Abstract
Chemotaxonomic and genetic properties were determined for 14 mycobacterial isolates identified as members of a newly described species Mycobacterium bohemicum. The isolates recovered from clinical, veterinary, and environmental sources were compared for lipid composition, biochemical test results, and sequencing of the 16S ribosomal DNA (rDNA) and the 16S-23S rDNA internal transcribed spacer (ITS) regions. The isolates had a lipid composition that was different from those of other known species. Though the isolates formed a distinct entity, some variations were detected in the features analyzed. Combined results of the phenotypic and genotypic analyses were used to group the isolates into three clusters. The major cluster (cluster A), very homogenous in all respects, comprised the M. bohemicum type strain, nine clinical and veterinary isolates, and two of the five environmental isolates. Three other environmental isolates displayed an insertion of 14 nucleotides in the ITS region; they also differed from cluster A in fatty alcohol composition and produced a positive result in the Tween 80 hydrolysis test. Among these three, two isolates were identical (cluster B), but one isolate (cluster C) had a unique high-performance liquid chromatography profile, and its gas liquid chromatography profile lacked 2-octadecanol, which was present in all other isolates analyzed. Thus, sequence variation in the 16S-23S ITS region was associated with interesting variations in lipid composition. Two of the isolates analyzed were regarded as potential inducers of human or veterinary infections. Each of the environmental isolates, all of which were unrelated to the cases presented, was cultured from the water of a different stream. Hence, natural waters are potential reservoirs of M. bohemicum.
In the past few years, species descriptions of several unclassified mycobacteria have been published, and more new species will certainly be identified in the future. In 1998, a novel species, Mycobacterium bohemicum, isolated from a patient with Down's syndrome and tuberculosis, was described by Reischl and coworkers (11). Only one additional report on this species has been presented so far (17). Despite its recent discovery, M. bohemicum seems to be more common than many of the other newly classified species. From 1989 through 1996, we isolated unidentifiable mycobacteria from specimens taken from four human patients and one goat. Isolates shown to be similar by means of gas liquid chromatography (GLC) analyses of cellular fatty acid and alcohol composition and biochemical testing (12) were also detected among environmental isolates grown from water taken from Finnish streams in 1990 (4). These isolates have recently been verified as M. bohemicum by partial sequencing of the 16S rRNA gene. In 1999, an additional three clinical isolates were identified as M. bohemicum.
We describe here the characteristics of Finnish M. bohemicum isolates originating from clinical, veterinary, and environmental sources, together with some descriptions of the sources of the isolates. Some minor but interesting differences were detected between the clinical and environmental isolates, and they are also discussed in this paper.
CASE REPORTS
Case report 1.
An 85-year-old farmer's wife, without an earlier history of skin disease, was admitted to the department of dermatology due to a chronic skin disorder of unknown origin. The first skin lesions had appeared on the dorsal sides of her hands approximately 1.5 years earlier and soon spread to cover both arms, legs, and the trunk. The eczema was resistant to the topical treatments that were given, including corticosteroids. When entering the hospital, she had nummular lesions with localized conglomerations covering most parts of her skin. Among these lesions, discrete scratched noduli were observed. Some of the lesions had bluish discoloration. Tentative clinical diagnoses included eczema nummulare, sarcoidosis, and livedo reticularis. Biopsy specimens taken for histopathological examinations in 1989 and 1990 (10) showed ulcerative dermatitis. No granuloma formation was detected, but two acid-fast bacilli were discovered in acid-fast staining. A parallel biopsy submitted for cultivation of mycobacteria grew M. bohemicum (1949/90) abundantly. Her chest radiography indicated no inflammatory changes suggestive of mycobacterial infection. The patient soon succumbed to cerebrovascular disturbances, and no antimycobacterial therapy was administered. M. bohemicum was regarded as a potential inducer of her skin disease; however, this possibility remained unverified owing to the rapid death of the patient.
Case report 2.
