Abstract
There is evidence that Staphylococcus aureus colonisation is linked to severity of atopic dermatitis. As no gold standard for S. aureus sampling on atopic dermatitis skin lesions exists, this study compared three commonly used methods. In addition, effectiveness of standard skin disinfection to remove S. aureus colonisation from these inflamed skin lesions was investigated. In 30 atopic dermatitis patients, three different S. aureus sampling methods, i.e. detergent scrubbing, moist swabbing and tape stripping, were performed on naïve and disinfected skin lesions. Two different S. aureus selective media, mannitol salt agar and chromID agar, were used for bacterial growing. Quantifying the S. aureus load varied significantly between the different sampling methods on naïve skin lesions ranging from mean 51 to 1.5 × 104 CFU/cm2 (p < 0.001). The qualitative detection on naïve skin was highest with the two detergent-based techniques (86% each), while for tape stripping, this value was 67% (all on chromID agar). In comparison, mannitol salt agar was less sensitive (p < 0.001). The disinfection of the skin lesions led to a significant reduction of the S. aureus load (p < 0.05) but no complete eradication in the case of previously positive swab. The obtained data highlight the importance of the selected sampling method and consecutive S. aureus selection agar plates to implement further clinical studies for the effectiveness of topical anti-staphylococcal antibiotics. Other disinfection regimes should be considered in atopic dermatitis patients when complete de-colonisation of certain skin areas is required, e.g. for surgical procedures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10096-021-04365-5.
Keywords: Atopic dermatitis, Staphylococcus aureus, Colonisation, Sampling methods, Selective media
Introduction
Staphylococcus aureus is one of the most common pathogens associated with serious infections in almost all parts of the body. It is a well-known highly virulent pathogen that can cause a variety of diseases in humans, for example skin infections which range from rather benign such as folliculitis to more serious infections like furunculosis with a high risk of spreading with blood which may lead to fatal consequences. Osteomyelitis, pneumonia or endocarditis caused by S. aureus can also be the source of bloodstream infections and a life-threatening sepsis. Apart from that, up to 50% of healthy individuals are temporarily and 10–20% persistently colonised with S. aureus on intact skin and mucosae in the nose, hands, axilla and perineum. Together with a number of cell-bound virulence factors, S. aureus may secrete 30 or more specific products such as exotoxins that interfere with the host defence. Due to these different exotoxins, S. aureus is causing a variety of different systemic toxin-mediated diseases, from serious like staphylococcal toxic shock syndrome up to the frequent and mainly self-resolving staphylococcal food poisoning [1]. Additionally, there is a growing body of evidence linking S. aureus colonisation and degree of severity of atopic dermatitis (AD) [2]. However, since the 1970s, there is evidence that patients with AD are more likely to be colonised with S. aureus ranging from 30% to nearly 100% [3–5]. Finally, a systematic literature search summarised and confirmed that patients with AD are significantly more often colonised with S. aureus than healthy controls (odds ratio 19.7; 95% confidence interval 10.8–35.8) [6]. The detection of S. aureus in all these summarised studies was predominantly performed with culture-based swab or scrub methods based on a 1965 published survey by Williamson and Kligman [7]. While recent studies have relied on culture-independent methods, like DNA sequencing methods [8–10], they are not able to quantitatively assess the absolute degree of colonisation of vital S. aureus on affected skin and their load reduction after antimicrobial treatment.
Although the commonly used non-invasive detection methods (detergent scrubbing (DS), moist swabbing (MS) and tape stripping (TS)) have been known for decades to detect and quantify the bacterial load from human skin [11], no gold standard for the sampling method of culture-based S. aureus detection on skin lesions from patients with AD exists so far. To establish the optimal non-invasive S. aureus sampling method, DS, MS and TS were compared in this prospective, single-centre study. Thereby, we set out to determine the most sensitive method for qualitative and quantitative detection of living S. aureus on skin lesions from patients with AD on naïve and disinfected skin. This is of high interest for future clinical studies of topical anti-staphylococcal agents to measure their antimicrobial activity as well as for clinical routine use.
In addition, the sensitivity of two different S. aureus selection agars for detecting S. aureus and the effectiveness of common skin disinfection to remove S. aureus from AD skin lesions were investigated.
