Abstract
During our survey into the diversity of woody litter fungi across the Greater Mekong Subregion, three rhytidhysteron-like taxa were collected from dead woody twigs in China and Thailand. These were further investigated based on morphological observations and multi-gene phylogenetic analyses of a combined DNA data matrix containing SSU, LSU, ITS, and tef1-α sequence data. A new species of Rhytidhysteron, R.xiaokongense sp. nov. is introduced with its asexual morph, and it is characterized by semi-immersed, subglobose to ampulliform conidiomata, dark brown, oblong to ellipsoidal, 1-septate, conidia, which are granular in appearance when mature. In addition to the new species, two new records from Thailand are reported viz. Rhytidhysterontectonae on woody litter of Betula sp. (Betulaceae) and Fabaceae sp. and Rhytidhysteronneorufulum on woody litter of Tectonagrandis (Lamiaceae). Morphological descriptions, illustrations, taxonomic notes and phylogenetic analyses are provided for all entries.
Keywords: Ascomycota, one new taxon, phylogeny, saprobic, taxonomy, Yunnan
Introduction
Hysteriaceae was introduced by Chevallier (1826) with Hysterium as the type genus, which was characterized by hysterothecial or apothecioidal, carbonaceous ascomata with a pronounced, longitudinal slit running the length of the long axis, 8-spored, clavate to cylindric asci with an ocular chamber as well as obovoid, clavate, ellipsoid or fusoid, hyaline to light- or dark brown, one to multi-septate or muriform, smooth-walled ascospores with or without a sheath (Boehm et al. 2009b; Hongsanan et al. 2020; Hyde et al. 2020a). In recent outlines of Dothideomycetes (Hongsanan et al. 2020; Pem et al. 2020; Wijayawardene et al. 2020), 14 genera have been accepted in Hysteriaceae.
Rhytidhysteron was introduced by Spegazzini (1881) to accommodate two species: Rhytidhysteronbrasiliense (type species) and R.viride in Patellariaceae (Clements and Shear 1931; Kutorga and Hawksworth 1997). Boehm et al. (2009a, b) transferred Rhytidhysteron from Patellariaceae to Hysteriaceae based on molecular data. Subsequent studies introduced more taxa and records in Rhytidhysteron with both morphological and molecular evidence (Thambugala et al. 2016; Doilom et al. 2017; Cobos-Villagrán et al. 2020; Dayarathne et al. 2020; de Silva et al. 2020; Hyde et al. 2020a, b; Mapook et al. 2020; Wanasinghe et al. 2021). Currently, 24 species are recognized in Rhytidhysteron (Species Fungorum 2021; Wanasinghe et al. 2021).
Rhytidhysteron species have been documented from a wide range of hosts in various countries such as Australia, Bermuda, Bolivia, Brazil, China, Colombia, Cuba, France, Hawaii, India, New Zealand, Thailand, Ukraine, USA, and Venezuela (Kutorga and Hawksworth 1997; de Silva et al. 2020). Most Rhytidhysteron species are identified as saprobes on woody-based substrates in terrestrial habitats as well as from mangrove wood in marine habitats (Thambugala et al. 2016; Kumar et al. 2019; Hyde et al. 2020a, b; Wanasinghe et al. 2021). However, they have also been reported as endophytes or weak pathogens on woody plants and seldom as human pathogens (Soto and Lucking 2017; de Silva et al. 2020). From a biotechnological perspective, Rhytidhysteron species have great potential for their commercial applications and in industry. In particular, interest in secondary metabolites has rekindled in recent years, for instance with the discovery of palmarumycins. The latter is a potential inhibitor of thioredoxin–thioredoxin reductase cellular redox systems, with potential antimicrobial and antifungal properties (Murillo et al. 2009). Other Rhytidhysteron species discovered from the Southeast Asian region, such as R.bruguierae (MFLUCC 17-1515) and R.chromolaenae (MFLUCC 17-1516) also showed antimicrobial activity against Mucorplumbeus (Mapook et al. 2020) and hence this demonstrates a potential biotechnological application.
The Greater Mekong Subregion (GMS) is regarded as a global biodiversity hotspot due to its widely varying environmental conditions. Accordingly, the GMS harbors a diverse array of numerous florae, fauna and microorganisms (Li et al. 2018). Woody litter microfungi is an overlooked group of fungi in GMS and based on previous fungal estimates, there is undoubtedly a large number of new species yet to be described from this region. Our ongoing studies into the diversity of microfungi of the GMS are actively contributing towards filling in the knowledge gap in fungal taxonomy, phylogeny, host association and ecological distribution of Rhytidhysteron species in this region (Luo et al. 2018; Bao et al. 2019; Dong et al. 2020; Hyde et al. 2020b; Monkai et al. 2020, 2021; Wanasinghe et al. 2020, 2021; Yasanthika et al. 2020). Our specific objectives of this study are as follows: 1) to describe a novel species of Rhytidhysteron with evidence from morphology and DNA sequence data; 2) to characterize (based on morphology and phylogeny) additional new records of Rhytidhysteron; 3) to investigate the phylogenetic relationships of our Rhytidhysteron samples based on DNA sequence analyses from rDNA and protein coding genes and update the taxonomy of Rhytidhysteron.
Materials and methods
Samples collection and morphological analyses
Woody litter samples were collected from China (Kunming, Yunnan Province) during the wet season (August 2019) and during the dry season (December 2019) collections were done in Thailand (Chiang Rai and Tak Provinces). Samples were brought to the laboratory in plastic Ziploc bags. Fungal specimens were then examined using a stereomicroscope (Olympus SZ61, China). Pure cultures were obtained via single spore isolation on potato dextrose agar (PDA) following the methods described in Senanayake et al. (2020). Cultures were incubated at 25 °C for one week in the dark. Digital images of the fruiting structures were captured with a Canon (EOS 600D) digital camera fitted to a Nikon ECLIPSE Ni compound microscope. Squash mount preparations were prepared to determine micro-morphology and free hand sections of sporocarps made to observe the shapes of ascomata/conidiomata and peridium structures. Measurements of morphological structures were taken from the widest part of each structure. When possible, more than 30 measurements were made. Measurements were taken using the Tarosoft (R) Image Frame Work program. Figures were processed using Adobe Photoshop CS6. Field data are presented in ‘Material examined’. Other details pertaining to good practices of morphological examinations were done following guidelines by Senanayake et al. (2020). New species are established based on recommendations proposed by Jeewon and Hyde (2016). Type specimens were deposited in the herbarium of the Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS). Ex-type living cultures were deposited at the Culture Collection of Mae Fah Luang University (MFLUCC) and Kunming Institute of Botany Culture Collection (KUMCC).
