Abstract
Population-based analysis of Mycobacterium tuberculosis transmission in Houston, Tex., over 5 years identified 377 patients infected with an isolate containing one to four copies of IS6110. The isolates were analyzed by spoligotyping and assigned to one of three major genetic groups based on nucleotide polymorphisms in codons katG 463 and gyrA 95. Prospectively obtained patient interviews were reviewed to assess epidemiologic links between apparently clustered patients. A total of 13 groups of isolates with the same IS6110 profile were identified, representing 326 of the 377 patients (86.5%; range 2 to 113 patients). In contrast, 28 groups of isolates containing 334 patients (88.6%) had the same spoligotype (range, 2 to 143 patients). Combination of IS6110 profile and spoligotype data identified 31 clusters with 300 patients (79.6%; range, 2 to 82 patients). All 377 isolates belonged to major genetic group 1 (77 patients) or genetic group 2 (300 patients); no major genetic group 3 isolates were identified. Among the 228 patients interviewed, 33 patients (14.5%) were directly linked to another patient in the same cluster. Possible epidemiologic links were also found among 11 patients. Moreover, many clusters consisted of individuals with the same ethnicity. In conclusion, we confirmed that IS6110 profiling and spoligotyping together provide enhanced molecular discrimination of M. tuberculosis isolates with low copy numbers of IS6110. Identification of epidemiologic links among some of the patients verified that the combination of these two methods reliably indexes tuberculosis transmission.
Characterization of Mycobacterium tuberculosis complex isolates by restriction fragment length polymorphism (RFLP) with the IS6110 element as a probe is the standard tool used in tuberculosis epidemiology studies. M. tuberculosis organisms usually contain several copies of IS6110, and the RFLP results are stable and reproducible, providing reliable differentiation of epidemiologically unrelated isolates. Moreover, large population-based studies have shown that identification of tuberculosis case clusters is significantly enhanced by IS6110 profiling compared to conventional contact tracing (1, 16, 22).
However, M. tuberculosis isolates containing five or fewer IS6110 copies cannot be reliably differentiated by the RFLP method (3), and in rare cases, the IS6110 element is missing from the genome altogether (21). M. tuberculosis strains with low copy numbers of IS6110 (hereafter in this work referred to as low-copy-number strains) have been more frequently isolated from Asian patients than from patients of European origin. For example, 56% of the strains collected from India and 33% of strains collected from Vietnam, Thailand, and Malaysia contained five or fewer IS6110 elements (5, 15), whereas the frequencies of low-copy-number isolates in Denmark and France were 11 and 8%, respectively (2, 10). In the United States, analysis of isolates from Texas, California, and Colorado found that 25% contained fewer than six IS6110 copies (23).
Other molecular characterization techniques, such as RFLP analysis of the polymorphic GC-rich repetitive sequence (PGRS) (13, 21) or spoligotyping, can to be used to differentiate the IS6110 low-copy-number isolates. Spoligotyping discriminates among M. tuberculosis isolates on the basis of genetic polymorphisms in the chromosomal direct repeat (DR) region consisting of identical DRs and unique spacer sequences (9, 11). Spoligotyping has been reported to be more sensitive than standard IS6110 profiling for subtyping M. tuberculosis isolates with five or fewer IS6110 copies (2, 7, 8, 13, 17). However, epidemiologic relationships among patients infected with IS6110 low-copy-number isolates and clustered by spoligotyping have not been systematically investigated in detail.
In order to address this issue, we characterized low-copy-number M. tuberculosis isolates obtained from patients in Houston by IS6110 profiling, spoligotyping, and major genetic group determination and investigated the epidemiologic links between patients in case clusters identified by these molecular methods.
MATERIALS AND METHODS
Bacterial isolates.
