Abstract
Fluorescent amplified-fragment length polymorphism (FAFLP) analysis, a high-resolution PCR-based genome fingerprinting method, was used to subtype Salmonella enterica serovar Enteritidis phage type 4. This single phage type is responsible for the majority of salmonellosis in Europe. Twenty strains isolated from nine outbreaks, five isolates from sporadic cases of human infection, four strains of poultry origin, and one laboratory-derived strain were comparatively studied by pulsed-field gel electrophoresis (PFGE) and FAFLP analysis. Following macrorestriction with XbaI, PFGE classified 73% of PT4 strains as a single type. FAFLP analysis was carried out with the primer pair EcoRI+0 and MseI+C, by simultaneously sampling 170 to 190 loci throughout the PT4 genome. Twenty-three FAFLP profiles, with 1 to 61 amplified-fragment differences, were found among the 30 strains. The index of discriminatory power of FAFLP analysis was 0.98, compared to 0.47 for PFGE. FAFLP analysis assigned genotypes to each PT4 outbreak, as well as sporadic PT4 infections, a significant development for the epidemiology and control of this zoonotic enteric pathogen.
Food poisoning with Salmonella enterica subsp. enterica is a major cause of human illness, and the most common serovar isolated is Enteritidis. One of the 60 phage types of Salmonella serovar Enteritidis (23), phage type 4 (PT4), has been responsible for the majority of salmonellosis in the United Kingdom and Europe since the early 1980s (18). A dramatic increase in such infections was observed in 1987 to 1988, when United Kingdom isolations of serovar Enteritidis increased from 6,858 to 15,427, 81% of which were due to PT4 (2, 28). Since Salmonella serovar Enteritidis PT4 infection is a zoonosis, major interventions have been aimed at the poultry reservoir. For example, between 1989 and 1993, nearly 2 million serovar Enteritidis-infected birds were compulsorily slaughtered. A feature of this serovar is its ability to infect the ovary and/or oviduct, providing a site where egg contents are contaminated (transovarian infection). Although contamination of eggs occurs sporadically rather than endemically (9), many human outbreaks have nonetheless involved grade-A table eggs (21). Human isolations of PT4 in the United Kingdom peaked in 1993 at 17,371 infections, declining in the last three years to 6,984 (provisional figure) in 1999 (30), due to the combination of improved infection control and hygiene at poultry breeding sites with vaccination of poultry against serovar Enteritidis.
Various genotyping methods have been applied, with limited success, to the epidemiological analysis of serovar Enteritidis PT4. The majority of isolates (70% of poultry isolates and 90% of human isolates) carry a 38-MDa virulence plasmid (14, 26), and since there is limited variation in the numbers and sizes of other plasmids, plasmid profiling is sometimes of use. However, extrachromosomal DNAs are subject to lateral transfer and spontaneous elimination. There are several other approaches based on restriction of genomic DNA with diverse endonucleases, followed by hybridization with probes. Again, these offer only limited differentiation of PT4. All PT4 strains belong to a single clonal line, which includes 11 other PTs as defined by insertion sequence IS200 profiling. However, the low copy number of IS200 in this serovar renders profiling ineffective for subtyping serovar Enteritidis (25). Similarly, ribotyping detects only four subtypes among PT4 strains (18). A modest improvement in discriminatory power is offered by macrorestriction with an infrequently cutting restriction enzyme(s) and pulsed-field gel electrophoresis (PFGE). Nine XbaI subtypes were detected among an epidemiologically diverse group of 39 PT4 strains (22). However, the majority of PT4 infections are caused by strains of a single XbaI pulsed-field profile (PFP) type, designated X1, which predominates among the type strains of the remaining Salmonella serovar Enteritidis phage types (23). In general, results from different molecular typing methods based on outer membrane proteins, lipopolysaccharides, multilocus enzyme electrophoresis, IS200 type, ribotype, and PFGE identify PT4 strains as members of one predominant clone. The inadequate discriminatory power even of a combination of the above techniques to subtype PT4 strains means that well-differentiated genotypes cannot be assigned to outbreaks or sporadic infections.
