Abstract
Background
: Viral diversity presents an ongoing challenge for diagnostic tests, which need to accurately detect all circulating variants. The Abbott Global Surveillance program monitors severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants and their impact on diagnostic test performance.
Objectives
: To evaluate the capacity of Abbott molecular, antigen, and serologic assays to detect circulating SARS-CoV-2 variants, including all current variants of concern (VOC): B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma) and B.1.617.2 (delta).
Study design
: Dilutions of variant virus cultures (B.1.1.7, B.1.351, B.1.429, B.1.526.1, B.1.526.2, B.1.617.1, B.1.617.2, P.1, R.1 and control isolate WA1) and a panel of N = 248 clinical samples from patients with sequence confirmed variant infections (B.1.1.7, B.1.351, B.1.427, B.1.429, B.1.526, B.1.526.1, B.1.526.2, P.1, P.2, R.1) were evaluated on at least one assay: Abbott ID NOW COVID-19, m2000 RealTime SARS-CoV-2, Alinity m SARS-CoV-2, and Alinity m Resp-4-Plex molecular assays; the BinaxNOW COVID-19 Ag Card and Panbio COVID-19 Ag Rapid Test Device; and the ARCHITECT/Alinity i SARS-CoV-2 IgG and AdviseDx IgM assays, Panbio COVID-19 IgG assay, and ARCHITECT/Alinity i AdviseDx SARS-CoV-2 IgG II assay.
Results
: Consistent with in silico predictions, each molecular and antigen assay detected VOC virus cultures with equivalent sensitivity to the WA1 control strain. Notably, 100% of all tested variant patient specimens were detected by molecular assays (N = 197 m2000, N = 88 Alinity m, N = 99 ID NOW), and lateral flow assays had a sensitivity of >94% for specimens with genome equivalents (GE) per device above 4 log (85/88, Panbio; 54/57 Binax). Furthermore, Abbott antibody assays detected IgG and IgM in 94–100% of sera from immune competent B.1.1.7 patients 15–26 days after symptom onset.
Conclusions
: These data confirm variant detection for 11 SARS-CoV-2 assays, which is consistent with each assay target region being highly conserved. Importantly, alpha, beta, gamma, and delta VOCs were detected by molecular and antigen assays, indicating that these tests may be suitable for widescale use where VOCs predominate.
Keywords: SARS-CoV-2, Molecular diagnostics, Variant of concern, Rapid antigen tests, Antibody assays
1. Background
As viruses continue to evolve, diagnostic tests must keep pace to ensure accurate detection of all circulating variants. While assay design can mitigate the impact of viral diversity, assay performance must be continually monitored through variant testing and molecular surveillance. To meet this challenge, the Abbott Global Surveillance program has been tracking the sequences of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants that have emerged throughout the pandemic, including variant of concern (VOC) lineages that have spread globally: B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma), and B.1.617.2 (delta). The B.1.1.7 lineage was first identified in the United Kingdom and carries spike mutations that have been linked to increased transmissibility, including N501Y [1], [2], [3]. The B.1.351 lineage was first identified in South Africa and has since spread globally, with several reports indicating that this variant can escape neutralizing antibodies [4], [5], [6]. The P.1 variant was first reported in Japan as a branch of the B.1.128 lineage, but was subsequently traced back to Brazil. The P.1 variant contains similar mutations in the spike gene as the B.1.1.7 and B.1.351 variants, suggesting a convergence of SARS-CoV-2 spike mutations that may increase transmissibility and risk of re-infection [7]. Similarly, the B.1.617.2 lineage first identified in India has been shown to be less susceptible to neutralizing antibodies and is linked to the highest transmissibility increases worldwide, likely due to the presence of L452R and P681R mutations in the spike protein [8], [9], [10]. Additional lineages have been identified as variants of interest (VOI) or under the ‘monitoring’ category by the World Health Organization (WHO) due to the presence of identical or similar mutations present in VOCs and increasing prevalence. While spike gene mutations are key hallmarks of VOC/VOI lineages, the presence of additional mutations throughout other regions of the genome warrant further examination for potential impact on diagnostic assay performance. With diagnostic tests designed to target viral regions that are expected to be well conserved, continued evaluation of those assumptions by sequence analysis and specimen testing is critical to ensure that diagnostic tests continue to keep pace with viral evolution throughout the SARS-CoV-2 pandemic.
2. Objective
The goal of this study was to evaluate the performance of Abbott SARS-CoV-2 molecular, antigen, and serological assays to detect SARS-CoV-2 variants and variant-specific antibodies.