Materials and methods
The protocol had been approved by the Ethical Board of the Medical University of Vienna (No. 2155/2016) and performed in accordance with the Declaration of Helsinki (1964), Good Clinical Practice guidelines of the European Commission and the Good Scientific Practice guidelines of the Medical University of Vienna. The study recruitment was conducted from February 2017 to March 2018 at the Department of Dermatology of the Medical University of Vienna during routine outpatient visits. All patients included were between 18 and 70 years old, male or female with signed and dated informed consent obtained and diagnosed with localised atopic dermatitis according to the Hanifin & Rajka criteria [12] (e.g. flexural eczema in a more or less symmetrical distribution on arms) with two individual lesions each covering an area between 40 and 300 cm2. The Eczema Area and Severity Index (EASI) score [13] has been routinely used at our clinic during the study period. Excluded were patients with a history of irritation following contact with the topical products Triton X-100, Octenidindihydrochlorid and/or 2-phenoxyethanol and/or systemic or topical treatment with antibiotics and/or use of antiseptic soaps at the tested skin lesion within 7 days before this study. For comparing three non-invasive sampling methods for detecting and quantifying S. aureus on skin lesions from AD, the sample size was calculated with at least 11 eligible S. aureus–positive patients for analysis of variance (based on www.statstodo.com). Because the S. aureus status was unknown at the beginning and AD lesions are colonised ranging from 30% to nearly 100% with S. aureus, we estimated a number of 30 AD patients with unknown S. aureus status to be sufficient for the purposes of this study.
Skin disinfection
Two localised AD skin lesions, e.g. flexural eczema in a more or less symmetrical distribution on arms of an area between 40 and 300 cm2, were randomly selected, one for disinfection while the other skin lesion remained naïve. First, one randomly selected skin lesion was disinfected with a one-time wipe disinfection with the antiseptic agent Octenisept® containing 0.1 g Octenidindihydrochlorid and 2 g 2-phenoxyethanol per 100 ml. After an impact time of at least 1 min, the three sampling methods were performed on a defined skin area (i.e. 4.8 cm2 for DS, 4.5 cm2 for MS and 3.8 cm2 for TS) on the disinfected and also on the naïve skin lesion.
Performing the non-invasive bacterial sampling methods
At the study visit, three non-invasive sampling methods were compared concomitantly. The techniques were always being performed in the same order for all subjects and both skin lesions.
Detergent scrub technique
A cylinder with a defined internal area of 4.8 cm2 was held firmly against the affected area and filled with 2.5 ml of sterile buffered non-ionic wash fluid (0.1% Triton X-100 in 0.075 M sodium phosphate buffer, pH 7.9) as initially described by Williamson and Kligman in 1965 [7]. The affected skin surface within the ring was rubbed firmly with a hard stick (Teflon policeman) for 1 min. After this, all the wash fluid was aspirated by a pipette from the inner cylinder and transferred to sterile tubes.
Moistened swab technique
A circle template with a defined internal area of 4.5 cm2 was applied to the affected area and the limited skin area swapped with a sterile cotton swap moistened with sterile buffered non-ionic wash fluid (0.1% Triton X-100 in 0.075 M phosphate buffer, pH 7.9) 3 times; the swap was then submerged in 1 ml wash fluid [14]. The swap buffer solution was vortexed mildly to release bacteria.
Tape stripping technique
A tape strip (D-SQUAME Standard Sampling Discs, CuDerm, USA) of a defined size of 3.8 cm2 was applied with gentle pressure to the lesional skin surface for 1 min by using a sterile gloved hand [15, 16]. After peeled off from the skin, the tape strip was transferred to a tube in a sterile way and submerged in wash fluid (0.1% Triton X-100 in 0.075 M phosphate buffer, pH 7.9). The tape strip containing wash fluid was vortexed gently.
Microbiological processing
After performing the three methods, all collected liquid samples were immediately processed (but not later than 4 h after collecting) at the microbiology laboratory. To detect the bacterial load (colony-forming units (CFU)/ml) at each method, a serial dilution method was performed according to the Clinical and Laboratory Standards Institute (appendix 27). After incubation at 37 °C over 24 h, the S. aureus–specific colonies were counted. S. aureus–specific colonies were defined according to the manufacturer’s instructions at mannitol salt agar (MSA; Becton Dickinson GmbH, Germany) as medium-sized yellow colonies with yellow surrounding medium and at S. aureus chromID (SAID; bioMérieux, France) green colonies. MSA were used for samples from all patients (n = 30) and SAID selection agar for samples from the 10th patient on continually (n = 21). The amount of bacterial load was indicated in CFU per cm2 of the investigated AD skin area. All S. aureus strains were routinely confirmed on species level by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF, Bruker Daltonics, Bremen, Germany).