DNA extraction, amplification and sequencing
Genomic DNA was extracted from the mycelium grown on PDA at 25–30 °C for one week using a Biospin Fungus Genomic DNA Extraction Kit (BioFlux Hangzhou, P. R. China). Three partial rDNA genes and a protein coding gene were processed in our study, including the small ribosomal subunit RNA (SSU) using the primer pair NS1/NS4 (White et al. 1990), internal transcribed spacer region (ITS) using the primer pair ITS5/ITS4 (White et al. 1990), large nuclear ribosomal subunit (LSU) using primer pair LR0R/LR5 (Vilgalys and Hester 1990), translation elongation factor 1-alpha gene (tef1-α) using primer pair 983F/2218R (Rehner and Buckley 2005). Amplification reactions were performed in a total volume of 25 μL of PCR mixtures containing 8.5 μL ddH2O, 12.5 μL 2X PCR MasterMix (TIANGEN Co., China), 2 μL DNA template and 1 μL of each primer. PCR thermal cycle program for SSU, LSU, ITS, and tef1-α were set as described in Wanasinghe et al. (2020). The PCR products were sent to the Qingke Company, Kunming City, Yunnan Province, China, for sequencing. Sequences were deposited in GenBank (Table 1).
Table 1.
GenBank accession numbers of sequences used for the phylogenetic analyses.
| Taxon | Strain number | GenBank accession numbers | Reference | |||
|---|---|---|---|---|---|---|
| SSU | LSU | ITS | tef1-α | |||
| Gloniopsiscalami | MFLUCC 15-0739 | KX669034 | NG_059715 | KX669036 | KX671965 | Hyde et al. (2016) |
| Gloniopsispraelonga | CBS 112415 | FJ161134 | FJ161173 | NA | FJ161090 | Boehm et al. (2009a) |
| Rhytidhysteronbruguierae | MFLUCC 18-0398T | MN017901 | MN017833 | NA | MN077056 | Dayarathne et al. (2020) |
| Rhytidhysteronbruguierae | MFLUCC 17-1515 | MN632463 | MN632452 | MN632457 | MN635661 | Mapook et al. (2020) |
| Rhytidhysteronbruguierae | MFLUCC 17 1511 | MN632465 | MN632454 | MN632459 | NA | Mapook et al. (2020) |
| Rhytidhysteronbruguierae | MFLUCC 17-1502 | MN632464 | MN632453 | MN632458 | MN635662 | Mapook et al. (2020) |
| Rhytidhysteronbruguierae | MFLUCC 17-1509 | MN632466 | MN632455 | MN632460 | NA | Mapook et al. (2020) |
| Rhytidhysteroncamporesii | HKAS 104277T | NA | MN429072 | MN429069 | MN442087 | Hyde et al. (2020a) |
| Rhytidhysteronchromolaenae | MFLUCC 17-1516T | MN632467 | MN632456 | MN632461 | MN635663 | Mapook et al. (2020) |
| Rhytidhysteronerioi | MFLU 16-0584T | NA | MN429071 | MN429068 | MN442086 | Hyde et al. (2020a) |
| Rhytidhysteronhongheense | KUMCC 20-0222T | MW264224 | MW264194 | MW264215 | MW256816 | Wanasinghe et al. (2021) |
| Rhytidhysteronhongheense | HKAS112348 | MW541831 | MW541820 | MW54182 | MW556132 | Wanasinghe et al. (2021) |
| Rhytidhysteronhongheense | HKAS112349 | MW541832 | MW541821 | MW541825 | MW556133 | Wanasinghe et al. (2021) |
| Rhytidhysteronhysterinum | EB 0351 | NA | GU397350 | NA | GU397340 | Boehm et al. (2009b) |
| Rhytidhysteronmagnoliae | MFLUCC 18-0719T | MN989382 | MN989384 | MN989383 | MN997309 | de Silva et al. (2020) |
| Rhytidhysteronmangrovei | MFLUCC 18-1113T | NA | MK357777 | MK425188 | MK450030 | Kumar et al. (2019) |
| Rhytidhysteronneorufulum | MFLUCC 13-0216T | KU377571 | KU377566 | KU377561 | KU510400 | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | GKM 361A | GU296192 | GQ221893 | NA | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | HUEFS 192194 | NA | KF914915 | NA | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | MFLUCC 12-0528 | KJ418119 | KJ418117 | KJ418118 | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | CBS 306.38 | AF164375 | FJ469672 | NA | GU349031 | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | MFLUCC 12-0011 | KJ418110 | KJ418109 | KJ206287 | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | MFLUCC 12-0567 | KJ546129 | KJ526126 | KJ546124 | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | MFLUCC 12-0569 | KJ546131 | KJ526128 | KJ546126 | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | EB 0381 | GU397366 | GU397351 | NA | NA | Thambugala et al. (2016) |
| Rhytidhysteronneorufulum | MFLUCC 21-0035 | MZ346025 | MZ346015 | MZ346020 | MZ356249 | This study |
| Rhytidhysteronopuntiae | GKM 1190 | NA | GQ221892 | NA | GU397341 | Mugambi et al. (2009) |
| Rhytidhysteronrufulum | MFLUCC 14-0577T | KU377570 | KU377565 | KU377560 | KU510399 | Thambugala et al. (2016) |
| Rhytidhysteronrufulum | EB 0384 | GU397368 | GU397354 | NA | NA | Boehm et al. (2009b) |
| Rhytidhysteronrufulum | EB 0382 | NA | GU397352 | NA | NA | Boehm et al. (2009b) |
| Rhytidhysteronrufulum | EB 0383 | GU397367 | GU397353 | NA | NA | Boehm et al. (2009b) |
| Rhytidhysteronrufulum | MFLUCC 12-0013 | KJ418113 | KJ418111 | KJ418112 | NA | de Silva et al. (2020) |
| Rhytidhysterontectonae | MFLUCC 13-0710T | KU712457 | KU764698 | KU144936 | KU872760 | Doilom et al. (2017) |
| Rhytidhysterontectonae | MFLUCC 21-0037 | MZ346023 | MZ346013 | MZ346018 | MZ356247 | This study |
| Rhytidhysterontectonae | MFLUCC 21-0034 | MZ346024 | MZ346014 | MZ346019 | MZ356248 | This study |
| Rhytidhysteronthailandicum | MFLUCC 14-0503T | KU377569 | KU377564 | KU377559 | KU497490 | Thambugala et al. (2016) |
| Rhytidhysteronthailandicum | MFLUCC 12-0530 | KJ546128 | KJ526125 | KJ546123 | NA | Thambugala et al. (2016) |
| Rhytidhysteronthailandicum | MFLU17-0788 | MT093495 | MT093472 | MT093733 | NA | de Silva et al. (2020) |
| Rhytidhysteronxiaokongense | KUMCC 20-0158 | MZ346021 | MZ346011 | MZ346016 | MZ356245 | This study |
| Rhytidhysteronxiaokongense | KUMCC 20-0160T | MZ346022 | MZ346012 | MZ346017 | MZ356246 | This study |
Ex-type strains are indicated with superscript “T”, and newly generated sequences are shown in bold. NA represents sequences that are unavailable in GenBank.
