Abstract
The coronavirus outbreak 2019, called COVID-19, which originated in Wuhan, negatively affected the lives of millions of people and many people died from this infection. To prevent the spread of the disease, which is still in effect, various restriction decisions have been taken all over the world. In addition, the number of COVID-19 tests has been increased to quarantine infected people. However, due to the problems encountered in the supply of RT-PCR tests and the ease of obtaining Computed Tomography and X-ray images, imaging-based methods have become very popular in the diagnosis of COVID-19. Therefore, studies using these images to classify COVID-19 have increased. This paper presents a classification method for computed tomography chest images in the COVID-19 Radiography Database using features extracted by popular Convolutional Neural Networks (CNN) models (AlexNet, ResNet18, ResNet50, Inceptionv3, Densenet201, Inceptionresnetv2, MobileNetv2, GoogleNet). The determination of hyperparameters of Machine Learning (ML) algorithms by Bayesian optimization, and ANN-based image segmentation are the two main contributions in this study. First of all, lung segmentation is performed automatically from the raw image with Artificial Neural Networks (ANNs). To ensure data diversity, data augmentation is applied to the COVID-19 classes, which are fewer than the other two classes. Then these images are applied as input to five different CNN models. The features extracted from each CNN model are given as input to four different ML algorithms, namely Support Vector Machine (SVM), k-Nearest Neighbors (k-NN), Naive Bayes (NB), and Decision Tree (DT) for classification. To achieve the most successful classification accuracy, the hyperparameters of each ML algorithm are determined using Bayesian optimization. With the classification made using these hyperparameters, the highest success is obtained as 96.29% with the DenseNet201 model and SVM algorithm. The Sensitivity, Precision, Specificity, MCC, and F1-Score metric values for this structure are 0.9642, 0.9642, 0.9812, 0.9641 and 0.9453, respectively. These results showed that ML methods with the most optimum hyperparameters can produce successful results.
Keywords: Bayesian Optimization, COVID-19 pandemic, Convolutional neural networks, Machine learning
1. Introduction
A novel coronavirus family called COVID-19 spread around the world, starting in China as a respiratory infection in late 2019. As a result of the rapid spread of this disease and its accompanying symptoms, the World Health Organization (WHO) announced COVID-19 as a pandemic. COVID-19 has caused severe economic problems for many countries since its emergence and has had a very negative impact on human life. Currently, worldwide, the total number of cases has exceeded 267.000.000 and the total number of deaths has exceeded 5.000.000 [1]. It has expanded to over 200 nations throughout the world. To counteract COVID-19, rulers and governments in various countries have implemented new policies and adopted new lifestyles [2]. While many medical experts work on vaccinations to prevent the virus, various treatments and medical procedures are being created to heal infected people and keep them from spreading the infection to others [3]. Although precautions are taken, the virus, which has undergone different mutations, unfortunately, becomes more resistant to increasingly developed treatment methods. This indicates that the infection may continue for a longer period of time. Today, studies such as early diagnosis of infection, reduction of its effects, and effects on human life are carried out by many researchers. Most previous studies aim to successfully identify people infected with COVID-19 as soon as possible so that infected people can be quarantined and treated quickly [4].
Initially, the common method for detecting COVID-19 was RT-PCR test kits. However, the use of medical images such as Chest X-Ray (CXR), ultrasound, computed tomography, and MRI became an important alternative way to diagnose infection. Among these, computed tomography and CXR have become very popular and are often used to detect lung disease abnormalities, including COVID-19 [5]. CXR is more accessible in hospitals worldwide than computed tomography, but for staging COVID-19 disease, early detection of disease changes, and monitoring progress, a computed tomography image is more precise than a CXR image of the chest [6,7]. Moreover, computed tomography images are a robust tool broadly used in medical imaging [8] and diagnosis [9] applications. However, it is not easy to classify the properties of community-acquired bacterial pneumonia as COVID-19 [10]. For medical analysis, computer science researchers have an important role in developing novel methods to support coronavirus diagnosis. In particular, Artificial intelligence (AI) is very important to develop solutions to support virus diagnosis. For this reason, AI methods have started to be used today to create automatic systems for the detection of COVID-19 [11].
Nowadays, with the emergence of AI-based Deep Learning (DL) models, large datasets can be processed simply. As in many fields of medicine, the most used DL model is Convolutional neural network (CNN) architectures. DL models perform both feature extraction and classification. Moreover, the extracted features can distinguish small details. This powerful feature extraction step makes DL superior to Machine Learning (ML) methods [12]. CNN extracts features with its structure consisting of deep and various layers and performs classification or regression similar to ML algorithm using these features. Numerous pre-designed CNN models such as VGG-Net, GoogLeNet, ResNet, Inceptionv3, DenseNet, and AlexNet have been applied many times in image classification and pattern recognition. Especially with Transfer Learning (TL), the use of these pre-trained models has increased. Thanks to TL, using the knowledge of the previously trained network, higher success models have been developed with a small number of training data [13].
CNN architectures extract features from the image according to the designed architecture and then classify them through fully connected layers. This layer performs the task of an ML algorithm. Therefore, successful classifications can be achieved by using the ML algorithm instead of this layer. In literature, various ML methods are employed due to their different advantages. It is important to identify the ML method that offers the best performance according to the application. The powerful algorithms commonly preferred in the literature are Naive Bayes (NB), Support Vector Machine (SVM), k-Nearest Neighbor (k-NN) and Decision Tree (DT). An unsupervised ML algorithm, k-NN [14] is an uncomplicated method to understand and use. Thanks to having high precision, the capability of dealing with high-dimensional data, and flexibility, the SVM has been found wide usage in bioinformatics [15]. DT [16] is a supervised ML algorithm that classifies big data similarly to the structure of a tree (leaves, branches, and nodes). Finally, NB [17] is a supervised ML algorithm that statistically classifies data using the Bayesian method.
In order to optimally implement the ML algorithm that is most compatible with the data and features, these ML algorithms must also be configured. Because the performance of any ML algorithm associated with the parameters in the algorithm affects accuracy. To get the most out of the ML algorithm, supervised or unsupervised all ML methods need to be configured before the training process. In this study, hyperparameter optimization (HO) [18] is applied to ML algorithms to achieve this. The hyperparameter search technique improves both the performance in the training step, the prediction accuracy, and the quality of the ML algorithm. Many algorithms, like Bayesian optimization, grid search, swarm optimization algorithm, etc. can be employed to perform in search of hyperparameter [19].
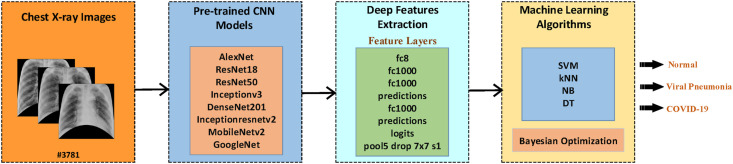
In the diagnostic process carried out in previous AI-based studies, raw images were used directly and no preprocessing was applied. However, pixels other than the lungs in the raw image should not affect the result. An image that includes only the lungs makes more sense for infection detection. To achieve this, the lung region must be segmented from the raw image. In this way, more successful diagnoses can be made if the input images containing only the region of the disease are combined with powerful estimation methods. This is the most important factor that motivates this work. In this research, studies on COVID-19 are investigated and a different study is presented using the COVID-19 Radiography Database [20]. First, in raw CXR chest images, to improve classification performance and prevent irrelevant parts such as patient ID, date-time texts, etc. from affecting the estimate, non-lung areas are eliminated, and for this, ANN lung segmentation is performed. The lung area of the original image is cropped by the segmentation process. Afterwards, to increase segmented image count, data augmentation is employed, thus providing data diversity. Then, features are extracted from each segmented image using state-of-the-art CNN models (AlexNet, ResNet18, ResNet50, Inceptionv3, Densenet201, Inceptionresnetv2, MobileNetv2, and GoogleNet). k-NN, SVM, DT, and NB methods are used to classify these features as COVID-19, Normal, and Viral Pneumonia. In order to obtain the most successful results from these methods, the hyperparameters of each method are determined with Bayesian optimization. As a result of the classification made using these hyperparameters, the results of the combination of eight different CNN models and four different ML methods are compared. Classification results are successful and at the end of the study, they are compared with previous similar studies. The high classification success and, the combination of state-of-the-art CNN models and optimized ML algorithms are the contributions of this study. The contributions of this study are summarized below.
