Abstract
Background
While emerging evidence indicates that N6-methyladenosine (m6A) regulators play crucial roles in cancer progression, their clinical significance in gastric cancer (GC) has thus far not been elucidated.
Methods
We investigated the expression of the m6A regulator genes and their prognostic potential in a large clinical cohort of 173 GC patients using qRT-PCR assays. In addition, we undertook a series of in-vitro and in-vivo functional studies to investigate the oncogenic role of FTO.
Results
GC patients with low expression of METTL3, METTL14, ALKBH5, WTAP and YTHDF1 demonstrated significantly poor OS, while patients with high FTO expression exhibited markedly worse OS. Furthermore, the cumulative risk-score derived from these gene panel also significantly associated with poor OS, with a corresponding hazard ratio of 5.47 (95% CI: 3.18–9.41, p < 0.0001). We observed that FTO expression was frequently upregulated in GC cell lines, with epithelial-mesenchymal-transition (EMT) features. FTO knockdown in HGC27 and AGS cells inhibited cell proliferation and migratory potential, while its overexpression in MKN28 cells resulted in enhanced proliferation and migration. Finally, confirming our in-vitro findings, FTO suppression led to significant tumour growth inhibition in a HGC27 xenograft model.
Conclusions
We demonstrate that m6A regulators may serve as promising prognostic biomarkers in GC. Our functional studies reveal that FTO is an important oncogene and may be a promising therapeutic target associated with EMT-alterations in gastric cancer.
Subject terms: Oncology, Molecular biology
Introduction
Gastric cancer (GC) is the fourth most frequent cause of mortality worldwide, accounting for about 8% of all cancers and 10% of cancer-related deaths [1, 2]. Although the survival benefit from surgical treatments as well as chemotherapy and chemoradiotherapy in GC patients is well recognised, the prognostic outcomes from this malignancy still remains poor [3]. The currently used prognostic biomarkers in GC patients are primarily clinicopathological, which are inadequate for robust prognostication due to their low overall accuracy [4]. Therefore, identification of critical factors that drive cancer progression and development of prognostic biomarkers for improving recurrence prediction and overall prognosis of GC patients are much needed.
RNA can be post-transcriptionally modified through acquisition of more than 150 chemical modifications [5, 6]. Among them, N [6]-methyladenosine (methylation of N6 adenosine; m6A) has been identified as one of the most abundant internal modifications in eukaryotic mRNA [7, 8]. The m6A modifications enrich around stop codons, in 3′ untranslated regions (UTR), within long internal exons, and also occur at 5′UTRs [9–11]. Epigenetic regulation of RNA through m6A methylation affects post-transcriptional fate of a large number of target mRNAs, which plays important roles in proliferation and differentiation. In fact, m6A dysregulation has a dual role as it leads to degradation of specific mRNAs, as well as facilitates elevated expression of certain oncogenes by inducing translation through different pathways. Moreover, low m6A tend to correlate with cell pluripotency, while m6A modifications continue to increase during cellular differentiation.
It is well known that there are several basic m6A regulators, which are involved in RNA metabolism including RNA stability, translation, splicing, transport and localisation [8, 12]. Among various categories of m6A regulators, the ‘writers’ consist of a multicomponent methyltransferase complex including methyltransferase-like 3 (METTL3), METTL14, and Wilms’ tumour 1-associating protein (WTAP1) [13, 14]. These m6A marks of mRNAs are erased by two RNA demethylases (‘erasers’), fat mass and obesity-associated protein (FTO) and alkylated DNA repair protein alkB homologue 5 (ALKBH5) proteins [15, 16]. Finally, these m6A methylated mRNAs are identified by “readers” represented by YTH family of proteins that preferentially binds to RNA containing m6A at the G [G>A] m6ACU consensus sequences [7].
Aberrant m6A modifications have been reported to play a crucial role in cancer progression across various malignancies, such as prostate, breast, pancreatic, gastric, glioblastoma, hepatocellular carcinoma (HCC), and acute myeloid leukaemia (AML) [17–23]. Recent functional studies have revealed that knockdown of METTL3 and METTL14 and overexpression of FTO promoted glioblastoma stem cell (GSC) self-renewal, pluripotency and enhanced tumour growth [17]. In breast cancer, hypoxia increases the expression of ALKBH5 which stabilises NANOG mRNA and promotes tumour formation [19, 20]. In acute myeloid leukaemia (AML), FTO enhances leukaemic oncogene-mediated cell transformation and leukemogenesis, through regulation of the expression of ASB2 and RARA by reducing m6A levels in these mRNA transcripts [21, 24]. Furthermore, with regards to the biomarker potential of m6A regulators, mutations and/or copy number variations in m6A regulators are reported to confer a worse prognosis in AML [22]. Furthermore in HCC, knock-down of METTL14 enhanced tumour metastatic potential in cell culture and animal model experiments, and down-regulation of METTL14 was associated with poor recurrence-free survival [23].
Recently, we have identified that a gene expression panel composed of basic m6A regulators in various malignancies including gastric, colorectal, breast, and ovarian cancer [25], and the risk scores derived from these m6A regulators significantly associated with poor prognosis in GC patients [25]. In order to confirm these intriguing findings, we assessed the association between the expression level of these m6A regulator genes and prognosis using an independent clinical cohort of GC patients. Furthermore, we investigated the oncogenic role of FTO using in a series of in-vitro and in-vivo experimental models. Our efforts led us to further validate the prognostic ability of m6A regulator genes in GC, and in particular, the clinical significance of FTO as a therapeutic target in GC patients.