Amplified-fragment length polymorphism (AFLP) analysis was originally described for characterization of plant genomes (27) using radioactively labeled primers for PCR amplification. The technique involves restriction of genomic DNA with two restriction enzymes, followed by ligation of restricted fragments to double-stranded oligonucleotide adapters. The latter process creates target sites for stringent primer annealing. Subsets of fragments are then amplified using selective or nonselective primers which have sequences complementary to the ligated adapter-restriction site. When modified for use with fluorophore-labeled primers, fluorescent AFLP (FAFLP) analysis displays, on an automated DNA sequencer, large numbers of DNA polymorphisms based upon multiple restriction sites throughout a bacterial genome. Amplified fragments (AFs) are sized accurately to within 1 bp, an important and novel feature in defining the genotypes of bacterial clones. High levels of reproducibility and discriminatory power in the definition of epidemiological clonality have been found for Streptococcus pyogenes (7, 8), Staphylococcus aureus (12, 13), Escherichia coli (4), and Mycobacterium tuberculosis (11). In this study, we investigated the capacity of FAFLP analysis to subtype the clone Salmonella serovar Enteritidis PT4 and to identify genotypes associated with outbreaks of this important human enteropathogen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Thirty strains of Salmonella serovar Enteritidis PT4 were studied (Table 1). Of these, 28 strains were isolated in England and Wales between 1967 and 1998, one was from a patient who had been infected in Spain, and the last was a laboratory-derived strain (5). Twenty-four strains were isolated from human infections, three were from chickens, one was from liquid egg, and one was from human food. Of the 24 strains from humans, 5 were from sporadic infections. The remaining 19 strains, chosen to represent epidemiologically defined outbreaks in specific locations in England and Wales, included multiple isolates from seven epidemiologically distinct outbreaks (outbreaks 3 to 9) which occurred between 1995 and 1998 and two single strains from outbreaks which occurred in 1993 (outbreaks 1 and 2). The strain from human food was thought to be associated with outbreak 9. All these strains were confirmed to be outbreak related by PFGE following XbaI digestion. The first nine strains in Table 1 represented the different PFP types identified in XbaI digests (22). These nine had no known epidemiological linkage and were isolated in England and Wales between 1967 and 1992.
TABLE 1.
Epidemiology and genotyping of Salmonella serovar Enteritidis PT4 isolates
| Strain | LEPb strain no. | Yr of isolation | Source and context | PFP typec | FAFLP profiled |
|---|---|---|---|---|---|
| 1 | E2187T | 1967 | Human, sporadic | X1 | A1 |
| 2 | P125678 | 1988 | Liquid egg | X2 | A2 |
| 3 | P104871 | 1987 | Human, sporadic | X3 | A3 |
| 4 | P125588 | 1988 | Chicken | X4 | A4 |
| 5 | P132344/0 | 1988 | Chicken | X5 | A5 |
| 6 | P4268400 | 1983 | Human, sporadic | X6 | A6 |
| 7 | P3012660 | 1992 | Human, sporadic | X7 | A7 |
| 8 | P3026660 | 1992 | Chicken | X8 | A8 |
| 9 | P132344/1 | 1988 | Lab derived | X9 | A9 |
| 10a | P3139460 | 1993 | Human, sporadic | X1 | A10 |
| 11 | P3116120 | 1993 | Human, outbreak 1 | X1 | A11 |
| 12 | P3115760 | 1993 | Human, outbreak 2 | X1 | A12 |
| 13 | P3776690 | 1995 | Human, outbreak 3 | X1 | A13 |
| 14 | P3776940 | 1995 | Human, outbreak 3 | X1 | A14 |
| 15 | P3780290 | 1995 | Human, outbreak 3 | X1 | A13 |
| 16 | P3874000 | 1995 | Human, outbreak 4 | X1 | A15 |
| 17 | P3874010 | 1995 | Human, outbreak 4 | X1 | A15 |
| 18 | P3908810 | 1996 | Human, outbreak 5 | X1 | A16 |
| 19 | P3908830 | 1996 | Human, outbreak 5 | X1 | A17 |
| 20 | P3908840 | 1996 | Human, outbreak 5 | X1 | A17 |
| 21 | P4214000 | 1997 | Human, outbreak 6 | X1 | A18 |
| 22 | P4223140 | 1997 | Human, outbreak 6 | X1 | A18 |
| 23 | P4305670 | 1997 | Human, outbreak 7 | X1 | A19 |
| 24 | P4306200 | 1997 | Human, outbreak 7 | X1 | A19 |
| 25 | P4656910 | 1998 | Human, outbreak 8 | X1 | A20 |
| 26 | P4661540 | 1998 | Human, outbreak 8 | X1 | A20 |
| 27 | P4662200 | 1998 | Human, outbreak 8 | X1 | A20 |
| 28 | P4931360 | 1998 | Human, outbreak 9 | X1 | A21 |
| 29 | P4937000 | 1998 | Food, outbreak 9 | X1 | A22 |
| 30 | P4940010 | 1998 | Human, outbreak 9 | X1 | A23 |
Strain 10 was isolated from a patient infected in Spain; all other strains were isolated in England and Wales.