-
•
ANN-based automatic lung segmentation to achieve powerful features,
-
•
Extracting features from pre-trained CNN models,
-
•
Determining the best ML algorithm parameters with Bayesian optimization,
-
•
With the proposed method, COVID-19 can be detected easily and with high accuracy.
The remainder of the article is organized as follows. In section 2, previous studies on COVID-19 are reviewed. The dataset, ANN-based image segmentation, data augmentation, feature extraction and Bayesian optimization are summarized in section 3. Performance evaluation, discussion of results and comparison with previous studies are explained in section 4. Finally, section 5 concludes the article and provides information about future work.
2. Related works
This section summarizes some of the previous state-of-the-art studies that have performed DL-based COVID-19 diagnosis using computed tomography or CXR images. A review study about this has been done by Lalmuanawma, Hussain and Chhakchhuak [21]. Similarly, Shi, Wang, Shi, Wu, Wang, Tang, He, Shi and Shen [22] detailed studies using AI techniques to diagnose COVID-19. Nayak, Nayak, Sinha, Arora and Pachori [23] compared AlexNet, ResNet-50, GoogleNet, MobileNet V2, VGG16, ResNet34, SqueezeNet, and Inception V3 models using CXR images for early detection of COVID-19 infection. Factors such as batch size, learning rate, number of epochs were taken into account to find the most suitable model. The results showed that ResNet34 outperformed other competing networks with 98.33% accuracy. Asif and Wenhui [24] applied CNN-based TL for COVID-19 diagnosis using CXR images. The images were given to the Inception-V3 model without any preprocessing step. The classification success of that proposed system was 96%. Wu, Hui, Niu, Li, Wang, He, Yang, Li, Li and Tian [25] proposed DL to detect COVID-19 patients using computed tomography images with maximal lung regions in axial, coronal, and sagittal views. After training a multi-view fusion model, the multi-view DL fusion model achieved 76% accuracy. Ardakani, Kanafi, Acharya, Khadem and Mohammadi [26] performed an AI technique using computed tomography images to detect COVID-19. They compared CNN models trained on computed tomography images with each other's and they discovered that Xception and ResNet-101 with accuracy rates of 99.02% and 99.51% respectively have better performance than the others. Other researchers who also used ResNet-18 classified computed tomography images for detection of COVID-19, Influenza viral pneumonia, normal subjects and obtained an accuracy of 86.7% [27]. Zhang, Liu, Shen, Li, Sang, Wu, Zha, Liang, Wang and Wang [28] developed an AI method with a large computed tomography dataset and this method differentiates COVID-19 and other types of viral pneumonia. They used the ResNet-18 model of CNN and their accuracy rate was 92.49%. Panwar, Gupta, Siddiqui, Morales-Menendez and Singh [29] presented a TL-based nCOVnet method based on DL to diagnose the COVID-19. They trained the patients' computed tomography images and tested them with an accuracy of 88.10%. A similar study including 3D deep CNN was called as DeCoVNet and presented to detect COVID-19 [30]. Nour, Cömert and Polat [2] advised SVM-based feature extractor with an accuracy of 98.97% on the CNN architecture model compared with other feature extractors such as k-NN and DT Elaziz, Hosny, Salah, Darwish, Lu and Sahlol [31] proposed an ML method with a modified Manta-Ray Foraging Optimization used for the selection of useful features from computed tomography images. Ezzat, Hassanien and Ella [32] introduced a method named GSA-DenseNet121-COVID-19. This method applied the pre-trained DenseNet121 model to detect COVID-19 by using the TL method. They applied the gravitational search algorithm (GSA) to select the optimal hyperparameters of the DenseNet121 model and ultimately achieved 98.38% success. Qaid, Mazaar, Al-Shamri, Alqahtani, Raweh and Alakwaa [33] have developed new hybrid models with DL and TL methods to detect COVID-19 using x-ray images. A COVID-19 Radiography database from Kaggle and data collected from a local hospital, Asir Hospital, were used in the training and testing of the designed hybrid models. It is reported that the proposed model did not make mistakes in detecting COVID-19, and the success was 97.8% in multi-class situations. Das, Kalam, Kumar and Sinha [34] presented a fully automated COVID-19 test model using CXR images in their study. For this purpose, the COVID-19 radiography dataset in the Kaggle repository was preferred. This dataset was used by grouping it into three classes as COVID-19 positive, other pneumonia infection and no infection. The performances of the used CNN models, VGG-16, and ResNet-50 were determined. In that study, it was stated that the VGG-16 model showed the best performance with an accuracy of 97.67%. Monshi, Poon, Chung and Monshi [35] stated that they optimized data augmentation and CNN hyperparameters to increase validation accuracy in COVID-19 detection using CXR. It is stated that this method increased the success of VGG-19 and ResNet-50 models. In addition, a new model was proposed based on EfficientNet-B0 and optimization called CovidXrayNet. It has been determined that this model has 95.82% accuracy when tested with data created with two different databases. Rajpal, Lakhyani, Singh, Kohli and Kumar [36] have considered the COVID-19 detection problem as a three-class classification problem. These classes were COVID-19, pneumonia and normal. They proposed a classification structure with features derived from the CNN network and a set of handpicked features. The proposed structure consisted of 3 parts. In the first part, 2048 parameters were obtained by using ResNet-50 with TL. In the second part, 64 features were selected from 252 handpicked features using PCA. In the third part, the features obtained in the previous two modules were combined and classified. Classification success was given as 0.979. Kumar, Tripathi, Satapathy and Zhang [37] proposed SARS-Net for the diagnosis of COVID-19 using CXR. In that study, the open-source COVIDx database containing CXR data was used. It has been demonstrated by quantitative analysis that the proposed architecture has higher success. It is stated that the proposed model has an accuracy of 97.60%.
It is possible to increase the number of similar previous studies summarized above. The difference of this study from the above studies is that it uses state-of-the-art CNN models together with optimized ML algorithms.
3. Methodology
This section provides information about the dataset used for COVID-19 detection. Later, it deals with the ANN-based lung segmentation, data augmentation, deep feature extraction and classification steps performed for the proposed method. Fig. 1 presents the block diagram of the proposed study. The detailed description of steps presented in the block diagram is explained in the following sections.
Fig. 1.
Block diagram of the proposed study.
