Abstract
A new repetitive DNA element was identified in an isolate of Leptospira interrogans serovar copenhageni from a patient in Salvador, Brazil. A Sau3A genomic library from this strain was constructed and screened for repetitive DNA elements. An insert of 438 bp (Rep1) from one library clone hybridized to multiple chromosomal DNA fragments resolved electrophoretically after digestion with BamHI, HindIII, and MfeI. A single oligonucleotide primer, designated iRepl, was designed to generate multiple PCR amplicons of various electrophoretic mobilities in a PCR typing method. The method distinguished strains belonging to the eight pathogenic and three saprophytic species of the genus Leptospira. Clinical isolates obtained during urban epidemics between 1996 and 1998 in Salvador, Brazil, were analyzed by this PCR method. Although the iRep1 primer was unable to discriminate strains among L. interrogans serovar copenhageni isolates, it was able to differentiate strains belonging to different species and serogroups of Leptospira identified in Salvador. This PCR-based method may provide a faster and less expensive alternative to serologic tests used in reference laboratories.
Leptospirosis, caused by the spirochete Leptospira, is considered an important reemerging infectious disease worldwide (6, 14, 30). Spirochetes have the ability to survive in a wide range of environmental reservoirs, including mammalian hosts, factors that combined with the great diversity of this organism to make leptospirosis the most widespread zoonosis in the world (6, 8, 10). In Salvador, Brazil, more than 300 cases of leptospirosis are identified each year during the rainy season, and 15% of them die (14). Because of the association of certain leptospira serogroups with severe disease manifestation and complications of leptospirosis, a test that can rapidly and easily distinguish serogroups and serovars during outbreak investigations is urgently needed.
Conventional identification and diagnosis of Leptospira are based on the serologic method of agglutination (6, 8, 16, 17, 23, 30). This method of classification is complicated by the extreme diversity of the genus, comprising 11 species organized into 31 serogroups and over 250 serovars based on their antigenic relatedness (6, 31–35). The basic taxon is the serovar (6), defined by the cross-agglutinin absorption test, a serologic method requiring the preparation of antisera and the maintenance of a large number of reference serovars in culture (4, 6, 7). Batteries of monoclonal antibodies raised against each isolated strain or reference serovar are also used for rapid diagnosis and identification, but are limited by the availability or access to these antibodies, which are usually confined to few reference laboratories (6). These tests are often complicated by the extensive cross-reactivity of the antisera to shared Leptospira serovar antigenic epitopes (3, 4, 6, 7, 9, 13). Because of this complexity of serologic identification, several laboratories have developed new classification techniques based on genetic heterogeneity among members of the genus Leptospira (3, 4, 7, 8, 9, 12, 19, 24).
Numerous PCR-based techniques such as random amplified polymorphic DNA, arbitrarily primed PCR, and the use of insertion sequences in PCR-based assays (IS1500 and IS1533) have been developed and evaluated based on typing of leptospiral reference strains (1–3, 6, 7–9, 12–19, 22, 27, 29, 31, 33, 34–37). Other molecular techniques such as restriction fragment length polymorphism (RFLP) and pulsed-field gel electrophoresis (PFGE) have been helpful in classifying leptospires at the serogroup level (4, 8, 12, 18, 23, 26, 28, 30, 32, 35), but are not easily applicable in outbreak investigations because of the time and expensive equipment and reagents required to do them.
In this report, we describe a new repetitive DNA sequence identified and cloned from a clinical isolate of Leptospira interrogans serovar copenhageni. This cloned DNA fragment was used to develop a simple PCR assay based on a single oligonucleotide (iRep1) that could rapidly differentiate leptospiral serogroups in an epidemiologic investigation during an urban epidemic in Salvador, Brazil.
MATERIALS AND METHODS
Leptospira strains.