Phylogenetic analyses
Representative species used in the phylogenetic analyses were selected based on previous publications (Thambugala et al. 2016; Mapook et al. 2020; Wanasinghe et al. 2021). Sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) and their accession numbers are listed in Table 1. The newly generated sequences in this study were assembled by BioEdit 7.0.9.0 (Hall 1999). Individual gene regions were separately aligned in MAFFT v.7 web server (http://mafft.cbrc.jp/alignment/server/) (Katoh et al. 2019). The alignments of each gene were improved by manually deleting the ambiguous regions and gaps, and then combined using BioEdit 7.2.3. Final alignments containing SSU, LSU, ITS, and tef1-α were converted to NEXUS format (.nxs) using CLUSTAL X (2.0) and PAUP v. 4.0b10 (Thompson et al. 1997; Swofford 2002) and processed for Bayesian and maximum parsimony analysis. The FASTA format was changed into PHYLIP format via the Alignment Transformation Environment (ALTER) online program (http://www.sing-group.org/ALTER/) and used for maximum likelihood analysis (ML).
ML was carried out in CIPRES Science Gateway v.3.3 (http://www.phylo.org/portal2/; Miller et al. 2010) using RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2014) with the GTRGAMMA substitution model and 1,000 bootstrap iterations. Maximum parsimony analysis (MP) was performed in PAUP v. 4.0b10 (Swofford 2002) with the heuristic search option and Tree-Bisection-Reconnection (TBR) of branch-swapping algorithm for 1,000 random replicates. Branches with a minimum branch length of zero were collapsed and gaps were treated as missing data (Hillis and Bull 1993). ML and MP bootstrap values (ML) ≥ 75% are given above each node of the phylogenetic tree (Fig. 1).
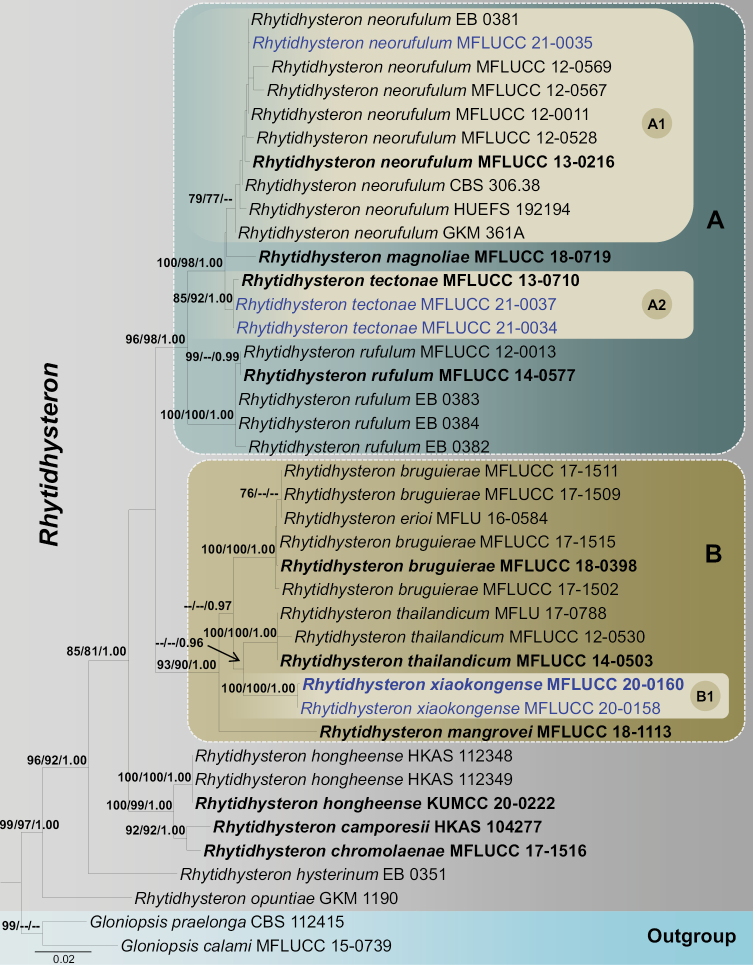
Figure 1.
RAxML tree based on a combined dataset of partial SSU, LSU, ITS, and tef1-α sequence analyses. Bootstrap support values for ML and MP equal to or higher than 75% and Bayesian PP equal to or greater than 0.95 are shown at the nodes. Hyphens (--) represent support values less than 75% / 0.95 BYPP. The ex-type strains are in bold and the new isolate in this study is in blue. The tree is rooted with Gloniopsiscalami (MFLUCC 15-0739) and G.praelonga (CBS 112415).
Bayesian analysis was executed in MrBayes v.3.2.2 (Ronquist et al. 2012). The model of evolution was estimated using MrModeltest v. 2.3 (Nylander et al. 2008) via PAUP v. 4.0b10 (Ronquist and Huelsenbeck 2003). The HKY+I for SSU; GTR+I+G for ITS, LSU and tef1-α were used in the final command. Markov chain Monte Carlo sampling (MCMC) in MrBayes v.3.2.2 (Ronquist et al. 2012) was used to determine posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002). Bayesian analyses of six simultaneous Markov chains were run for 2,000,000 generations and trees were sampled every 200 generations (resulting in 10,001 total trees). The first 25% of sampled trees were discarded as part of the burn-in procedure, the remaining 7,501 trees were used to create the consensus tree, and the average standard deviation of split frequencies was set as 0.01. Branches with Bayesian posterior probabilities (BYPP) ≥ 0.95 are indicated above each node of the phylogenetic tree (Fig. 1). Phylogenetic trees were visualized in FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/; Rambaut 2012). The tree was edited using Microsoft PowerPoint before being, then saved in PDF format and finally converted to JPG format using Adobe Illustrator CS6 (Adobe Systems, USA). The finalized alignments and trees were deposited in TreeBASE, submission ID: TB2:S28620 (http://purl.org/phylo/treebase/phylows/study/TB2:S28620).
Results
Phylogenetic analysis
The phylogenetic analysis was conducted using 38 strains in Rhytidhysteron, and two outgroup taxa viz. Gloniopsiscalami (MFLUCC 15-0739) and G.praelonga (CBS 112415) in Pleosporales (Table 1). The aligned sequence matrix comprised four gene regions (SSU: 1018 bp, LSU: 891 bp, ITS: 742 bp and tef1-α: 953 bp) and a total of 3,604 characters (including gaps), of which 3,095 characters were constant, 161 variable characters were parsimony-uninformative and 348 characters were parsimony-informative. The Kishino-Hasegawa test shows length = 928 steps with CI = 0.696, RI = 0.846, RC = 0.589 and HI = 0.304. The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of -10181.226009. The matrix had 723 distinct alignment patterns, with 26.6% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.242390, C = 0.244261, G = 0.276352, T = 0.236997; substitution rates AC = 1.457846, AG = 2.708684, AT = 1.298658, CG = 0.909442, CT = 6.323746, GT = 1.00; gamma distribution shape parameter α = 0.02.