3.1. The used database
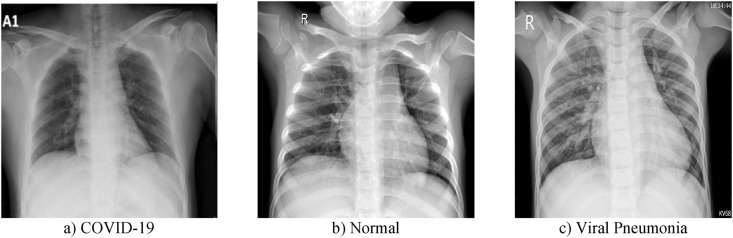
In this application, a public dataset that contains the CXR images, which are taken from the posterior-to-anterior chest, is utilized [20]. Collaborative medical doctors and university professors from Bangladesh, Qatar, Malaysia, and Pakistan created the COVID-19 Radiography Database by collecting CXR images. This dataset is in three classes as COVID-19, Viral Pneumonia and Normal. In Table 1 , the classes in the database and the number of CXR images in these classes are given. For this dataset, CXR images collected from various sources were combined. Some CXR images of this dataset are shown in Fig. 2 .
Table 1.
Number of samples classes in the COVID-19 Radiography Database.
| Class | COVID-19 | Normal | Viral Pneumonia |
|---|---|---|---|
| Number of CXR images | 219 | 1341 | 1345 |
| Total | 2905 | ||
Fig. 2.
Some raw CXR images of COVID-19 Radiography Database.
3.2. ANN-based segmentation of lungs
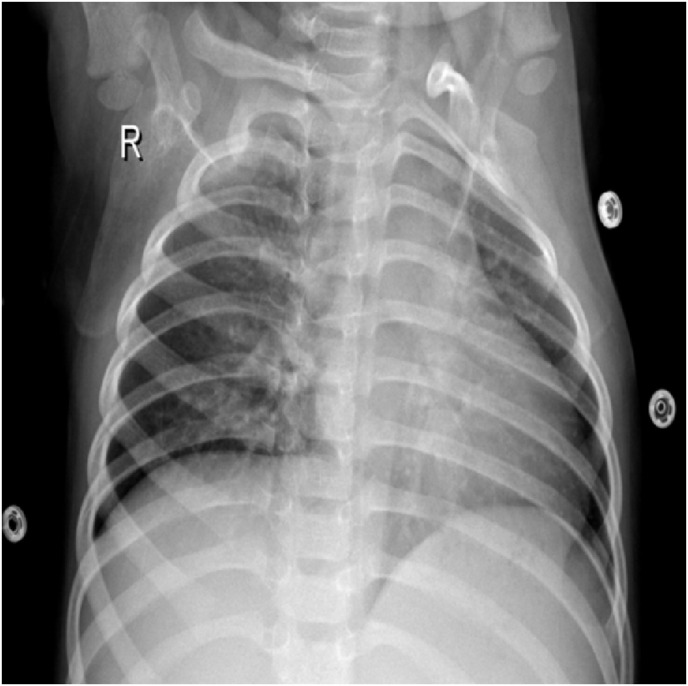
For AI applications, segmentation of meaningful regions in the image is very important for tasks such as object recognition, pattern recognition, etc. Otherwise, noise patterns or unrelated blobs in the CXR scans may pose inaccurate forecasts. The raw CXR images used in this study also contain different types of noises, as shown in Fig. 3 . The segmentation process is applied to exclude these noises from the AI algorithm.
Fig. 3.
A CXR Image sample that contains irrelevant patterns or noises.
For the COVID-19 diagnosis with CXR images, the region of interest needs to be the lungs in the chest image. Therefore, the usage of lungs area masked images will make AI possible to conduct a more precise prediction. However, the CXR images of patients are not always perfectly aligned. Therefore, the lung positions change from one CXR image to another. That's why, for images with only the lung part, each image should be segmented individually. This is not a logical solution as the dataset used contains 2905 images. Therefore, the segmentation step should be performed automatically. ANN-based automated segmentation is recommended to perform prediction based only on the lung part in CXR images [38].
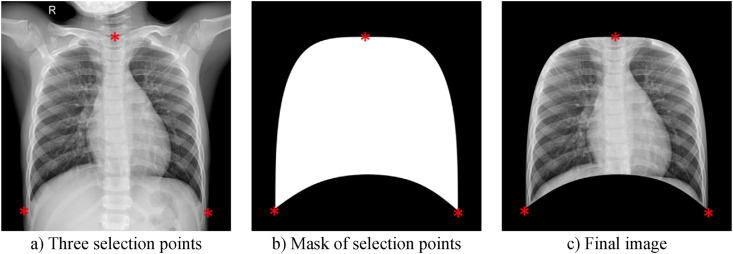
To provide automatic segmentation, 50 images from each class in the dataset are given as input to ANN. For each of these images, the three points (up, left, and right) are determined manually as seen in Fig. 4 (a). Next, according to these points, the mask presented in Fig. 4(b) is obtained. In the last step, the raw image is cropped according to the mask. Thus, the final image ready for AI input is obtained as shown in Fig. 4(c).
Fig. 4.
Manually selected points, mask image, and segmented lung image.
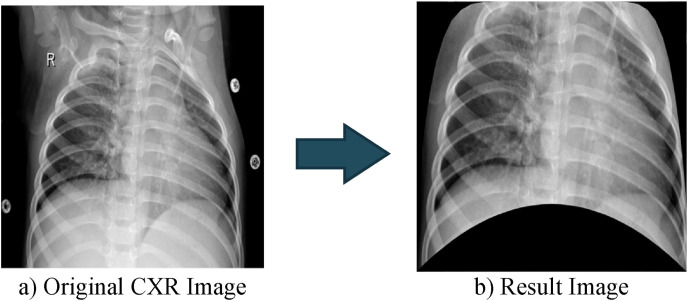
After 50 randomly selected images from each class are manually converted to the resulting image (see Fig. 4 (c)), ANN-based automatic segmentation is carried out to generate these result images. The CXR image with a size of 1024 × 1024 has been converted to a vector (one-dimensional matrix). During the training of the ANN, while this vector is employed as input, the manually determined points are used as the target. The Levenberg-Marquardt algorithm is used for training ANN. The designed neural network contains a single hidden layer of 100 neurons. ANN output consists of the x and y coordinates of three manually selected selection points, that is, it contains 6 neurons. The training process is completed with 94% accuracy [38]. To determine the mask creation coordinates, All the remaining CXR images are applied to the trained ANN. By using the coordinate values obtained at the end of ANN, cropping is performed. Fig. 5 shows the raw and result image of an image obtained by ANN-based segmentation. Fig. 5 shows the resulting image produced by applying ANN-based segmentation to the CXR image in Fig. 3.
Fig. 5.
Cropped image obtained by ANN-based segmentation.
3.3. Data augmentation
A large dataset must be used to ensure successful classification with DL. However, in many applications, the number of data that can be accessed is limited. In such cases, successful network training is performed by increasing the number of data in the computer environment [38,39]. As seen in Table 1, there is an imbalance in the number of images belonging to the classes in the dataset. COVID-19 is the class with the least number of images with 219 images. In this study, rotation data augmentation method is applied to COVID-19 class images to eliminate this imbalance. Since the number of images belonging to the other two classes is sufficient, there is no need for data augmentation. Rotation produces the same image at different angles to increase the variety of data available. This provides stronger training. The features of the deep network, trained with more variety and more images, better represent the data.
In this step, images with COVID-19 class are increased 4 times. Consequently, the total number of COVID-19 classified images reaches 1095 and the total number of data reaches 3781. This process is implemented to the cropped images produced at the end of ANN-based segmentation, and each image is rotated randomly between 0 and 360° (see Fig. 6 ).
Fig. 6.
Rotation operation.