Statistics
The statistical significance (p < 0.05) of the measured difference of the quantitative detection of S. aureus with the three different sampling methods was determined by the Friedman test; furthermore, the difference on naïve versus disinfected AD skin and on the two different S. aureus selection agar plates was determined by the Wilcoxon test. The statistical significance (p < 0.05) of the different qualitative S. aureus detection (yes/no) by two S. aureus selective media was estimated by the McNemar test.
Results
In a total number of 30 patients, each suffering from AD, the three different non-invasive sampling methods (DS, MS, TS) were successfully performed on a disinfected and symmetrically distributed naïve AD skin lesion. The included patients with AD display a representative sample of a larger population of AD patients with mainly moderate disease according to the EASI score. Patient characteristics and disease severity are summarised in Table 1.
Table 1.
Patient characteristics
| Patients, n (female) | 30 (16) |
| Age, years; median (minimum–maximum) | 31.1 (19–69) |
| EASIa score, n (%) | |
|
0.1–1.0 (almost clear) 1.1–7.0 (mild) 7.1–21.0 (moderate) 21.1–50.0 (severe) 50.1–72.0 (very severe) |
0 3 (10%) 22 (73%) 5 (17%) 0 |
aEczema Area and Severity Index score
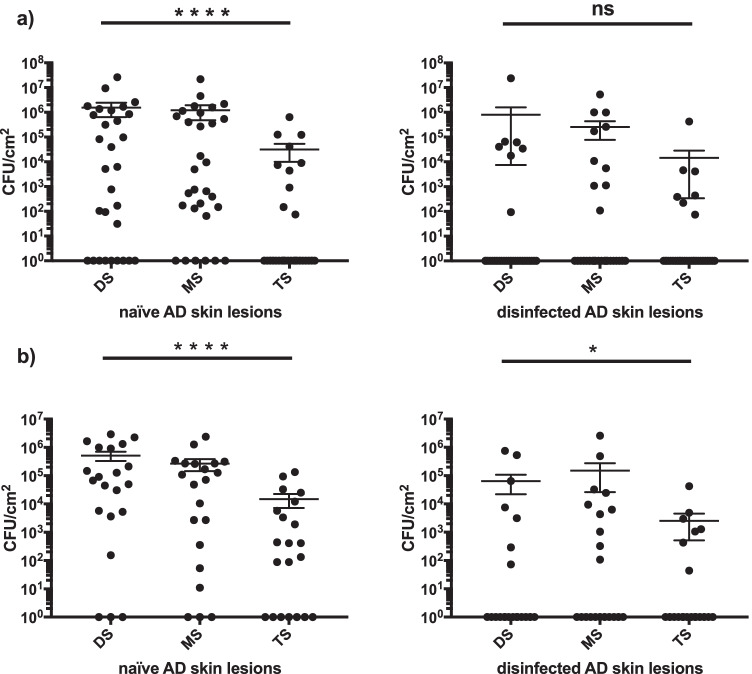
The sampling methods differed significantly in quantitative detection of S. aureus on naïve skin lesions (p < 0.001). The TS technique was notably less effective on naïve skin lesions compared to detergent-based methods like MS and DS, with the latter two showing no significant difference. After standardised disinfection, colonisation with S. aureus was significantly reduced (p < 0.05) independent of the detection method and selective media. Nevertheless, S. aureus was not fully eradicated and 47.6% were still colonised with S. aureus, as detected with MS, the most sensitive method. The results of the quantitative detection of S. aureus are summarised in Table 2 and Fig. 1.
Table 2.