Methods
Study design and patient specimens
We enrolled a cohort of 173 gastric cancer patients who underwent surgery between January 2006 and December 2012 at the Mie University Hospital, for the clinical validation of m6A regulators. Each tumour sample was fixed in 10% phosphate-buffered formalin and embedded in paraffin. Information on patient demographics and clinicopathological characteristics are provided in Supplementary Table 1. A written informed consent was obtained from all patients, and the Institutional Review Boards of Mie University Hospital (IRB number: H2019-197).
RNA isolation and quantitative real time PCR
Total RNA from formalin-fixed paraffin-embedded (FFPE) specimens of GC patients were extracted using the AllPrep DNA/RNA FFPE Kit (Qiagen, Valencia, CA), and RNA from cancer cell-lines was extracted using the miRNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) synthesis was performed using 500 ng total RNA with High Capacity cDNA Reverse Transcription Kit (Invitrogen, Carlsbad, CA). Quantitative real-time polymerase chain reaction (qRT-PCR) assays for the expression of each transcripts were performed by SensiFAST SYBR (Bioline, London, UK) using the QuantStudioTM 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA). The β-actin gene was used as an internal control for data normalisation. The relative expression levels of each target transcripts were calculated using the 2−ΔCT method. The sequences of each PCR primer in the current study are provided in Supplementary Table 2.
FTO knockdown and overexpression in GC cell lines
GC cell lines HGC27, AGS, and MKN28 were purchased from the American Type Culture Collection (Manassas, VA). All cell lines were grown in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco, Carlsbad, CA) with 10% foetal bovine serum and 1% penicillin and streptomycin and cultured at 37 °C in a humidified incubator at 5% CO2. The PLKO.1 lentivirus vector bearing control or FTO cDNA sequences (sh-FTO) were used to perform stable knockdown of FTO. These plasmids were kindly provided by Professor Chuan He’s Laboratory. Briefly, 2.24 μg psPAX2, 0.76 μg pMD2.G and 1.5 μg shFTO or control were co-transfected into HEK-293T using Lipofectamine 2,000 (Invitrogen, Carlsbad, CA). The lentivirus particles were harvested at 48 and 72 h after transfection. For transduction, the lentivirus particles were directly added to HGC27 and AGS cells along with 10 ug/ml Polybrene (MilliporeSigma, Burlington, MA) and incubated at 37 °C for 48–72 h.
Overexpression of FTO
The FTO overexpression plasmid was also kindly provided by Professor Chuan He’s Laboratory. Each FTO expression construct was transfected into MKN28 cells (2000 ng per 1 million cells) using Lipofectamine 2,000 (Invitrogen, Carlsbad, CA). For all transfections, empty pcDNA vector was used as a control. Forty-eight hours after transfection of these plasmids, the transfected MKN28 cells were collected for further experiments.
Western blotting
Total cellular protein from HGC27 and AGS were collected at 48-h post transduction, while MKN28 cells were extracted following 96-h post transfection. Proteins were extracted using a RIPA Lysis and Extraction Buffer (ThermoFisher Scientific, Waltham, MA) containing 0.1% protease inhibitor cocktail (ThermoFisher Scientific). Twenty μg of each protein was loaded on sodium dodecyl sulfate-polyacrylamide gels (12%), followed by electrophoresis, and immediate transfer of the proteins to a nitrocellulose membrane using a transfer buffer (25 mM Tris, pH 8.3, 192 mM glycine with 20% methanol). Nitrocellulose membranes were incubated in TBS-T (Tris-NaCl buffer (TBS) with 0.1% Tween 20) containing 5% dry milk for 1 h at room temperature for nonspecific binding blocking. The blots were incubated overnight at 4 °C with the primary antibodies in TBS-T containing 1% bovine serum albumin (BSA). Lists of the primary antibodies that were used are shown in Supplementary Table 3. The blots were washed five times for 5 m each with TBS-T at room temperature, incubated for 60 m in species-appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Dallas, TX) in TBS-T containing 1% BSA, washed five times, and incubated on Super-Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) for 2 m. All protein bands on the membranes were visualised using ChemiDocTMMP Imaging system (ver 5.2.1, BioRad Laboratories Inc, Hercules, CA). B-actin (Sigma-Aldrich) was used as a reference protein. Band intensity was quantified using Image J ver. 1.52 (Bethesda, MD) [26], and shown as a ratio to B-actin band intensity.
MA2 (FTO inhibitor)
Meclofenamic acid (MA)/MA2 is a non-steroidal-anti-inflammatory-drug approved by the US Food and Drug Administration (FDA) known for its inhibition of prostaglandin synthesis, cyclooxygenase enzymes and lipoxygenases [27], as well as the inhibition of FTO [28]. In-vitro functional studies performed by Huang et al, demonstrated that MA/MA2 specifically competed with FTO binding for the m6A-containing nucleic acids, and HeLa cells treated with MA/MA2 resulted in the upregulation of m6A modification levels in mRNAs [29]. In our study, MA2 compound was kindly provided by Professor Cai-Guang Yang’s Laboratory.