3.4. Deep feature extraction and Bayesian Optimization
Fig. 7 summarizes the steps described in this section. The segmented and augmented CXR images obtained as a result of the previous steps are given as input to eight different CNN models for feature extraction. CNN architectures consist of three different layers. These are convolutional layers, pooling layers, and fully connected layers. When these layers come together in different combinations, CNN architecture is formed. With the convolution layers, the input images are subjected to the convolution process and the features related to the input image are formed at the output. The pooling layer further reduces the number of parameters by downsampling the output from the convolution result. Finally, fully connected layers allow us to separate the data into various classes after feature extraction, that is, it does what an ML (e.g. ANN) algorithm does. Different CNN models differentiate these layers by combining them with different techniques, combinations and rules [40]. For example, what makes ResNet different and special is that it uses residual blocks to transmit residual values to the next layers to avoid the vanishing gradient problem [41].
Fig. 7.
Block diagram representation of feature extraction, optimization, and classification steps.
As seen in Table 2 , each CNN model has a specific input size. Therefore, the image sizes are adjusted for each model. After the input images are given to the models, convolutional processes, pooling, Rectified Linear Unit (ReLU), etc. processes are implemented to these images. These processes continue until the feature layer within the relevant model is reached. Deep features of CNN models are extracted from a specific layer of each model. The feature layer name of each model is shown in Table 2 and Fig. 7. The layers used for feature extraction can produce a different number of outputs depending on the CNN model. All of the feature extraction layers used in our study extract 1000 features (see Table 2). 85% of the extracted features are separated for train and 15% for test. Classification is carried out using the features occurring in feature layers.
Table 2.
State-of-the-art CNN model features.
| CNN models | Alexnet | Resnet18 | Resnet50 | Inceptionv3 | Densenet201 | Inceptionresnetv2 | GoogleNet | MobileNetv2 |
|---|---|---|---|---|---|---|---|---|
| Input Size | 227 × 227 | 224 × 224 | 224 × 224 | 299 × 299 | 224 × 224 | 299 × 299 | 224 × 224 | 224 × 224 |
| Feature Layer | fc8 | fc1000 | fc1000 | predictions | fc1000 | predictions | pool5-drop_7 × 7_s1 | logits |
| Number of Extracted Features | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 | 1000 |
The features from the feature layer are given to ML algorithms for classification. For ML methods to provide maximum classification accuracy, it is of great importance to determine the parameters that affect the classification accuracy. Therefore, the best parameters should be determined according to the features used. In our application, relevant parameters (hyperparameters) are selected by employing Bayesian optimization in the training step for each ML algorithm. Hyperparameter types differ according to the ML algorithm. Hyperparameters are distance and the number of neighbors for k-NN; box constraints (C), coding method, kernel scale for SVM; minimum leaf size for DT, kernel width, and distribution type for NB. The values of these parameters calculated by Bayesian optimization are presented in Table 3 . Then, the training and testing processes are conducted while the ML algorithm has these parameter values. In the training phase, the stopping criterion is determined as making a certain number of iterations for each ML algorithm. Bayesian optimization is a hyperparameter search technique. For this reason, it is an effective technique that is frequently applied among HO methods. The general expression for this optimization method is as in Eq. (1).
| (1) |
Table 3.
Hyperparameters of ML algorithm obtained as a result of Bayesian Optimization.
| ML alg. | Hyperparameters | CNN models used for feature extraction |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Alexnet | Resnet18 | Resnet50 | Inceptionv3 | Densenet201 | Inceptionresnetv2 | GoogleNet | MobileNetv2 | ||
| SVM | Coding method | onevsall | onevsall | onevsone | onevsone | onevsone | onevsone | onevsall | onevsall |
| Box constraints (C) | 0.0010143 | 0.10313 | 0.0046093 | 0.0027652 | 0.003083 | 0.0010105 | 0.0010391 | 0.0023347 | |
| Kernel scale | 0.16082 | 1.1931 | 0.2569 | 0.43568 | 0.77278 | 0.32909 | 0.082927 | 0.66399 | |
| Total elapsed time(s) | 4834.7835 | 4949.4327 | 5109.2594 | 5298.5672 | 5436.9534 | 5314.5984 | 5273.306 | 5380.0478 | |
| k-NN | Number of neighbors | 3 | 3 | 3 | 3 | 6 | 4 | 3 | 4 |
| Distance | correlation | seuclidean | seuclidean | spearman | seuclidean | spearman | seuclidean | spearman | |
| Total elapsed time(s) | 81.6571 | 82.5329 | 86.2578 | 87.9510 | 95.5283 | 89.1256 | 86.6401 | 94.8055 | |
| NB | Distribution type | kernel | kernel | kernel | kernel | kernel | kernel | kernel | kernel |
| Kernel width | 0.051574 | 0.058872 | 0.058477 | 0.067775 | 0.070217 | 0.059119 | 0.063142 | 0.060272 | |
| Total elapsed time(s) | 2589.5847 | 2714.706 | 2730.5094 | 2930.5064 | 3010.9512 | 2906.2571 | 2760.4442 | 3006.6945 | |
| DT | Minimum leaf size | 4 | 12 | 2 | 6 | 3 | 8 | 8 | 10 |
| Total elapsed time(s) | 85.2036 | 86.1918 | 88.0563 | 89.2318 | 90.6345 | 88.8135 | 83.7987 | 89.5203 | |
This Eq. (1) finds the value of that maximizes the function. The components of that denotes a hyperparameter can be continuous reals, integers, or categorical, meaning a discrete set of names. represents an objective function also called a Gaussian Process (GP) model to try to minimize on a validation set. represents the search space for the hyperparameter type. Bayesian optimization is derived from Bayes' theorem. In Bayes' theorem, the inference is made by making use of prior knowledge. Bayes' theorem is presented in Eq. (2). In Eq. (2), the posterior distribution is directly proportional () to the product of likelihood () and a priori distribution (). refers to the prediction performed, called “belief” [42,43].
| (2) |
In ML algorithms, the objective function is expensive to find the optimal hyperparameter in the search space. In these cases, a simpler surrogate model of the objective function is created to solve the optimization problem more cost-effectively. In short, the model used to approximate the objective function is called the surrogate model. Also, it would be costly to use all the function values to find the point that would give the maximum value of x as in Eq. (1). More optimal is that the function approaches areas where it is most likely optimal. For this, an iterative approach is often recommended. Bayesian Optimization incorporates prior models into the space of possible posterior functions, that is, it applies Bayesian-based iterative optimization. Many probability models can be used in the Bayesian optimization algorithm, but the GP is the most popular. Therefore, in this study, GP is determined as a surrogate model for objective functions in Bayesian optimization. GP is a stochastic process that collects random variables, usually indexed by space or time. Any finite collection of these variables is distributed according to a multivariate Gaussian distribution [44]. GP can be expressed as a mean function () and a covariance () as in Eq. (3). As an example, the response of this function at the input and the output y is shown in Eq. (4). Here denotes noise with Gaussian distribution (Eq. (5)) [45]. The GP is updated by adding the found sampling point ) to the previous samples.
| (3) |
| (4) |
| (5) |
Bayesian optimization also uses the acquisition function ( for Eq. (1)), which moves sampling to the area where improvement is possible. The acquisition function is optimized over the GP, and the sampling point is located. The acquisition function, Expected Improvement (EI), used in this study can be defined as in Eq. (6). Where represents the value of the best sample.