3. Study design
In silico analysis – Sequences containing lineage-defining mutations were obtained from GISAID [11] for in silico analysis by customized application of the NextClade tool (clades.nextstrain.org) to compare individual assay target sequences for Abbott ID NOW COVID-19, m2000 RealTime SARS-CoV-2, Alinity m SARS-CoV-2, and Alinity m Resp-4-Plex molecular assays; the BinaxNOW COVID-19 Ag Card and Panbio COVID-19 Ag Rapid Test Device; and the ARCHITECT/Alinity i SARS-CoV-2 IgG and AdviseDx IgM assays, Panbio COVID-19 IgG assay, and ARCHITECT/Alinity i AdviseDx SARS-CoV-2 IgG II assay. Mismatches were considered significant if they were present in >75% of lineage sequences and reduced the target region nucleotide identity or amino acid homology to less than 90%.
Virus cultures – A panel of virus cultures were received from BEI Resources: B.1.1.7 (NR-54,011), B.1.351 (NR-54,008, NR-54,009), P.1 (NR-54,982), B.1.617.1 (NR-55,486), B.1.617.2 (NR-55,611), and the A lineage USA-WA1/2020 control (NR-52,881). Virus cultures were also generated by inoculating Vero cells with remnant patient specimens for the following lineages: R.1, B.1.429, B.1.1.7, B.1.526.1, B.1.617.1, and B.1.617.2. Culturing conditions were followed as previously described [12]. All virus stocks were titered using a calibrated, quantitative research lab developed test (LDT) on the m2000 instrument to calculate genome equivalents per ml (GE/ml) as previously described [13].
Remnant patient specimens - Anonymized remnant patient nasopharyngeal, oropharyngeal, mid-turbinate, and nasal swab samples in viral transport media (VTM) or saline were collected at the point of discard from patients receiving care at Guys’ and St Thomas’ Hospital in London (UK Research Ethics Committee 20/SC/0310), Montefiore Medical Center (Einstein IRB, 2018–9587), Medical University of South Carolina (IRB-III, Medical University of South Carolina, Pro00107968), Rush University Medical Center (Rush IRB, 20,120,410-IRB05), University of California San Francisco (UCSF Institutional Review Board, 11–05,519), Universidade Federal do Rio de Janeiro (CAAE, 30,161,620.0.0000.5257), Institut de Recherche en Santé de Surveillance Epidémiologique et de Formation (Ethical Committee of the Ministry of Health of Senegal 000,129/MSAS/CNERS), and University of Kwa-Zulu Natal (University of KwaZulu–Natal Biomedical Research Ethics Committee, BREC/00,001,510/2020). A panel of remnant viral transport media (VTM) specimens collected in Florida was also purchased from SLR (BioMed IRB, 06,012,006). All specimens were initially positive for SARS-CoV-2 by an RT-PCR assay and subsequently titered using a calibrated, quantitative research assay on the m2000 instrument to calculate genome equivalents per ml (GE/ml) [13] with the exception of a panel of N = 31 South African specimens that were exclusively tested with the Seegene Allplex 2019-nCoV qualitative assay and have a neat viral load reported as a cycle threshold (Ct) value.
Next generation sequencing and variant classification – Specimens were sequenced using one of three previously described methods: (1) the ARTIC protocol [14] on Nanopore instruments, (2) unbiased metagenomic library amplification with custom xGen probe enrichment and sequencing on Illumina instruments [15], (3) metagenomic enrichment sequencing using spiked primers on Illumina instruments [16]. Genomes were deposited into GISAID with references indicated in Supplemental Table 1, analyzed as described in the in silico section above, and assigned lineage classifications by the Pangolin tool [17].
Molecular, antigen, and antibody testing – For molecular experiments, virus cultures and clinical specimens were heat inactivated at 65 °C for 30 min and then diluted in PBS or Alinity m multi-Collect solution (Abbott Molecular Diagnostics, part number 09N19–001) prior to testing. Specimen and virus culture samples were tested according to the manufacturer's instructions for m2000 RealTime SARS-CoV-2, Alinity m SARS-CoV-2, and Alinity m Resp-4-Plex molecular assays. For ID NOW testing, specimens and virus culture samples were directly loaded into the sample receiver (50–200 μl).
For rapid antigen testing, virus cultures and clinical specimens were tested without any inactivation in a biosafety level 3 laboratory. Dilutions were prepared in assay elution buffer or storage buffer. In Panbio experiments, samples were directly pipetted onto the device (1–50 #x03BC;l), followed immediately by 5 drops of elution buffer. For BinaxNOW experiments, 4 drops of elution buffer were placed in the card prior to addition of sample (1–50 μl) and insertion of the swab. All subsequent steps were followed for each test according to the manufacturer's instructions.