Cell proliferation assay
The MTT (3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide) assay was used to evaluate the cell proliferation ability. Cells were plated in 96-well dishes at a density of 500–1000 cells/well in IMDM with 10% FBS and antibiotics. Cell proliferation in each group (naive, sh-ctrl, sh-FTO, pcDNA, and OE-FTO) was measured at 24, 48, 72, and 96 h after seeding. Thereafter, 10 µl of 5 mg/ml MTT (Thiazolyl Blue Tetrazolium Bromide; Sigma-Aldrich, St Louis, MO) reagent was added to the cell-culture medium, incubated for 4 h, removed the medium, and crystals were completely dissolved with 100 µl DMSO (Sigma-Aldrich). Thereafter The light absorbance OD570 reading was detected using Tecan Infinite 200 PRO (TECAN, Mannedorf, Switzerland). Experiments in each group were performed at least with three technical replicates.
Cell migration assay
5 × 104 cells/well with FBS negative IMDM were plated in Corning Transwell polycarbonate membrane cell culture inserts (3422, Sigma-Aldrich) for migration assays. 500 μl of IMDM + 10% FBS media were added to the bottom chamber of the migration plate to induce chemotaxis. Cells were allowed to migrate for 16–36 h (HGC27: 16 h, AGS: 36 h, MKN28: 24 h) and stained using the three-step stain (3300, ThermoFisher Scientific) according to the manufacturer’s instruction. The stained cells were observed and counted under the microscope at least three times in each group. Number of migrated cells were calculated using Image J software, and the percentage of migrated cells in each field (X100) was calculated.
Wound-healing assay
0.5×10^6 cells were plated in 6-well dishes with IMDM + 10% FBS + 1% penicillin/streptomycin. The cells were allowed to attach overnight, and a straight line was scratched using a pipet tip in each well. Images were taken immediately (0 h) and 16 h (for HGC27 cells), or 24 h (for AGS and MKN28 cells) after forming the scratch. Distance migrated by the cells was calculated as a percentage of the width of the gap at 16/24 h compared to that of 0 h. The average of three technical replicates were plotted.
Xenograft experiments
Seven-week-old male athymic nude mice (Envigo, Houston, TX) were kept under controlled conditions of light and fed ad libitum. 1 × 106 HGC27 cells in each group (naive, sh-ctrl, sh-FTO, and MA2 treatment group) were suspended in matrigel matrix (BD Biosciences) and injected subcutaneously into the left flank using 27-gauge needle (n = 10 in each group). About Five days after injection, subcutaneous tumours became palpable. In MA2 treatment group, we started intratumoral injection of 5 nmol of MA2 once a week for 4 weeks, as reported previously [30]. Tumour size was daily measured by calipers for 28 days, and tumour volume was calculated using the following formula: 1/2 (length × width × height). All mice were sacrificed on day 28, collected tumour mass, and calculated the tumour weight. The animal protocol was approved by the Institutional Animal Care and Use Committee, Baylor Scott & White Research Institute, Dallas, Texas.
Statistical analysis
Comparison of target gene expression between two independent groups were analysed using the two tailed Mann–Whitney U test (D’Agostino-Pearson omnibus normality test was used to determine Gaussian distribution of target gene expression) or two-tailed t-test. To analyse correlations between individual target genes and various clinicopathological features, as well as in multivariate analysis, Youden’s index was used to dichotomise patients into high and low expression groups. The differences between groups were analysed by the Fisher’s exact test. Overall survival (OS) was defined as the period from the date of GC diagnosis to the date of last follow up, and recurrence free survival (RFS) was defined as the period from the date of GC diagnosis to the date of recurrence (recurrence positive cases) or the date of last follow up (recurrence negative cases). Analysis for OS and RFS were performed by Log-rank test by dichotomising patients using the Youden’s index for OS and RFS in individual target gene and combined gene panel. Risk scores composed of combined target gene panel and various clinical risk factors which remained in univariate analysis were also generated by Cox proportional hazard model for OS and RFS in order to analyse whether each of the predictors affected the outcome after adjusting for known confounders. As for in-vitro and in-vivo experiments, statistical comparisons were determined by unpaired t-test when such comparisons were made between two groups, and one-way ANOVA with Tukey’s post hoc tests in cases of multiple comparisons. A p-value of <0.05 was considered statistically significant. All statistical analyses were performed using the Medcalc statistical software V.16.2.0 (Medcalc Software bvba, Ostend, Belgium), JMP software 10.0.2 (SAS Institute, Cary, NC), and GraphPad Prism V7.0 (GraphPad Software, San Diego, CA).
Results
Evaluation of the prognostic ability of m6A regulators in gastric cancer patients
In order to evaluate the overall prognostic significance of the m6A regulator genes, we investigated their expression in a clinical cohort composed of stage 1–4 GC patients by qRT-PCR assays. Association between various clinicopathological factors and the expression of individual m6A genes are shown in Supplementary Table 4. In advanced stage patients, the expression of FTO was significantly upregulated, while the expression of other m6A regulators (METTL3, METTL14, ALKBH5, WTAP, and YTHDF1) were significantly downregulated. We thereafter examined whether the expression of these m6A regulators is associated with OS by performing Kaplan–Meier survival analysis. Patients with high expression of FTO associated with obviously worse OS than those with low expression (p < 0.0001, Supplementary Fig. 1). On the other hand, patients with low expression of METTL3, METTL14, ALKBH5, WTAP, and YTHDF1 were associated with significantly poorer OS than those with high expression (p = 0.002, 0.0002, 0.0008, 0.0001, and 0.01, respectively, Supplementary Fig. 1). Furthermore, patients with low expression of YTHDF2 showed a trend for worse OS than those with high expression (p = 0.09, Supplementary Fig. 1). Collectively, these data suggest that the expression levels of individual m6A regulators are significantly associated with OS in GC patients.