| (6) |
4. Results and discussion
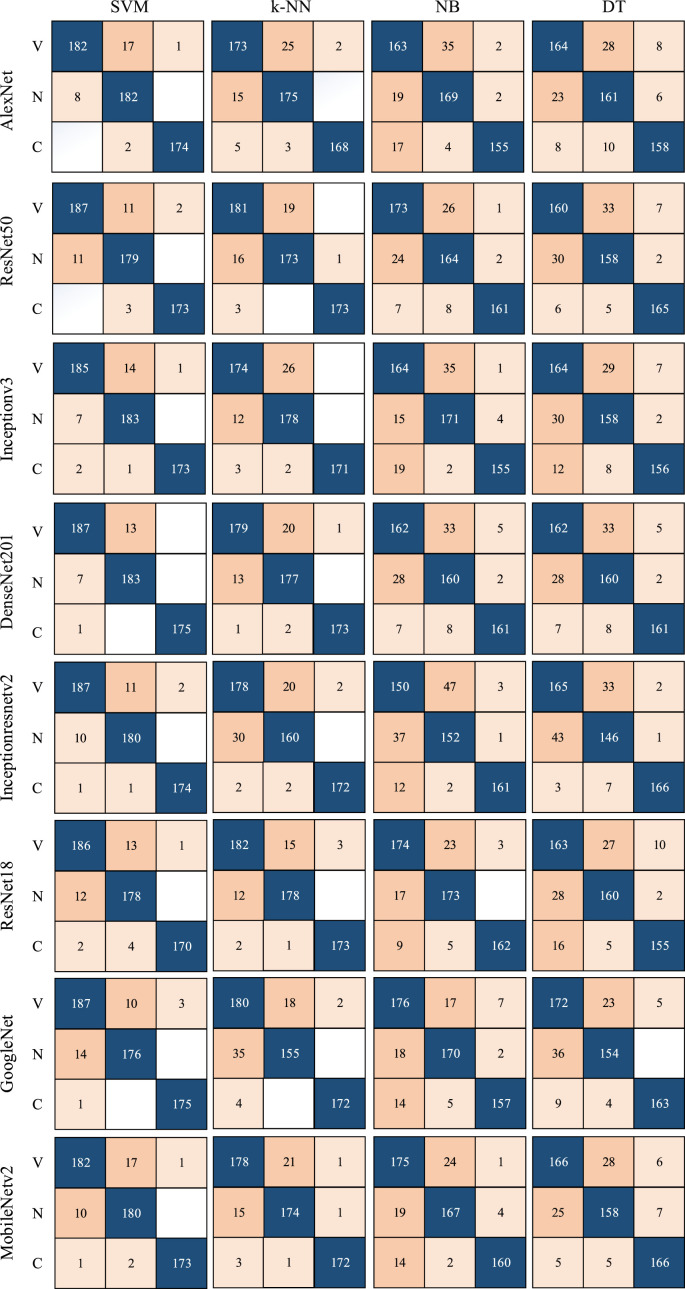
Segmented lung images are classified using 1000 features, each extracted from eight different popular CNN models. For experimental applications where DL algorithms are implemented, a laptop with 16 GB RAM, Intel Core i7 CPU, and NVIDIA GeForce GTX 1050 is used. In the training step, the parameters that affect the accuracy of each ML method are also optimized by Bayesian optimization. Therefore, as a result of the training, both the trained network and the model's hyperparameters, as seen in Table 3, are found. The determined hyperparameters are not changed during the training and testing steps. The features of each model are classified using four different ML methods. The confusion matrices determined depending on the result of the classification are given in Fig. 8 . Additionally, various metrics calculated using the confusion matrix are also calculated and presented for performance measurement. The formulas of used metrics are presented in Eq. (7) - Eq. (12). The values obtained using these formulas and showing the results of this study are given in Table 4 .
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
Fig. 8.
Confusion matrices of state-of-the-art CNN models according to ML algorithms.
Table 4.
The calculated metrics of state-of-the-art CNN models according to ML algorithms.
| Model | ML | Accuracy (%) | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|---|
| AlexNet | SVM | 95.05 | 0.9522 | 0.9750 | 0.9526 | 0.9519 | 0.9273 |
| k-NN | 91.17 | 0.9135 | 0.9553 | 0.9156 | 0.9140 | 0.8698 | |
| NB | 86.04 | 0.8617 | 0.9292 | 0.8688 | 0.8639 | 0.7945 | |
| DT | 85.34 | 0.8550 | 0.9261 | 0.8562 | 0.8554 | 0.7817 | |
| ResNet18 | SVM | 94.35 | 0.9443 | 0.9713 | 0.9457 | 0.9448 | 0.9163 |
| k-NN | 94.17 | 0.9433 | 0.9705 | 0.9430 | 0.9431 | 0.9136 | |
| NB | 89.93 | 0.9003 | 0.9489 | 0.9042 | 0.9017 | 0.8512 | |
| DT | 84.45 | 0.8459 | 0.9213 | 0.8496 | 0.8475 | 0.7691 | |
| ResNet50 | SVM | 95.23 | 0.9534 | 0.9759 | 0.9535 | 0.9534 | 0.9293 |
| k-NN | 93.11 | 0.9328 | 0.9650 | 0.9334 | 0.9331 | 0.8981 | |
| NB | 87.99 | 0.8810 | 0.9391 | 0.8860 | 0.8830 | 0.8225 | |
| DT | 85.34 | 0.8564 | 0.9258 | 0.8569 | 0.8565 | 0.7824 | |
| Inceptionv3 | SVM | 95.58 | 0.9570 | 0.9777 | 0.9574 | 0.9570 | 0.9348 |
| k-NN | 92.40 | 0.9261 | 0.9615 | 0.9282 | 0.9264 | 0.8887 | |
| NB | 86.57 | 0.8669 | 0.9320 | 0.8730 | 0.8687 | 0.8020 | |
| DT | 84.45 | 0.8460 | 0.9213 | 0.8506 | 0.8479 | 0.7695 | |
| DenseNet201 | SVM | 96.29 | 0.9642 | 0.9812 | 0.9642 | 0.9641 | 0.9453 |
| k-NN | 93.46 | 0.9365 | 0.9669 | 0.9371 | 0.9365 | 0.9036 | |
| NB | 85.34 | 0.8556 | 0.9258 | 0.8589 | 0.8569 | 0.7830 | |
| DT | 85.34 | 0.8556 | 0.9258 | 0.8589 | 0.8569 | 0.7830 | |
| Inceptionresnetv2 | SVM | 95.58 | 0.9570 | 0.9776 | 0.9569 | 0.9569 | 0.9345 |
| k-NN | 90.11 | 0.9031 | 0.9496 | 0.9051 | 0.9038 | 0.8539 | |
| NB | 81.80 | 0.8216 | 0.9076 | 0.8273 | 0.8239 | 0.7319 | |
| DT | 84.28 | 0.8455 | 0.9201 | 0.8497 | 0.8473 | 0.7677 | |
| MobileNetv2 | SVM | 94.52 | 0.9468 | 0.9723 | 0.9473 | 0.9467 | 0.9193 |
| k-NN | 92.58 | 0.9277 | 0.9624 | 0.9281 | 0.9278 | 0.8903 | |
| NB | 88.69 | 0.8877 | 0.9426 | 0.8921 | 0.8894 | 0.8326 | |
| DT | 86.57 | 0.8683 | 0.9323 | 0.8672 | 0.8677 | 0.8000 | |
| GoogleNet | SVM | 95.05 | 0.9519 | 0.9749 | 0.9517 | 0.9517 | 0.9267 |
| k-NN | 89.58 | 0.8977 | 0.9468 | 0.9021 | 0.8987 | 0.8470 | |
| NB | 88.87 | 0.8889 | 0.9437 | 0.8925 | 0.8903 | 0.8345 | |
| DT | 86.40 | 0.8656 | 0.9308 | 0.8712 | 0.8676 | 0.7993 |
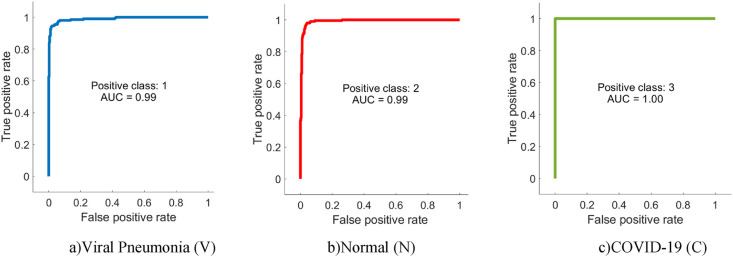
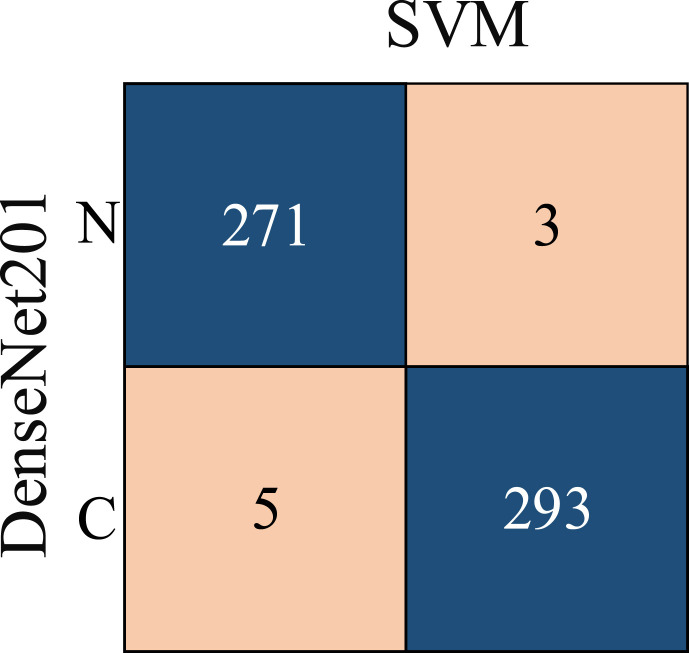
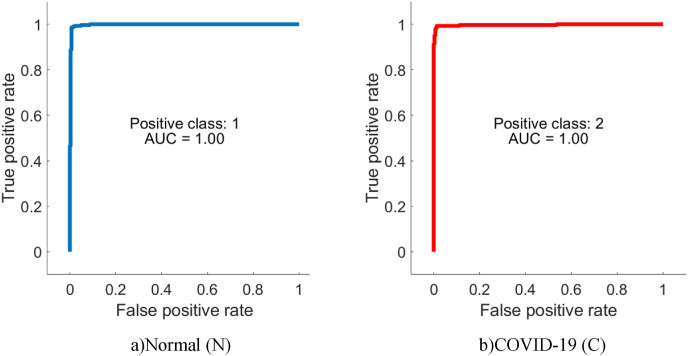