Quantitative detection of S. aureus by three different sampling methods collected on naïve and disinfected atopic dermatitis skin lesions and cultivated on two different S. aureus selective media
| S. aureus selective media | Sampling methods | CFUa × 104/cm2 Mean (SD) |
CFUa × 104/cm2 Median (percentile 25; 75) |
||
|---|---|---|---|---|---|
| Naïve skin | Disinfected skin | Naïve skin | Disinfected skin | ||
| MSA (n = 30) | DS | 155.4 (495.2) | 80.6 (437.3) | 0.5 (0; 92.2) | 0 (0; 0.002) |
| MS | 120.3 (398) | 25.8 (98.6) | 0.2 (0.01; 75.5) | 0 (0; 0.2) | |
| TS | 3.2 (11.8) | 1.5 (7.8) | 0 (0; 0.2) | 0 (0; 0.002) | |
| SAID (n = 21) | DS | 51.9 (85.2) | 6.5 (19.5) | 6.8 (0.5; 95.8) | 0 (0; 0.2) |
| MS | 26.9 (55.9) | 15.1 (57.1) | 7.1 (0.02; 27) | 0 (0; 0.8) | |
| TS | 1.5 (3.5) | 0.3 (0.9) | 0.04 (0; 0.9) | 0 (0; 0.08) | |
aColony-forming units of S. aureus
Fig. 1.
Quantitative detection of S. aureus by using different detection methods and selective media (mean and standard error of the mean values). a Quantitative detection of S. aureus by using selective media MSA (n = 30). b Quantitative detection of S. aureus by using selective media SAID (n = 21). Abbreviations: ****p < 0.0001; *p < 0.05; ns = p > 0.05; MSA, mannitol salt agar; CFU, colony-forming units of Staphylococcus aureus; SAID, Staphylococcus aureus chromID; DS, detergent scrubbing; MS, moist swabbing; TS, tape stripping; AD, atopic dermatitis
The most sensitive sampling method for S. aureus detection on naïve and disinfected AD skin lesions was the MS technique in combination with chromogenic SAID selective media where the detected rate of colonisation was 85.7% on naïve skin lesions respectively and 47.6% on disinfected skin lesions compared with the lowest rate (33.3% respectively 19.1%) by using the TS technique in combination with MSA. The SAID selective media was more sensitive for the quantitative detection of S. aureus compared to MSA (p < 0.001). All results of qualitative detection of S. aureus are summarised in Tables 3 and 4 and in Fig. 2. The direct comparison of bacterial load (CFU) detected on MSA and SAID plates from samples collected with the three different sampling methods from naïve AD skin lesions showed on both agar plates a significant difference (supplementary Fig. 1). On disinfected AD skin lesions, the difference in bacterial density (CFU) between the collection techniques was significant only on SAID in direct comparison (supplementary Fig. 2).
Table 3.
Comparison of two different S. aureus selective media with cultivated samples of three different sampling methods collected on naïve and disinfected skin lesions
| S. aureus positive | |||||||
|---|---|---|---|---|---|---|---|
| MSA (n = 21) | SAID (n = 21) | ∆a SAID − MSA | |||||
|
Atopic dermatitis skin lesions |
Sampling methods | n | % | n | % | n | % |
| Naïve | DS | 13 | 61.9 | 18 | 85.7 | 5 | + 23.8 |
| MS | 16 | 76.2 | 18 | 85.7 | 2 | + 9.5 | |
| TS | 7 | 33.3 | 14 | 66.7 | 7 | + 33.4 | |
| Disinfected | DS | 3 | 14.3 | 7 | 33.3 | 4 | + 19.0 |
| MS | 6 | 28.6 | 10 | 47.6 | 4 | + 19.0 | |
| TS | 4 | 19.1 | 7 | 33.3 | 3 | + 14.2 | |
| ∆a naïve − disinfected | DS | 10 | − 47.6 | 11 | − 52.4 | ||
| MS | 10 | − 47.6 | 8 | − 38.1 | |||
| TS | 3 | − 14.1 | 7 | − 33.4 | |||
aDifference
Table 4.
Direct comparison of the qualitative detection of S. aureus from non-invasive skin bacteria sampling by two different S. aureus selective media
| Mannitol salt agar | Total | |||
|---|---|---|---|---|
| S. aureus positive |
S. aureus negative |
|||
| chromID agar | S. aureus positive | 40 | 34 | 74 |
| S. aureus negative | 9 | 43 | 52 | |
| Total | 49 | 77 | 126 | |
Fig. 2.