Strains used in this study were supplied by the World Health Organization/Food and Agriculture Organization (WHO/FAO) Collaborating Center for Reference and Research on Leptospirosis, Amsterdam, The Netherlands, or the Brazilian National Leptospirosis Reference Laboratory, or purchased from the American Type Culture Collection. Leptospiral strains were isolated from patients and captured rodents during sequential urban epidemics in Salvador, Brazil, between 1996 and 1998 (14). All isolates were propagated in liquid Ellinghausen-McCullough-Johnson-Harris medium (Difco Laboratories, Detroit, Mich.) at 29°C.
Genomic library.
Purified chromosomal DNA from L. interrogans serovar copenhageni strain L1-130 isolated from a patient was partially digested with Sau3A (New England Biolabs, Beverly, Mass.) and ligated to BamHI-digested pQE-30 vector (Qiagen, Valencia, Calif.). Escherichia coli DH5α cells were transformed by electroporation with the recombinant plasmids and grown on Luria-Bertani agar plates supplemented with ampicillin (100 μg/ml).
Screening the genomic library and DNA hybridization analysis.
The DNA insert in the Sau3A library clones was amplified by PCR with the T3 and T7 primer set. Amplified inserts of 500 to 1,000 bp were size selected after agarose gel electrophoresis, purified with the QIAquick nucleotide removal kit (Qiagen), and labeled with the DIG-end digoxigenin (Dig) labeling kit (Roche Molecular Biochemicals, Indianapolis, Ind.).
Approximately 4 μg of genomic DNA from strain L. interrogans copenhageni L1-130 was digested with restriction enzymes BamHI, MfeI, and HindIII and resolved at 20 V on a 1% agarose (Gibco) gel in 1 × TAE (40 mM Tris acetate, 2 mM EPTA) overnight. DNA was extracted according to the protocol provided by the supplier (Qiagen), with the blood and cell culture kit from 500 ml of 7-day cultures of Leptospira cells. Genomic DNA fragments were denatured and blotted onto positively charged nylon membranes (Roche Molecular Biochemicals) according to the method described by Southern (25). Both prehybridization (1 h) and hybridization (overnight) were performed under low-stringency conditions of 40°C and repeated at high-stringency conditions of 60°C in Dig Easy hybridization solution (Roche). After hybridization with Dig-labeled PCR products, the membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.01% sodium dodecyl sulfate (SDS) for 5 min at room temperature and twice in 0.1× SSC–0.01% SDS at 40°C and then at 60°C for 15 min. Membranes were exposed for 25 min to Biomax BL film (Eastman Kodak, Rochester, N.Y.) for the detection of chemiluminescent products.
PCR amplification and DNA extraction.
DNA from 3-day leptospiral culture pellets was resuspended in distilled water to an approximate concentration of 108 organisms/ml, boiled for 15 min to kill the organism and release the DNA, vortexed, and centrifuged at 14,000 × g for 15 min. Samples were stored at −80°C in 15 μl single-use aliquots.
PCR was carried out with the GeneAmp PCR System 9600 (Perkin-Elmer, Norwalk, Conn.). The PCR mix included 10 to 100 ng of DNA, 100 pmol of primers, 2.5 mM MgCl2 (Gibco BRL, Grand Island, N.Y.), 1× PCR buffer (Gibco), 200 μM deoxynucleoside triphosphates (Roche Molecular Biochemicals), and 1 U of recombinant Taq polymerase (Gibco) in a final volume of 50 μl. Temperature cycles for the amplification were 94°C for 5 min, 94°C for 30 s, 50°C for 1.5 min, and 72°C for 4 min, with a final extension time of 7 min after a total of 35 cycles. Sample dye (0.25% bromophenol blue, 0.25% xylene cyanol FF, and 30% glycerol in water) was added to the PCR products, which were resolved for 1 h at 100 V on a 1.5% agarose gel in 1× TAE. PCR assays were performed in triplicate, and products were visualized by the UV illumination of an ethidium bromide-stained gel. DNA from L. interrogans serovar copenhageni strain Winjberg was used as a positive control, while blank samples lacking DNA were used as negative controls.
Sequence analysis.