A 2-year-old female Rocky Mountain goat, which had lived its whole life in a zoo, was euthanized due to an acute total lameness of the right forelimb and severe disability of the right elbow joint. In a postmortem examination at the National Veterinary and Food Research Institute, Helsinki, Finland, the goat was found to be in a good bodily condition. There were bruises around the right shoulder, and the capsule of the right elbow joint was discovered to be ruptured. Some mesenteric lymph nodes were found to be slightly enlarged, and on the cut surface of the enlarged areas, several 1- to 4-mm-diameter grayish foci were detected by the naked eye. Histopathological stainings verified granulomatous lymphadenitis with numerous acid-fast bacilli. M. bohemicum grew in culture on Löwenstein-Jensen medium. It was concluded that the goat had verified mycobacterial mesenteric lymphadenitis due to M. bohemicum at the time of the traumatic disease, which led to its euthanasia.
MATERIALS AND METHODS
Bacterial strains.
The clinical isolates of 1989 to 1993 (21910/89, 1949/90, HO365/91, and 926/93) were recovered from sputum (n = 3) and skin biopsy (n = 1) specimens. The specimens were processed by conventional procedures and cultured in egg-based media, and biopsy specimens were also cultured in liquid media as described in detail earlier (10). More recent isolates (2267/99, 7981/99, and 3739-2/99) were grown in the MGIT 960 system (Becton Dickinson Microbiology Systems, Cockeysville, Md.) as described elsewhere in detail (6). The veterinary isolate (3448/96) from a specimen taken during the postmortem biopsy of a mesenterial lymph node of a Rocky Mountain goat (Oreamnos americanus) at the National Veterinary and Food Research Institute was grown in Löwenstein-Jensen medium. The environmental isolates (E170-2, E445, E590, E743, and E744) were recovered in a study of stream waters, as described in detail by Iivanainen et al. (4). After the initial identification, the isolates were stored in 7H9 broth at −80°C. The M. bohemicum reference strain (DSM 44277T) was kindly provided for the present study by J. Kaustova (Ostrava, Czech Republic). A German clinical isolate (2938/96), which was a gift from L. Naumann (Regensburg, Germany), was also included in the study. The type strain of Mycobacterium interjectum (ATCC 51457T), a species easily confused with M. bohemicum, was also included in the analyses.
Lipid analyses.
The isolates were cultured on mycobacteria 7H11 agar supplemented with Middlebrook OADC enrichment (Difco, Detroit, Mich.) at 30°C (environmental isolates) or 36°C (clinical isolates). Fatty acid methyl esters were produced by acid methanolysis (5). Analyses of lipid composition, including fatty acids, fatty alcohols, and mycolic acid cleavage products, were performed using a Perkin-Elmer (Norwalk, Conn.) AutoSystem gas chromatograph. Peaks were identified using a Hewlett-Packard model G1800A gas chromatograph (Palo Alto, Calif.) equipped with an electron ionization detector as described in detail elsewhere (12).
The cell wall mycolic acids were analyzed by high-performance liquid chromatography (HPLC) using a previously described method (1, 14). The type strain (DSM 44277T) and all isolates except for the three recent ones (2267/99, 7981/99, and 3739-2/99) were subjected to this HPLC analysis. In addition, isolates 926, 1949, E744, and M. interjectum (ATCC 51457T) were analyzed for mycolic acids using one-dimensional thin-layer chromatography (TLC) as described in detail (18).
Biochemical tests.
The isolates were tested at 36°C for urease, arylsulfatase (day 10), pyrazinamidase, semiquantitative catalase, nitrate reduction, and Tween 80 hydrolysis as described earlier (12, 13).
16S rDNA and ITS region sequencing.
Amplification of the complete 16S rRNA gene and sequencing of the amplified DNA fragments were performed as described previously (7). The internal transcribed spacer (ITS) region between 16S and 23S ribosomal DNAs (rDNAs) was amplified, and the amplification products were sequenced as described in detail earlier (13).
Environmental analyses.
Characteristics of the drainage areas and the chemical, physical, and microbiological qualities of the natural water reservoirs from which the environmental M. bohemicum isolates were obtained were analyzed as described in detail earlier (4).