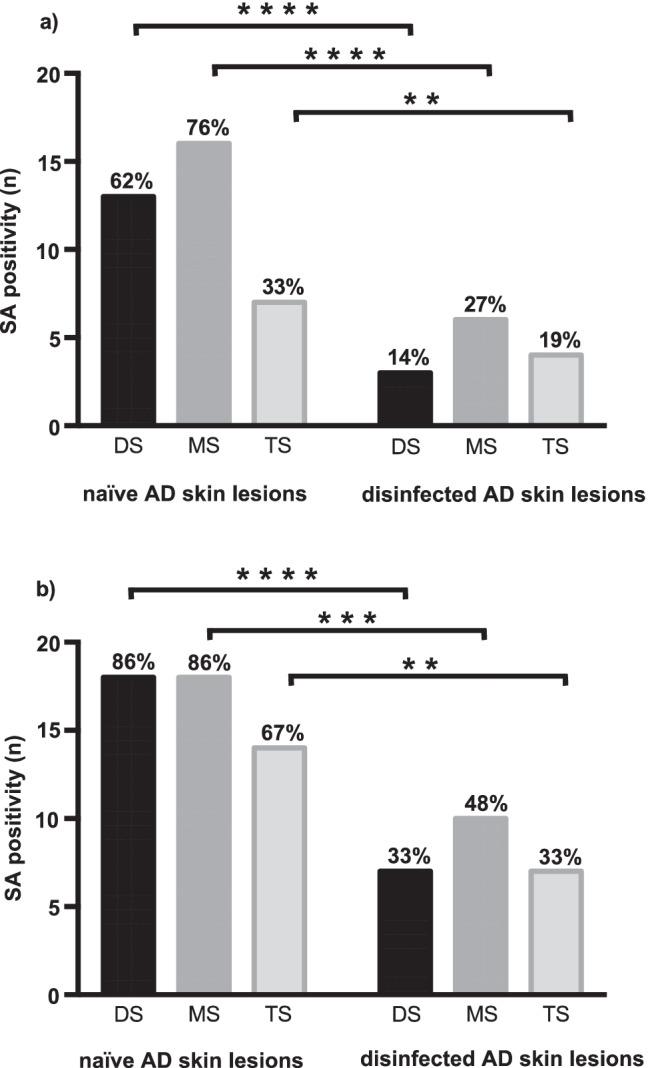
Qualitative detection of S. aureus by using different detection methods and selective media. a Qualitative detection of S. aureus by using selective media MSA (n = 21). b Qualitative detection of S. aureus by using selective media SAID (n = 21). Abbreviations: ****p < 0.0001; ***p < 0.001; **p < 0.1; SA, S. aureus; MSA, mannitol salt agar; SAID, S. aureus chromID; DS, detergent scrubbing; MS, moist swabbing; TS, tape stripping; AD, atopic dermatitis
Discussion
Atopic dermatitis is a frequent skin disease with an observed association of increased S. aureus colonisation [6]. Nevertheless, there is currently no defined gold standard for the sampling for culture to detect living S. aureus on skin lesions from AD patients and other forms of chronic skin conditions. Therefore, the three most promising non-invasive S. aureus sampling methods, two detergent-based (DS, MS) and one tape stripping (TS) methods, were compared in this study [11]. At first, the culture-based qualitative detection of S. aureus and the change of the viable S. aureus load after disinfection on AD skin lesions of patients with predominantly moderate disease according to the EASI score were investigated by these three different sampling methods. Results for these two main endpoints clearly differed between the sampling methods used and show the importance of the selected method in the clinical setting and research use before and after disinfection or antimicrobial treatment.
In detail, the highest sensitivity to detect S. aureus culture based on AD skin lesions and the persistence of S. aureus after disinfection was found with the detergent-based methods (DS or MS) in combination with the SAID selection agar. The sensitivity results differed clearly on naïve AD skin lesions, from 86% (used DS or MS method with SAID plates) to 33.3% (used TS method with MSA plates). Therefore, the prevalence of S. aureus colonisation was up to 86% on investigated naïve AD skin lesions where the density of bacteria on the skin lesions is naturally the highest, while the pooled prevalence of S. aureus colonisation reported in a systematic review of 81 studies was 70% (95% confidence interval 0.66–74). However, in a subgroup analysis from severe AD, the prevalence was comparable to our results [6] although our patients were mainly suffering from moderate AD (73% according to EASI score). S. aureus could be detected in all AD severity levels. The used EASI score, along with the two scores SCORAD and POEM, is one of the best-validated outcome measures for atopic dermatitis [17] and over the last years well established at our clinic to estimate the severity of atopic dermatitis. The data collected shows how different the culture results can be depending on the sampling method. Naïve skin lesions were clearly defined without topical or systemic antibiotics or local antimicrobial active soaps, nor was any sampling performed under cortisone or dermatological local therapy in the course of a (usually initial) evaluation of an episode of atopic dermatitis. The eradication rate of S. aureus after the used disinfection procedure reduced the bacterial load significantly independent from the used sampling method or S. aureus selection agar plates but surprisingly was not fully effective on AD skin lesions against S. aureus in a major proportion of patients. An octenidine-based antiseptic agent was chosen because it is approved for use on skin and mucous membranes and has good bactericidal activity against S. aureus (comparable to chlorhexidine) and good tolerability also on sensitive skin [18]. According to the used sampling method and S. aureus selection agar, the persistence rates were 48% (38% difference compared to non-disinfected skin) using the MS method and SAID selective media to 19% (14% difference compared to non-disinfected skin) using the TS method and MSA at disinfected AD skin lesions (Table 3). The antiseptic agent was applied only once for an impact time of at least 1 min, which may have been too short for effective decolonisation or should be repeated several times. However, the difference in the detection of living S. aureus before and after therapy is important, for example, in clinical trials to demonstrate the efficacy of new antiseptic agents to reduce the S. aureus load.