The DNA fragment that hybridized to multiple Leptospira DNA fragments was sequenced by dideoxynucleotide chain termination reactions with the ABI Prism 310 genetic analyzer (Perkin Elmer). Nucleic acid sequences were analyzed by BLAST nucleotide similarity search at the National Center for Biotechnology Information. Primers were designed using Amplify Software (University of Wisconsin, Madison, Wis.).
RESULTS
Screening the genomic library and identifying the DNA repetitive element.
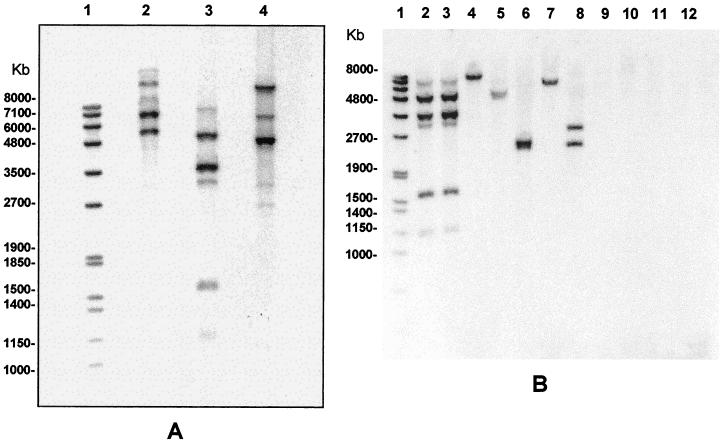
An L. interrogans serovar copenhageni strain isolated from a patient during an urban epidemic in Salvador, Brazil, was used to construct the genomic library. On initial screening, PCR amplification of the genomic library identified 36 clones containing inserts of 500 to 1,000 bp. Of these, 35 clones hybridized to only one or two BamHI DNA fragments (data not shown). One clone, pRep1, hybridized to five BamHI fragments, indicating that this DNA insert contained a multicopy element (Fig. 1A). Fragments varied in size based on the restriction enzyme. Southern blot analysis of BamHI or HindIII-digested L1-130 DNA demonstrated high-molecular-weight fragments of between 3,000 and 8,000 bp, while that for MfeI-digested L1-130 DNA generated five fragments between 1,500 and 6,000 bp (Fig. 1). Sequence analysis of the pRep1 DNA insert revealed a 438-bp AT-rich sequence, designated Rep1 (Fig. 2), with one 348-bp open reading frame (accession number AF303218). The EMBL and GenBank databases identified no sequences similar to that of Rep1.
FIG. 1.
Distribution of the repetitive element among reference strains representing 11 species of the genus Leptospira. (A) Genomic DNA from a clinical isolate of L. interrogans serovar copenhageni was digested with BamHI (lane 2), MfeI (lane 3), and HindIII (lane 4). (B) Genomic DNA from leptospiral species was digested with BamHI, separated on a 0.7% agarose gel electrophoresis, transferred to a positively charged nylon membrane, and hybridized with Dig-labeled pRep1. Genomic DNA was analyzed from the following species: L. interrogans serovar copenhageni strain Winjberg (lane 2), L. interrogans serovar copenhageni strain L1-130 (lane 3), L. borgpetersenii serovar castelloni strain Castellon 3 (lane 4), L. weilii serovar celledoni strain Celledoni (lane 5), L. noguchi serovar panama strain CZ 214 K (lane 6), L. santarosai serovar shermani strain 1342 K (lane 7), L. kirshneri serovar grippotyphosa strain Moskva V (lane 8), L. inadai serovar lyme strain 10 (lane 9), L. fainei serovar hurstbridge strain BUT6 (lane 10), L. meyeri serovar ranarum strain Ranae (lane 11), and L. biflexa serovar patoc strain Patoc1 (lane 12). The Dig-labeled molecular weight marker set VII is represented in lane 1.
FIG. 2.
Sequence analysis of the repetitive element identified from a genomic library of L. interrogans serovar copenhageni strain L1-130. Structural features of the repetitive element are shown including the location of primer iRepl used for PCR genotyping. iRep1 primer is shown as the reverse complement of the target sequence.
