggio Policlinico of Milan and approved by the local review board Study Polimmune.
Method details
Sequencing of breakthrough individuals
Participants were enrolled after SARS-CoV-2 positive RT-PCR results that showed S-gene dropout or delay using a primer set that predates the S probe set in the "TaqPath" test kit sold by Thermo Fisher. Specifically, the Delta S 157-158 deletion causes S-gene dropout in our diagnostic RT-qPCR assay. The RT-qPCR assay uses probes for human RNase P and SARS-CoV-2 Orf1b and S gene. S-gene dropout or delay was identified using the following individual sample criteria: 1) internal control RNase P amplification was present, 2) Orf1b amplification was present, and 3) S-gene specific probes showed no amplification or a delay of >5 cycles compared to Orf1b due to the 156-157 deletion in delta. Three samples (278C, 284C, and 286C) included in this analysis were either not sequenced due to low volumes or failed sequencing due to low viral loads, but according to outbreaks.info over 90% of all cases in Washington state at the time were Delta SARS-CoV-2.
SARS-CoV-2 genome sequencing was then conducted using a targeted enrichment approach. SARS-CoV-2 was detected using a laboratory-developed test (LDT) or research assay. For the LDT, SARS-CoV-2 detection was performed using real-time RT-PCR with a probe sets targeting Orf1b and S with FAM fluor (Life Technologies 4332079 assays # APGZJKF and #APXGVC4) multiplexed with an RNaseP probe set with VIC or HEX fluor (Life Technologies A30064 or IDT custom) each in duplicate on a QuantStudio 6 instrument (Applied Biosystems). RNA from positive specimens was converted to cDNA using random hexamers and reverse transcriptase (Superscript IV, Thermo) and a sequencing library was constructed using the Illumina TruSeq RNA Library Prep for Enrichment kit (Illumina). The sequencing library was enriched for SARS-CoV-2 using the Twist Respiratory Virus Research Panel (Twist). Libraries were sequenced on a MiSeq or NextSeq instruments. The resulting reads were assembled against the SARS-CoV-2 reference genome Wuhan-Hu-1/2019 (Genbank accession MN908947) using the bioinformatics pipeline https://github.com/seattleflu/assembly. Consensus sequences were deposited to Genbank and GISAID (Bedford et al., 2020). Sequence quality was determined using Nextclade version 1.0.0-alpha.8 (https://clades.nextstrain.org/). Lineage was assigned using the Pangolin COVID-19 Lineage Assigner version 3.1.11 (https://pangolin.cog-uk.io/) (Paredes et al., 2021)
Roche anti-N analysis
0.5mL serum/plasma samples were tested in CAP/CLIA-accredited clinical laboratory using the FDA-authorized Roche Elecsys Anti-SARS-CoV-2 for anti-nucleocapsid antibodies using manufacturer’s established positivity cut-off index of ≥1.0 (Theel et al., 2021).
Recombinant protein expression and purification
The SARS-CoV-2 Hexapro S (Hsieh et al., 2020), SARS-CoV S 2P (Pallesen et al., 2017; Walls et al., 2020b), HKU1 S2P, and OC43 S (Tortorici et al., 2019) ectodomains were produced in Expi293F Cells (ThermoFisher Scientific) grown in suspension using Expi293 Expression Medium (ThermoFisher Scientific) at 37°C in a humidified 8% CO2 incubator with constant rotation at 130 rpm. Cells grown to a density of 3 million cells per mL were transfected using the ExpiFectamine 293 Transfection Kit (ThermoFisher Scientific) and cultivated for four days prior to supernatent harvest. SARS-CoV-2 Hexapro S, SARS-CoV S2P, and HKU1 S2P ectodomains were purified from clarified supernatants using a HisTrapHP column (Cytiva) and washed with 10 column volumes of 25 mM sodium phosphate pH 8.0 and 150 mM NaCl before elution on a gradient up to 500 mM imidazole. OC43 S, fused to a strep tag, supernatant was clarified by 10 minute centrifugation at 800xg and brought to 100 mM Tris-HCL pH 8, 150 mM NaCl, and 18.1 mL/liter biotin blocking solution (BioLock). After a 20-minute incubation, supernatant was further centrifuged at 10,000xg for 20 minutes. Supernatant was then bound to a 1 mL StrepTrap HP column (Cytiva) and washed with 10 column volumes of 100 mM Tris-HCl, 150 mM NaCl, and 1 mM EDTA pH 8 before elution in wash buffer supplemented with 2.5 mM desthiobiotin. Purified protein was buffer exchanged into 20 mM Tris-HCl pH 8.0 and 100 mM NaCl, concentrated using 100 kDa MWCO centrifugal filters (Amicon Ultra) to 1-2 mg/mL, and flash frozen.