The analysis was performed as part of an ongoing, population-based tuberculosis epidemiology study in Houston, Tex. During the 5-year study period, from October 1994 to September 1999, 502 patients were identified that were infected with an M. tuberculosis isolate having zero to four IS6110 copies. This number represented 20.3% (502 of 2,478) of the total number of patients studied. During this time period, 85% of culture-positive tuberculosis (TB) patients reported in Houston were enrolled in our study, and the average yearly TB case rate was 19.7 per 100,000 members of the population. Isolates from 383 (76.3%) patients were available for this analysis. Only one isolate per patient was included. A total of 377 patients were infected with an isolate with one to four IS6110 copies (low-copy-number isolates), and six patients were infected with an isolate having no copies of IS6110 (zero-copy-number isolates). The specimen source was pulmonary in 296 (77.3%) cases, extrapulmonary in 50 (13.1%) cases, and unknown in 37 (9.7%) cases. If the patient had both pulmonary and extrapulmonary isolates, the extrapulmonary isolate was used in this study.
DNA methods.
DNA extraction and IS6110 RFLP analysis were performed by an internationally standardized protocol (19). The IS6110 profiles were analyzed with the BioImage (Ann Arbor, Mich.) Whole Band Analysis program, version 3.2. Spoligotyping was performed with a commercially available kit (Isogen Bioscience BV, Maarssen, The Netherlands) according to the instructions of the manufacturer. The isolates were assigned to one of three principal genetic groups on the basis of nucleotide polymorphism at codon 463 and 95 of the genes encoding the catalase-peroxidase and A subunit of DNA gyrase, respectively (18). Isolates lacking IS6110 elements were also characterized by PCR-based sequencing of a 360-bp segment of the hsp65 gene as described previously (12). A subset of the low-copy-number isolates was also analyzed by the double-repetitive-element PCR (DRE-PCR) according to a previously published protocol (6, 14). The DRE-PCR method is based on PCR amplification of genomic segments located between the IS6110 and PGRS repetitive sequences. A cluster was defined as two or more patients infected with an M. tuberculosis isolate having the same IS6110 profile and spoligotype.
Patient interviews.
Prospectively obtained interviews were available from 228 of the 300 (76.0%) patients that were clustered by the combination of the IS6110 profiling and spoligotyping results. The extensive questionnaire inquired about demographic information; living situation; use of tobacco, alcohol, and illicit drugs; sexual preferences and related habits; and patient medical history, including possible location of exposure to M. tuberculosis.
Statistical analysis.
A chi-square analysis was performed on the effect of clustering and major genetic grouping by the country of origin. A P value of ≤0.05 was considered statistically significant.
RESULTS
IS6110 profiling.
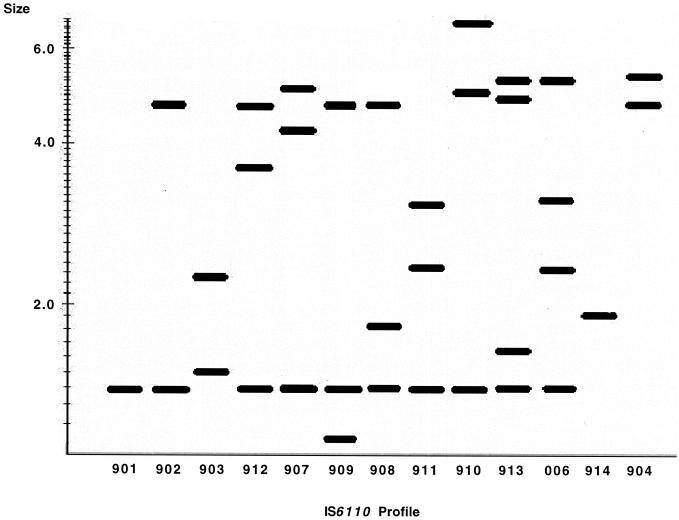
Analysis of the 377 low-copy-number isolates by IS6110 profiling alone revealed 64 distinct profiles. Fifty-one isolates had a unique profile, while 326 (86.5%) belonged to one of 13 groups (Fig. 1) consisting of 2 to 113 patients.
FIG. 1.
Schematic representation of the IS6110 profiles of the 13 IS6110 groups obtained from the clustered low-copy-number M. tuberculosis isolates.
Spoligotyping.
A total of 72 spoligotypes was identified among the 377 isolates. Forty-four of the 377 isolates had a unique spoligotype, while for 334 isolates (88.6%) 2 to 143 patients shared 1 of 28 spoligotypes. One Mycobacterium bovis isolate and four M. bovis BCG isolates were identified based on characteristic spoligotypes (11). The most common spoligotypes observed were S12 (n = 143) and S3 (n = 43) (17).