LEP, Laboratory of Enteric Pathogens.
Macrorestriction profiles were obtained with XbaI following PFGE (22).
FAFLP profiles were obtained with the EcoRI+0 and MseI+C primer pair.
All strains were identified as PT4 by phage typing, in accordance with the Salmonella serovar Enteritidis phage typing scheme (29). They included the type strain of PT4, E2187, isolated in 1967. Strains were cultured aerobically at 37°C for 18 to 24 h on nutrient agar plates, and preserved for reference on Protect bacterial beads (Technical Service Consultants Ltd., Heywood, Lancashire, United Kingdom) at −70°C, after suspensions were made in nutrient broth with 10% (vol/vol) glycerol.
Standard nucleic acid extraction and PFGE.
Genomic DNA was extracted from 18- to 24-h Salmonella plate cultures by the cetyltrimethylammonium bromide (CTAB) method (31). Briefly, cell growth was scraped off the plate culture and washed in 1× Tris-EDTA buffer (pH 8.0). The cells were resuspended in 500 μl of Tris-EDTA buffer, 30 μl of 10% sodium dodecyl sulfate, and 3 μl of 10-mg/ml proteinase K. The sample was mixed thoroughly and incubated at 37°C for 1 h. To this suspension was added 100 μl of 5 M NaCl and 80 μl of CTAB-NaCl solution (31). The sample was incubated at 65°C for 10 min. This was followed by a chloroform-isoamyl alcohol (24:1) extraction, a phenol-chloroform extraction, and a repeat chloroform-isoamyl alcohol extraction. The DNA was precipitated with isopropanol, washed with 70% ethanol, and air dried. The concentration of DNA was estimated using a spectrophotometer (Beckman DU 640) by standard methods (24). All strains were subjected to PFGE following macrorestriction with XbaI as previously described (22, 23). The ramping and electrophoresis conditions used for PFGE were 10 to 100 s at 4.8 V cm−1 for 64 h on 1.2% agarose gels.
FAFLP analysis.
FAFLP analysis was performed on 500 ng of DNA which had been extracted from each strain and digested with endonucleases EcoRI and MseI. Restriction fragments were ligated to double-stranded adapters as described previously (8). The forward primer, a nonselective 5-carboxyfluorescein-labeled EcoRI primer (EcoRI+0; 5′-GACTGCGTACCAATTC-3′), was used in all reactions. The reverse primer, a nonlabeled MseI primer, had an extra selective base at the 3′ end (MseI+A, MseI+T, MseI+G, or MseI+C; 5′-GATGAGTCCTGAGTAAX-3′, where X is the selective base). PCRs and touchdown PCR conditions were as described previously (6, 8). FAFLP analysis products were separated on an ABI 377 automated DNA sequencer using Premix Long Ranger 5% polyacrylamide gel solution (FMC BioProducts, Vallensbaek Strand, Denmark) as described previously (6, 8). Each FAFLP reaction mixture was loaded with an internal size marker (GeneScan-2500 labeled with red fluorescent dye 6-carboxy-x-rhodamine). The running buffer was 1× TBE, and the electrophoresis conditions were 2.0 kV at 51°C for 10 h. The well-to-read distance was 48 cm.
Fragment analysis.
Fluorescent AFs obtained on the acrylamide sequencing gel were sized with GeneScan 3.0 software (Perkin-Elmer Corp., Norwalk, Conn.). Gel displays were transformed into electropherograms, which were visually inspected for polymorphisms, i.e., the presence or absence of AFs. The software Genotyper (Perkin-Elmer Corp.) was used to generate a table in a binary matrix format. Dice coefficients of similarity were calculated with in-house software. Cluster analysis was performed by UPGMA (NEIGHBOR program by PHYLIP), and the dendrogram was displayed with TreeView (20).
RESULTS
Macrorestriction profiles and PFGE.
Nine XbaI macrorestriction profiles (PFP types) were observed among the 30 PT4 strains studied. Each profile consisted of 11 to 15 fragments. The majority of strains (73%), including the type strain E2187 isolated in 1967, shared a single XbaI profile, X1. Eight other profiles were unique to single strains (Table 1). The numerical index of discriminatory power, or D value (15), for PFGE subtyping of PT4 was calculated as 0.47.
FAFLP analysis.