The detergent-based methods were significantly better to detect S. aureus on AD skin lesions. The buffered non-ionic detergent (0.1% Triton X-100 in 0.075 M sodium phosphate buffer, pH 7.9) first described by Williamson and Kligman [7] used for the scrubbing and swabbing method as sampling fluid was well tolerated without any side effects or skin irritations.
Remarkable are the weak results of the TS method to detect viable S. aureus on pathological AD skin lesions with high bacterial load. This is in contrast to previous results for skin microbiome by next-generation sequencing and culture-based studies, where even the culture-based study showed a greater number and wider variety of viable skin bacteria by the TS than the swabbing method [19]. However, compared to our study, the sample size was small (n = 7 versus 30), the skin was always healthy, the detergent buffer differed and Staphylococcus epidermidis was the lead detected bacterium; S. aureus was only detected once.
Sellotape stripping is in general an interesting method and easy to perform [19]. Only one tape strip per sample was used as is common in daily practice; this can be seen as a limitation of the study results. Another limitation could be that not all bacteria are successfully transferred from the foil by submerged in the washing fluid and vortexed gently. However, this is generally a disadvantage of the method, because in case of transfer of the tape strip directly on the agar plate also not all bacteria are successfully transferred to the agar medium or by leaving the tape strip in situ on the agar medium, the oxygen supply for the growth of aerobic and facultatively anaerobic bacteria such as S. aureus is inhibited and this could also falsify the results for the detection of growing S. aureus. A known disadvantage of the tape stripping technique is certainly that only bacteria on the skin surface are collected and the bacteria in the depth of the skin structure are not captured [16]. We suspect because 25% of the cutaneous bacteria population is localised in the depths of the skin structure and hair follicles [20] and S. aureus is found in the upper and the lower layers of the epidermis [21] and hair follicles [22] which may be a reason for the poor results with this particular pathogen of this method.
As expected, the DS method was highly effective to collect quantitative S. aureus from skin as has already been demonstrated by its widespread use for research purposes [11]. However, it was the most complex sampling method studied in our series. In addition, it was slightly unpleasant for the patient due to the necessary firm rubbing with a Teflon policeman for 1 min at the sensitive AD skin lesion as compared to the others. Although for detection of superficial skin bacteria this method is still considered as gold standard for the specific qualitative and quantitative detection of S. aureus on AD skin lesions, it was only equally effective to the less complex and less unpleasant MS method. Different kinds of swabs are often used routinely as a sampling method on intact or damaged skin. Generally, swab-based sampling methods for skin bacteria have a poor reputation among researchers mainly because of improper use of dry swabs [11]. For standardised MS performed with a non-ionic detergent on a defined skin area for detection of S. aureus on lesional AD skin, our study nevertheless showed excellent results. In addition, the handling of swabs is easy and quick and the defined skin area is simply marked with a sterile ring as a template. Former studies showed that swab-based methods collect mainly bacteria from the superficial skin layer and scrub-based methods collect the superficial skin cells and the associated microbes [8]. Indeed, we cannot rule out that for other settings a difference in detecting S. aureus in AD skin lesions might exist between the MS and DS methods.