All serological testing was conducted with patient sera according to the manufacturer's instructions for ARCHITECT/Alinity i SARS-CoV-2 IgG and AdviseDx IgM assays, Panbio COVID-19 IgG assay, and ARCHITECT/Alinity i AdviseDx SARS-CoV-2 IgG II assay.
4. Results
In silico analysis of 746,345 sequences from 45 SARS-CoV-2 lineages revealed no significant mutations of concern for the performance of Abbott assays targeting the RNA-dependent RNA polymerase (RDRP) region (ID NOW COVID-19, m2000 RealTime SARS-CoV-2, Alinity m SARS-CoV-2, Alinity m Resp-4-Plex), nucleocapsid region (m2000 RealTime SARS-CoV-2, Alinity m SARS-CoV-2, Alinity m Resp-4-Plex, BinaxNOW COVID-19 Ag Card, Panbio COVID-19 Ag Rapid Test Device, ARCHITECT/Alinity i SARS-CoV-2 IgG, and Panbio COVID-19 IgG), and spike region (ARCHITECT/Alinity i AdviseDx SARS-CoV-2 IgM, ARCHITECT/Alinity i AdviseDx SARS- CoV-2 IgG II) (Supplemental Table 2). To evaluate these predictions for Abbott's high throughput molecular assays, dilution series of heat-inactivated B.1.1.7, B.1.351, and P.1 virus cultures were tested on m2000 RealTime SARS-CoV-2, Alinity m SARS-CoV-2, Alinity m Resp-4-Plex (Table 1 ). All dilutions were detected in the expected ranges previously observed with other strains in both tissue culture infectious dose (TCID50) and genome equivalents (GE) units [18, 19]. However, considerable variability was observed in the ratio between GE quantitated with a standard curve and the calculated median TCID50 for each virus isolate stock (Table 2 ). Notably, the GE/TCID50 ratios were 23 to 102-fold higher on average for the B.1.1.7 culture than either of the B.1.351 stocks or the P.1 stock. Therefore, GE units are more reliable for comparisons between virus culture stocks and were utilized for all subsequent experiments.
Table 1.
m2000 and Alinity m SARS-CoV-2 assay results with B.1.1.7, B.1.351, and P.1 virus culture dilutions.
| Sample ID+ | TCID50/mL | Log GE/mL* | RealTime SARS-CoV-2 | Alinity m SARS-CoV-2 | Alinity m Resp-4-Plex | |
|---|---|---|---|---|---|---|
| B.1.1.7 variant | BEI NR-54,011 Dilution 1 | 28 | 6.25 | Detected | Detected | Detected |
| BEI NR-54,011 Dilution 2 | 2.8 | 5.19 | Detected | Detected | Detected | |
| BEI NR-54,011 Dilution 3 | 0.28 | 4.00 | Detected | Detected | Detected | |
| BEI NR-54,011 Dilution 4 | 0.028 | 2.85 | Detected | Detected | Detected | |
| B.1.351 variant | BEI NR-54,008 Dilution 1 | 28 | 4.63 | Detected | Detected | Detected |
| BEI NR-54,008 Dilution 2 | 2.8 | 3.83 | Detected | Detected | Detected | |
| BEI NR-54,008 Dilution 3 | 0.28 | 2.51 | Detected | Detected | Detected | |
| BEI NR-54,008 Dilution 4 | 0.028 | 1.27 | Detected | Detected | Detected | |
| BEI NR-54,009 Dilution 1 | 28 | 4.35 | Detected | Detected | Detected | |
| BEI NR-54,009 Dilution 2 | 2.8 | 3.27 | Detected | Detected | Detected | |
| BEI NR-54,009 Dilution 3 | 0.28 | 2.11 | Detected | Detected | Detected | |
| BEI NR-54,009 Dilution 4 | 0.028 | 0.73 | Detected | Detected | Detected | |
| P.1 variant | BEI NR-54,982 Dilution 1 | 28 | 4.56 | Detected | Detected | Detected |
| BEI NR-54,982 Dilution 2 | 2.8 | 3.45 | Detected | Detected | Detected | |
| BEI NR-54,982 Dilution 3 | 0.28 | 2.39 | Detected | Detected | Detected | |
| BEI NR-54,982 Dilution 4 | 0.028 | 1.32 | Detected | Detected | Detected |
Log genome equivalents (GE)/mL were calculated from a standard curve plot of TCID50 vs GE with an R2 value of 0.99. Four out of four replicates were detected for all dilutions tested.
Table 2.