FAFLP analysis was performed on the set of PT4 DNAs with a nonselective forward primer (EcoRI+0) and one of four selective reverse primers, MseI+A, MseI+T, MseI+G, or MseI+C. In these experiments (data not shown), the combination of EcoRI+0 and MseI+C was found to be the most effective primer pair for epidemiological subtyping of PT4. FAFLP analysis gel data using this primer pair consisted of AFs in the size range 60 to 1,000 bp. Only fragments with sizes between 60 and 550 bp were included in the final analysis, since the accuracy of sizing above the latter value decreases to approximately ±2.0 bp, compared to ±1.0 bp below it. The number of AFs generated in the chosen size range was 170 to 190 per profile. Among all profiles, 87 AFs were polymorphic; the remainder were common.
Of the 30 strains included in this study, epidemiological data assigned 20 to nine different outbreaks. For three of those outbreaks, two isolates were analyzed from each, while for four outbreaks, three isolates were analyzed from each (Table 1). Single strains were analyzed from each of two further outbreaks. The 10 remaining strains, including the PT4 type strain, had been isolated between 1967 and 1993 from humans, chickens, and liquid egg (Table 1).
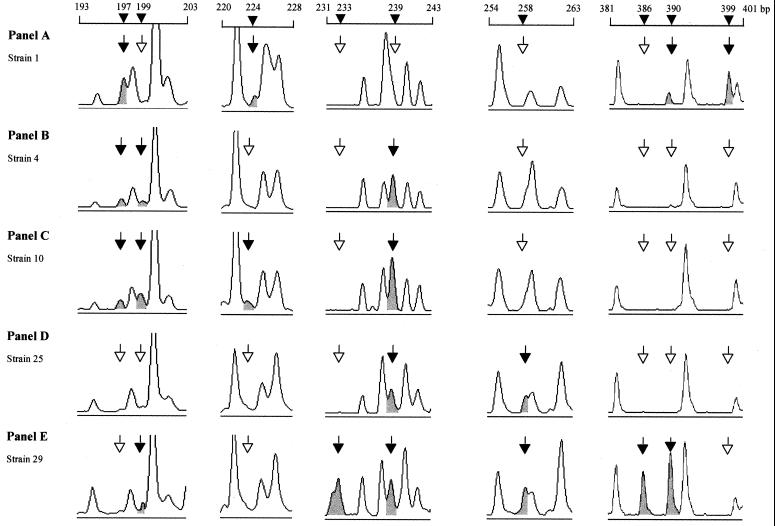
Twenty-three distinct FAFLP profiles were detected among the 30 strains. Seventeen strains had unique profiles. The number of AF differences between individual profiles ranged from 1 to 61. The type strain of PT4, E2187, had the most divergent FAFLP profile (Fig. 1A), with 48 to 61 AF differences from the other strains. Two other strains with very divergent FAFLP profiles were strain 29 (Fig. 1E), with 17 to 61 AF differences from other strains, and strain 7, with 19 to 33 AF differences from the others. All nonoutbreak strains exhibited unique FAFLP profiles (A2 to A10) (Table 1). The index of discriminatory power of FAFLP analysis for PT4 with the EcoRI+0 and MseI+C primer pair was thus 0.98.
FIG. 1.
FAFLP electropherograms showing polymorphisms within five Salmonella serovar Enteritidis PT4 strain genomes. Electropherograms were derived with GeneScan 3.0 software and show examples of windows of polymorphisms within the FAFLP profiles obtained with EcoRI+0 and MseI+C. Panels A to E correspond to profiles A1, A4, A10, A20, and A22, respectively (Table 1). The solid arrows and peaks indicate an AF characteristic of that profile (sizes are indicated in base pairs). Open arrows indicate the absence of a polymorphic fragment from that profile.