From Table 4, it is possible to observe that ML methods using hyperparameters calculated by Bayesian optimization successfully classify the features extracted from each CNN model. When the models are compared, it is understood that the highest accuracy is achieved with DenseNet201. Among ML algorithms, the strongest classification is carried out with SVM. The highest classification accuracy is calculated as 96.29% as a result of extracting features from segmented images via DenseNet and classification with SVM. The Sensitivity, Precision, Specificity, MCC, and F1-Score values for the DenseNet-SVM structure are 0.9642, 0.9642, 0.9812, 0.9641 and 0.9453, respectively. In addition, ROC curves are shown in Fig. 9 , which indicates that this DenseNet-SVM structure has a strong ability to discriminate between classes. Also, as seen in Table 4, these values are higher than the values obtained with other CNN models. In the classification made with SVM, kNN, NB and DT in all CNN models, the average accuracy values are 95.21%, 92.07%, 86.90%, and 85.27% respectively. This indicates that the structure has a more successful overall prediction and unbiased classification.
Fig. 9.
ROC curves of the DenseNet-SVM structure.
The comparison between this study and some previous studies in terms of classification accuracy is shown in Table 5 . Accordingly, the proposed method is comparable to previous state-of-the-art studies. There are two important contributions to the higher success of this study compared to other studies. First, the lungs are selected by ANN-based segmentation from the raw image and given to CNN models. The second is to determine the hyperparameters of the ML algorithms fed by the features extracted from the CNN models by Bayesian Optimization. Since this study uses only segmented lungs instead of using raw CXR images, the classification success has increased. However, this segmentation process can be further developed to provide a more accurate diagnosis. DL-based segmentation methods may be more effective for this.
Table 5.
The comparison of the proposed study and previous studies.
| Previous Studies | Methodologies | Accuracy (%) |
|---|---|---|
| Wang, Wong [46] | DL | 92.30 |
| Brunese, Mercaldo, Reginelli and Santone [47] | DL & TL | 97.00 |
| Afshar, Heidarian, Naderkhani, Oikonomou, Plataniotis and Mohammadi [48] | DL & Capsule network | 95.70 |
| Ismael and Şengür [49] | DL & SVM | 94.70 |
| Chowdhury, Rahman, Khandakar, Mazhar, Kadir, Mahbub, Islam, Khan, Iqbal and Al-Emadi [20] | DL & TL | 97.94 |
| Farooq, Hafeez [50] | DL & TL | 96.20 |
| Hemdan, Shouman and Karar [51] | VGG19 | 90.00 |
| Ucar, Korkmaz [52] | Bayes & SqueezeNet | 98.30 |
| Apostolopoulos, Mpesiana [53] | DL & TL | 93.48 |
| Xu, Jiang, et al. [54] | ResNet & Location Attention | 86.70 |
| Ozturk, Talo, Yildirim, Baloglu, Yildirim and Acharya [55] | DarkCovidNet | 87.02 |
| Rahimzadeh, Attar [56] | Xception - ResNet50V2 | 91.40 |
| Narin, Kaya and Pamuk [57] | DL & TL | 98.00 |
| Asif and Wenhui [24] | DL & TL | 96.00 |
| Nour, Cömert and Polat [2] | DL & ML | 98.97 |
| Wang, Xiao, Li, Zhang, Lu, Hou and Liu [58] | ResNet & Feature Pyramid Network | 94.00 |
| Khan, Shah and Bhat [59] | DL & TL | 95.00 |
| Momeny et al. [60] | DL & AlexNet | 77.60 |
| Rahman et al. [61] | HOG & DL | 96.04 |
| Shadin et al. [62] | DL & InceptionV3 | 85.94 |
| Monshi et al. [35] | DL & CovidXRayNet | 95.82 |
| Singh et al. [63] | DL & LeNet-5/ResNet-50 | 87.00 |
| Proposed Method | DL & ML | 96.29 |
The robustness and applicability of the proposed method in this study are also tested on a different dataset. The COVID-19 positive CXR images are collected from the open-source Covid-Chestxray-Dataset that is firstly introduced by Cohen et al. [64]. This dataset contains both CXR images and Computed Tomography images. Due to the concept of our article the Computed Tomography images are removed from the dataset. In the Covid-Chestxray-Dataset, all the CXR images that are taken from patients with COVID-19, other infected, no infection and non-commented, are combined in a single directory. The CXR images in this folder are associated with the disease via a list file. According to this list, the relevant CXR images have been selected. Due to the low number of disease samples other than COVID-19, only COVID-19 CXR are taken from this dataset. The number of CXR images taken from this database is 476. The number of non-COVID samples in the Covid-Chestxray-Dataset is quite low. Therefore, non-COVID CXR images are taken from a different dataset called ChexPert dataset. The ChexPert dataset is introduced by Irvin et al. [65]. 1913 non-COVID-19 CXR in the ChexPert dataset are added to the new database. As a result, a new two-class dataset consisting of a total of 2389 images, 476 COVID-19 and 1913 non-COVID-19, is obtained.