In our study, we could also demonstrate that the choice of the S. aureus selection agar is an important issue. Overall, MSA was not without difficulties in identifying S. aureus due to difficulties in differentiating between S. aureus and coagulase-negative staphylococci why S. aureus strains were always confirmed by MALDI-TOF. The growing S. aureus colony resulted in yellow colouration of the agar in an area bigger than the diameter of the single colony, which resulted in a partly insufficient discrimination when the bacterial load was high and the single colonies were too close together despite using the highest dilution level. Therefore, after the first 9 patients, the SAID selective media were used, a chromogenic agar medium known to be highly sensitive and specific for detecting staphylococci and specifically identifying S. aureus [23]. The two different S. aureus selection agar plates showed significant differences by detecting S. aureus. Interestingly, MSA was significantly less sensitive regarding qualitative detection of living S. aureus compared to SAID agar as shown in detail in Table 4. Similar results were previously described with S. aureus chromogenic agars using clinical samples, i.e. from nasal swab specimens [24, 25] and food [26]. The importance of the choice of the selection agar was also shown by a subanalysis in direct comparison of the same sample on both plates, where on low bacterial load (on disinfected AD skin lesions) a significant difference between the sampling methods could be shown only with SAID plates (see supplementary Figs. 1 and 2).
Conclusion
The obtained data highlight the importance of the selected sampling method and consecutive S. aureus selection agar plates. To implement further clinical studies for the effectiveness of topical anti-staphylococcal antibiotics and to study the links between S. aureus colonisation and AD, it is essential to standardise the used sampling technique during the whole study and for all investigators. The obtained data in our study clearly suggest the use of detergent-based methods like scrubbing or swabbing. A chromogenic S. aureus selection agar additionally improves the results. Due to the convenient handling and less sampling associated burden for the AD patient, the standardised moistened swab technique on a defined skin area should be considered the standard sampling method for S. aureus.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
HL, CB, MOAS and MZ designed the research. CB, TQ and MW performed the experiments. ANP and SE performed the microbiological assays. HL, MK and MZ analysed the data. HL wrote the main manuscript and all authors reviewed and commented on the manuscript. The whole study was performed under supervision by MZ.
Funding
Open access funding provided by Medical University of Vienna.
Data availability
The data generated and analysed in this study are available from the authors on request.
Declarations
Ethics approval
The Ethical Board of the Medical University of Vienna (No. 2155/2016) approved the study.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/17/2022
Missing Open Access funding information has been added in the Funding Note.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;20(339):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep. 2015;15:65. doi: 10.1007/s11882-015-0567-4. [DOI] [PubMed] [Google Scholar]
- 3.Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol. 1974;90:525–530. doi: 10.1111/j.1365-2133.1974.tb06447.x. [DOI] [PubMed] [Google Scholar]
- 4.Park H-Y, Kim C-R, Huh I-S, Jung M-Y, Seo E-Y, Park J-H, et al. Staphylococcus aureus colonization in acute and chronic skin lesions of patients with atopic dermatitis. Ann Dermatol. 2013;25:410–416. doi: 10.5021/ad.2013.25.4.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsui K, Nishikawa A, Suto H, Tsuboi R, Ogawa H. Comparative study of Staphylococcus aureus isolated from lesional and non-lesional skin of atopic dermatitis patients. Microbiol Immunol. 2000;44:945–947. doi: 10.1111/j.1348-0421.2000.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 6.Totté JEE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SGMA. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. 2016;175:687–695. doi: 10.1111/bjd.14566. [DOI] [PubMed] [Google Scholar]
- 7.Williamson P, Kligman AM. A new method for the quantitative investigation of cutaneous bacteria. J Invest Dermatol. 1965;45:498–503. doi: 10.1038/jid.1965.164. [DOI] [PubMed] [Google Scholar]