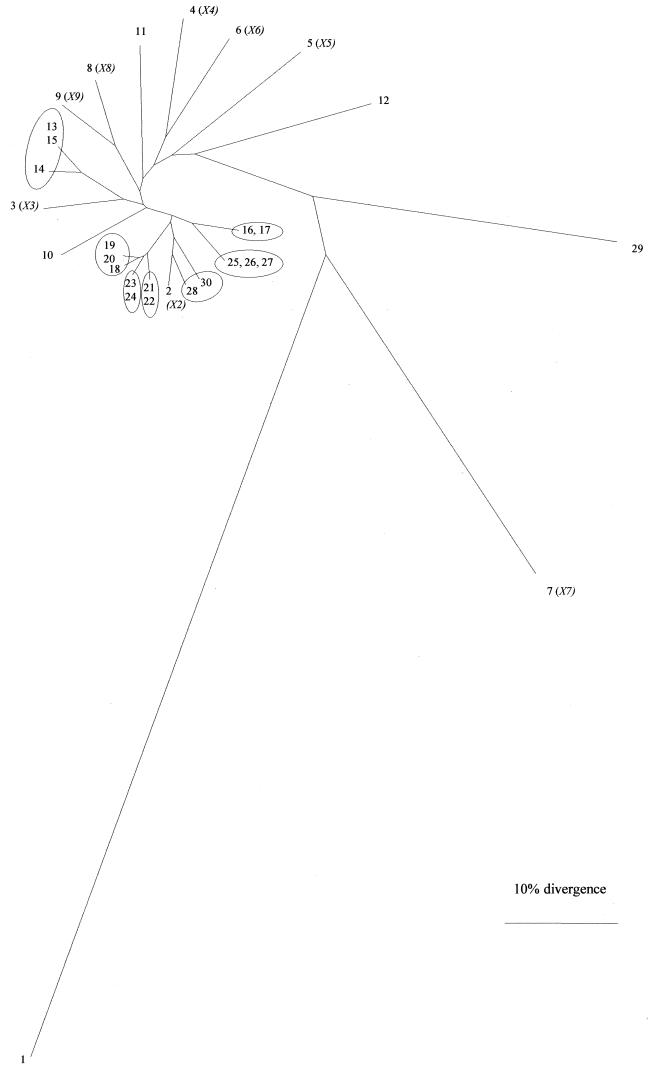
A dendrogram (Fig. 2) was derived from FAFLP analysis data by UPGMA cluster analysis (see Materials and Methods) and exhibited distinctive features. The type strain of PT4 was greatly divergent from all but the two other strains described above, as well as from most of the nonoutbreak ones (Fig. 2). All strains from outbreaks 4 to 9 were found in the same area of the tree. The nonoutbreak strains (numbered 2 to 9 in Table 1) having distinct PFGE profiles were characterized by unique FAFLP profiles, as was a strain isolated from a patient infected in Spain, which had the same PFGE profile as all the outbreak strains.
FIG. 2.
Genetic relationships between Salmonella serovar Enteritidis PT4 strains. The dendrogram was derived from FAFLP data by UPGMA and the PHYLIP and TreeView programs (20). Each branch represents a different FAFLP profile. All strains (Table 1) were PFP type X1 unless otherwise shown in parentheses (Table 1).
Cluster analysis of outbreak strains.
Most of the strains, which epidemiological information had suggested to be part of different outbreaks, were loosely clustered in the FAFLP dendrogram (Fig. 2). Both isolates from outbreak 4 (strains 16 and 17) shared FAFLP profile A15, branching next to the single profile shared by the three isolates from outbreak 8 (Fig. 2) (strains 25, 26, and 27; profile A20). Profiles A15 and A20 differed from each other by AFs of 182, 212, and 297 bp. Two of the three isolates from outbreak 9 (strains 28 and 30) differed from each other by two AFs (108 and 164 bp). However, strain 29, a food isolate, which was suggested by epidemiological data to be part of outbreak 9, was widely separated from these two by 17 AF differences (profile A22). Strains 21 and 22 from outbreak 6 shared one FAFLP profile, A18. Similarly, profile A19 was shared by strains 23 and 24 from outbreak 7. Profiles A18 and A19 differed by AFs of 108 and 306 bp. Of the three isolates from outbreak 5, strains 19 and 20 shared profile A17, while A16 differed by the absence of a 199-bp AF.
The three isolates from outbreak 3 clustered on a branch separate from the other outbreak profiles (with 4 to 14 AF differences from all other outbreak isolates studied). Two of these (strains 13 and 15) shared FAFLP profile A13; the other (strain 14) had a profile which differed by AFs of 112 and 182 bp. The single strains representing outbreaks 1 and 2 had unique FAFLP profiles, each with 4 to 15 AF differences from those of the other outbreak strains. In summary, characterization of nine “outbreak genotypes” could be made on the basis of the 22 polymorphic AFs shown in Table 2.
TABLE 2.