Copy number and distribution of the Rep1 element among Leptospira species.
RFLP patterns for the 11 species of Leptospira varied based on Southern blot analysis with the Rep1 element (Fig. 1B). Four leptospiral species, L. borgpetersenii, L. weilii, L. noguchi, and L. santarosai, displayed one copy of Rep1, while L. kirshneri displayed two copies. The other four species tested, L. inadai. L. fainei, L. meyeri, and L. biflexa, contained no Rep1 elements based on the DNA hybridization results. L. interrogans copenhageni Winjberg reference strain and L. interrogans copenhageni L1-130 displayed identical patterns based on five chromosomal fragments. These results indicate that the Rep1 element not only contains a repetitive sequence but also allows the differentiation of species based on a distinct pattern (Fig. 1B).
Differentiation of Leptospira reference strains by PCR-based typing methods.
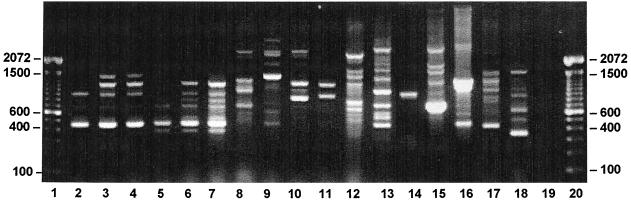
Electrophoretic band patterns resulting from PCR-amplified products of 46 leptospiral reference strains were generated with the use of the iRep1 primer, a single 21-mer oligonucleotide based on a sequence within the Rep1 element (Fig. 2). Each of the 11 species revealed a distinct pattern. Among 17 strains tested representing 16 serovars, the iRep1 assay distinguished 14 unique banding patterns (Fig. 3). Further comparison of L. interrogans serovar hardjo and L. borgpetersenii serovar hardjo revealed identical patterns, although they belonged to different species.
FIG. 3.
iRep1 PCR-based molecular typing of reference strains from the genus Leptospira. DNA from boiled culture pellets was used in PCRs with the iRep1 primer for L. interrogans serovar autumnalis strain Akiyami A (lane 2), serovar icterohaemorrhagiae strain RGA (lane 3), serovar copenhageni strain Winjberg (lane 4), serovar wolfii strain 3705 (lane 5), and serovar hardjo strain Hardjoprajitno (lane 6); L. borgpetersenii serovar hardjo strain Lely 607 (lane 7), serovar sejroe strain M84 (lane 8), and serovar castellonis strain Castellon 3 (lane 9); L. weilii serovar celledoni strain Celledoni (lane 10); L. noguchi serovar panama strain CZ 214 K (lane 11); L. santarosai serovar shermani strain 1342 (lane 12); L. kirshneri serovar grippotyphosa strain Moskva V (lane 13); L. wolbachi serovar codice strain CDC (lane 14); L. inadai serovar lyme strain 10 (lane 15); L. fainei serovar hurstbridge strain BUT6 (lane 16); L. meyeri serovar ranarum strain ranae (strain 17); and L. biflexa serovar patoc strain Patoc1 (lane 18). A negative control sample without DNA is shown in lane 19. The positions of the 100-bp size markers are represented in lanes 1 and 20.
Three distinct banding patterns differentiated the three serovars hardjo, sejroe, and castellonis belonging to the species L. borgpetersenii. Among 17 serovars tested, five L. interrogans serovars (automnalis, copenhageni, icterohaemorrhagiae, wolfii, and hardjo) displayed similar yet unique patterns. L. interrogans serovar icterohaemorrhagiae strain RGA and serovar copenhageni strain Winjberg displayed identical patterns.
Application of iRep1-based PCR to epidemiological investigation of leptospirosis.