Nucleotide sequence accession numbers.
The ITS region sequence of M. bohemicum type strain DSM 44277T and the 16S rDNA ITS region sequences of the isolates E590 and E743 have been submitted to the EMBL with the accession numbers AJ277282, AJ277283, and AJ277284, respectively.
RESULTS
Lipid analyses.
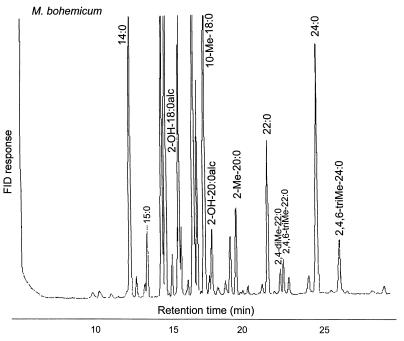
In GLC analysis, the type strain of M. bohemicum is characterized by the presence of 2-octadecanol (2-OH-18:0alc), 2-eicosanol (2-OH-20:0ac), 2-methyl-eicosanoic acid (2-Me-20:0), 2,4-dimethyl-docosanoic acid (2,4-diMe-22: 0), 2,4,6-trimethyl-docosanoic acid (2,4,6-triMe-22:0), and 2,4,6-trimethyl-tetracosanoic acid (2,4,6-triMe-24:0) in addition to the fatty acid markers regarded as typical of mycobacteria (9). This combination of markers is different from the profiles described for other known species. All of the isolates analyzed in this study produced highly similar profiles. However, minor variations were detected, and these variations made it possible to divide the isolates into three GLC clusters. All of the clinical isolates, the veterinary isolate, and two of the five environmental isolates (E445 and E170-2) had a GLC profile that was identical to that of the type strain (Fig. 1) (cluster A). The three other environmental isolates, which were otherwise similar, lacked 2-eicosanol (2-OH-20:0alc) (cluster B), or 2-eicosanol and 2-octadecanol (2-OH-18:0alc) (cluster C [isolate E590]) (Table 1).
FIG. 1.
GLC profile of M. bohemicum DSM 44277T. For definitions of marker designations, see Results.
TABLE 1.
Characteristics of M. bohemicum isolates from various sources compared with type strains of M. bohemicum and M. interjectum
| Clustera and strain or isolate(s) by source (n) | Fatty markers
|
Mycolic acids
|
Pigment | Growth at 42°Cb | Nitrate reductionb | Semiquantitative catalase (45 mm foam)b | Tween hydrolysisb | Arylsulfatase (10 d)b | Ureaseb | Pyrazinamidaseb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18:0alc | 20:0alc | 2-Me-20:0 | 2,4-diMe- 22:0 | 2,4,6-triMe- 22:0 | 2,4,6-triMe- 24:0 | 26:0 | TLC | HPLC | |||||||||
| A. M. bohemicum DSM 44277Tc | 5.3 | 1.8 | 1.7 | 0.6 | 0.9 | 1.7 | 0 | NDd | Typical | Yellow | + | − | + | − | − | + | + |
| A. Clinical isolates (8)e | 5.1 ± 0.8f | 1.4 ± 0.4 | 3.0 ± 1.2 | 1.1 ± 0.5 | 1.6 ± 1.0 | 2.2 ± 0.9 | 0 | α, metoxy, keto, dicarboxy | Typicalg | Yellow | + | v | + | − | v | + | − |
| A. Veterinary isolate (1) | 5.6 | 1.4 | 3.7 | 1.3 | 2.1 | 2.1 | 0 | ND | Typical | Yellow | + | − | + | − | +w | + | − |
| Environmental isolates | |||||||||||||||||
| A. E170-2, E445 | 5.5–6.0h | 0.7–1.4 | 2.9–3.6 | 0.8–1.0 | 1.1–1.2 | 2.0–2.7 | 0 | ND | Typical | Yellow | + | v | + | − | + | + | − |
| B. E743, E744 | 4.5–4.9h | 0 | 1.9–3.1 | 0.2–0.6 | 0.5–1.9 | 1.0–1.5 | 0 | α, metoxy, keto, dicarboxy | Typical | Yellow | + | v | + | + | + | v | + |
| C. E590 | 0 | 0 | 2.5 | 0.5 | 7.5 | 1.6 | 0 | ND | Atypical | Yellow | + | + | + | + | + | + | + |
| M. interjectum ATCC 51457Tc | 5.5 | 1.8 | 4.1 | 2.3 | 0.0 | 2.2 | 5.1 | α, keto, dicarboxy | ND | Yellow | − | + | + | + | − | + | + |
Clustering (A, B, and C) was based on ITS sequences, GLC fatty acids, and HPLC.