- 8.Grice EA, Kong HH, Renaud G, Young AC, NISC Comparative Sequencing Program. Bouffard GG, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tauber M, Balica S, Hsu C-Y, Jean-Decoster C, Lauze C, Redoules D, et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol. 2016;137:1272–1274.e3. doi: 10.1016/j.jaci.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 10.Kong HH, Andersson B, Clavel T, Common JE, Jackson SA, Olson ND, et al. Performing skin microbiome research: a method to the madness. J Investig Dermatol. 2017;137:561–568. doi: 10.1016/j.jid.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eady EA. Handbook of non-invasive methods and the skin. CRC Press, Taylor & Francis Group; 2006. Sampling the bacteria of the skin; pp. 457–466. [Google Scholar]
- 12.Hanifin JM, Rajka G (1980) Diagnostic features of atopic dermatitis. Acta Derm Venereol (Stockh) 92(suppl):44–47. 10.2340/00015555924447
- 13.Hanifin JM, Thurston M, Omoto M, Cherill R, Tofte SJ, Graeber M. The Eczema Area and Severity Index (EASI): assessment of reliability in atopic dermatitis. EASI Evaluator Group. Exp Dermatol. 2001;10:11–18. doi: 10.1034/j.1600-0625.2001.100102.x. [DOI] [PubMed] [Google Scholar]
- 14.Keyworth N, Millar MR, Holland KT. Swab-wash method for quantitation of cutaneous microflora. J Clin Microbiol. 1990;28:941–943. doi: 10.1128/JCM.28.5.941-943.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinkus H. Examination of the epidermis by the strip method of removing horny layers. I. Observations on thickness of the horny layer, and on mitotic activity after stripping. J Invest Dermatol. 1951;16:383–6. doi: 10.1038/jid.1951.45. [DOI] [PubMed] [Google Scholar]
- 16.Brown E, Wenzel RP, Hendley JO. Exploration of the microbial anatomy of normal human skin by using plasmid profiles of coagulase-negative staphylococci: search for the reservoir of resident skin flora. J Infect Dis. 1989;160:644–650. doi: 10.1093/infdis/160.4.644. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt J, Langan S, Williams HC, European Dermato-Epidemiology Network What are the best outcome measurements for atopic eczema? A systematic review. J Allergy Clin Immunol. 2007;120:1389–98. doi: 10.1016/j.jaci.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Krishna BVS, Gibb AP. Use of octenidine dihydrochloride in meticillin-resistant Staphylococcus aureus decolonisation regimens: a literature review. J Hosp Infect. 2010;74:199–203. doi: 10.1016/j.jhin.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Ogai K, Nagase S, Mukai K, Iuchi T, Mori Y, Matsue M, et al. A comparison of techniques for collecting skin microbiome samples: swabbing versus tape-stripping. Front Microbiol. 2018;9:2362. doi: 10.3389/fmicb.2018.02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange-Asschenfeldt B, Marenbach D, Lang C, Patzelt A, Ulrich M, Maltusch A, et al. Distribution of bacteria in the epidermal layers and hair follicles of the human skin. Skin Pharmacol Physiol. 2011;24:305–311. doi: 10.1159/000328728. [DOI] [PubMed] [Google Scholar]
- 21.Hanssen A-M, Kindlund B, Stenklev NC, Furberg A-S, Fismen S, Olsen RS, et al. Localization of Staphylococcus aureus in tissue from the nasal vestibule in healthy carriers. BMC Microbiol. 2017;5(17):89. doi: 10.1186/s12866-017-0997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Broeke-Smits NJP, Kummer JA, Bleys RLAW, Fluit AC, Boel CHE. Hair follicles as a niche of Staphylococcus aureus in the nose; is a more effective decolonisation strategy needed? J Hosp Infect. 2010;76:211–214. doi: 10.1016/j.jhin.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Perry JD, Rennison C, Butterworth LA, Hopley ALJ, Gould FK. Evaluation of S. aureus ID, a new chromogenic agar medium for detection of Staphylococcus aureus. J Clin Microbiol. 2003;41:5695–8. doi: 10.1128/jcm.41.12.5695-5698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Souza HA, Baron EJ. BBL CHROMagar Staph aureus is superior to mannitol salt for detection of Staphylococcus aureus in complex mixed infections. Am J Clin Pathol. 2005;123:806–808. doi: 10.1309/FVHR-F3GR-LEQX-GBAG. [DOI] [PubMed] [Google Scholar]
- 25.Han Z, Lautenbach E, Fishman N, Nachamkin I. Evaluation of mannitol salt agar, CHROMagar Staph aureus and CHROMagar MRSA for detection of meticillin-resistant Staphylococcus aureus from nasal swab specimens. J Med Microbiol. 2007;56:43–46. doi: 10.1099/jmm.0.46777-0. [DOI] [PubMed] [Google Scholar]
- 26.Kim H-J, Oh S-W. Performance comparison of 5 selective media used to detect Staphylococcus aureus in foods. Food Sci Biotechnol. 2010;19:1097–1101. doi: 10.1007/s10068-010-0155-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analysed in this study are available from the authors on request.