ELISA
For anti-S ELISA, 50 μL of 2-6 μg/mL S was plated onto 384-well Nunc Maxisorp (ThermoFisher) plates in PBS and sealed overnight at 4°C. The next day plates were washed 4 × in Tris Buffered Saline Tween (TBST- 20mM Tris pH 8, 150mM NaCl, 0.1% Tween) using a plate washer (BioTek) and blocked with Casein (ThermoFisher) for 1 h at 37°C. Plates were washed 4 × in TBST and 1:5 serial dilutions of human sera or plasma were made in 50 μL TBST starting at 1:10, 1:50, or 1:250 and incubated at 37°C for 1 h. Plates were washed 4 × in TBST, then anti-human (Invitrogen) horseradish peroxidase-conjugated antibodies were diluted 1:5,000 and 50 μL added to each well and incubated at 37°C for 1 h. Plates were washed 4 × in TBST and 50 μL of TMB (SeraCare) was added to every well for 5 min at room temperature. The reaction was quenched with the addition of 50 μL of 1 N HCl. Plates were immediately read at 450 nm on a VarioSkanLux plate reader (ThermoFisher) and data plotted and fit in Prism (GraphPad) using nonlinear regression sigmoidal, 4PL, X is log(concentration) to determine EC50 values from curve fits. Where the curve did not reach an OD450 of 4, a constraint of OD450 4 was placed on the upper bounds of the fit.
VSV pseudovirus production
G614 SARS-CoV-2 S (Crawford et al., 2020), Delta S, Beta S, Omicron S, and SARS-CoV S pseudotyped VSV viruses were prepared as described previously (McCallum et al., 2021a; Walls et al., 2021). Briefly, HEK293T cells in DMEM supplemented with 10% FBS, 1% PenStrep seeded in 10-cm dishes were transfected with the plasmid encoding for the corresponding S glycoprotein using lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. One day post-transfection, cells were infected with VSV(G∗ΔG-luciferase) (Kaname et al., 2010) and after 2 h were washed five times with DMEM before adding medium supplemented with anti-VSV-G antibody (I1- mouse hybridoma supernatant, CRL- 2700, ATCC). Virus pseudotypes were harvested 18-24 h post-inoculation, clarified by centrifugation at 2,500 x g for 5 min, filtered through a 0.45 μm cut off membrane, concentrated 10 times with a 30 kDa cut off membrane, aliquoted and stored at -80°C.
VSV pseudovirus neutralization
VeroE6-TMPRSS2 (Lempp et al., 2021) were cultured in DMEM with 10% FBS (Hyclone), 1% PenStrep and 8 μg/mL puromycin (to ensure retention of TMPRSS2) with 5% CO2 in a 37°C incubator (ThermoFisher). Cells were trypsinized using 0.05% trypsin and plated to 40,000 cells/well. The following day, cells were checked to be at 80% confluence. In an empty half-area 96-well plate, a 1:3 serial dilution of sera was made in DMEM and diluted pseudovirus was then added and incubated at room temperature for 30-60 min before addition of the sera-virus mixture to the cells at 37°C. 2 hours later, 40 μL of a DMEM solution containing 20% FBS and 2% PenStrep was added to each well. After 17-20 hours, 40 μL/well of One-Glo-EX substrate (Promega) was added to the cells and incubated in the dark for 5-10 min prior to reading on a BioTek plate reader. Measurements were done at least in duplicate using distinct batches of pseudoviruses and one representative experiment is shown. Relative luciferase units were plotted and normalized in Prism (GraphPad). Nonlinear regression of log(inhibitor) versus normalized response was used to determine IC50 values from curve fits. Normality was tested using the D’agostino-Pearson test and in the absence of a normal distribution, kruskal-wallis tests were used to compare two groups to determine whether differences reached statistical significance. Fold changes were determined by comparing individual animal IC50 and then averaging the individual fold changes for reporting.
Quantification and statistical analysis
8-15 subjects were included in each group as detailed in Table S1. Experiments were all done in at least biologic duplicates. Normality was tested using the D’agostino-Pearson test and in the absence of a normal distribution, Kruskal Wallis tests were used to compare two groups to determine whether differences reached statistical significance. Significance is indicated with stars: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001 with non-significant groups are not shown. Fold changes were determined by comparing individual animal IC50 and then averaging the individual fold changes for reporting. Where significance is not shown in the figure itself, it is presented in Table S2.