Grading of results: +, positive; −, negative; +w, weak positive; v, variable.
Results of the present study.
ND, not done.
Seven isolates from Finland and one from Germany; see Materials and Methods.
Percent of total peak area (mean ± SD).
Five of eight isolates analyzed; see Materials and Methods.
Percent of total peak area (range).
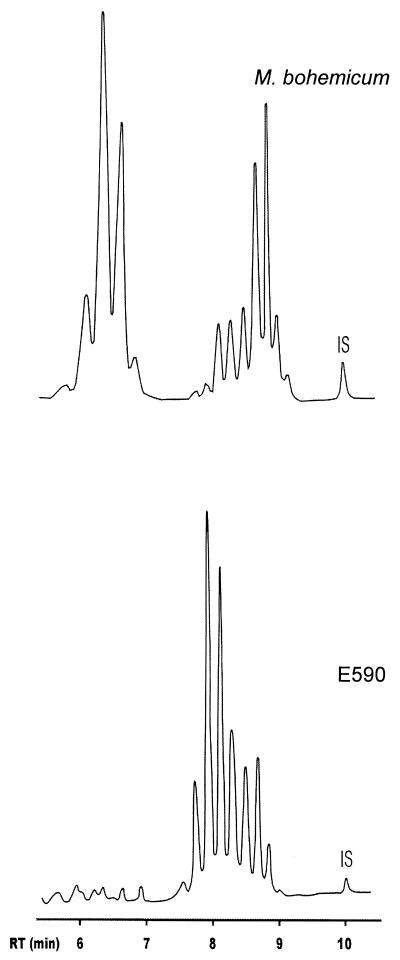
The TLC analysis for mycolic acids verified the presence of α-, metoxy-, keto-, and dicarboxy-mycolates in all three of the M. bohemicum isolates analyzed. In further mycolic acid analyses using HPLC, all but 1 of the 12 isolates examined, including the M. bohemicum type strain, had an identical mycolic acid profile characterized by a major cluster of peaks eluting between 6 and 7 min and a second cluster eluting between 8 and 9.5 min (Fig. 2). One environmental isolate (E590), on the other hand, was characterized by a unique cluster of peaks between 7.5 and 9 min. Thus, it differed distinctly in its mycolic acid composition from the other isolates described here (Fig. 2) as well as from any known species.
FIG. 2.
Representative HPLC profiles of M. bohemicum (type strain) and of the isolate E590. IS, internal standard.
Biochemical characteristics.
The results of biochemical testing are presented in Table 1. All isolates were smooth, bright yellow, and grew at 42°C. They were positive for semiquantitative catalase, and all but one (E744) was also positive for urease. In contrast to the isolates belonging to GLC cluster A, the isolates of cluster B (E743 and E744) and cluster C (E590) hydrolyzed Tween 80.
16S rDNA and ITS region sequencing.
All of the isolates included in the study had 16S rDNA sequences that were highly similar to that of the M. bohemicum type strain (GenBank accession number U84502) (11). Compared to the type strain 16S rDNA sequence, there were four nucleotide differences detected in isolates E743 and E744 and three in isolate E590. All other isolates showed 100% similarity. In ITS sequencing, all but three environmental isolates had an identical ITS sequence, which also was detected in the M. bohemicum type strain. These three environmental isolates (i.e., E590, E743, and E744) had an insertion of 14 nucleotides in the ITS region sequence (Fig. 3). This insertion was identical in two of the isolates (cluster B) and included a one-nucleotide difference in the third isolate (E590; cluster C).