Genotypes of outbreak strains defined by FAFLP analysis with EcoRI+0 and MseI+C primers
| Strain | Source | FAFLP profile | Sizes of polymorphic AFa (bp) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | Outbreak 1 | A11 | 114 | 164 | 182 | 199 | 212 | 274 | 297 | 306 | 359 | |||||||||||||
| 12 | Outbreak 2 | A12 | 66 | 74 | 87 | 108 | 112 | 164 | 182 | 199 | 212 | 215 | 275 | 276 | 285 | 297 | 366 | 390 | 525 | |||||
| 13, 15 | Outbreak 3 | A13 | 66 | 108 | 112 | 114 | 164 | 212 | 274 | 297 | 306 | 359 | 525 | |||||||||||
| 14 | Outbreak 3 | A14 | 66 | 108 | 114 | 164 | 182 | 212 | 274 | 297 | 306 | 359 | 525 | |||||||||||
| 16, 17 | Outbreak 4 | A15 | 66 | 74 | 108 | 112 | 114 | 164 | 274 | 276 | 297 | 359 | 525 | |||||||||||
| 18 | Outbreak 5 | A16 | 66 | 74 | 108 | 112 | 114 | 164 | 182 | 212 | 274 | 276 | 297 | 306 | 310 | 359 | 525 | |||||||
| 19, 20 | Outbreak 5 | A17 | 66 | 74 | 108 | 112 | 114 | 164 | 182 | 199 | 212 | 274 | 276 | 297 | 306 | 310 | 359 | 525 | ||||||
| 21, 22 | Outbreak 6 | A18 | 66 | 74 | 112 | 114 | 164 | 182 | 199 | 212 | 274 | 276 | 297 | 306 | 310 | 359 | 525 | |||||||
| 23, 24 | Outbreak 7 | A19 | 66 | 74 | 108 | 112 | 114 | 164 | 182 | 199 | 212 | 274 | 276 | 297 | 310 | 359 | 525 | |||||||
| 25, 26, 27 | Outbreak 8 | A20 | 66 | 74 | 108 | 112 | 114 | 164 | 182 | 212 | 274 | 275 | 359 | 525 | ||||||||||
| 28 | Outbreak 9 | A21 | 66 | 74 | 87 | 108 | 112 | 114 | 164 | 182 | 199 | 212 | 274 | 276 | 297 | 359 | 525 | |||||||
| 29 | Outbreak 9 | A22b | 66 | 74 | 87 | 112 | 114 | 182 | 199 | 212 | 274 | 276 | 285 | 359 | 366 | 390 | 525 | |||||||
| 30 | Outbreak 9 | A23 | 66 | 74 | 87 | 108 | 114 | 182 | 199 | 212 | 274 | 276 | 297 | 359 | 525 | |||||||||
AFs are present in some strains and absent in others; a unique combination of AFs defines an outbreak genotype.
FAFLP profile A22 (strain 29) was characterized by the presence of unique AFs of 134, 233, 250, 251, 252, 314, 372, 374, and 386 bp, in addition to the polymorphic fragments listed above, and the absence of AFs of 116 and 136 bp, which are present in all other isolates.
DISCUSSION
Radioactive and fluorescent AFLP methods have been applied to genotyping of Salmonella enterica, essentially to the level of serovar discrimination only. Aarts et al. (1) assigned reproducible serovar profiles based on an EcoRI primer with two selective nucleotides at the 3′ end and a nonselective MseI primer. Lindstedt et al. (16) examined 97 strains belonging to seven Salmonella serovars, including seven serovar Enteritidis strains, by FAFLP typing on a capillary sequencer. In this system, the primer pairs EcoRI+0-MseI+C and XbaI+0-MseI+0 exhibited discriminatory power equivalent only to that of PFGE. Since the strains in that study were not phage typed and lacked associated epidemiological markers, and since the range of AFs was not reported, the applicability of this interesting report to the serovar Enteritidis problem is limited. Nair et al. (19) compared FAFLP analysis with PFGE for genotype analysis of 30 strains of Salmonella serovar Typhi, finding a higher index of discriminatory power (D = 0.88) for FAFLP analysis than for PFGE (D = 0.74), though the difference was not as marked as in the present report.
In this report, we describe a highly discriminatory FAFLP analysis of Salmonella serovar Enteritidis PT4, comparing it with PFGE for the analysis of nine independent outbreaks and a collection of strains causing sporadic infection. PFGE is currently the best generally available method for molecular subtyping of Salmonella serovar Enteritidis PT4. Powell et al. (22) found nine distinct PFGE subtypes among 39 strains, the majority (77%) sharing the same XbaI subtype. Lukinmaa et al. (17), using XbaI, SpeI, and NotI endonucleases for macrorestriction and PFGE, found the same XbaI PFGE subtype for 33 of 43 strains (75%) in their study. These findings have been supported by extensive fingerprinting of PT4 strains from cases of human infection in England and Wales over the last seven years (Laboratory of Enteric Pathogens, unpublished data).