First, lungs are selected from the raw images in the new dataset, using ANN-based segmentation. Due to the imbalance between COVID-19 positive and normal CXR images, data augmentation techniques applied in the first dataset are applied to COVID-19 positive images. Thus, the number of COVID-19 positive images is 1904. As a result, the new two-class dataset consists of 3817 CXR images. Then, the features are extracted using the DenseNet201 model so that the new dataset can be tested with the DenseNet-SVM model, which has the highest accuracy. 85% of 3817 data containing 1000 features are used for training and the remaining 15% for testing. Finally, classification was made with the Bayesian optimization-based SVM model and 98.60% accuracy is obtained. The Sensitivity, Precision, Specificity, MCC, and F1-Score values for the DenseNet-SVM structure are 0.9891, 0.9819, 0.9832, 0.9855 and 0.9720, respectively. The confusion matrix obtained with the DenseNet-SVM model is shown in Fig. 10 . The receiver operating characteristic (ROC) curves of the test dataset are also shown in Fig. 11 . The results show that the model proposed in this study is successful and powerful in the diagnosis of COVID-19.
Fig. 10.
Confusion matrices of DenseNet-SVM structure.
Fig. 11.
ROC curves of the test dataset.
5. Conclusion
Early detection of the COVID-19 pandemic is important to prevent the spread and early treatment. This study easily diagnoses COVID-19 using CXR images. First, the raw CXR images are segmented by using ANN. In this way, masked CXR images with only the lungs part are considered for the detection of COVID-19. After that, images of the COVID-19 class are augmented to ensure data diversity. In the next step, the segmented and augmented images are applied as input to state-of-the-art CNN models. These CNN models are used for feature extraction only. 85% of the extracted features are separated for train and 15% for test. The features extracted from each model are given to four different ML methods for classification. These ML methods contain parameters that affect classification accuracy. In order to ensure the most accurate classification, each ML parameter is optimized by Bayesian Optimization in the training step. Test data are classified in the test step by using the hyperparameters found as a result of the optimization.
As a result of the classification, it is seen that the features extracted from different CNN models are successfully classified with four ML algorithms. However, the most successful classification is realized as 96.29% with DenseNet-SVM structure. The results obtained with SVM are higher than the other three ML algorithms. The average accuracy of eight different CNN models based on SVM is 95.21%. The results show that this proposed study is robust for COVID-19 diagnosis. In addition, to prove this experimentally, the most successful DenseNet-SVM structure is applied on a different dataset and 98.60% accuracy is achieved.
The shortcoming in the studies so far is generally to give the whole scan image directly as input to the network. Also, the number of studies using optimization to improve the prediction performance of the network is limited. It is clearly seen from results when compared with previous studies with the same dataset, lung segmentation and hyperparameter optimization of ML methods create a significant difference in increasing system performance. As a result of these, this study contributes in terms of achieving high classification accuracy and proposing the combination of state-of-the-art CNN models and optimized ML algorithms.
This study proposes an end-to-end learning structure, so this model does not require any manual feature extraction. COVID-19 infection is automatically detected using CXR images. Thus, a secure and fast decision support system is provided to assist expert radiographs. This reduces the radiologists' workload and prevents misdiagnosis. The methodological significance/contribution of this study can be discussed in three steps. First, the segmentation of lung regions in CXR images is one of the most important factors affecting the accuracy of this study. Due to the patient's condition during CXR imaging, it is not always possible to match the patient's chest position to the same location on the scan. In addition, there are always some informative symbols on the scan that possibly affects the ML performance negatively. The extracted features based on these symbols may cause faulty learning of the AI used for diagnosis. Therefore, other parts should not affect the diagnosis, except the lung. To avoid this situation, ANN-assisted lung segmentation methods are proposed. In this way, AI focuses only on features in the lung region. Therefore, this provides a stronger basis for diagnosis. Secondly, this study presents a comprehensive comparison based on DL and ML. It extracts features from eight frequently preferred CNN models and classifies these features with four different ML methods. The results obtained prove that different combinations of DL and ML show high performance for the diagnosis of COVID-19. In this sense, it provides a great benefit to researchers for future studies. Finally, the third step aims to increase the success of DL-ML combinations by optimizing the ML methods used. In this context, thanks to Bayesian optimization, the improvement of classification results is provided.
Although this study provides high performance in the diagnosis of COVID-19, it also has limitations. This proposed method, which gives high classification accuracy in the COVID-19 Radiography Database dataset, may give lower accuracy in a different dataset. Because the scan images in different datasets differ from each other due to the label, noise, etc. reasons they contain. In order to overcome this problem, AI should be trained by using scan images taken at different times and places. In addition to the diversity of the data, the distribution among the classes in the data is also important. The imbalance between the numbers of classes negatively affects training. The difference in data augmentation methods used to eliminate this imbalance also changes the accuracy of classification. As another limitation, although the Bayesian optimization applied in our study increases the ML classification successes, the processing time is too high. Therefore, parameter optimization slows down the diagnosis speed.
Considering all these limitations, in future works, experiments will be carried out using more and more various scan images. It is aimed to use different optimization techniques that are more suitable in terms of calculation time. To extract more robust features, it is planned to combine the features extracted from different CNN models, and then determine the most effective ones among these many features by feature selection algorithms. Thus, the accuracy of the diagnosis of COVID-19 can be increased and more reliable results can be produced.
Declaration of competing interest
The authors of this study declare that they have no conflict of interest.
References
- 1.E.C.f.D.P.a.C. (ECDC) 2020. COVID-19 Situation Update Worldwide. as of 18 October 2020. [Google Scholar]
- 2.Nour M., Cömert Z., Polat K. Applied Soft Computing; 2020. A Novel Medical Diagnosis Model for COVID-19 Infection Detection Based on Deep Features and Bayesian Optimization; p. 106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Awadhi A.M., Alsaifi K., Al-Awadhi A., Alhammadi S. Death and contagious infectious diseases: impact of the COVID-19 virus on stock market returns. J. Behav. Exp. Finance. 2020;27:100326. doi: 10.1016/j.jbef.2020.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islam M., Karray F., Alhajj R., Zeng J. 2020. A Review on Deep Learning Techniques for the Diagnosis of Novel Coronavirus (Covid-19) arXiv preprint arXiv:2008.04815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canayaz M., MH-COVIDNet Diagnosis of COVID-19 using deep neural networks and meta-heuristic-based feature selection on X-ray images. Biomed. Signal Process Control. 2021;64:102257. doi: 10.1016/j.bspc.2020.102257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W.-Y., Lo C.S.Y., Lui M.M.-S., Lee J.C.Y., Chiu K.W.-H., Chung T. 2020. Frequency and Distribution of Chest Radiographic Findings in COVID-19 Positive Patients, Radiology; p. 201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S., Lim S., Kim C., Wi S., Kwon T., Youn W.S., Lee S.H., Kang B.S., Cho S. Enhancement of soft-tissue contrast in cone-beam CT using an anti-scatter grid with a sparse sampling approach. Phys. Med. 2020;70:1–9. doi: 10.1016/j.ejmp.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 8.J.W. Bulte, M.M. Modo, Design and Applications of Nanoparticles in Biomedical Imaging, Springer2017.