Topologies of the phylogenetic trees under ML, MP and BI criteria recovered for each gene dataset were visually compared, and the overall tree topology was similar to those obtained from the combined dataset (Figure 1). Our analyzed molecular data generated phylogeny of Rhytidhysteron species was consistent with those of Wanasinghe et al. (2021). The maximum likelihood tree generated based on sequence analysis of the combined (ribosomal DNA: SSU, LSU and ITS; and protein coding gene: tef1-α) dataset recovered three major monophyletic clades within Rhytidhysteron (A-C, Figure 1) and two basal lineages viz. R.hysterinum (EB 0351) and R.opuntiae (GKM 1190). Clade A comprises Rhytidhysteronmagnoliae, R.neorufulum, R.rufulum and R.tectonae with 96% ML, 98% MP and 1.00 BYPP support values.
One of our new isolates, MFLUCC 21-0035 grouped with another nine Rhytidhysteronneorufulum strains (CBS 306.38, EB 0381, GKM 361A, HUEFS 192194, MFLUCC 12-0011, MFLUCC 12-0528, MFLUCC 12-0567, MFLUCC 12-0569, MFLUCC 13-0216, MFLUCC 21-0035). However, this relationship is not statistically supported in Bayesian analysis, retrieving 79% and 77% support values in ML and MP, respectively (sub clade A1, Figure 1). Rhytidhysteronmagnoliae (MFLUCC 18-0719) constitutes an independent lineage and is a sister taxon to others in sub clade A1, and this was not statistically supported.
Two newly generated sequences MFLUCC 21-0034 and MFLUCC 21-0037 grouped with the type strain of Rhytidhysterontectonae (MFLUCC 13-0710) as a monophyletic clade within Clade A (subclade A2, Figure 1). This association was supported by 85% ML, 92% MP and 1.00 BYPP bootstrap values (subclade A2, Figure 1). Five strains of Rhytidhysteronrufulum (EB 0382, EB 0383, EB 0384, MFLUCC 12-0013, MFLUCC 14-0577) constitute another strongly monophyletic group basal to Clade A.
Two of our newly generated sequences, Rhytidhysteronxiaokongense (KUMCC 20-0158, KUMCC 20-0160), grouped with R.bruguierae (MFLUCC 17-1511, MFLUCC 17-1502, MFLUCC 17-1509, MFLUCC 17-1515, MFLUCC 18-0398), R.erioi (MFLU 16-0584), R.mangrovei (MFLUCC 18-1113) and R.thailandicum (MFLU 17-0788, MFLUCC 12-0530, MFLUCC 14-0503). These taxa form a monophyletic clade (Clade B) in Rhytidhysteron with 93% ML, 91% MP and 1.00 BYPP bootstrap values. Within this clade (Clade B), Rhytidhysteronxiaokongense (KUMCC 20-0158 and KUMCC 20-0160) clusters together (subclade B1) with high bootstrap values (100% ML, 100% MP and 1.00 BYPP) and is sister to Rhytidhysteronthailandicum. However, the latter relationship was only supported by BI analysis with 0.96 BYPP.
Rhytidhysteroncamporesii (HKAS104277), R.chromolaenae (MFLUCC 17-1516) and R.hongheense (HKAS112348, HKAS112349, KUMCC 20-0222) grouped as a monophyletic clade. This relationship is statistically supported with 100% ML, 99% MP and 1.00 BYPP values (Figure 1). Rhytidhysteronhysterinum (EB 0351) and R.opuntiae (GKM 1190) nested as basal lineages in Rhytidhysteron (Figure 1).
Taxonomy
. Rhytidhysteron xiaokongense
G.C. Ren & K.D. Hyde sp. nov.
87692543-D03E-5EF3-BDED-7D2E30016BF7
558453
Facesoffungi Number No: FoF09903
Figure 2.
Rhytidhysteronxiaokongense (HKAS 112728, holotype) a, b conidiomata on natural wood surface c sections through conidioma d ostiolar neck e conidioma wall f–h conidiogenous cells and developing conidia i–m conidia n germinated conidium o, p culture characters on PDA (o = above, p = reverse). Scale bars: 100 μm (c, d); 50 μm (e); 15 μm (f–h); 10 μm (i–m); 20 μm (n); 25 mm (o, p).
Etymology.
The species epithet reflects the location where the species was collected.
Holotype.
HKAS 112728.
Diagnosis.
Similar to R.hysterinum and R.rufulum, but differs in some conidial features.
Description.
Saprobic on woody litter of Prunus sp. Sexual morph Undetermined. Asexual morphConidiomata 448–464 × 324–422 µm (x̄ = 454 × 378 μm, n = 5), solitary, scattered, semi-immersed in the host, black, unilocular, subglobose to ampulliform. Ostioles 178–227 × 166–234 µm (x̄ = 205 × 198 μm, n = 6), central, short papillate. Conidiomata wall 30–40 μm thick, 4–6 layers, reddish-brown to dark brown cells of textura angularis. Conidiogenous cells 5–8 × 3–6 µm (x̄ = 6.8 × 4.5 μm, n = 10), subglobose or ellipsoidal, hyaline, smooth, forming in a single layer over the entire inner surface of the wall, discrete, producing a single conidium at the apex. Conidia 20–25 × 8–10 μm (x̄ = 22 × 9 μm, n = 20), hyaline to yellowish-brown when immature, becoming brown to dark brown at maturity, oblong to ellipsoidal, with rounded ends, straight to slightly curved, aseptate when immature, becoming 1-septate when mature, with granular appearance, slightly constricted at septa.
Habitat and distribution.
Known to inhabit woody litter of Prunus sp. (Yunnan, China) (this study).
Material examined.
China, Yunnan Province, Kunming city, Xiaokong Mountain (25.171311°N, 102.703690°E), on dead wood of Prunus sp. (Rosaceae), 21-Dec-2019, G.C. Ren, KM18 (HKAS 112728, holotype), ex-type living culture KUMCC 20-0160; KM17 (HKAS 112727, paratype), ex-paratype living culture KUMCC 20-0158.
Notes.
Rhytidhysteronxiaokongense is similar to R.hysterinum and R.rufulum in having black, unilocular, subglobose conidiomata and dark brown, 1-septate conidia. However, some of the conidia features in these species are different: R.xiaokongense has oblong to ellipsoidal conidia with rounded ends, whereas the conidia of R.rufulum and R.hysterinum have a truncated base with a pore in the middle of the septum (Samuels and Müller 1979). In the phylogenetic analyses, R.xiaokongense is distinct from R.rufulum and R.hysterinum and is more closely related to R.thailandicum. Rhytidhysteronxiaokongense has 1-septate, dark brown, oblong to ellipsoidal conidia, while R.thailandicum has globose to subglobose, hyaline conidia (Thambugala et al. 2016). The sequence data from both mycelium and fruiting bodies confirms that single spore isolation was successfully performed.