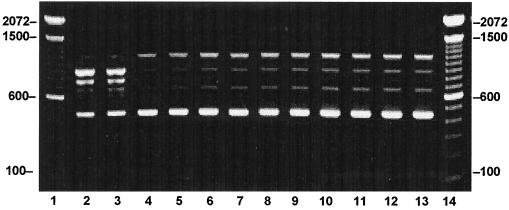
To assess relatedness among strains, we analyzed 22 clinical and 5 captured rodent (Rattus norvegicus) isolates with primer iRep1. Twenty-two of the clinical isolates and all five rodent isolates displayed a four-band pattern identical to that of the L. interrogans copenhageni Winjberg reference strain (Fig. 4). However, one of the clinical isolates, L1-133, revealed a unique pattern. Compared to the reference bank strains from WHO, L1-133 had a pattern identical to that of L. interrogans canicola strain Hond Utrecht.
FIG. 4.
iRep1 PCR-based molecular typing of human and rat Leptospira strains isolated from an urban epidemic in Salvador, Brazil. DNA from boiled Leptospira culture pellets was amplified with the iRep1 primer. The following samples were identified as L. interrogans serovar copenhageni except for L1-133, shown in lane 3 (L. interrogans serovar canicola). The following clinical isolates of L. interrogans serovar copenhageni were evaluated: L1-130 (lane 5), L1-212 (lane 6), L8-38 (lane 7), L8-118 (lane 8), and L8-163 (lane 9). Rat isolates included R1-15 (lane 10), R1-98 (lane 11), and R1-147 (lane 12), and R1-152 (lane 13). Lanes 2 and 4 represent reference strains L. interrogans serovar canicola strain Hond Utrecht IV and L. interrogans serovar copenhageni strain Winjberg, respectively. The positions of the 100-bp size markers are represented in lanes 1 and 14.
DISCUSSION
This report describes the identification and cloning of a new repetitive DNA sequence from a clinical isolate of L. interrogans serovar copenhageni. This cloned repetitive element was used in the development of a rapid and specific PCR assay to distinguish among serogroups and serovars during an urban epidemic that occurred between 1996 and 1998 in Salvador, Brazil (14). The zoonosis caused by leptospiral species has a worldwide distribution, with recent outbreaks reported from Nicaragua (36), Brazil (14), and parts of the United States (30). The 1996 epidemic of leptospirosis in Salvador, Brazil, had a mortality rate of 15% (14). Leptospira species have been associated with specific animal reservoirs (6, 14, 30, 31, 33, 34, 35), such as serovars icterohaemorrhagiae and copenhageni, which are most commonly associated with domestic rats worldwide. Rapid identification of the etiologic agents in outbreaks and the differentiation of leptospiral serovars are critical in preventing high mortality associated with certain serogroups. Although this new PCR-based assay was not able to distinguish among serovars within the same serogroups, it was able to differentiate the 11 species and 16 serogroups of Leptospira.
Leptospira species are heterogeneous and are further grouped into 31 serogroups and over 250 serovars (1, 2, 4, 6–11, 14, 18, 20, 21, 27, 30–36). These spirochetes are taxonomically classified by genetic techniques such as DNA-DNA hybridization (2, 3, 6, 21, 35), but in many laboratories, diagnosis and classification rely on serologic methods based on the microscopic agglutination test. In reference laboratories, methods such as the cross-absorption agglutinin test and the use of monoclonal antibodies allow serovar identification (6, 7). Due to the elaborate and complex nature of these classification methods and confusion with nomenclature, most clinical laboratories have had to rely on other techniques to distinguish leptospiral serogroups, particularly those associated with human disease.
The iRep1 PCR typing assay has several advantages compared to other methods used to classify leptospires. First, the use of a single oligonucleotide primer eliminates the need for difficult and time-consuming techniques such as maintenance of reference serum batteries, dark-field microscopy (3, 6, 7, 16, 17, 23), and preparation of homologous rabbit antiserum used for the serologic assays. Second, iRep1 PCR was specific enough to detect leptospiral DNA from cultures that became contaminated with other bacteria, a frequent problem in the tropical environment. Third, iRep1 PCR was inexpensive compared to serological techniques and other genetic approaches in the identification of Leptospira. Unlike PCR-based typing assays, RFLP, Southern blot analysis, and PFGE require expensive reagents and equipment (4, 6, 8, 9, 12, 16, 18). Other PCR-based methods have been reported, and the discovery of IS1500 and IS1533 DNA elements (34, 36) has allowed the development of repetitive-element PCR assays. Unlike iRep1 PCR, which uses only one oligonucleotide primer to amplify DNA, the insertion sequence PCR requires two inverted primers (34, 36).