Combination of IS6110 profiling and spoligotyping.
Combination of the IS6110 profile and spoligotyping data identified 31 clusters with 300 of the 377 patients (79.6%). The cluster size varied from 2 to 82 patients (mean, 9.7; median, 4) (Table 1). Isolates assigned to the two largest clusters (999.008 and 999.016) were also analyzed by the DRE-PCR method. All isolates in both of these clusters had the same DRE-PCR profile.
TABLE 1.
Characteristics of the clusters identified by combination of IS6110 profile and spoligotype
| Clustera | IS6110 copy no. | Genetic groupb | IS6110 profilea | Spoligotypec | No. of patients | No. of linksd | Patient characteristic(s)e |
|---|---|---|---|---|---|---|---|
| 999.001 | 1 | 1 | 901 | S10 | 13 | 0 | Vietnamese |
| 999.002 | 1 | 1 | 901 | S20 | 2 | 0 | |
| 999.003 | 1 | 1 | 901 | S22 | 3 | 0 | Vietnamese |
| 999.004 | 1 | 1 | 901 | S55 | 5 | 0 | |
| 999.005 | 1 | 1 | 901 | S101 | 2 | 0 | |
| 999.006 | 1 | 1 | 901 | S162 | 3 | 0 | Vietnamese |
| 999.007 | 2 | 2 | 902 | S3 | 12 | 0 | Hispanic |
| 999.008 | 2 | 2 | 902 | S12 | 82 | 6 | Black, jail, homeless, drug use, 7 cross-contaminants |
| 999.009 | 2 | 2 | 902 | S52 | 7 | 0 | Black, homosexual, HIV positive |
| 999.010 | 2 | 2 | 902 | S60 | 2 | 0 | |
| 999.011 | 2 | 2 | 902 | S63 | 2 | 0 | Hispanic |
| 999.012 | 2 | 1 | 903 | S11 | 6 | 4 | White |
| 999.013 | 2 | 2 | 904 | S19 | 2 | 0 | Vietnamese |
| 999.014 | 3 | 2 | 909 | S3 | 3 | 0 | |
| 999.015 | 3 | 2 | 907 | S3 | 2 | 0 | |
| 999.016 | 3 | 2 | 908 | S12 | 30 | 0 | Black, jail |
| 999.017 | 3 | 2 | 909 | S15 | 7 | 2 | Black, homosexual, HIV positive |
| 999.018 | 3 | 2 | 909 | S21 | 4 | 0 | White |
| 999.019 | 3 | 2 | 909 | S53 | 3 | 0 | Black, homeless, jail, prison |
| 999.020 | 3 | 2 | 910 | S18 | 3 | 0 | |
| 999.021 | 3 | 2 | 911 | S54 | 7 | 0 | Black |
| 999.022 | 3 | 2 | 912 | S242 | 3 | 0 | Hispanic, jail, prison, homeless |
| 999.023 | 4 | 2 | 913 | S3 | 8 | 0 | |
| 999.024 | 4 | 2 | 006 | S3 | 14 | 0 | Hispanic |
| 999.025 | 4 | 2 | 006 | S25 | 26 | 14 | Black |
| 999.026 | 4 | 2 | 006 | S26 | 4 | 0 | |
| 999.027 | 4 | 2 | 006 | S27 | 8 | 2 | Hispanic |
| 999.028 | 4 | 2 | 006 | S29 | 8 | 0 | |
| 999.029 | 1 | 1 | 914 | BCG | 4 | 0 | |
| 999.030 | 3 | 2 | 910 | S12 | 2 | 0 | |
| 999.031 | 3 | 2 | 912 | S12 | 23 | 5 | Six cross-contaminants |
| Total | 300 | 33 |
U.S.- and foreign-born patients.
Of the 377 patients infected with low-copy-number isolates 241 (63.9%) were U.S. born, 129 (34.2%) were foreign born, and the birthplace was unknown for 7 (1.9%) individuals. The majority of foreign-born patients originated from Mexico (n = 57) and Vietnam (n = 45). Comparison of clustering and place of birth is shown in Table 2.