. Rhytidhysteron tectonae
Doilom & K.D. Hyde, Fungal Diversity. 82: 107–182 (2017)
A040E115-BF8E-5F4A-B91B-BCF1737464B4
551964
Facesoffungi number No: FoF01849
Figure 3.
Rhytidhysterontectonae (HKAS 115533) a, bHysterothecium on wood c vertical section through hysterothecia d exciple e pseudoparaphyses f–i immature and mature asci j ocular chamber. k–r immature and mature ascospores s Germinating ascospore t, u culture characters on PDA (t = above view, u = reverse view). Scale bars: 300 μm (c); 50 μm (d); 30 μm (e); 50 μm (f–i); 10 μm (j–r); 15 μm (s); 25 mm (t, u).
Description.
Saprobic on decaying wood. Sexual morphHysterothecia 550–950 µm long, 450–600 µm high, 400–500 diam. (x̄ = 800 × 500 × 450 µm, n = 5), semi-immersed to superficial, scattered, apothecial, erumpent from the substrate, dark brown to black, coriaceous, elongate with a longitudinal slit. Exciple 70–110 µm (x̄ = 90 µm, n = 15), thick-walled, composed of brown to dark brown cells of textura globulosa to angularis. Hamathecium comprising 1–2 μm wide, numerous, septate, branched, pseudoparaphyses. Asci 170–200 × 10–12 μm (x̄ = 190 × 11, n = 15), 8-spored, bitunicate, cylindrical, with short pedicel, rounded at the apex, with an ocular chamber. Ascospores 25–29 × 8–10 µm (x̄ = 27 × 9 µm, n = 20), uniseriate, hyaline to brown, 1–3-septate, smooth-walled, ellipsoidal to fusoid, straight or curved, rounded to slightly pointed at both ends, guttulate. Asexual morph Undetermined.
Habitat and distribution.
Known to inhabit dead branches of Tectonagrandis, Betula sp. (Betulaceae) and Fabaceae sp (Thailand) (Doilom et al. 2017; this study).
Material examined.
Thailand, Chiang Rai Province, Mae Yao District, on dead woody twigs of Betula sp. (Betulaceae), 23-Sep-2019, G.C. Ren, MY09 (HKAS 115533), living culture MFLUCC 21-0037; Thailand, Chiang Rai Province, Mae Fah Luang University, on dead woody twigs of Fabaceae, 5-Jul-2019, G.C. Ren, RMFLU19001 (HKAS 115532), living culture MFLUCC 21-0034.
Notes.
Rhytidhysterontectonae was introduced by Doilom et al. (2017) based on morphological and phylogenetic analyses from dead branches of Tectonagrandis in Thailand. Based on our phylogenetic analysis of the combined SSU, LSU, ITS, and tef1-α sequence data, our collections (MFLUCC 21-0034 and MFLUCC 21-0037) cluster with the strain of R.tectonae (MFLUCC 13-0710) with 85% ML, 92% MP, 1.00 PP bootstrap support (Figure 1). Our collection shares similar morphological features with R.tectonae (MFLU 14-0607). However, our new collection has smaller hysterothecia (800 × 500 × 450 μm vs 2175 × 585 × 523 μm) and longer asci (190 μm vs 155 μm) in comparison to the type. Based on morphological characteristics and phylogenetic analysis, we introduce MFLUCC 21-0034 and MFLUCC 21-0037 as new host records of R.tectonae from decaying wood of Betula sp. and Fabaceae sp. in Thailand.
. Rhytidhysteron neorufulum
Thambug. & K.D. Hyde, Cryptog. Mycol. 37(1): 110 (2016)
52DFAE6A-942F-58F9-944E-34EB9C0FC19E
551865
Facesoffungi number No: FoF01840
Figure 4.
Rhytidhysteronneorufulum (HKAS 115534) a, bHysterothecium on wood c vertical section through hysterothecia d exciple e pseudoparaphyses f–h immature asci and mature asci i–m immature ascospores and mature ascospores n germinating ascospore o, p culture characters on PDA (o = above view, p = reverse view). Scale bars: 1000 μm (a, b); 200 μm (c); 15 μm (d); 20 μm (e); 50 μm (f–h); 10 μm (i–m); 20 μm (n); 20 mm (o, p).
Description.
Saprobic on decaying wood of Tectonagrandis. Sexual morphHysterothecia 1400–2100 μm long, 350–500 μm high, 600–1000 μm diam. (x̄ = 1780 × 400 × 700 μm, n = 5), superficial, black, solitary to aggregated, coriaceous, smooth, elliptical or irregular in shape, elongated with a longitudinal slit. Exciple 75–115μm (x̄ = 90, n = 20) wide, composed of several layers of brown to dark brown, thick-walled cells of textura angularis. Hamathecium 2–3.5 μm wide, dense, septate pseudoparaphyses, constricted at the septum, filiform, pale-yellow pigmented, forming epithecium above the asci and enclosed in a gelatinous matrix. Asci 190–260 × 13–18 μm (x̄ = 230 × 16 μm, n = 10), 8-spored, bitunicate, clavate to cylindrical, with a short furcate pedicel, apically rounded, without a distinct ocular chamber. Ascospores 36–44 × 11–17 μm (x̄ = 41 × 13 μm, n = 30), uni-seriate, yellowish to brown, with 1–3-septa, ellipsoidal to fusiform, slightly rounded or pointed at both ends, constricted at the central septum, with granular appearance. Asexual morph Undetermined.
Habitat and distribution.
Bursera sp (Mexico), Heveabrasiliensis and Tectonagrandis (Thailand) (Thambugala et al. 2016; Cobos-Villagran et al. 2020; this study).
Material examined.
Thailand, Tak Province, Mogro District, Amphoe Umphang, on dead woods of Tectonagrandis (Lamiaceae), 20-Aug-2019, G.C. Ren, T203 (HKAS 115534), living culture MFLUCC 21-0035.
Notes.
Rhytidhysteronneorufulum was introduced by Thambugala et al. (2016) based on both morphological and phylogenetic analyses of a combined dataset of LSU, SSU and tef1-α sequence data. Thambugala et al. (2016) accounted R.neorufulum (MFLUCC 13-0216) from decaying woody stems and twigs in Thailand. Our new collection shares similar morphology to that of the type description of Rhytidhysteronneorufulum (MFLUCC 13-0216) in having superficial, coriaceous, elliptical or irregular, elongated hysterothecia with a longitudinal slit, bitunicate, cylindrical, short furcate pedicel asci and yellowish to brown, ellipsoidal to fusiform ascospores with 1–3-septa (Thambugala et al. 2016). However, our new collection has larger asci (190–260 × 13–18 μm vs 185–220 × 9.5–13 μm) and ascospores (36–44 × 11–17 μm vs 19–31 × 8–13 μm) in comparison to the type of Rhytidhysteronneorufulum (MFLUCC 13-0216). The multi-gene phylogenetic analysis based on combined SSU, LSU, ITS, and tef1-α sequence data showed that our collection is related to Rhytidhysteronneorufulum (Figure 1).