Interestingly, the iRep1 PCR assay yielded electrophoretic bands from strains belonging to serogroups that did not have DNA fragments that hybridized to the 348-bp repetitive element (Fig. 1). The short primer sequence used for the PCR may anneal to similar sequences in the genomes of strains that lack the full repetitive element sequence. This would also explain the large number of PCR products obtained from strains that harbor the repetitive element.
One disadvantage of the iRep1 PCR assay was its inability to distinguish among isolates at the serovar level. However, it may still be helpful in discriminating serogroups from different animal reservoirs during an outbreak. Inability to genetically differentiate organisms at the serovar level by other techniques has been described (33, 36). Both IS1500- and IS1533-based PCR typing methods were unable to differentiate serovars (they were able to differentiate serogroups) among clinical isolates during an outbreak in Nicaragua (34, 36). Similarly, we were unable to distinguish the 22 isolates typed as serovar copenhageni based on their iRep1 PCR patterns.
The iRep1 PCR assay found similarities among many of the isolates from the urban epidemic in Salvador, Brazil. All 27 cultured isolates, both human and rat, were identified as belonging to the L. interrogans serovar copenhageni group, with the exception of isolate L1-133. This isolate was typed as L. interrogans serovar canicola based on the reference strain pattern. Standard serologic methods were used to confirm the serovar status of clinical isolates. Interestingly, the iRep1 PCR found that two serovar hardjo isolates from two different species (L. interrogans and L. borgpetersenii) produced identical electrophoretic banding patterns. Others have reported high similarity of the lipopolysaccharide biosynthetic loci (rfb) of these subtypes, suggesting horizontal acquisition of a large segment of the loci by one strain from another (5). Such an observation made by this PCR-based typing system further validates its usefulness. This rapid typing method may therefore be applied in places with limited resources to assist clinical management of leptospirosis.
ACKNOWLEDGMENTS
We thank Patrícia Guimarães de Oliveira, Fernanda Pinheiro Carvalho, and Hygia Guerreiro for their assistance in isolating and maintaining strains from patients and captured rodents; Ruud Hartskeerl and Marga Goris for supplying the reference battery of Leptospira serovars; Sabine Ehrt and Sanqwei Lu for their support and critical advice; and Brendan Flannery for advice during the preparation of the manuscript.
This work was supported by the Oswaldo Cruz Foundation/Brazilian Ministry of Health (0250.250.415), Brazilian National Research Council (52.1229/98-7, 30.0861/96-6, 35.0052/95-6, and Pronex 4196086200), the Fogarty Program in International Research and Training in Emerging Infectious Diseases (TW00905, TW00919, and AI30639), and a KO8 Award from the National Institute of Allergy and Infectious Diseases (AI01605).