TABLE 2.
Comparison of clustering and place of birth among patients infected with low-copy-number isolates
| Birth | No. (%)
|
P | ||
|---|---|---|---|---|
| All patients | Clustered | Nonclustered | ||
| U.S. born | 241 (63.9) | 218 (72.7) | 23 (29.8) | <0.01 |
| Foreign born | 129 (34.2) | 75 (25.0) | 54 (70.1) | <0.01 |
| Unknown | 7 (1.9) | 7 (2.3) | 0 (0.0) | 0.35a |
| Total | 377 | 300 (79.6) | 77 (20.4) | |
Fisher's exact, two-tailed test.
Major genetic group designation.
Seventy-seven isolates (20.4%) and eight clusters were assigned to major genetic group 1, and 300 isolates (79.6%) and 23 clusters belonged to major genetic group 2 (18). No group 3 isolates were identified among the 377 isolates. A comparison of major genetic group and place of birth is shown in Table 3. Of note, significantly more Vietnamese patients were infected with an isolate belonging to major genetic group 1 (37 of 45; 82.2%), whereas significantly more Mexican patients were infected with major genetic group 2 isolates (51 of 57; 89.5%) (P < 0.01).
TABLE 3.
Comparison of major genetic group and place of birth among patients infected with low-copy-number isolates
| Birth | No. (%)
|
P | ||
|---|---|---|---|---|
| All patients | Group 1a | Group 2a | ||
| U.S. born | 241 (63.9) | 13 (16.9) | 228 (76.0) | <0.01 |
| Foreign born | 129 (34.2) | 61 (79.2) | 68 (22.7) | <0.01 |
| Unknown | 7 (1.9) | 3 (3.9) | 4 (1.3) | 0.15b |
| Total | 377 | 77 (20.4) | 300 (79.6) | |
Defined by Sreevatsan et al. (18).
Fisher's exact, two-tailed test.
Proven or potential epidemiologic links.
Among the 228 patients interviewed, only 69 (30.3%) individuals had a history of contact with a known TB patient. A direct epidemiologic link, as defined by known person-to-person contact between two patients in the same cluster, was found between 33 (14.5%) patients in six clusters (Table 1). For example, in cluster 999.025, 14 of the 26 patients were epidemiologically linked. This cluster consisted mainly of black patients living in two neighborhoods on the north and the south side of Houston that were linked by a public bus route. In addition, 11 patients in each of three clusters (999.009, 999.019, and 999.022) shared similar TB risk factors even though direct links between the patients could not be identified. For one of the three clusters, patients were human immunodeficiency virus (HIV)-positive homosexual men, and for two clusters the patients had a history of homelessness and incarceration. Seven patients in cluster 999.008 and six patients in cluster 999.031 were identified as laboratory cross-contaminants. Overall, for 18 of the 31 clusters, the majority (51 to 100%) of the patients belonged to the same ethnic group. Four clusters consisted mainly of Vietnamese patients, five of Hispanic patients, two of white patients, and seven of black patients. No epidemiologic links were identified between patients whose isolates were not clustered.
Zero-copy-number isolates.
Six patients were infected with M. tuberculosis isolates that did not hybridize with the standard, right-side IS6110 probe, suggesting that the isolates did not contain IS6110. PCR-based sequencing of the hsp65 gene confirmed that the isolates belonged to the M. tuberculosis complex. Four different spoligotypes were obtained from the six isolates. The four spoligotypes had similar patterns (lacking spacers 19 to 41) but have not been previously identified (Table 4). Two U.S.-born patients were infected with M. tuberculosis isolates with the spoligotype arbitrarily designated S295, whereas two of the four Vietnamese patients were infected with isolates with the spoligotype arbitrarily designated S296. However, no direct epidemiologic links were found between the patients sharing the same spoligotype. All zero-copy-number isolates belonged to major genetic group 1.
TABLE 4.
Spoligotypes of the IS6110 zero-copy-number M. tuberculosis isolates
Arbitrary designation.
Black squares denote hybridization with the spacer probe; open squares denote lack of hybridization.