Comparison of GE*/TCID50 Ratios.
| B.1.1.7 variant(NR-54,011) | B.1.351 variant (NR-54,008) | B.1.351 variant (NR-54,009) | P.1 variant(NR-54,982) | Fold difference in GE/TCID | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dilution | TCID50/mL | Mean log GE/mL | GE/TCID50 ratio | Mean log GE/mL | GE/TCID50 ratio | Mean log GE/mL | GE/TCID50 ratio | Mean log GE/mL | GE/TCID50 ratio | RatioNR-54,011/ NR-54,008 | Ratio NR-54,011/ NR-54,009 | Ratio NR-54,011/ NR-54,982 |
| 1 | 28 | 6.25 | 65,189 | 4.63 | 1540 | 4.35 | 809 | 4.56 | 1296 | 42 | 81 | 50 |
| 2 | 2.8 | 5.19 | 55,963 | 3.83 | 2454 | 3.27 | 672 | 3.45 | 1020 | 23 | 83 | 55 |
| 3 | 0.28 | 4.00 | 36,169 | 2.51 | 1193 | 2.11 | 471 | 2.39 | 913 | 30 | 77 | 40 |
| 4 | 0.028 | 2.85 | 25,294 | 1.27 | 674 | 0.73 | 248 | 1.32 | 755 | 38 | 102 | 33 |
| Average | NA | NA | 45,654 | NA | 1465 | NA | 550 | NA | 996 | 31 | 83 | 46 |
*Genome equivalents (GE)/mL were calculated from a standard curve plot of TCID50 vs GE with an R2 value of 0.99.
To further evaluate core lab molecular assays for performance with variant lineages, a control virus culture (lineage A, WA1) and B.1.526.1, B.1.526.2, B.1.617.1, B.1.617.2, P.1, and R.1 virus culture stocks were diluted to 4–8 log GE/ml and tested on the m2000 and Alinity m assays (Supplemental Table 3). Consistent with variant detection for B.1.1.7, B.1.351, and P.1 lineages in Table 1, all additional virus cultures were also detected by m2000 and Alinity m (Supplemental Table 3). Furthermore, serial dilutions in the range of 2.5–4 log GE/test (approximately Ct 26–31) were tested on ID NOW for all variant virus cultures and the lineage A control. All variant virus cultures and the control strain were detected at all dilutions tested with ID NOW, indicating excellent variant detection as low as 2.5 log GE/test with heat inactivated virus stocks (Supplemental Table 3).
To evaluate variant detection by rapid antigen assays, virus culture dilution series (B.1.1.7, B.1.351, B.1.429, B.1.526.1, B.1.526.2, B.1.617.1, B.1.617.2, P.1, R.1, and WA1) ranging from 4 to 6 log GE/test (approximately Ct 19–25) were prepared without inactivation and tested on Panbio and BinaxNOW. Variant virus cultures were detected with equivalent sensitivity as the WA1 control for both Panbio and BinaxNOW at 4.5 log GE/test or lower. These results are consistent with recent Panbio Ag evaluations of the B.1.1.7 and B.1.351 lineages [20, 21] and BinaxNOW evaluation of the B.1.1.7, B.1.351, P.1, and B.1.617.2 lineages [22].
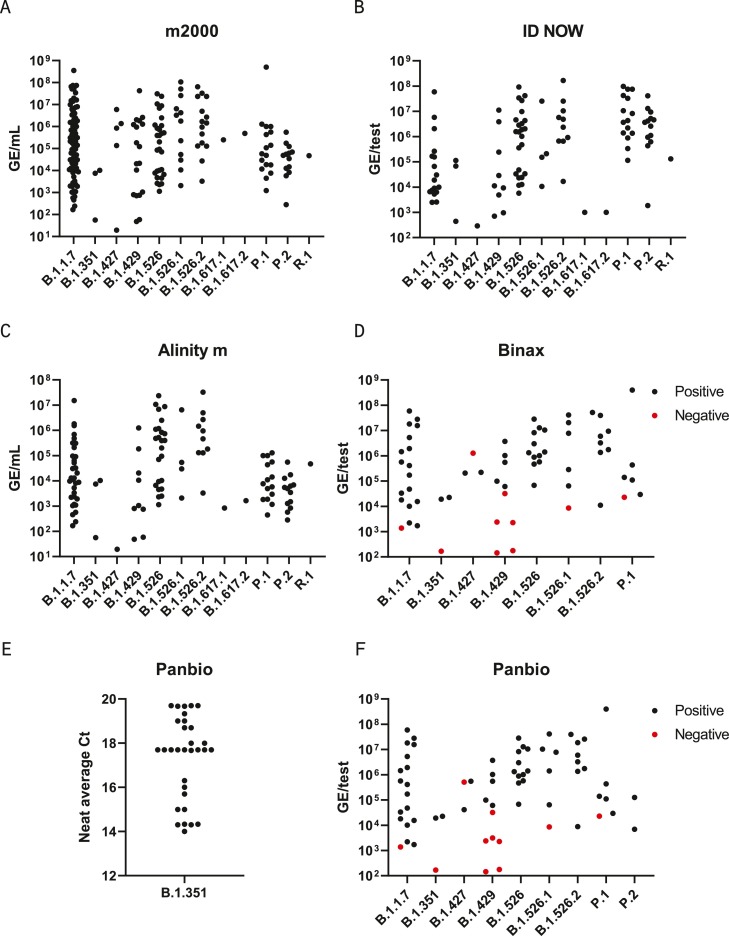
Further evaluation was performed with a panel of N = 228 remnant clinical upper respiratory specimens with sequence-confirmed VOC/VOI lineages present. Remnant specimens were collected in the United States (New York, Illinois, Wisconsin, Florida, South Carolina, California), Brazil, Senegal, United Kingdom, and South Africa for sequencing by viral whole genome next generation sequencing (NGS) before quantitation with the calibrated m2000 assay (N = 197) or a qualitative RT-PCR assay (N = 31) (Fig. 1 ). The full panel consisted of 12 variant lineages, including all four VOCs (Fig. 1). Due to limited sample