REFERENCES
- 1.Baranton G, Old I G. The spirochetes: a different way of life. Bull Inst Pasteur. 1995;93:63–95. [Google Scholar]
- 2.Boursaux-Eude C, Saint Girons I, Zuerner R. Leptospira genomics. Electrophoresis. 1998;19:589–592. doi: 10.1002/elps.1150190421. [DOI] [PubMed] [Google Scholar]
- 3.Brown P D, Levett P N. Differentiation of Leptospira species and serovars by PCR-restriction endonuclease analysis, arbitrary primed PCR and low-stringency PCR. J Med Microbiol. 1997;46:173–181. doi: 10.1099/00222615-46-2-173. [DOI] [PubMed] [Google Scholar]
- 4.Corney B G, Colley J, Graham G C. Simplified analysis of pathogenic leptospiral serovars by random amplified polymorphic DNA fingerprinting. J Med Microbiol. 1997;46:927–932. doi: 10.1099/00222615-46-11-927. [DOI] [PubMed] [Google Scholar]
- 5.de la Peña-Moctezuma A, Bulach D M, Kalambaheti T, Adler B. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol Lett. 1999;177:319–326. doi: 10.1111/j.1574-6968.1999.tb13749.x. [DOI] [PubMed] [Google Scholar]
- 6.Faine S, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis. 2nd ed. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 7.Feresu S B, Bolin C B, Korver H, Terpstra W J. Classification of leptospires of the pyrogenes serogroup isolated from cattle in Zimbabwe by cross-agglutinin absorption and restriction fragment length polymorphism analysis. Int J Bacteriol. 1994;44:541–546. doi: 10.1099/00207713-44-3-541. [DOI] [PubMed] [Google Scholar]
- 8.Gerritsen M A, Smits M A, Olyhoek T. Random amplified polymorphic DNA fingerprinting for rapid identification of leptospiras of serogroup Sejroe. J Med Microbiol. 1995;42:336–339. doi: 10.1099/00222615-42-5-336. [DOI] [PubMed] [Google Scholar]
- 9.Gravekampp C, van de Kemp H, Frazen M, Carrington D, Schoone G J, van Eys G J J M, Everard C O R, Hartskeerl R A, Terpstra W J. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J Gen Microbiol. 1993;139:1691–1700. doi: 10.1099/00221287-139-8-1691. [DOI] [PubMed] [Google Scholar]
- 10.Haake D A, Chao G, Zuerner R L, Barnett J K, Barnett D, Mazel M, Matsunaga J, Levett P N, Bolin C A. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. J Clin Microbiol. 2000;68:2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann J L. Genomic techniques for identification of leptospira strains. Pathol Biol. 1993;41:943–950. [PubMed] [Google Scholar]
- 12.Herrmann J L, Bellenger E, Perolat P, Baranton G, Saint Girous I. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol. 1992;30:1696–1702. doi: 10.1128/jcm.30.7.1696-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann J L, Baril C, Bellenger E, Perolat P, Baranton G, Saint Girons I. Genome conservation in isolates of Leptospira interrogans. J Bacteriol. 1991;173:7582–7588. doi: 10.1128/jb.173.23.7582-7588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko A I, Reis M G, Dourado C R, Johnson W D, Riley L W the Salvador Leptospirosis Study Group. Urban epidemic of severe leptospirosis in Brazil. Lancet. 1999;354:820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence J G, Dykhuizen D E, DuBose R F, Hartl D L. Phylogenetic analysis using insertion sequence fingerprinting in Escherichia coli. Mol Biol Evol. 1989;6:1–14. doi: 10.1093/oxfordjournals.molbev.a040531. [DOI] [PubMed] [Google Scholar]
- 16.Letocart M, Baranton G, Perolat P. Rapid identification of pathogenic Leptospira species (Leptospira interrogans, L. borgpetersenii, and L. kirshneri) with species-specific DNA probes produced by arbitrary primed PCR. J Clin Microbiol. 1997;35:248–253. doi: 10.1128/jcm.35.1.248-253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meriens F, Amouriaux P, Perolat P, Baranton G, Saint Girons I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol. 1992;30:2219–2224. doi: 10.1128/jcm.30.9.2219-2224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacciarini M L, Savio M L, Tagliabue S, Rossi C. Repetitive sequences cloned from Leptospira interrogans serovar hardjo genotype hardjoprajitno and their application to serovar identification. J Clin Microbiol. 1992;30:1243–1249. doi: 10.1128/jcm.30.5.1243-1249.