Key to asexual morphs of Rhytidhysteron species
| 1 | Asexual morph has two types of conidia | 2 |
| – | Asexual morph has only one type of conidia | 3 |
| 2 | Comprising paraphyses | R.hysterinum |
| – | Paraphyses are absent | R.rufulum |
| 3 | Diplodia-like conidia | R.xiaokongense |
| – | Aposphaeria-like conidia | R.thailandicum |
Discussion
Rhytidhysteron is one of the first genera that trainee mycologists working on microfungi find in nature, as the hysterothecia are conspicuous (Hyde et al. 2020a). Species also easily germinate in culture and can easily be sequenced (Hyde et al. 2020a). Thus, it is even more remarkable that we found a new species in this study, indicating we are far from finding all species in this genus, and that more collections need be done on other continents (Hyde et al. 2020c). Most of Rhytidhysteron species are saprobes, which are essential for ecosystems functioning in terrestrial habitats and are commonly recognized as key biotic agents of wood decomposition, playing a vital role in carbon and nitrogen cycling in arid ecosystems, soil stability, plant biomass decomposition, and endophytic interactions with plants (Lustenhouwer et al. 2020; Dossa et al. 2021). Furthermore, Rhytidhysteron species have numerous antimicrobial and antifungal applications (Murillo et al. 2009; Mapook et al. 2020), and the discovery of new species provides new resources for future applied research in the field of biotechnology and industry.
Since the genus was established in 1881, a total of 24 species have been found to date, and the most commonly encountered species are Rhytidhysteronneorufulum and R.rufulum, so it might be difficult for mycologists to find new species within Rhytidhysteron. Rhytidhysteron is mainly identified via its sexual morph (Dayarathne et al. 2020; de Silva et al. 2020; Hyde et al. 2020a, b; Mapook et al. 2020; Wanasinghe et al. 2021). The asexual morphs of Rhytidhysteron have been reported as aposphaeria-like or diplodia-like, including R.hysterinum and R.rufulum (Samuels and Müller 1979). Thambugala et al. (2016) confirmed the asexual-sexual morph connection for R.thailandicum by aposphaeria-like asexual morphs forming in culture on PDA. Herein, we found a diplodia-like asexual morph of Rhytidhysteron from woody litter of Prunus sp. in China. In comparison to the occurrence of the sexual morph of Rhytidhysteron, asexual morphs seldom form under natural conditions. The discovery of this new species provides an important reference for the study of the asexual morphs of Rhytidhysteron. Moreover, findings from this study further enrich GMSRhytidhysteron species diversity.
In our phylogenetic analyses, the new species, Rhytidhysteronxiaokongense was basal to R.thailandicum (Fig. 1). Although species in Rhytidhysteron are morphologically similar, our new species is an asexual form of the species found in nature, so it is easy to distinguish from other speices excluding the asexual forms of R.hysterinum, R.rufulum and R.thailandicum. Rhytidhysteronxiaokongense shares similar morphological characters to R.hysterinum and R.rufulum in having black, unilocular, subglobose conidiomata and dark brown, 1-septate conidia but conidial features differ (Samuels and Müller 1979). Rhytidhysteronthailandicum can be differentiated from R.xiaokongense with respects to its globose to subglobose, hyaline conidia (Thambugala et al. 2016). To further support the establishment of the new taxon as proposed by Jeewon and Hyde (2016), we examined the nucleotide differences within the ITS regions (ITS1-5.8S-ITS2) gene region. Comparison of the 507 nucleotides across the ITS regions reveals 39 bp (7.7%) differences between Rhytidhysteronthailandicum and R.xiaokongense.
Rhytidhysteron species are widely distributed throughout the globe (de Silva et al. 2020); however, they appear to be particularly abundant in Asia, where they are well studied. There is an abundance of species and collections in the Greater Mekong Subregion (China and Thailand), such as R.brasiliense, R.camporesii, R.chromolaenae, R.erioi, R.hongheense, R.hysterinum, R.magnoliae, R.mangrovei, R.neorufulum, R.tectonae and R.thailandicum (Thambugala et al. 2016; Doilom et al. 2017; Soto-Medina et al. 2017; Kumar et al. 2019; Cobos-Villagran et al. 2020; Dayarathne et al. 2020; de Silva et al. 2020; Hyde et al. 2020a; Mapook et al. 2020; Wanasinghe et al. 2021). We provide morphological and phylogenetic data for three species of Rhytidhysteron collected from the Greater Mekong Subregion: one new species, Rhytidhysteronxiaokongense, as a geographical record from China, two new host records of R.tectonae from woody litter of Betula sp and Fabaceae sp, and one new host record of R.neorufulum from woody litter of Tectonagrandis. Based on our current work and that of past studies (de Silva et al. 2020; Hyde et al. 2020a, b; Mapook et al. 2020; Wanasinghe et al. 2021), it is clear that species within Rhytidhysteron are likely cosmopolitan and not host-specific, with evidence of the same species being found on a number of different hosts. Importantly, the morphology of a single species sometimes shows slight variations under different environmental conditions, geographical regions, hosts and different life modes (Senanayake et al. 2020). It is therefore crucial to collect more species of Rhytidhysteron across different geographic regions and hosts, obtain more cultures and sequence data, and describe their morphology to improve knowledge of taxonomy and phylogeny.
Supplementary Material
Acknowledgements
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDA2602020). We thank the support from the National Natural Science Foundation of China (NSFC32001296). We also would like to thank the Thailand Research Fund for the grant entitled Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (No. RDG6130001). Dhanushka Wanasinghe thanks the CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2021FYB0005), the Postdoctoral Fund from Human Resources and Social Security Bureau of Yunnan Province and the National Science Foundation of China, High-End Foreign Experts” in the High-Level Talent Recruitment Plan of Yunnan Province (2021) and Chinese Academy of Sciences (grant no. 41761144055) for financial support. Austin G. Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing.
Citation
Ren G-C, Wanasinghe DN, Jeewon R, Monkai J, Mortimer PE, Hyde KD, Xu J-C, Gui H (2022) Taxonomy and phylogeny of the novel rhytidhysteron-like collections in the Greater Mekong Subregion. MycoKeys 86: 65–85. https://doi.org/10.3897/mycokeys.86.70668
Funding Statement
Kunming Institute of Botany, Chinese Academy of Science,the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDA2602020).
Contributor Information
Dhanushka N. Wanasinghe, Email: dnadeeshan@gmail.com.