volume, most specimens were diluted before molecular and antigen testing and many did not have sufficient volume for testing on every assay. Furthermore, specimens that had been previously inactivated at the collection site were excluded from antigen testing due to destruction of the nucleocapsid structure. Amongst the samples comprising the variant clinical specimen panel, N = 197 were tested with m2000, N = 116 were tested with Alinity m, and N = 99 were tested with ID NOW. Each molecular assay detected 100% of all variants tested across a wide range of viral loads (Fig. 2 ). Likewise, most variant specimens with >4 log GE/test were detected by both antigen assays, with Panbio detecting 85/88 (96.6%) and BinaxNOW detecting 54/57 (94.7%) above 4 log GE/test. Within the subset panel of N = 31 B1.351 specimens with qualitative viral loads available, 100% were detected by Panbio after 2–15 fold dilution. Three specimens (B.1.427, B.1.429, B.1.526.1) were missed by both antigen assays, although variant mutations are unlikely to be the cause since other specimens in these lineages were detected at similar or lower viral loads and no mutations were present in the assay target regions. Two of the three were near the 4 log GE/test level and the third was tested at a viral load of approximately 6 log GE/test after multiple freeze/thaw cycles, which may have impacted the integrity of the specimen.
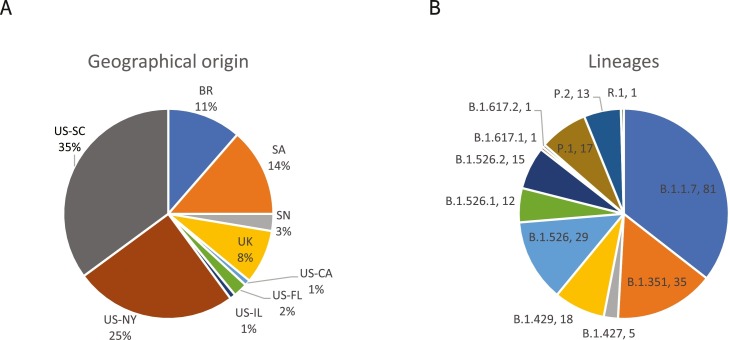
Fig. 1.
Characteristics of clinical specimen panel. The percentage of N = 228 total remnant respiratory samples collected from each of the indicated countries (country two letter codes, BR – Brazil, SA – South Africa, SN – Senegal, UK – United Kingdom, US – United States) and US states (CA – California, FL – Florida, IL – Illinois, NY – New York, SC – South Carolina) are shown in panel A. The total number of specimens (N) for each lineage in the panel of 228 remnant respiratory samples are shown in panel B.
Fig. 2.
Clinical variant specimen panel. Remnant clinical specimens from patients with sequence-confirmed VOI/VOC infections were tested on at least one diagnostic assay –m2000 RealTime SARS-CoV-2 (A), Abbott ID NOW COVID-19 (B), Alinity m SARS-CoV-2 (C), the BinaxNOW COVID-19 Ag Card (D) and Panbio COVID-19 Ag Rapid Test Device (E-F). Genome equivalents (GE).
To evaluate IgG and IgM antibody assays, a panel of remnant sera were collected for 20 of the B.1.1.7 infections that had already been sequenced and tested on molecular and antigen tests (Table 3 ). All sera were collected 15–26 days post symptom onset, and three immunocompromised patients (#2, #4, and #10) were included in the panel. Antibodies for patients #2 and #4 were detected by four assays (ARCHITECT and Alinity i IgM and IgG II), but not by ARCHITECT, Alinity i, or Panbio IgG assays. Patient #10 had cancer and did not have a detectable antibody response at 15 days post symptom onset by any assay. Excluding immunocompromised patients, antibodies were detected in 100% (17/17) of patients by ARCHITECT/Alinity i IgM and IgG II assays and were detected in 94% (16/17) of patients by ARCHITECT/Alinity i, and Panbio IgG assays (Table 3).