Acknowledgments
We thank Hideki Tani (University of Toyama) for providing the reagents necessary for preparing VSV pseudotyped viruses. We thank Antonio Lanzavecchia for coordinating SARS-CoV-2 naive blood donors. We thank Christina Lockwood and Lea Starita for their assistance in providing the sequencing data for participant identification. This study was supported by the National Institute of Allergy and Infectious Diseases (DP1AI158186 and HHSN272201700059C to D.V.), a Pew Biomedical Scholars award (D.V.), Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund (D.V.), Fast Grants (D.V.), and the Bill and Melinda Gates Foundation (OPP1156262 to D.V.). D.V. is an Investigator of the Howard Hughes Medical Institute.
Author contributions
A.C.W., J.E.B., and D.V. conceived the project. A.C.W. and D.V. designed the experiments and supervised this study. K.R.S., M.J.N., A.J., J.E.B., and C.S. carried out ELISAs. A.C.W., K.R.S., M.J.N., E.C., and D.C. cloned and coordinated pseudovirus genes, produced pseudoviruses, and ran pseudovirus neutralization assays. J.E.B., A.J., and M.M. expressed and purified proteins. E.A.G., E.J.D.-A., and A.G. coordinated and ran Roche anti N experiments. N.F., J.L., and H.Y.C. coordinated the collection of clinical samples and provided them. A.C.W., K.R.S., M.J.N., A.J., C.S., J.E.B., and D.V. analyzed the data. A.C.W. and D.V. wrote the manuscript with input from all authors.
Declaration of interests
The Veesler laboratory has received an unrelated sponsored research agreement from Vir Biotechnology. A.C.W. and D.V. are named as inventors on patent applications filed by the University of Washington for SARS-CoV-2 and sarbecovirus receptor-binding domain nanoparticle vaccines. E.C. and D.C. are employees of Vir Biotechnology and may hold shares in Vir Biotechnology. H.Y.C. is a consultant for Merck, Pfizer, Ellume, and the Bill and Melinda Gates Foundation and has received support from Cepheid and Sanofi-Pasteur. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Published: January 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.01.011.
Supplemental information
Data and code availability
Data generated in this study are available upon request.
References
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Greninger A.L., Roychoudhury P., Starita L.M., Famulare M., Huang M.L., Nalla A., Pepper G., Reinhardt A., Xie H., et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.E., Walls A.C., Joshi A., Sprouse K.R., Stewart C., Tortorici M.A., Franko N.M., Logue J.K., Mazzitelli I.G., Tiles S.W., et al. SARS-CoV-2 spike conformation determines plasma neutralizing activity. bioRxiv. 2021 doi: 10.1126/sciimmunol.adf1421. bioRxiv:10.1101/2021.12.19.473391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021 doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B.K., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P., et al. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020;28:465–474.e4. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.-M., Zeng Q., Tahan S., Droit L., et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.-J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A.-R.Y., Brown C.M., Mcmahan K., Yu J., Liu J., Jacob-Dolan C., Chandrashekar A., Tierney D., Ansel J.L., Rowe M., et al. Immune responses in fully vaccinated individuals following breakthrough infection with the SARS-CoV-2 delta variant in Provincetown, Massachusetts. medRxiv. 2021 doi: 10.1126/scitranslmed.abn6150. medRxiv:2021.10.18.21265113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Nason M.C., Flach B., Gagne M., O’Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:4593–4595. doi: 10.1016/j.cell.2021.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Torortici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv. 2021 doi: 10.1016/j.cell.2021.12.033. medRxiv:10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Feldmann F., Zhao H., Curiel D.T., Okumura A., Tang-Huau T.-L., Case J.B., Meade-White K., Callison J., Chen R.E., et al. A single intranasal dose of chimpanzee adenovirus-vectored vaccine protects against SARS-CoV-2 infection in rhesus macaques. Cell Rep. Med. 2021;2:100230. doi: 10.1016/j.xcrm.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaname Y., Tani H., Kataoka C., Shiokawa M., Taguwa S., Abe T., Moriishi K., Kinoshita T., Matsuura Y. Acquisition of complement resistance through incorporation of CD55/decay-accelerating factor into viral particles bearing baculovirus GP64. J. Virol. 2010;84:3210–3219. doi: 10.1128/JVI.02519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Richardson S.I., Moyo-Gwete T., Hermanus T., Tincho M.B., Benede N., Manamela N.P., Baguma R., Makhado Z., Ngomti A., et al. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe. 2021;29:1611–1619.e5. doi: 10.1016/j.chom.