Heng Gui, Email: guiheng@mail.kib.ac.cn.
References
- Bao DF, Hyde KD, Luo ZL, Su HY, Nalumpang S. (2019) Minutisphaeraaquaticum sp. nov. increases the known diversity of Minutisphaeraceae. Asian Journal of Mycology 2: 306–314. 10.5943/ajom/2/1/21 [DOI] [Google Scholar]
- Barr ME. (1990) Rhytidhysteronopuntiae. Memoirs of the New York Botanical Garden 62: e72.
- Boehm EW, Mugambi GK, Miller AN, Huhndorf SM, Marincowitz S, Spatafora JW, Schoch CL. (2009a) A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Studies in Mycology 64: 49–83. 10.3114/sim.2009.64.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EWA, Schoch CL, Spatafora JW. (2009b) On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycological Research 113: 461–479. 10.1016/j.mycres.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Chevallier FF. (1826) Flore générale des environs de Paris, vol I. Ferra Librairie-Editeur, Paris.
- Clements FE, Shear CL. (1931) The Genera of Fungi; Hafner Publishing Co. : New York, NY, USA, 632 pp. [Google Scholar]
- Cobos-Villagrán A, Hernández-Rodríguez C, Valenzuela R, Villa-Tanaca L, Calvillo-Medina RP, Mateo-Cid LE, Martínez-Pineda M, Raymundo T. (2020) The genus Rhytidhysteron (Dothideomycetes, Ascomycota) in Mexico. Acta Botanica Mexicana 127: e1675. 10.21829/abm127.2020.1675 [DOI]
- Dayarathne MC, Jones EBG, Maharachchikumbura SSN, Devadatha B, Sarma VV, Khongphinitbunjong K, Chomnunti P, Hyde KD. (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11: 1–188. 10.5943/mycosphere/11/1/1 [DOI] [Google Scholar]
- De Silva NI, Tennakoon DS, Thambugala KM, Karunarathna SC, Lumyong S, Hyde KD. (2020) Morphology and multigene phylogeny reveal a new species and a new record of Rhytidhysteron (Dothideomycetes, Ascomycota) from China. Asian Journal of Mycology 3: 295–306. 10.5943/ajom/3/1/4 [DOI] [Google Scholar]
- Doilom M, Dissanayake AJ, Wanasinghe DN, Boonmee S, Liu JK, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EHC, Hyde KD. (2016) Micro fungi on Tectonagrandis (teak) in Northern Thailand. Fungal Diversity 82: 107–182. 10.1007/s13225-016-0368-7 [DOI] [Google Scholar]
- Dong W, Wang B, Hyde KD, McKenzie EHC, Raja HA, Tanaka K, Abdel-Wahab MA, Abdel-Aziz FA, Doilom M, Phookamsak R, Hongsanan S, Wanasinghe DN, Yu XD, Wang GN, Yang H, Yang J, Thambugala KM, Tian Q, Luo ZL, Yang JB, Miller AN, Fournier J, Boonmee S, Hu DM, Nalumpang S, Zhang H. (2020) Freshwater Dothideomycetes. Fungal Diversity 105: 319–575. [DOI] [Google Scholar]
- Dossa GGO, Yang YQ, Hu W, Paudel E, Schaefer D, Yang YP, Cao KF, Xu JC, Bushley KE, Harrison RD. (2021) Fungal succession in decomposing woody debris across a tropical forest disturbance gradient. Soil Biology and Biochemistry 155: e108142. 10.1016/j.soilbio.2021.108142 [DOI]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie EHC, Sarma VV, Boonmee S, Lücking R, Bhat DJ, Liu NG, Tennakoon DS, Pem D, Karunarathna A, Jiang SH, Jones EBG, Phillips AJL, Manawasinghe IS, Tibpromma S, Jayasiri SC, Sandamali DS, Jayawardena RS, Wijayawardene NN, Ekanayaka AH, Jeewon R, Lu YZ, Dissanayake AJ, Zeng XY, Luo ZL, Tian Q, Phukhamsakda C, Thambugala KM, Dai DQ, Chethana KWT, Samarakoon MC, Ertz D, Bao DF, Doilom M, Liu JK, Pérez-Ortega S, Suija A, Senwanna C, Wijesinghe SN, Konta S, Niranjan M, Zhang SN, Ariyawansa HA, Jiang HB, Zhang JF, Norphanphoun C, de Silva NI, Thiyagaraja V, Zhang H, Bezerra JDP, Miranda-González R, Aptroot A, Kashiwadani H, Harishchandra D, Sérusiaux E, Aluthmuhandiram JVS, Abeywickrama PD, Devadatha B, Wu HX, Moon KH, Gueidan C, Schumm F, Bundhun D, Mapook A, Monkai J, Chomnunti P, Suetrong S, Chaiwan N, Dayarathne MC, Yang J, Rathnayaka AR, Bhunjun CS, Xu JC, Zheng JS, Liu G, Feng Y, Xie N. (2020) Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11: 1553–2107. 10.5943/mycosphere/11/1/13 [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu NG, Abeywickrama PD, Mapook A, Wei D, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao DF, Li J, Samarakoon MC, Chaiwan N, Lin CG, Phutthacharoen K, Zhang SN, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang HB, Yang J, Zeng M, Huanraluek N, Liu JK, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang SK, Thiyagaraja V, Lu YZ, Jayawardena RS, Dong W, Yang EF, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, da Silva Santos AC, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu J, Sheng J. (2020a) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Hyde KD, de Silva N, Jeewon R, Bhat DJ, Phookamsak R, Doilom M, Boonmee S, Jayawardena RS, Maharachchikumbura SSN, Senanayake IC, Manawasinghe IS, Liu NG, Abeywickrama PD, Chaiwan N, Karunarathna A, Pem D, Lin CG, Sysouphanthong P, Luo ZL, Wei DP, Wanasinghe DN, Norphanphoun C, Tennakoon DS, Samarakoon MC, Jayasiri SC, Jiang HB, Zeng XY, Li JF, Wijesinghe SN, Devadatha B, Goonasekara ID, Brahmanage RS, Yang EF, Aluthmuhandiram JVS, Dayarathne MC, Marasinghe DS, Li WJ, Dissanayake LS, Dong W, Huanraluek N, Lumyong S, Liu JK, Karunarathna SC, Jones EBG, Al-Sadi AM, Xu JC, Harishchandra D, Sarma VV. (2020b) AJOM new records and collections of fungi: 1–100. Asian Journal of Mycology 3: 22–294 10.5943/ajom/3/1/3 [DOI] [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ, Bhunjun CS, Calabon MS, Jiang HB, Lin CG, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena RS, Liu JK, Maharachchikumbura SSN, Phukhamsakda C, Phookamsak R, Al-Sadi AM, Naritsada Thongklang N, Wang Y, Gafforov Y, Jones EBG, Lumyong S. (2020c) The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 103: 219–271. 10.1007/s13225-020-00458-2 [DOI]
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, Santiago ALCMA, Drechsler-Santos ER, Senanayake I, Tanaka K, Tenna-koon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu HX, Ynag J, Zeng XY, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XZ, Lücking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen TC, Xu JC, Zhang ZK, Zhao YC, Zhou JL, Zhu L. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, McKenzie EHC, Jeewon R, Phillips AJL, Bhat DJ, Wanasinghe DN, Liu JK, Lu YZ, Kang JC, Xu J, Karunarathna SC. (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10: 1–186. 10.5943/mycosphere/10/1/1 [DOI] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in bioinformatics 20: 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Cheewangkoon R, Thambugala KM, Jones GEB, Brahmanage RS, Doilom M, Jeewon R, Hyde KD. (2019) Rhytidhysteronmangrovei (Hysteriaceae), a new species from mangroves in Phetchaburi Province, Thailand. Phytotaxa 401: 166–178. 10.11646/phytotaxa.401.3.2 [DOI] [Google Scholar]