Table 3.
SARS-CoV-2 Serology Assay Results with B.1.1.7 Clinical Samples.
| Sample ID | ARCH IgM (Index)+ | Alinity IgM (Index)+ | ARCH IgG (Index)+ | Alinity IgG (Index)+ | ARCH IgG II (AU/mL)+ | Alinity IgG II (AU/mL)+ | Panbio IgG Result | Days post symptom onset |
|---|---|---|---|---|---|---|---|---|
| Patient 1 | 18.36 | 18.68 | 6.76 | 6.97 | 13,359.5 | 11,595.4 | Positive | 26 |
| Patient 2 | 1.22 | 1.26 | 0.22 | 0.22 | 178.5 | 189.5 | Negative | 20 |
| Patient 3 | 11.68 | 12.05 | 5.62 | 6.01 | 5288.1 | 6570.1 | Positive | 19 |
| Patient 4 | 11.71 | 11.04 | 0.8 | 0.87 | 149.4 | 153.7 | Negative | 25 |
| Patient 5 | 6.47 | 6.58 | 5.29 | 5.44 | 5472.9 | 5468.1 | Positive | 20 |
| Patient 6 | 1.31 | 1.32 | 6.6 | 6.58 | > 40,000.0 | > 40,000.0 | Positive | 16 |
| Patient 7 | 14.16 | 13.51 | 6.8 | 6.98 | 12,034.7 | 11,191.5 | Positive | 22 |
| Patient 8 | 7.41 | 7.2 | 6.39 | 6.36 | 14,489.9 | 12,718.3 | Positive | 21 |
| Patient 9 | 15.15 | 14.86 | 4.73 | 4.82 | 2745.6 | 2603.7 | Positive | 20 |
| Patient 10 | 0.03 | 0.04 | 0.02 | 0.02 | 0.9 | 0.2 | Negative | 15 |
| Patient 11 | 1.27 | 1.22 | 0.22 | 0.23 | 186.4 | 186.8 | Negative | 20 |
| Patient 12 | 26.79 | 26.67 | 6.62 | 6.6 | 22,392.8 | 19,367.7 | Positive | 23 |
| Patient 13 | 5.81 | 6.06 | 4.31 | 4.33 | 1255.4 | 1183.3 | Positive | 19 |
| Patient 14 | 88.25 | 88.59 | 6.67 | 6.42 | 6811.4 | 6250 | Positive | 16 |
| Patient 15 | 88.27 | 88.14 | 6.66 | 6.58 | 6815.2 | 6587.5 | Positive | 21 |
| Patient 16 | 88.87 | 89.11 | 6.39 | 6.5 | 6924.4 | 6401.6 | Positive | 18 |
| Patient 17 | 2.09 | 2.2 | 6.23 | 6.19 | 1626.4 | 1630.5 | Positive | 17 |
| Patient 18 | 91.31 | 87.56 | 6.62 | 6.55 | 7459.3 | 6330 | Positive | 19 |
| Patient 19 | 4.56 | 4.58 | 7.36 | 7.31 | 2345.6 | 2274.9 | Positive | 19 |
| Patient 20 | 4.55 | 4.61 | 7.18 | 7.53 | 2491.1 | 2236.1 | Positive | 20 |
An index value of ≥1.4 is positive for IgG assays, ≥1.0 is positive for IgM assays, a result of ≥50 AU/ml per mL is positive and <22 AU/mL is not reported in a clinical setting for IgG II assays.
5. Discussion
Viral diversity will continue to challenge diagnostic tests as new SARS-CoV-2 strains arise and spread globally. To prevent potentially undetected outbreaks of new variant lineages, regular evaluation of diagnostic tests must be conducted to challenge assays with emerging and widely circulating strains, especially those that have been linked to increased transmissibility and immune escape [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. While in silico analyses did not predict impact to detection by Abbott molecular, antigen, and antibody assays for any of the 45 lineages examined, these predictions must be continually tested. The data herein confirmed that Abbott COVID-19 molecular, antigen, and serological assays effectively detect circulating SARS-CoV-2 VOC/VOI infections and B.1.1.7-specific antibodies.