The D value (15) of PFGE for our set of PT4 strains was less than half that of FAFLP analysis. Seventy-three percent of the strains, including the 20 from the nine outbreaks in the study, were included in a single XbaI PFP type. PFGE was unable to resolve epidemiological clonality for PT4 outbreaks whereas FAFLP analysis yielded 23 distinct profiles (compare Table 1 with Fig. 2). The FAFLP profile of the PT4 type strain E2187 differed in approximately one-third of the 180 AFs constituting its profile from those of the 21 other strains sharing its PFP type. This provides a striking example of the relative resolving power of the two techniques. With one exception, all strains found in one area of the tree (delimited by the clusters of strains 19, 20, and 18 and strains 16 and 17 in Fig. 2) were from six outbreaks. The exception, strain 2, was included in the study as representative of PFP type X2. It had a unique FAFLP profile, A2, which differed even from those of the most closely related strains (strains 28 and 30) by four and six AF differences, respectively. FAFLP analysis encompasses a much greater number of data points than PFGE, with each profile sampling in a nonbiased manner approximately 1.2% of the Salmonella serovar Enteritidis genome information found at The Institute for Genomic Research website (http://salmonella.life.uiuc.edu/).
FAFLP analysis was able to group isolates from four outbreaks into unique profiles (A15, A18, A19, and A20). Profiles A18 and A19 differed by two AFs (Table 2) but were designated as distinct outbreak genotypes on the basis of epidemiological context. For three other outbreaks, a pair of closely related profiles having one or two AF differences (A13 and A14; A16 and A17; and A21 and A23) were found within one outbreak genotype (Table 2). Thus, the definition of “outbreak genotype” should be made on the basis of a combination of FAFLP analysis and epidemiological context. Similar contextual considerations are applicable to the establishment of genetic relationship by PFGE in the USA PulseNet program for surveillance of E. coli O157 (E. Ribot, personal communication).
With the exception of one isolate from outbreak 9, there was complete concordance between the FAFLP analysis and epidemiological data. The exception (strain 29) was isolated from a food sample, whereas strains 28 and 30 were from human infections. From the epidemiological evidence, and because it shows the same PFGE profile, strain 29 was considered to be part of the same outbreak, which occurred in northwest London in 1998. However, its FAFLP profile (A22), which differed from the profile of each of the two human isolates by 17 AFs (Table 2 and Fig. 2), suggests that strain 29 did not belong to outbreak 9; this is an important finding, since the food involved had been one of those suspected as a source of this outbreak. The substantial number of differential data points, the reproducibility of FAFLP profiles, and the precision of AF sizing by the automated sequencer result in a significant qualitative improvement in definition of epidemiological clonality.
The findings of this study for PT4 parallel those for group A streptococci, where strains of the invasive serotype M1 subclone, homogeneous by all other molecular methods including combined PFGE data with three endonucleases, were readily assigned by FAFLP analysis to 17 subtypes. Eisenstein (10) noted that as the discriminatory power of a technique increases, the number of genetic differences found increases, and the appearance of clonality recedes. Additionally, FAFLP analysis can be modeled on and related back to the whole genome sequence (3, 11), which is approaching completion for Salmonella serovar Enteritidis at the time of writing (information can be found at http://salmonella.org). In summary, we have shown that FAFLP analysis resolves considerable genetic microheterogeneity among strains of Salmonella serovar Enteritidis PT4. We have demonstrated that this enteropathogenic “clone” in fact constitutes a rapidly evolving clone complex, within which genotypes can be readily assigned both to outbreaks and to sporadic infections.
ACKNOWLEDGMENT
We thank Linda Ward for phage typing the strains.
REFERENCES
- 1.Aarts H J, van Lith L A, Keijer J. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett Appl Microbiol. 1998;26:131–135. doi: 10.1046/j.1472-765x.1998.00302.x. [DOI] [PubMed] [Google Scholar]
- 2.Advisory Committee on the Microbiological Safety of Food. Report on Salmonella in eggs. London, United Kingdom: Her Majesty's Stationery Office; 1993. [Google Scholar]
- 3.Arnold C, Metherell L, Clewley J P, Stanley J. Predictive modelling of fluorescent AFLP: a new approach to the molecular epidemiology of E. coli. Res Microbiol. 1999;150:33–44. doi: 10.1016/s0923-2508(99)80044-8. [DOI] [PubMed] [Google Scholar]
- 4.Arnold C, Metherell L, Willshaw G, Maggs A, Stanley J. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J Clin Microbiol. 1999;37:1274–1279. doi: 10.1128/jcm.37.5.1274-1279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chart H, Threlfall E J, Rowe B. Virulence of Salmonella enteritidis phage type 4 is related to the possession of a 38 MDa plasmid. FEMS Microbiol Lett. 1989;49:299–303. doi: 10.1016/0378-1097(89)90057-8. [DOI] [PubMed] [Google Scholar]