DISCUSSION
The aim of this study was to evaluate spoligotyping as a technique for subtyping IS6110 low-copy-number isolates cultured in Houston, and to investigate the epidemiologic links between patients in case clusters identified by the molecular methods.
Clustering and epidemiologic links.
The number of low-copy-number M. tuberculosis clusters increased from 13 when examined by IS6110 profiling alone to 31 after using a combination of IS6110 profiling and spoligotyping results. However, the clustering percentage only decreased from 86.5 to 79.6%. This clustering percentage is much higher than that observed in a similar study performed in Denmark, where the clustering percentage decreased from 83 to 55% (2), and higher than the 59% clustering observed in Houston overall (data not shown).
A review of patient interviews and contact investigation reports revealed that only 33 patients (14.5%) were directly linked to another patient in the same cluster. However, possible epidemiologic links were also found among 11 patients in three clusters, and 13 patients were found to be laboratory cross-contaminants. Moreover, many clusters consisted mainly of individuals with the same ethnicity. We thus believe that the clustering results obtained by combining IS6110 profile and spoligotype represent true clustering. However, clustering of M. tuberculosis isolates measured by DNA clonality does not always indicate recent transmission but may represent transmission of strains endemic to the area. Since some clusters consisted of foreign-born patients only, it is possible that the clustering results reflect reactivation of a latent infection with a common organism endemic in the country of origin of the patients. An analysis of a large sample of common M. tuberculosis isolates from these countries could verify this hypothesis. 
The combination of the methods used may have overestimated clustering of the low-copy-number isolates in some cases. Two of the largest clusters (999.008 and 999.016) had related IS6110 profiles (902 and 908 [Fig. 1]) that shared two IS6110 elements and had the same spoligotype, S12, but no epidemiologic links were identified between members of the two clusters. We have previously shown that this spoligotype is the most common spoligotype among low-copy-number M. tuberculosis isolates cultured in Houston (17). However, when the large clusters were analyzed by an additional typing method, DRE-PCR, they were not further divided, suggesting that these two clusters are distinct. Nevertheless, it is possible that in some cases clustering results could be further improved by using the PGRS RFLP method or by analyzing the isolates with the new spoligotyping test that contains additional spacer probes (20).
U.S.- and foreign-born patients.
Studies performed in Denmark and France show that the majority of the patients infected with low-copy-number isolates were foreign born (2, 10). In this study, 63.9% of the isolates originated from U.S.-born patients. However, the proportion of U.S.-born patients infected with low-copy-number isolates was slightly lower than the 70.3% that was observed in Houston overall (data not shown). Low-copy-number isolates from foreign-born patients were significantly less likely to be clustered than isolates from U.S.-born patients (P < 0.01).
Major genetic groups.
Sreevatsan et al. (18) have shown that M. tuberculosis complex isolates can be differentiated into three major genetic groups based on DNA polymorphism in codons katG 463 and gyrA 95. They proposed that group 1 isolates are evolutionarily older and have subsequently evolved to group 2 and 3 organisms (18). Our results show that major genetic group 1 isolates were significantly more prevalent among patients of Asian origin, whereas the majority of Mexican and U.S.-born patients were infected with isolates belonging to major genetic group 2. These data add to the concept that M. tuberculosis isolates of different genetic groups are prevalent in different areas of the world. No major genetic group 3 isolates were identified in our study, confirming the previous observation that low-copy-number M. tuberculosis isolates belong to evolutionarily older genetic groups 1 and 2 (18).
Zero-copy-number isolates.
The first isolate lacking IS6110 was identified in 1993 from an Indian patient (21), and thus far, about 15 such isolates have been reported in the literature, originating mainly from Asian patients (4, 5, 7, 13, 23). These isolates can be identified by other molecular typing methods such as spoligotyping (13). We identified six zero-copy-number isolates from four Vietnamese patients and two U.S.-born patients. The six isolates were divided into four spoligotypes that had similar patterns, suggesting that they may have evolved from a common ancestor. Our data also confirmed that the DR region appears to be present in genomes of all M. tuberculosis isolates, even in those that lack IS6110 (7, 13). All zero-copy-number isolates belonged to major genetic group 1, suggesting that they represent evolutionarily older M. tuberculosis isolates (18).
Conclusions.