FIG. 3.
Alignment of 16S-23S rDNA ITS sequences including the type strain of M. bohemicum, a patient isolate (2267/99), and two environmental isolates (E743 and E590). Dots indicate identity, and hyphens represent alignment gaps.
Environmental reservoirs of M. bohemicum.
The environmental isolates were detected in water samples from five different streams. The sampling sites were located in rural areas and were not downstream of any known source of animal or human contamination. Three of the sampling sites were clustered on the western coast of Finland. The area is former sea bottom with acidic sulfide soils, and the stream waters in this area are characterized by high levels of anions and cations (e.g., Cl−, F−, SO42−, Ca, Mg, Na, and K) (8). The other two sampling sites were located in inland regions with high peat coverage (60 and 70%). The concentration of M. bohemicum (100 to 200 CFU/liter) was low compared to the total concentrations of mycobacteria in these water samples (970 to 5,800 CFU/liter). As indicated by color (250 to 600 mg of Pt/liter) and chemical oxygen demand (69 to 180 mg of KMnO4/liter), all five water samples contained a high amount of organic matter. The pH in the waters ranged from 5.3 to 6.3.
Clinical significance of isolates.
The Finnish clinical isolates were recovered from seven patients, and the veterinary isolate was from a Rocky Mountain goat. In five of the patients, only a single isolate was detected in a minimum of three sputum specimens, and it was regarded as clinically insignificant. In a 52-year-old woman, M. bohemicum (21910/89) was isolated from sputum specimens in two consecutive years (1988 and 1989). In 1991 and 1992, she was found to harbor Mycobacterium avium complex in several sputum specimens. The patient has been lost to further follow-up. In one of the seven patients (case 1) and in the goat (case 2), M. bohemicum was isolated from biopsy specimens and was regarded as an inducer of clinical infection.
DISCUSSION
Published data on M. bohemicum are limited to the original species description and a very recent case report (16, 17). We isolated M. bohemicum from several sources during the 1990s, including clinical, veterinary, and environmental specimens. Although our present data indicate that its recovery from sputum specimens is likely to be insignificant, M. bohemicum should be listed among potential mycobacterial pathogens causing soft tissue infections in humans and animals. The genetic, biochemical, and lipid characteristics of the isolates from different sources, when compared to the published species description, showed minor variations in biochemical test reactivity, lipid composition, and ITS region sequences.
In 16S rDNA sequencing, the isolates were highly similar to the type strain. On the other hand, interesting variations were detected in the environmental isolates in the sequencing of the ITS region. Differences in both the genetic and GLC lipid compositions divided the studied isolates into the same three main entities. Two of the five environmental isolates as well as the clinical and veterinary isolates included in GLC cluster A were similar to the type strain (Table 1). In contrast, differences including a 14-nucleotide insertion were detected in the ITS region of the other three environmental isolates (GLC clusters B and C). Except for this insertion, these environmental isolates differed from the type strain by only 4 to 5 nucleotides within the ITS region. It is interesting that cluster C (isolate E590) differed appreciably from all other isolates also studied in HPLC analysis.