Subsequently, to elucidate whether the m6A regulator expression profiles can also be utilised for recurrence prediction, we evaluated their expression in stage 1–3 GC patients who had undergone curative surgery. Association between clinicopathological factors and the expression of individual m6A genes are shown in Supplementary Table 5. In line with the stage 1–4 patients, the expression of FTO was significantly upregulated, while the expression of most of other m6A regulators (METTL3, METTL14, ALKBH5, and WTAP) were significantly downregulated in patients with recurrence. We next evaluated by Kaplan–Meier analysis whether the expression of these m6A regulators was associated with recurrence free survival (RFS). In line with the OS data, patients with high expression of FTO associated with significantly poorer RFS than those with low expression (p < 0.0001, Supplementary Fig. 2). On the other hand, patients with low expression of METTL3, METTL14, ALKBH5, and WTAP were associated with significantly poorer OS than those with high expression (p = 0.03, 0.01, 0.006, and 0.02, respectively, Supplementary Fig. 2). Taken together, these data indicate that the expression levels of individual m6A regulators are also significantly associated with RFS in GC patients with curative intent.
The five gene m6A classifier exhibited significantly superior prognostic ability independent of clinicopathological factors
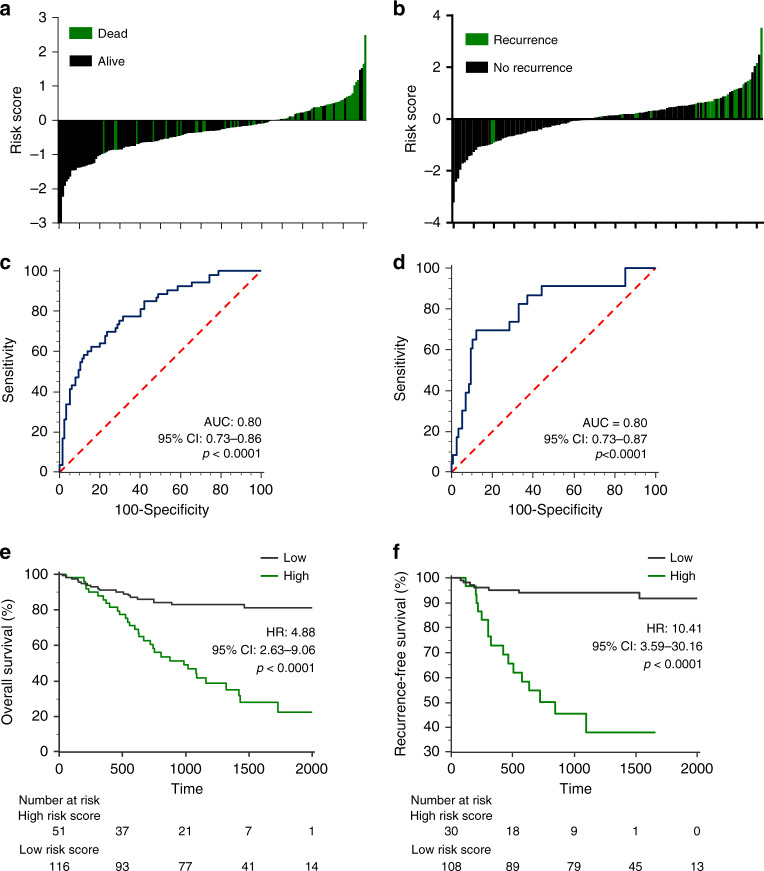
Next, we evaluated whether the combined expression profiles of m6A regulator genes further improves the prognostic ability in GC. We selected five of the seven m6A regulator genes as these five m6A regulators (FTO, METTL3, METTL14, ALKBH5, and WTAP) significantly distinguished both OS and RFS at single gene level. A multivariate cox-proportional hazard model was used to combine the 5-gene signature for OS and RFS prediction. The waterfall plots derived from the 5-gene cox-model, distinguished patients with poor OS (Fig. 1a) and RFS (Fig. 1b) with high accuracy. The risk scores derived from the 5-gene Cox model achieved an area under the curve (AUC) of 0.80 with a p < 0.0001 for OS and RFS prediction (Fig. 1c and d). Subsequently, Kaplan–Meier analysis revealed that patients with high-risk scores associated with significantly worse OS than those with low-risk scores with corresponding hazard ratio (HR) of 4.88 (p < 0.0001, Fig. 1e). As for RFS as well, patients with high risk scores associated with significantly worse RFS with corresponding HR of 10.41 (p < 0.0001, Fig. 1f). Taken together, these results demonstrate that the risk scores derived from the 5-gene m6A classifier offer robust prognostic potential in GC patients.
Fig. 1. Prognostic significance of the five gene m6A classifier in GC patients.
a, b The waterfall plot representing dead/alive (a) or recurrent/non-recurrent patients (b) based on the risk score derived from the five gene m6A classifier. c, d Receiver operating characteristic (ROC) curve for detection of dead/alive patients (c) and recurrent/non-recurrent patients (d) based on the five gene m6A classifier. (e, f Kaplan–Meier analysis for overall survival (OS; e) and recurrence free survival (RFS; f) between two groups derived from the five gene m6A classifier. HR hazard ratio, AUC area under curve.