- Kutorga E, Hawksworth DL. (1997) A re-assessment of the genera referred to the family Patellariaceae (Ascomycota). Systema Ascomycetum 15: 1–110. [Google Scholar]
- Li H, Guo J, Karunarathna SC, Ye L, Xu J, Hyde KD, Mortimer PE. (2018) Native Forests Have a Higher Diversity of Macrofungi Than Comparable Plantation Forests in the Greater Mekong Subregion. Forests 9: e402. 10.3390/f9070402 [DOI]
- Luo Z, Hyde KD, Bhat DJ, Jeewon R, Maharachchikumbura SSN, Bao DF, Li WL, Su XJ, Yang XY, Su HY. (2018) Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycological Progress 17: 511–530. 10.1007/s11557-018-1377-6 [DOI] [Google Scholar]
- Lustenhouwer N, Maynard DS, Bradford MA, Lindner DL, Oberle B, Zanne AE, Crowther TW. (2020) A trait-based understanding of wood decomposition by fungi. Proceedings of the National Academy of Sciences 117: 11551–11558. 10.1073/pnas.1909166117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkai J, Boonmee S, Ren GC, Wei DP, Phookamsak R, Mortimer PE. (2020) Distoseptisporahydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 459: 093–107. 10.11646/phytotaxa.459.2.1 [DOI] [Google Scholar]
- Monkai J, Wanasinghe DN, Jeewon R, Promputtha I, Phookamsak R. (2021) Morphological and phylogenetic characterization of fungi within Bambusicolaceae: introducing two new species from the Greater Mekong Subregion. Mycological Progress 20: 721–732. 10.1007/s11557-021-01694-9 [DOI] [Google Scholar]
- Mapook A, Hyde KD, McKenzie EHC, Jones EBG, Bhat DJ, Jeewon R, Stadler M, Samarakoon MC, Malaithong M, Tanunchai B, Buscot F, Wubet T, Purahong W. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaenaodorata (Siam weed). Fungal Diversity 101: 1–175. 10.1007/s13225-020-00444-8 [DOI] [Google Scholar]
- Murillo C, Albertazzi FJ, Carranza J, Lumbsch HT, Tamayo G. (2009) Molecular data indicate that Rhytidhysteronrufulum (ascomycetes, Patellariales) in Costa Rica consists of four distinct lineages corroborated by morphological and chemical characters. Mycological Research 113: 405–16. 10.1016/j.mycres.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Mugambi GK, Huhndorf SM. (2009) Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7: 453–464. 10.1017/S147720000999020X [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Niranjan M, Tiwari S, Baghela A, Sarma VV. (2018) New records of Ascomycetous fungi from Andaman Islands, India and their molecular sequence data. Current Research in Environmental & Applied Mycology 8: 331–350. 10.5943/cream/8/3/5 [DOI] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. (2008) AWTY: a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- Pem D, Hongsanan S, Doilom M, Tibpromma S, Wanasinghe DN, Dong W, Ningguo L, Phookamsak R, Phillips AJL, Jeewon R, Hyde KD. (2019) https://www.dothideomycetes.org: An online taxonomic resource for the classification, identification, and nomenclature of Dothideomycetes. Asian Journal of Mycology 2: 287–297. 10.5943/ajom/2/1/19 [DOI] [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree [Accessed 1 May 2020]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. 10.1080/15572536.2006.11832842 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels GJ, Müller E. (1979) Life-history studies of Brazilian ascomycetes. 7. Rhytidhysteronrufulum and the genus Eutryblidiella. Sydowia 32: 277–292. [Google Scholar]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Nguyen TT, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Jayawardena RS, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang MM. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Soto EM, Lücking R. (2017) A new species of Rhytidhysteron (Ascomycota: Patellariaceae) from Colombia, with a provisional working key to known species in the world Artículo original. Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales 41: 59–63. 10.18257/raccefyn.423 [DOI] [Google Scholar]
- Spegazzini C. (1881) Fungi argentini additis nonnullis brasiliensibus montevideensibusque. Pugillus quartus (Continuacion). An Soc Cient Argent 12: 174–189. [Google Scholar]
- Species Fungorum (2021) Species Fungorum. http://www.speciesfungorum.org/Names/Names.asp
- Stamatakis A. (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland, 56(9): 1776–1778. 10.1111/j.0014-3820.2002.tb00191.x [DOI] [Google Scholar]
- Thambugala KM, Hyde KD, Eungwanichayapant PD, Romero AI, Liu ZY. (2016) Additions to the Genus Rhytidhysteron in Hysteriaceae. Cryptogamie, Mycologie 37: 99–116. 10.7872/crym/v37.iss1.2016.99 [DOI] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Wijayawardene NN, Xu JC, Cheewangkoon R, Mortimer PE. (2020) Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS ONE 15: e0235855. 10.1371/journal.pone.0235855 [DOI] [PMC free article] [PubMed]
- Wanasinghe DN, Mortimer PE, Xu J. (2021) Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaeaviscosa in Honghe (China). Journal of Fungi 7: e180. 10.3390/jof7030180 [DOI] [PMC free article] [PubMed]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A Guide to Methods and Applications. Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk PM, Kolaříková K, Raja HA, Radek R, Papp V, Dima V, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska JZ, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Lateef AA, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu J, Wang Y, Tian F, Alvarado P, Li DW, Kušan I, Matočec N, Mešić A, Tkalčec Z, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCMA, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Silva-Filho AGS, Gentekaki E, Liu P, Cavender JC, Kang Y, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Kuhnert E, Grossart HP, Thines M. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11: 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Yasanthika E, Dissanayake LS, Wanasinghe DN, Karunarathna SC, Mortimer PE, Samarakoon BC, Monkai J, Hyde KD. (2020) Lonicericolafuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan Province, China. Phytotaxa 446: 103–113. 10.11646/phytotaxa.446.2.3 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 1–4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.