Slowly growing mycobacteria can be identified correctly by means of GLC by using an identification system which recognizes both fatty acid, fatty alcohols, and mycolic acid cleavage products (3, 5). Our results on M. bohemicum verified these earlier experiences. The characteristic markers of M. bohemicum were secondary alcohols (2-OH-18:0alc; 2-OH-20:0alc) and methyl-branched fatty acids (2-Me-20:0; 2,4-diMe-22:0; 2,4,6-triMe-22:0; 2,4,6-triMe-24:0). These branched-chain fatty acids elute after eicosanoic acid (20:0). Thus, M. bohemicum can be identified by GLC only if fatty acids with carbon chain lengths between 20 and 26, derived from mycolic acids, are included in the analysis. The GLC profile of the scotochromogenic M. interjectum closely resembles that of M. bohemicum. However, M. bohemicum lacks the hexacosanoic acid that is present in significant amounts (5.1% of total area) in M. interjectum. Mycobacterium malmoense is another species with branched-chain fatty acids, 2-Me-20:0, 2,4-diMe-22:0, and 2,4,6-triMe-24:0 (5, 12). This slowly growing species is characterized by the presence of a high amount of hexacosanoic acid, and because it is nonpigmented, it can easily be separated from M. bohemicum.
The HPLC profile of M. bohemicum is similar to but not identical with M. avium complex and Mycobacterium scrofulaceum, from which it differs by the retention times of the major peaks (14). In the present study, all but one of the isolates had identical HPLC profiles. One environmental isolate (E590) which was found to be highly similar to other isolates by 16S rDNA sequencing had a completely different HPLC chromatogram. This indicates that either M. bohemicum is a species characterized by two different mycolic acid profiles, as already reported to be the case with Mycobacterium gordonae and M. interjectum (2, 15), or E590 belongs to a different taxon, undistinguishable from M. bohemicum on the basis of 16S rDNA sequencing. In the TLC mycolic acid analysis, M. bohemicum contained α-, keto-, metoxy-, and dicarboxy-mycolates. This combination is unknown in any slow-growing mycobacterial species described, but is found in Mycobacterium komossense (19).
Variations in biochemical reactivity were detected among the tested isolates, and some of the results obtained also differed from the earlier species description (11). Nowadays, most new species are described on the basis of genetic analyses of a small number of strains, and biochemical data have also been collected from a limited number of strains. Increasing the number of isolates that are analyzed allows detection of natural variations in biochemical properties, as shown here.
M. bohemicum is not common in the Finnish environment. Only five strains of M. bohemicum (0.7%) were recovered among 757 isolates from 53 water samples. These water samples contained medium to high total mycobacterial concentrations. Yet, natural waters, either directly or via water distribution systems, are potential reservoirs of this recently recognized species, as well as of other species detected as colonizers or pathogens in clinical or veterinary samples.
ACKNOWLEDGMENTS
This work was partly funded by the Foundation of Finnish Antituberculous Association and The Finnish Cultural Foundation of Northern Savo (P.T.) and Oskar Öflund's Foundation (S.S.).
We express our gratitude to J. Kaustova and L. Naumann for the strains supplied. We also thank M. Viljanen for providing us with one of the early M. bohemicum isolates.
REFERENCES
- 1.Butler W R, Thibert L, Kilburn J O. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J Clin Microbiol. 1992;30:2698–2704. doi: 10.1128/jcm.30.10.2698-2704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cage G D. High-performance liquid chromatography patterns of Mycobacterium gordonae mycolic acids. J Clin Microbiol. 1992;30:2402–2407. doi: 10.1128/jcm.30.9.2402-2407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou S, Chedore P, Kasatiya S. Use of gas chromatographic fatty acid and mycolic acid cleavage product determination to differentiate among Mycobacterium genavense, Mycobacterium fortuitum, Mycobacterium simiae, and Mycobacterium tuberculosis. J Clin Microbiol. 1998;36:577–579. doi: 10.1128/jcm.36.2.577-579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iivanainen E K, Martikainen P J, Väänänen P K, Katila M-L. Environmental factors affecting the occurrence of mycobacteria in brook waters. Appl Environ Microbiol. 1993;59:398–404. doi: 10.1128/aem.59.2.398-404.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jantzen E, Tangen T, Eng J. Gas chromatography of mycobacterial fatty acids and alcohols: diagnostic applications. APMIS. 1989;97:1037–1045. doi: 10.1111/j.1699-0463.1989.tb00515.x. [DOI] [PubMed] [Google Scholar]
- 6.Katila M-L, Katila P, Erkinjuntti-Pekkanen R. Accelerated detection and identification of mycobacteria with MGIT 960 and COBAS AMPLICOR systems. J Clin Microbiol. 2000;38:960–964. doi: 10.1128/jcm.38.3.960-964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koukila-Kähkölä P, Springer B, Böttger E C, Paulin L, Jantzen E, Katila M-L. Mycobacterium branderi sp. nov., a new potential human pathogen. Int J Syst Bacteriol. 1995;45:549–553. doi: 10.1099/00207713-45-3-549. [DOI] [PubMed] [Google Scholar]
- 8.Lahermo, P., P. Väänänen, T. Tarvainen, and R. Salminen. 1996. Geochemical atlas of Finland, part 3. Environmental geochemistry—stream waters and sediments. Geological Survey of Finland, Espoo, Finland. Geochemical atlas of Finland, part 3. English abstract.)