- 9.Li B., Li X., Wang Y., Han Y., Wang Y., Wang C., Zhang G., Jin J., Jia H., Fan F. Diagnostic value and key features of computed tomography in Coronavirus Disease 2019. Emerg. Microb. Infect. 2020;9:787–793. doi: 10.1080/22221751.2020.1750307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobi A., Chung M., Bernheim A., Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin. Imag. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96:106691. doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos, Solit. Fractals. 2020;140:110122. doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang F., Qi Z., Duan K., Xi D., Zhu Y., Zhu H., Xiong H., He Q. A comprehensive survey on transfer learning. Proc. IEEE. 2020;109:43–76. [Google Scholar]
- 14.Gou J., Ma H., Ou W., Zeng S., Rao Y., Yang H. A generalized mean distance-based k-nearest neighbor classifier. Expert Syst. Appl. 2019;115:356–372. [Google Scholar]
- 15.A. Ben-Hur, J. Weston, A User's Guide to Support Vector Machines, Data Mining Techniques for the Life Sciences, Springer2010, pp. 223-239.
- 16.Brijain M., Patel R., Kushik M., Rana K. 2014. A Survey on Decision Tree Algorithm for Classification. [Google Scholar]
- 17.Huang Y., Li L. 2011 IEEE International Conference on Cloud Computing and Intelligence Systems, IEEE. 2011. Naive Bayes Classification Algorithm Based on Small Sample Set; pp. 34–39. [Google Scholar]
- 18.Bergstra J., Bengio Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012;13:281–305. [Google Scholar]
- 19.Yao Y., Cao J., Ma Z. International Conference on Service-Oriented Computing, Springer. 2018. A cost-effective deadline-constrained scheduling strategy for a hyperparameter optimization workflow for machine learning algorithms; pp. 870–878. [Google Scholar]
- 20.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N. 2020. Can AI Help in Screening Viral and COVID-19 Pneumonia? arXiv preprint arXiv:2003.13145. [Google Scholar]
- 21.Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos, Solit. Fractals. 2020:110059. doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev. Biomed. Eng. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 23.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process Control. 2021;64:102365. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asif S., Wenhui Y. Automatic detection of COVID-19 using X-ray images with deep convolutional neural networks and machine learning. medRxiv. 2020 doi: 10.1101/2020.05.01.20088211. [DOI] [Google Scholar]
- 25.Wu X., Hui H., Niu M., Li L., Wang L., He B., Yang X., Li L., Li H., Tian J. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur. J. Radiol. 2020:109041. doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J. 2020. A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia. Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos, Solit. Fractals. 2020:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. 2020:1–13. [Google Scholar]
- 31.Elaziz M.A., Hosny K.M., Salah A., Darwish M.M., Lu S., Sahlol A.T. New machine learning method for image-based diagnosis of COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. 2021;98:106742. doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qaid T.S., Mazaar H., Al-Shamri M.Y.H., Alqahtani M.S., Raweh A.A., Alakwaa W. Hybrid deep-learning and machine-learning models for predicting COVID-19. Comput. Intell. Neurosci. 2021;2021 doi: 10.1155/2021/9996737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das A.K., Kalam S., Kumar C., Sinha D. TLCoV-An automated Covid-19 screening model using Transfer Learning from chest X-ray images. Chaos, Solit. Fractals. 2021;144:110713. doi: 10.1016/j.chaos.2021.110713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monshi M.M.A., Poon J., Chung V., Monshi F.M. CovidXrayNet: optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Comput. Biol. Med. 2021;133:104375. doi: 10.1016/j.compbiomed.2021.104375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajpal S., Lakhyani N., Singh A.K., Kohli R., Kumar N. Using handpicked features in conjunction with ResNet-50 for improved detection of COVID-19 from chest X-ray images. Chaos, Solit. Fractals. 2021;145:110749. doi: 10.1016/j.chaos.2021.110749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A., Tripathi A.R., Satapathy S.C., Zhang Y.-D., SARS-Net COVID-19 detection from chest x-rays by combining graph convolutional network and convolutional neural network. Pattern Recogn. 2022;122:108255. doi: 10.1016/j.patcog.2021.108255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. CNN-based transfer learning–BiLSTM network: a novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021;98:106912. doi: 10.1016/j.asoc.2020.106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aslan M.F., Sabanci K., Durdu A. A CNN-based novel solution for determining the survival status of heart failure patients with clinical record data: numeric to image. Biomed. Signal Process Control. 2021;68:102716. [Google Scholar]
- 40.O'Shea K., Nash R. 2015. An Introduction to Convolutional Neural Networks. arXiv preprint arXiv:1511.08458. [Google Scholar]
- 41.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 42.Aslan M.F., Durdu A., Yusefi A., Sabanci K., Sungur C. A tutorial: mobile robotics, SLAM, bayesian filter, keyframe bundle adjustment and ROS applications. Robot Oper. Syst. 2021:227–269. [Google Scholar]
- 43.Wu J., Chen X.-Y., Zhang H., Xiong L.-D., Lei H., Deng S.-H. Hyperparameter optimization for machine learning models based on Bayesian optimization. J. Electron. Sci. Technol. 2019;17:26–40. [Google Scholar]
- 44.Frazier P.I. 2018. A Tutorial on Bayesian Optimization. arXiv preprint arXiv:1807.02811. [Google Scholar]
- 45.Bishnoi S., Ravinder R., Grover H.S., Kodamana H., Krishnan N.A. Scalable Gaussian processes for predicting the optical, physical, thermal, and mechanical properties of inorganic glasses with large datasets. Mater. Adv. 2021;2:477–487. [Google Scholar]
- 46.Wang L., Wong A. 2020. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Progr. Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. 2020. Covid-caps: A Capsule Network-Based Framework for Identification of Covid-19 Cases from X-Ray Images. arXiv preprint arXiv:2004.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2020;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farooq M., Hafeez A. 2020. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 51.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 52.Ucar F., Korkmaz D. COVIDiagnosis-net: deep Bayes-SqueezeNet based diagnostic of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J. 2020. A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahimzadeh M., Attar A. 2020. A New Modified Deep Convolutional Neural Network for Detecting COVID-19 from X-Ray Images. arXiv preprint arXiv:2004.08052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., Liu X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recogn. 2020;110:107613. doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Progr. Biomed. 2020:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Momeny M., Neshat A.A., Hussain M.A., Kia S., Marhamati M., Jahanbakhshi A., Hamarneh G. Learning-to-augment strategy using noisy and denoised data: improving generalizability of deep CNN for the detection of COVID-19 in X-ray images. Comput. Biol. Med. 2021;136:104704. doi: 10.1016/j.compbiomed.2021.104704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahman M.M., Nooruddin S., Hasan K., Dey N.K. HOG+ CNN Net: diagnosing COVID-19 and pneumonia by deep neural network from chest X-Ray images. Sn Comput. Sci. 2021;2:1–15. doi: 10.1007/s42979-021-00762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shadin N.S., Sanjana S., Lisa N.J. 2021 International Conference on Information Technology (ICIT), IEEE. 2021. COVID-19 diagnosis from chest X-ray images using convolutional neural network (CNN) and InceptionV3; pp. 799–804. [Google Scholar]
- 63.Singh S., Sapra P., Garg A., Vishwakarma D.K. 2021 5th International Conference on Computing Methodologies and Communication (ICCMC), IEEE. 2021. CNN based covid-aid: covid 19 detection using chest X-ray; pp. 1791–1797. [Google Scholar]
- 64.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. Covid-19 Image Data Collection: Prospective Predictions Are the Future. arXiv preprint arXiv:2006.11988. [Google Scholar]
- 65.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K. Proceedings of the AAAI Conference on Artificial Intelligence. 2019. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison; pp. 590–597. [Google Scholar]