Curation of specimen panels for SARS-CoV-2 diagnostic assay variant performance evaluations comes with many challenges. In this study, we have utilized virus cultures and frozen remnant clinical specimens, which each have limitations. Neither are the intended sample type for rapid molecular or antigen tests, yet they are the only suitable option since primary fresh samples have not been sequenced yet to identify whether a variant is present. However, freeze/thaw cycles and extended storage conditions reduce intact virus levels in sequenced specimens. Antigen tests utilizing lateral flow technology, such as Panbio and BinaxNOW, are especially sensitive to the impacts of specimen storage because they rely on complete solubilization of a sample with an intact antigen, which can easily be impacted by components of storage media and aggregation during freeze/thaw cycles. Furthermore, inactivation procedures, such as heat or addition of detergents, have been shown to impact nucleocapsid detection [12]. Fortunately, the effects of storage conditions can be compensated by a second viral load measurement at the time of variant performance testing. This approach was taken in the present study and confirmed differences between initial diagnostic Ct and post-sequencing Cts, although the qualitative molecular result remained positive at both measurements [23]. Even after compensating for changes in viral load, specimen integrity can still impact performance if nucleocapsid antigen is compromised, which is not accounted for in a viral load measurement. For instance, three specimens of three different lineages were negative at above a threshold of 4 log GE/test on lateral flow tests despite detection of other clinical specimens from these lineages (B.1.427, B.1.429, and B.1.526.1) at equivalent or lower viral loads (Fig. 2) Therefore, the loss of detection for these three specimens was likely due to specimen integrity issues and not any of the mutations present in these lineages. Detection of the B.1.429 and B.1.526.1 virus cultures at 4.5 log GE/test by both Panbio and Binax and complete sequence conservation in the assay target regions also supports this conclusion.
The effects of storage conditions are easier to control with contrived samples, such as virus cultures. However, a caveat of utilizing virus cultures is the potential introduction of new mutations that are selected during culture. To avoid this issue in our study, virus stocks were only used after 1 passage in cell culture and sequences were confirmed in the genes targeted by each assay. Virus culture conditions can also result in widely different titers for each stock as demonstrated in Table 2, making standardization of units for comparing stocks very critical.
Variant-specific antibody specimens are especially difficult to evaluate due to a lack of contrived samples and the rarity of sequenced infections that are linked with blood sampling after recovery. Even in cases when variant infections have been linked to followup blood collections, the potential for pre-existing antibodies to prior non-variant infections or vaccination cannot always be ruled out. Nonetheless, antibody assays that utilize a recombinant antigen with multiple epitopes are less prone to the potential impact of viral diversity due to the redundant nature of polyclonal immune responses as illustrated by the detection of spike antibodies in the sera of B.1.1.7 patients (Table 3). This observation is consistent with recent studies reporting that polyclonal antibodies in convalescent plasma bind the spike protein of SARS-CoV-2 B1.1.7 and B.1.351 variants with similar binding kinetics despite variable capacity to neutralize the virus [24, 25]. Future studies must be conducted to evaluate antibody detection in patients who have recovered from other variant infections besides B.1.1.7.
Variant lineages will continue to emerge as the SARS-CoV-2 pandemic progresses, with the pace at which they arise dependent upon the total number of infections and the mutation rate. For context, SARS coronaviruses maintain a higher rate of replication fidelity during replication due to an encoded proofreading mechanism in their RDRP [26]. As a result, SARS-CoV-2 variants are typically defined by a pattern of mutations totaling <30 nucleotides (0.1% divergence), which is much lower than the divergence of 30% or more between HCV genotypes, for example [11, 27, 28]. Just as viral surveillance has ensured that diagnostic tests for divergent viruses like HIV and HCV keep pace with diverse circulating strains, continued monitoring of SARS-CoV-2 variants should also serve as a proactive mechanism for maintaining a high level of accuracy for diagnostic tests.
Declaration of Competing Interest
MAR, AO, BH, CL, XL, MGB, TVM, AM, GSO, and GC are employees and shareholders of Abbott Laboratories.This work was funded by Abbott Laboratories.
Acknowledgements
This study was funded by Abbott Laboratories. We thank Dr Stephen Kovacs, Dr Richard Roth, Dr Shihai Huang, Ana Vallari, Dr Dan Toolsie, Kara Nordin, Dr Luis Gonzales, Dr Carsten Buenning, Xuan Yue, Antonio Covelli, Frank Mazur, Dylan Johnston, and Dr Jim Badciong for technical support in conducting the study.The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/PHC658/2021 (Lineage B.1.617.2; Delta Variant), NR-55611, contributed by Dr. Richard Webby and Dr. Anami Patel.The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/PHC658/2021 (Lineage B.1.617.2; Delta Variant), NR-55611, contributed by Dr. Richard Webby and Dr. Anami Patel.The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/CA-Stanford-15_S02/2021 (Lineage B.1.617.1; Kappa Variant), NR-55486, contributed by Dr. Mehul Suthar and Dr. Benjamin Pinsky.The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/South Africa/KRISP-K005325/2020, NR-54009, contributed by Alex Sigal and Tulio de Oliveira.The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/South Africa/KRISP-EC-K005321/2020, NR-54008, contributed by Alex Sigal and Tulio de Oliveira.The following reagent was deposited by Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA/CA_CDC_5574/2020, NR-54011.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105080.