Our study confirmed that IS6110 profiling and spoligotyping together provide an enhanced molecular discrimination method of low-copy-number M. tuberculosis isolates relative to IS6110 profiling or spoligotyping alone. Identification of epidemiologic links among some of the patients verified that the combination of the two methods reliably indexes tuberculosis transmission. In addition, all low-copy-number M. tuberculosis isolates were found to belong to the evolutionarily older major genetic groups 1 and 2.
ACKNOWLEDGMENTS
We thank Thanh Tung Bui, Pandora Davis, Lorretta Jackson, Weldon Mauney, Yuly Orozco, and Tony Prejean for conducting the patient interviews; Saif Shere for technical assistance; and Heather Tooker for assistance with graphics.
The study was supported by NIH Public Health Service grant DA-09238 to J.M.M.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Bauer J, Andersen A B, Kremer K, Miorner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol. 1999;37:2602–2606. doi: 10.1128/jcm.37.8.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burman W J, Reves R R, Hawkes A P, Rietmeijer C A, Yang Z, El-Hajj H, Bates J H, Cave M D. DNA fingerprinting with two probes decreases clustering of Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:1140–1146. doi: 10.1164/ajrccm.155.3.9117000. [DOI] [PubMed] [Google Scholar]
- 4.Das S, Chan S L, Allen B W, Mitchison D A, Lowrie D B. Application of DNA fingerprinting with IS986 to sequential mycobacterial isolates obtained from pulmonary tuberculosis patients in Hong Kong before, during and after short-course chemotherapy. Tuber Lung Dis. 1993;74:47–51. doi: 10.1016/0962-8479(93)90068-9. [DOI] [PubMed] [Google Scholar]
- 5.Das S, Paramasivan C N, Lowrie D B, Prabhakar R, Narayanan P R. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, South India. Tuber Lung Dis. 1995;76:550–554. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 6.Friedman C R, Stoeckle M Y, Johnson W D, Jr, Riley L W. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J Clin Microbiol. 1995;33:1383–1384. doi: 10.1128/jcm.33.5.1383-1384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goguet de la Salmonière Y-O, Li H M, Torrea G, Bunchoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal M, Saunders N A, van Embden J D A, Young D B, Shaw R J. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groenen P M A, Bunschoten A E, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis: application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez M C, Vincent V, Aubert D, Bizet J, Gaillot O, Lebrun L, Le Pendeven C, Le Pennec M P, Mathieu D, Offredo C, Pangon B, Pierre-Audigier C. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J Clin Microbiol. 1998;36:486–492. doi: 10.1128/jcm.36.2.486-492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapur V, Li L-L, Hamrick M R, Plikaytis B B, Shinnick T M, Telenti A, Jacobs W R, Jr, Banerjee A, Cole S, Yuen K Y, Clarridge III J E, Kreiswirth B N, Musser J M. Rapid Mycobacterium species assignment and unambigous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch Pathol Lab Med. 1995;119:131–138. [PubMed] [Google Scholar]
- 13.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montoro E, Valdivia J, Cardoso-Leão S. Molecular fingerprinting of Mycobacterium tuberculosis isolates obtained in Havana, Cuba, by IS6110 restriction fragment length polymorphism analysis and by the double-repetitive-element PCR method. J Clin Microbiol. 1998;36:3099–3102. doi: 10.1128/jcm.36.10.3099-3102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park Y-K, Bai G-H, Kim S-J. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the Western Pacific region. J Clin Microbiol. 2000;38:191–197. doi: 10.1128/jcm.38.1.191-197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 17.Soini H, Pan X, Amin A, Graviss E A, Siddiqui A, Musser J M. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol. 2000;38:669–676. doi: 10.1128/jcm.38.2.669-676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Embden J D A, van Gorkom T, Kremer K, Jansen R, van der Zeijst B A M, Schouls L M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaganehdoost A, Graviss E A, Ross M W, Adams G J, Ramaswamy S, Wanger A, Frothingham R, Soini H, Musser J M. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J Infect Dis. 1999;180:1245–1251. doi: 10.1086/314991. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Barnes P F, Chaves F, Eisenach K D, Weis S E, Bates J H, Cave M D. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]