- 9.Luquin M, Ausina V, López Calahorra F, Belda F, García Barceló M, Celma C, Prats G. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J Clin Microbiol. 1991;29:120–130. doi: 10.1128/jcm.29.1.120-130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattila J O, Katila M-L, Vornanen M. Slowly growing mycobacteria and chronic skin disorders. Clin Infect Dis. 1996;23:1043–1048. doi: 10.1093/clinids/23.5.1043. [DOI] [PubMed] [Google Scholar]
- 11.Reischl U, Emler S, Horak Z, Kaustova J, Kroppenstedt R M, Lehn N, Naumann L. Mycobacterium bohemicum sp. nov., a new slow-growing scotochromogenic mycobacterium. Int J Syst Bacteriol. 1998;48:1349–1355. doi: 10.1099/00207713-48-4-1349. [DOI] [PubMed] [Google Scholar]
- 12.Torkko P, Suutari M, Suomalainen S, Paulin L, Larsson L, Katila M-L. Separation among species of Mycobacterium terrae complex by lipid analyses: comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol. 1998;36:499–505. doi: 10.1128/jcm.36.2.499-505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torkko P, Suomalainen S, Iivanainen E, Suutari M, Tortoli E, Paulin L, Katila M-L. Mycobacterium xenopi and related organisms isolated from stream waters in Finland and description of Mycobacterium botniense sp. nov. Int J Syst Evol Microbiol. 2000;50:283–289. doi: 10.1099/00207713-50-1-283. [DOI] [PubMed] [Google Scholar]
- 14.Tortoli E, Bartoloni A. High-performance liquid chromatography and identification of mycobacteria. Rev Med Microbiol. 1996;7:207–219. [Google Scholar]
- 15.Tortoli E, Kirschner P, Bartoloni A, Burrini C, Manfrin V, Mantella A, Scagnelli M, Scarparo C, Simonetti M T, Böttger E C. Isolation of an unusual mycobacterium from an AIDS patient. J Clin Microbiol. 1996;34:2316–2319. doi: 10.1128/jcm.34.9.2316-2319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tortoli E, Kirschner P, Springer B, Bartoloni A, Burrini C, Mantella A, Scagnelli M, Scarparo C, Simonetti M T, Böttger E C. Cervical lymphadenitis due to an unusual mycobacterium. Eur Clin Microbiol Infect Dis. 1997;16:308–311. doi: 10.1007/BF01695636. [DOI] [PubMed] [Google Scholar]
- 17.Tortoli E, Bartoloni A, Manfrin V, Mantella A, Scarparo C, Böttger E. Cervical lymphadenitis due to Mycobacterium bohemicum. Clin Infect Dis. 2000;30:210–211. doi: 10.1086/313600. [DOI] [PubMed] [Google Scholar]
- 18.Vincent Lévy-Frébault V, Portaels F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42:315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 19.Yassin A F, Binder C, Schaal K P. Identification of mycobacterial isolates by thin-layer and capillary gas-liquid chromatography under diagnostic routine conditions. Zentbl Bakteriol. 1993;278:34–48. doi: 10.1016/s0934-8840(11)80277-x. [DOI] [PubMed] [Google Scholar]