In univariate analysis, along with the 5-gene m6A classifier, several clinicopathological factors showed significant association with poor OS and RFS (Table 1). Subsequent multivariate analysis identified that along with the 5-gene m6A classifier, older age, positive lymph node metastasis, hepatic metastasis, and distant metastasis, as independent risk factors for poorer OS (Table 1). With regards to RFS, multivariate analysis revealed that along with the 5-gene m6A classifier, larger tumour size was associated with worse RFS (Table 1). Collectively, we have demonstrated that the 5-gene m6A classifier is an independent risk factor of both worse OS and RFS in GC patients.
Table 1.
Univariate and multivariate cox proportional hazard analysis for overall survival and recurrence free survival in GC patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
| OS (n = 167) | ||||||
| Age ≥ 69 | 2.85 | 1.62–5.21 | 0.0002 | 3.72 | 1.84–7.86 | 0.0002 |
| Male | 1.67 | 0.91–3.32 | 0.1 | |||
| Macroscopic type 3 or 4 | 5.46 | 3.11–9.98 | <0.0001 | 1.79 | 0.81–4.12 | 0.15 |
| Tumour size >35 mm | 2.95 | 1.64–5.64 | 0.0002 | 1.12 | 0.51–2.48 | 0.78 |
| Poorly differentiated histology | 1.55 | 0.90–2.70 | 0.11 | |||
| T Stage greater than T4 | 7.06 | 3.97–13.13 | <0.0001 | 1.83 | 0.78–4.45 | 0.17 |
| Lymph node metastasis positive | 8.35 | 4.15–19.17 | <0.0001 | 4.26 | 1.54–13.67 | 0.004 |
| Lymphatic invasion positive | 4.3 | 1.99–11.24 | <0.0001 | 0.45 | 0.12–1.65 | 0.22 |
| Venous invasion positive | 4.05 | 2.32–7.32 | <0.0001 | 0.63 | 0.27–1.43 | 0.27 |
| Hepatic metastasis positive | 16.85 | 6.63–37.59 | <0.0001 | 6.17 | 2.30–14.93 | 0.0007 |
| Peritoneal metastasis positive | 7.67 | 3.89–14.19 | <0.0001 | 2.05 | 0.92–4.46 | 0.08 |
| Distant metastasis positive | 6.04 | 2.99–11.34 | <0.0001 | 6.57 | 2.64–16.27 | <0.0001 |
| Combined m6A panel high risk | 5.21 | 2.97–9.42 | <0.0001 | 2.24 | 1.15–4.49 | 0.02 |
| RFS (n = 138) | ||||||
| Age ≥69 | 2.48 | 1.07–6.16 | 0.03 | 1.14 | 0.40–3.40 | 0.81 |
| Male | 1.28 | 0.55–3.33 | 0.58 | |||
| Macroscopic type 3 or 4 | 5.57 | 2.44–13.4 | <0.0001 | 0.95 | 0.35–2.75 | 0.92 |
| Tumour size >35 mm | 4.68 | 1.87–14.19 | 0.0007 | 3.11 | 1.05–10.66 | 0.04 |
| Poorly differentiated histology | 4.1 | 1.63–12.41 | 0.002 | 1.41 | 0.48–4.87 | 0.55 |
| T Stage greater than T4 | 12.79 | 5.29–35.54 | <0.0001 | 2.07 | 0.63–7.58 | 0.24 |
| Lymph node metastasis positive | 19.79 | 5.79–123.84 | <0.0001 | 4.49 | 0.76–45.57 | 0.1 |
| Lymphatic invasion positive | 14.29 | 3.00–255.91 | <0.0001 | 1.47 | 0.14–34.10 | 0.76 |
| Venous invasion positive | 5.34 | 2.31–13.28 | <0.0001 | 1.52 | 0.51–4.71 | 0.46 |
| Combined m6A panel high risk | 11.62 | 4.81–31.32 | <0.0001 | 6.46 | 2.21–21.00 | 0.0005 |
HR hazard ratio, CI confidence interval.
Bold indicates a statistically significant.