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Bielak D.A., Carreño J.M., Chernet R.L., et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 2021;384:1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempp F.A., Soriaga L.B., Montiel-Ruiz M., Benigni F., Noack J., Park Y.J., Bianchi S., Walls A.C., Bowen J.E., Zhou J., et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- Levine-Tiefenbrun M., Yelin I., Alapi H., Katz R., Herzel E., Kuint J., Chodick G., Gazit S., Patalon T., Kishony R. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat. Med. 2021;27:2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- Li B., Deng A., Li K., Hu Y., Li Z., Xiong Q., Liu Z., Guo Q., Zou L., Zhang H., et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. medRxiv. 2021 doi: 10.1038/s41467-022-28089-y. medRxiv:2021.07.07.21260122v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu J., Johnson B.A., Xia H., Ku Z., Schindewolf C., Widen S.G., An Z., Weaver S.C., Menachery V.D., et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv. 2021 doi: 10.1016/j.celrep.2022.110829. bioRxiv:2021.08.12.456173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Efficacy of the ChAdOx1 nCoV-19 COVID-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Schäfer A., Leist S.R., De la Cruz G., West A., Atochina-Vasserman E.N., Lindesmith L.C., Pardi N., Parks R., Barr M., et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., De Marco A., Franko N., Tilles S.W., Logue J., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes M.I., Lunn S.M., Famulare M., Frisbie L.A., Painter I., Burstein R., Roychoudhury P., Xie H., Mohamed Bakhash S.A., Perez R., et al. Associations between SARS-CoV-2 variants and risk of COVID-19 hospitalization among confirmed cases in Washington State: a retrospective cohort study. medRxiv. 2021 doi: 10.1093/cid/ciac279. medRxiv:2021.09.29.21264272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry H., Bruton R., Stephens C., Brown K., Amirthalingam G., Otter A., Hallis B., Zuo J., Moss P. Differential immunogenicity of BNT162b2 or ChAdOx1 vaccines after extended-interval homologous dual vaccination in older people. Immun. Ageing. 2021;18:34. doi: 10.1186/s12979-021-00246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat S., Rikhtegaran Tehrani Z., Logue J., Newman M., Frieman M.B., Harris A.D., Sajadi M.M. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS-CoV-2. JAMA. 2021;325:1467–1469. doi: 10.1001/jama.2021.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2021 doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.-.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., Maccamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A.M., Taha T.Y., Tabata T., Chen I.P., Ciling A., Khalid M.M., Sreekumar B., Chen P.-Y., Hayashi J.M., Soczek K.M., et al. Rapid assessment of SARS-CoV-2 evolved variants using virus-like particles. Science. 2021;374:1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.S., Collier A.-R.Y., Liu J., Yu J., Wan H., McMahan K., He X., Jacob-Dolan C., Chandrashekar A., Sellers D., et al. Ad26.COV2.S or BNT162b2 boosting of BNT162b2 vaccinated individuals. medRxiv. 2021 medRxiv:10.1101/2021.12.02.21267198. [Google Scholar]
- Tan C.-W., Chia W.-N., Young B.E., Zhu F., Lim B.-L., Sia W.-R., Thein T.-L., Chen M.I.-C., Leo Y.-S., Lye D.C., et al. Pan-Sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel E.S., Johnson P.W., Kunze K.L., Wu L., Gorsh A.P., Granger D., Roforth M.M., Jerde C.R., Lasho M., Andersen K.J., et al. SARS-CoV-2 serologic assays dependent on dual-antigen binding demonstrate diverging kinetics relative to other antibody detection methods. J. Clin. Microbiol. 2021;59:e0123121. doi: 10.1128/JCM.01231-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.-J., Bosch B.-J., Rey F.A., de Groot R.J., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., Kim W., Kalaidina E., Goss C.W., Rauseo A.M., Schmitz A.J., Hansen L., Haile A., Klebert M.K., Pusic I., et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature. 2021;595:421–425. doi: 10.1038/s41586-021-03647-4. [DOI] [PubMed] [Google Scholar]
- Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies. Nat. Med. 2021 doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Miranda M.C., Schäfer A., Pham M.N., Greaney A., Arunachalam P.S., Navarro M.-J., Tortorici M.A., Rogers K., O’Connor M.A., et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447.e16. doi: 10.1016/j.cell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2020;183:1732. doi: 10.1016/j.cell.2020.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Zost S.J., Jang H., Huang Y., Allen J.D., Case J.B., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions and monocytes for optimal therapeutic protection. bioRxiv. 2020 doi: 10.1016/j.cell.2021.02.026. bioRxiv:2020.12.28.424554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B., Whitener B., VanBlargan L.A., Hassan A.O., Shrihari S., Liang C.-Y., Karl C.E., Mackin S., Chen R.E., Kafai N.M., et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. bioRxiv. 2021 doi: 10.1126/scitranslmed.abm3302. bioRxiv:2021.08.25.457693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this study are available upon request.