Appendix. Supplementary materials
References
- 1.Investigation of novel SARS-COV-2 variant Variant of concern 202012/01. Public Health England. 2020 [Google Scholar]
- 2.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. MedRxiv. 2021 [Google Scholar]
- 3.Faria N.R., Rambaut A., Suchard M.A., Baele G., Bedford T., Ward M.J., et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond M., Chen R., Xie X., Case J., Zhang X., VanBlargan L., et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res. Sq. 2021 doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv. 2021 [Google Scholar]
- 6.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 [Google Scholar]
- 7.Faria N., Claro I.M., Candido D., Franco L.A.M., Andrade P.S., Coletti T.M., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. 2021.
- 8.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. EuroSurveillance. 2021:26. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B1.617 by vaccine and convalescent serum. Cell. 2021 doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 11.Elbe S., Data Buckland-Merrett G. disease and diplomacy: gISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg M.G., Israeli E., Quaco E., Cloherty G.A., Hemken P.M. Tergitol-15-S-9 inactivates SARS-CoV-2 and boosts immunoassay signals. BioTechniques. 2021;71:370–375. doi: 10.2144/btn-2021-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg M.G .ZW., Lucic D., Degli-Angeli E.J., Anderson M., Forberg K., Olivo A., Sheikh F., Toolsie D., Greninger A.L., Cloherty G.A., Coombs R.W., Berry G.J. Development of the RealTime SARS-CoV-2 quantitative Laboratory Developed Test and correlation with viral culture as a measure of infectivity. J. Clin. Virol. 2021 doi: 10.1016/j.jcv.2021.104945. Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Artic Network. SARS-CoV-2 (2020). https://artic.network/ncov-2019.
- 15.Forberg K.O.G, Meyer T.V., Mowerman I., Mohaimani A., Faron M., Jennings C., Landay A.L., Goldstein Y., Fox A., Berg M.G., and Cloherty G.A. SNP and Phylogenetic Characterization of Low Viral Load SARS-CoV-2 Specimens By Target Enrichment. Front Vir. 2021.
- 16.Deng X., Gu W., Federman S., du Plessis L., Pybus O.G., Faria N.R., et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Áine O'Toole ES, Anthony Underwood, Ben Jackson, Verity Hill, J.T. McCrone, Chris Ruis, Khali Abu-Dahab, Ben Taylor, Corin Yeats, Louis du Plessis, David Aanensen, Eddie Holmes, Oliver Pybus, Andrew Rambaut. pangolin: lineage assignment in an emerging pandemic as an epidemiological tool. githubcom/cov-lineages/pangolin](githubcom/cov-lineages/pangolin. 2021. [DOI] [PMC free article] [PubMed]
- 18.Fung B., Gopez A., Servellita V., Arevalo S., Ho C., Deucher A., et al. Direct Comparison of SARS-CoV-2 Analytical Limits of Detection across Seven Molecular Assays. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilarowski G., Lebel P., Sunshine S., Liu J., Crawford E., Marquez C., et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiaa802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SARS-CoV-2 Lateral Flow Antigen Tests Evaluation of VUI-20201201. Public Health England. 2020 [Google Scholar]
- 21.Akingba O.L., Sprong K., Hardie D.R. Field performance evaluation of the PanBio rapid SARS-CoV-2 antigen assay in an epidemic driven by 501Y.v2 (lineage B.1.351) in the Eastern Cape, South Africa. medRxiv. 2021 doi: 10.1016/j.jcvp.2021.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frediani J.K., Levy J.M., Rao A., Bassit L., Figueroa J., Vos M.B., et al. Multidisciplinary assessment of the Abbott BinaxNOW SARS-CoV-2 point-of-care antigen test in the context of emerging viral variants and self-administration. Sci. Rep. 2021;11:14604. doi: 10.1038/s41598-021-94055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skalina K.A., Goldstein D.Y., Sulail J., Hahm E., Narlieva M., Szymczak W., et al. Extended storage of SARS-CoV-2 nasopharyngeal swabs does not negatively impact results of molecular-based testing across three clinical platforms. J. Clin. Pathol. 2020 doi: 10.1136/jclinpath-2020-206738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., et al. Sensitivity of infectious SARS-CoV-2 B1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021 doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 26.Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodcroft E. 2021.
- 28.Petruzziello A., Marigliano S., Loquercio G., Cozzolino A., Cacciapuoti C. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World J. Gastroenterol. 2016;22:7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.