Stable knockdown of FTO suppresses oncogenic function in GC cells
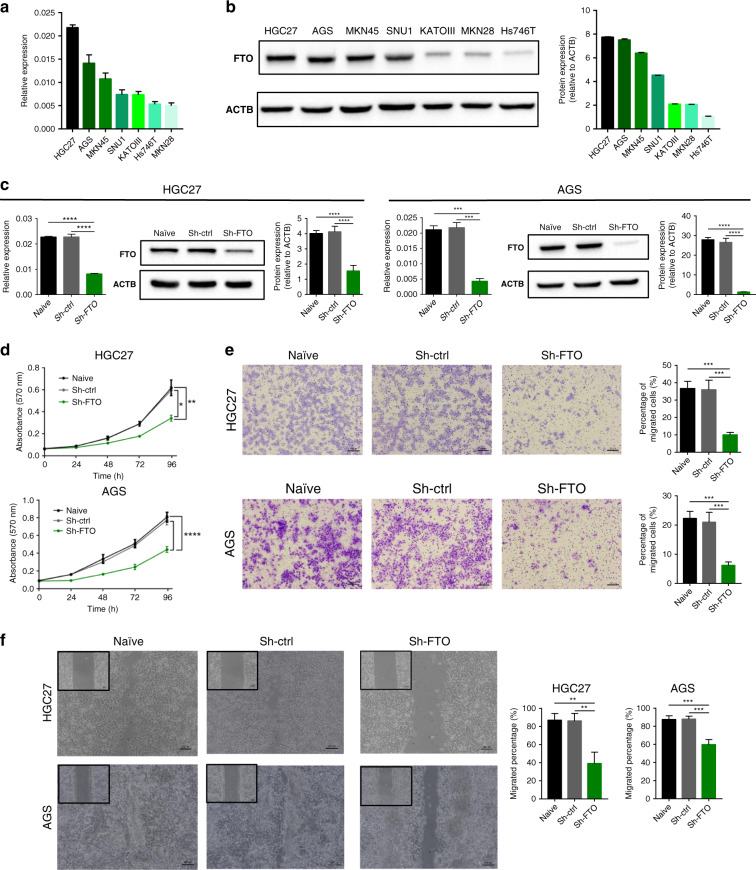
Considering that FTO was highly upregulated compared to other m6A regulators, we next performed functional experiments to investigate the oncogenic role of FTO in GC cell lines. We first evaluated the expression of FTO in various GC cells at mRNA and protein levels, which led to the selection of HGC27 and AGS cells that showed highest FTO expression as candidates for FTO knockdown studies (Fig. 2a and b). Following the stable knockdown of FTO, expression of FTO in HGC27 and AGS cells was confirmed by qRT-PCR and western blotting. In both the cell lines, we were able to achieve successful knockdown of the FTO (Fig. 2c).
Fig. 2. Stable knockdown of FTO suppressed cell proliferation and migration in gastric cancer cells.
a FTO mRNA expression in seven GC cell lines. b Western blotting image of FTO (left) and its relative signal intensity (right) in seven GC cell lines. c Stable knockdown of FTO by shRNA effectively decreased its both mRNA and protein expression level in HGC27 and AGS cells. d Cellular proliferation rate evaluated by MTT assay of sh-FTO cells compared to control cells in HGC27 (top) and AGS cell lines (bottom). e Representative image of cells that migrated through the permeable membrane of a transwell migration plate (left) and quantification of migrated cells (right) of sh-FTO and control cells in HGC27 and AGS cell lines. f Left: Representative image of monolayer of post scratch in HGC27 (16 h post scratch) and AGS cell lines (24 h post scratch). Right: Quantification of migrated cells in 16 h/24 h post scratch compared to 0 h post scratch. *p < 0.05, **p < 0.01, ***p < 0.001, and ***p < 0.0001 by one-way ANOVA test.
Subsequently, we performed additional assays to evaluate the effects of FTO suppression on the tumorigenicity in these two-cell lines. First, we performed MTT (3-4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay to evaluate the alteration of cell proliferation ability between the cells with and without FTO knockdown. As expected, knockdown of FTO expression significantly inhibited cell proliferation in both cell-lines following FTO stable knockdown compared with cells transduced with the control vector (Fig. 2d). Thereafter, we performed migration assays and wound-healing assays to interrogate the effects of FTO suppression on tumour cell migration potential. Interestingly, the cellular migratory capability was also significantly inhibited in both cell-lines following FTO stable knockdown compared with cells transduced with the control vector (Fig. 2e, and f). Collectively, these results suggest that FTO regulates not only cell proliferation, but also cell migration ability of GC cells.
FTO inhibition by MA2 suppressed oncogenic function in HGC27 and AGS cells
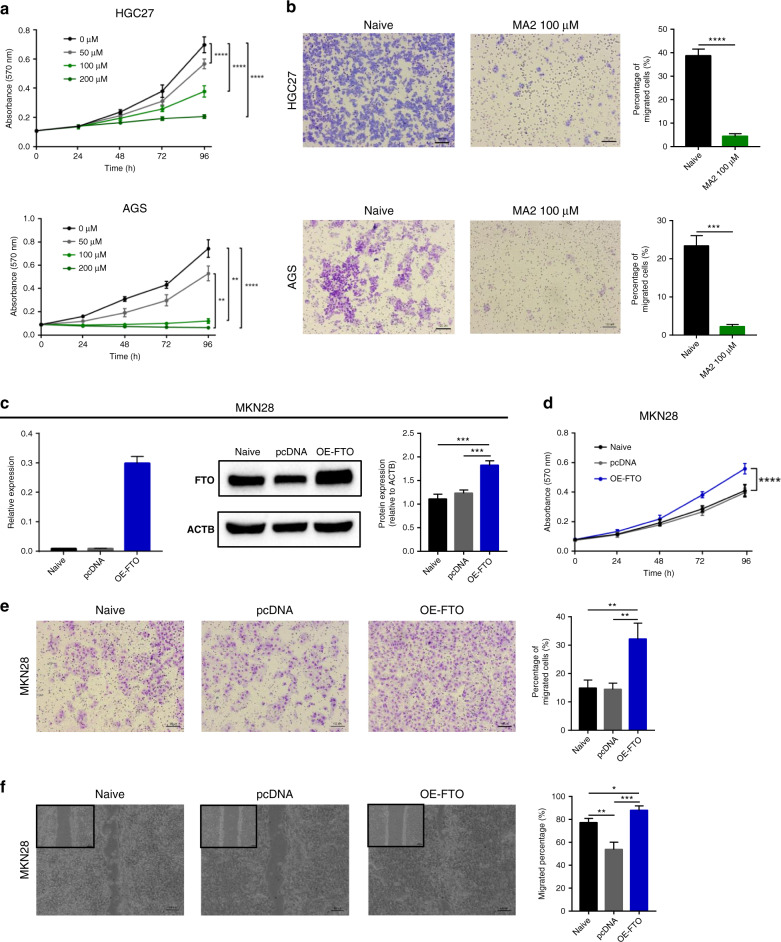
Next, we investigated the efficacy of MA2 which is reported as an FTO inhibitor [26, 27] through a series of functional assays. First, we performed MTT assays to investigate the effect of MA2 on cell proliferation. In line with the FTO knock down data, cell proliferation ability was indeed inhibited in a dose-dependent manner in both HGC27 and AGS cells (Fig. 3a). Subsequently, we performed cell migration assays to investigate the effects of MA2 on cell migratory ability. As expected, the migratory potential was also inhibited in both cell-line with 100 μM MA2 treatment compared with cells without treatment (Fig. 3b).