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perolat P, Merien F, Ellis W A, Baranton G. Characterization of Leptospira isolates from serovar hardjo by ribotyping arbitrary primed PCR, and mapped restriction site polymorphisms. J Clin Microbiol. 1994;32:1949–1957. doi: 10.1128/jcm.32.8.1949-1957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralph D, McClelland M. Phylogenetic evidence for horizontal transfer of an intervening sequence between species in a spirochete genus. J Bacteriol. 1994;176:5982–5987. doi: 10.1128/jb.176.19.5982-5987.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramadass P, Jarvis B D W, Corner R J, Penny D, Marshall R B. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992;42:215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- 22.Rantakokko-Jalava K, Nikkari S, Jalava J, Eerola E, Skurnik M, Meurman O, Ruuskanen O, Alanen A, Kotilainen E, Toivanen P, Kotilainen P. Direct amplification of rRNA genes in diagnosis of bacterial infections. J Clin Microbiol. 2000;38:32–39. doi: 10.1128/jcm.38.1.32-39.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero E C, Billerbeck A E C, Lando V S, Camargo E D, Souza C C, Yasuda P H. Detection of Leptospira DNA in patients with aseptic meningitis by PCR. J Clin Microbiol. 1998;36:1453–1455. doi: 10.1128/jcm.36.5.1453-1455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savio M L, Rossi C, Fusi P, Tagliabue S, Pacciarini M L. Detection and identification of Leptospira interrogans serovars by PCR coupled with restriction endonuclease analysis of amplified DNA. J Clin Microbiol. 1994;32:935–941. doi: 10.1128/jcm.32.4.935-941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi Y, Masami K, Satoru Y, Masahito F. Repetitive sequence of Leptospira interrogans serovar icterohaemorrhagiae strain Ictero No.1: a sensitive probe for demonstration of Leptospira interrogans strains. Microbiol Immunol. 1999;43:669–678. doi: 10.1111/j.1348-0421.1999.tb02455.x. [DOI] [PubMed] [Google Scholar]
- 27.Thiermann A B, Handsaker A L, Mosely S L, Kingscote B. New method for classification of leptospiral isolates belonging to serogroup Pomona by restriction endonuclease analysis: serovar kennewicki. J Clin Microbiol. 1985;21:585–587. doi: 10.1128/jcm.21.4.585-587.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. J Clin Microbiol. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinetz J M, Glass G E, Flexner C E, Mueller P, Kaslow D C. Sporadic urban leptospirosis. Ann Intern Med. 1996;125:794–798. doi: 10.7326/0003-4819-125-10-199611150-00002. [DOI] [PubMed] [Google Scholar]
- 31.Woo T H S, Patel B K C, Smythe L D, Symonds M L, Norris M A, Dohnt M F. Comparison of two PCR methods for rapid identification of Leptospira genospecies interrogans. FEMS Microbiol Lett. 1997;155:169–177. doi: 10.1111/j.1574-6968.1997.tb13874.x. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda B H, Steigerwalt A G, Sulzer K R, Kaufmann A F, Rogers F, Brenner D J. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol. 1987;37:407–415. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 33.Zuerner R L. Genomic structure, organization, and variation in Leptospira. J Cell Biochem. 1993;17E:309. [Google Scholar]
- 34.Zuerner R L, Alt D, Bolin C A. IS1533-based PCR assay for the identification of Leptospira interrogans sensu lato serovars. J Clin Microbiol. 1995;33:3284–3289. doi: 10.1128/jcm.33.12.3284-3289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuerner R L, Bolin C A. Nucleic acid probe characterizes Leptospira interrogans serovars by restriction fragment length polymorphisms. Vet Microbiol. 1990;24:355–366. doi: 10.1016/0378-1135(90)90183-v. [DOI] [PubMed] [Google Scholar]
- 36.Zuerner R L, Bolin C A. Differentiation of Leptospira interrogans isolates by IS1500 hybridization and PCR assays. J Clin Microbiol. 1997;35:2612–2617. doi: 10.1128/jcm.35.10.2612-2617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuerner R L, Ellis W A, Bolin C A, Montgomery J M. Restriction fragment length polymorphisms distinguish Leptospira borgpetersenii serovar hardjo type hardjo-bovis isolated from different geographical locations. J Clin Microbiol. 1993;31:578–583. doi: 10.1128/jcm.31.3.578-583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]