- 6.Desai M, Efstratiou A, George R, Stanley J. High-resolution genotyping of Streptococcus pyogenes serotype M1 isolates by fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol. 1999;37:1948–1952. doi: 10.1128/jcm.37.6.1948-1952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai M, Tanna A, Efstratiou A, George R, Clewley J, Stanley J. Extensive genetic diversity among clinical isolates of Streptococcus pyogenes serotype M5. Microbiology. 1998;144:629–637. doi: 10.1099/00221287-144-3-629. [DOI] [PubMed] [Google Scholar]
- 8.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eijdokun O O, Killalea D, Cooper M, Holmyard S, Cross A, Kemp C. Four linked outbreaks of Salmonella enteritidis phage type 4 infection—the continuing egg threat. Commun Dis Public Health. 2000;3:95–100. [PubMed] [Google Scholar]
- 10.Eisenstein B I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis. 1990;161:595–602. doi: 10.1093/infdis/161.4.595. [DOI] [PubMed] [Google Scholar]
- 11.Goulding J N, Stanley J, Saunders N, Arnold C. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:1121–1126. doi: 10.1128/jcm.38.3.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grady R, Desai M, O'Neill G, Cookson B, Stanley J. Genotyping of epidemic methicillin-resistant Staphylococcus aureus phage type 15 isolates by fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol. 1999;37:3198–3203. doi: 10.1128/jcm.37.10.3198-3203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grady R, O'Neill G, Cookson B, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of the MRSA epidemic. FEMS Microbiol Lett. 2000;187:27–30. doi: 10.1111/j.1574-6968.2000.tb09131.x. [DOI] [PubMed] [Google Scholar]
- 14.Helmuth R, Schroeter A. Molecular typing methods for S. enteritidis. Int J Food Microbiol. 1994;21:69–77. doi: 10.1016/0168-1605(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 15.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstedt B A, Heir E, Vardund T, Kapperud G. Fluorescent amplified-fragment length polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J Clin Microbiol. 2000;38:1623–1627. doi: 10.1128/jcm.38.4.1623-1627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukinmaa S, Schildt R, Rinttila T, Siitonen A. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J Clin Microbiol. 1999;37:2176–2182. doi: 10.1128/jcm.37.7.2176-2182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendoza M C, Landeras E. Molecular epidemiological methods for differentiation of Salmonella enterica serovar Enteritidis strains. In: Saeed A M, editor. Salmonella enterica serovar Enteritidis in humans and animals. Ames: Iowa State University Press; 1999. pp. 125–140. [Google Scholar]
- 19.Nair S, Schreiber E, Thong K, Pang T, Altwegg M. Genotypic characterization of Salmonella typhi by amplified fragment length polymorphism fingerprinting provides increased discrimination as compared to pulsed-field gel electrophoresis and ribotyping. J Microbiol Methods. 2000;41:35–43. doi: 10.1016/s0167-7012(00)00148-2. [DOI] [PubMed] [Google Scholar]
- 20.Page R D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 21.Poppe C. Epidemiology of Salmonella enterica serovar Enteritidis. In: Saeed A M, editor. Salmonella enterica serovar Enteritidis in humans and animals. Ames: Iowa State University Press; 1999. pp. 3–18. [Google Scholar]
- 22.Powell N G, Threlfall E J, Chart H, Rowe B. Subdivision of Salmonella enteritidis PT4 by pulsed-field gel electrophoresis: potential for epidemiological surveillance. FEMS Microbiol Lett. 1994;119:193–198. doi: 10.1111/j.1574-6968.1994.tb06888.x. [DOI] [PubMed] [Google Scholar]
- 23.Ridley A M, Threlfall E J, Rowe B. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1998;36:2314–2321. doi: 10.1128/jcm.36.8.2314-2321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Stanley J, Baquar N. Phylogenetics of Salmonella enteritidis. Int J Food Microbiol. 1994;21:79–87. doi: 10.1016/0168-1605(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 26.Threlfall E J, Hampton M D, Chart H, Rowe B. Use of plasmid profile typing for surveillance of Salmonella enteritidis phage type 4 from humans, poultry and eggs. Epidemiol Infect. 1994;112:25–31. doi: 10.1017/s0950268800057381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall P G, Ward L R. Epidemiology of Salmonella enterica serovar Enteritidis phage type 4 in England and Wales. In: Saeed A M, editor. Salmonella enterica serovar Enteritidis in humans and animals. Ames: Iowa State University Press; 1999. pp. 19–25. [Google Scholar]
- 29.Ward L R, de Sa J D, Rowe B. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward L R, Threlfall J, Smith H R, O'Brien S J. Salmonella enteritidis epidemic. Science. 2000;287:1753–1754. doi: 10.1126/science.287.5459.1753d. [DOI] [PubMed] [Google Scholar]
- 31.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1987. [DOI] [PubMed] [Google Scholar]