Fig. 3. FTO inhibitor (MA2) suppressed oncogenic role and overexpression of FTO enhanced the tumour progression in GC cell lines.
a Cellular proliferation rate was inhibited in a dose-dependent manner in MA2-treated cells compared to un-treated cells in HGC27 (top) and AGS cell lines (bottom). b Representative image of migration assay (left) and quantification of migrated cells (right) of MA2-treated cells compared to un-treated cells in HGC27 and AGS cell lines. c Transient overexpression (OE) of FTO significantly increased both mRNA and protein expression level in MKN28 cells. d Cellular proliferation rate was increased in OE-FTO cells compared to control cells in MKN28 cell line. e Representative image of migration assay (left) and quantification of migrated cells (right) of OE-FTO and control cells. f Left: Representative image of monolayer of 24 h post scratch in MKN28 cell line. Right: Quantification of migrated cells in 24 h post scratch compared to 0 h post scratch in MKN28 cell line. *p < 0.05, **p < 0.01, ***p < 0.001, and ***p < 0.0001 by two-tails-t-test, or by one-way ANOVA test.
Overexpression of FTO enhances proliferation and migratory potential in MKN28 cells
Next, we performed transient overexpression of FTO in MKN28 cells which showed lowest endogenous FTO expression (Fig. 2a). Forty-eight hours after transfection, the expression of FTO in MKN28 cells was confirmed by qRT-PCR and western blotting, and we observed that the FTO expression level was significantly overexpressed in these cells (Fig. 3c). Thereafter, we performed MTT assays to compare cell proliferation ability between the cells with and without FTO overexpression. We observed consistent results, in which MKN28 cells transfected with FTO overexpression plasmid showed significantly higher cell proliferation ability compared with cells transfected with the control plasmid (Fig. 3d). Next, we performed migration assays and wound-healing assays to assess the effects of FTO overexpression on tumour cell migration potential. As expected, the migration potential in FTO overexpression group was significantly higher compared with cells transfected with the control plasmid (Fig. 3e and f). Collectively, these results strengthened the observation that the oncogenic function of FTO especially in proliferative and migratory potential in multiple GC cell lines.
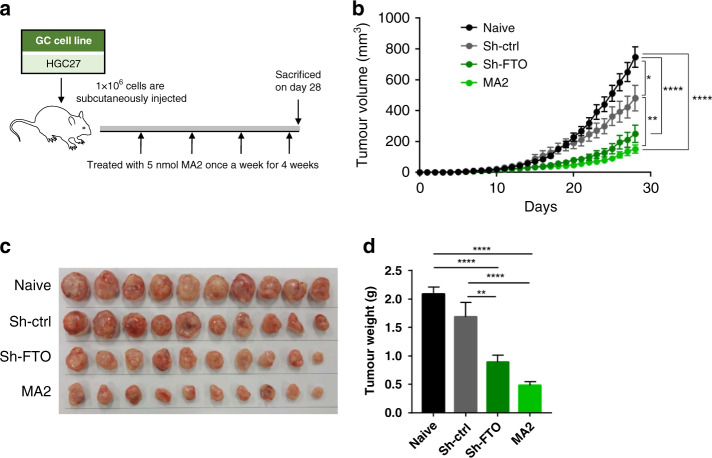
FTO stable knockdown and FTO inhibition suppress the tumour growth in HGC27 xenograft model
Next, in addition to in-vitro studies, we also performed an in-vivo xenograft animal model study to investigate whether FTO is involved in GC cell growth. We again used HGC27 cells, which belong to the epithelial-mesenchymal-transition (EMT) subtype, and performed subcutaneous injection of HGC27 cells to observe the daily tumour volume alteration in four groups (1.naive, 2.sh-ctrl; cells transfected with control vector, 3.sh-FTO; cells transfected with FTO stable knockdown vector, and 4.MA2 treatment group). In the MA2 treatment group, we performed intratumoral injection with 5 nmol of MA2 once a week for four weeks (Fig. 4a). Intriguingly, while tumour volume in the naive and sh-ctrl group continued to grow linearly, tumour growth in sh-FTO group and MA2 treatment groups was significantly attenuated (Fig. 4b). On day 28, we sacrificed all the mice, and tumours collected at end point also showed significant difference in tumour weights between the groups, which was in line with the daily tumour volume data (Fig. 4c and d). Overall, consistent with our in-vitro studies, we were able to successfully demonstrate that FTO has an oncogenic role in an animal model as well.
Fig. 4. Stable knockdown of FTO and inhibition by MA2 resulted in tumour growth inhibition in HGC27 xenografts.
a The schematic diagram of the in-vivo experiment and MA2 treatment in HGC27 xenografts. b Time course tumour volume alteration in each group. c Representative image of collected xenograft tumours. d Average weight of collected xenograft tumour between groups. *p < 0.05, **p < 0.01, and ***p < 0.0001 by one-way ANOVA test.
FTO is associated with the alteration of epithelial-mesenchymal-transition (EMT) related genes
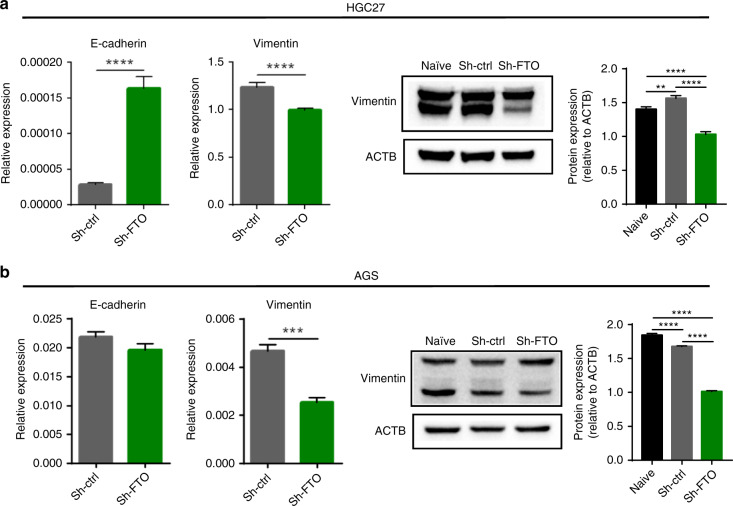
Our previous in-silico study revealed that the deregulation of m6A regulators demonstrated strong associations with the activation of EMT pathway in multiple human malignancies including gastric cancer [25]. Having observed the tumorigenic potential of FTO and its association with tumour cell migration, we next examined the expression EMT regulated genes upon FTO knockdown. The mRNA expression level of E-cadherin was significantly higher in sh-FTO group of HGC27 cells (Fig. 5a and b). On the other hand, vimentin expression level was significantly downregulated in sh-FTO group of both cell lines (Fig. 5a and b). The results of western blot confirmed that protein expression level of vimentin was downregulated in sh-FTO group of both cell lines (Fig. 5a and b). These data suggest that FTO might play an oncogenic role via EMT pathway.
Fig. 5. FTO inhibition resulted in the modulation of core of epithelial-mesenchymal-transition related genes in GC cell lines.
a, b Left: mRNA expression levels of E-cadherin and vimentin. Right: western blotting image of FTO and vimentin, and its relative signal intensity of sh-FTO cells compared to control cells in HGC27 (a) and AGS cell line (b). *p < 0.05, **p < 0.01, and ***p < 0.0001 by two-tails-t-test, or by one-way ANOVA test.
Discussion
Recent studies have demonstrated the role of aberrant m6A modifications in cancer progression across various malignancies [17–23]. Especially among them, decreased m6A RNA methylation levels induced by FTO or ALKBH5 often exert oncogenic functions in various cancers [17, 18, 24, 31, 32]. We previously identified a 7-gene m6A classifier and demonstrated its ability in predicting prognosis across multiple cancers including gastric cancer [25]. We were also intrigued by the fact that the m6A gene-derived high-risk patients showed activation of epithelial-mesenchymal pathway (EMT) [25]. Here we have systematically investigated the prognostic ability of m6A genes in an independent cohort of gastric cancer patients and elucidated the functional role of FTO in gastric cancer progression.
Several previous studies also suggested the oncological importance and functional role of m6A regulators including FTO in gastric cancer [33–41]; however, currently most of these findings are based on in-silico data analysis. In this study, we performed a series of investigations consisting of clinical evaluations, in-vitro and in-vivo experimental models, and we demonstrated several additional findings that were not reported in the previous studies. In addition to the FTO knock-down and overexpression studies in cells, we also confirmed the oncological functional role of FTO using an animal model derived from HGC27 cells, which possesses a strong EMT phenotype. Furthermore, we also demonstrated the effectiveness of MA2 (FTO inhibitor) both in cultured cells and an animal model. Our results indicated that the FTO knock-down or inhibition altered EMT potential. Collectively, our study strengthened and added further findings as for the oncogenic role of FTO to the previous study in gastric cancer.
EMT is a fundamental developmental process that is reactivated in wound healing and a variety of diseases including cancer where it induces the development of metastasis, resistance to treatment, and generation and maintenance of cancer stem cells [42, 43]. There are various epigenetic modifications that are associated with modulation of EMT; DNA methylation, histone acetylation, RNA m6A and m5C RNA methylation, and reprogramming of the epigenome by transcription factors or non-coding RNAs [44]. Indeed, for MALAT1 which is a major lncRNA associated with EMT potential, two major m6A sites were found at consensus sites in two hairpin stem structures in several human cell lines like breast cancer [45]. The consequence of this modification would destabilise these secondary structures and would modify interactions with RNA binding proteins.
In this study, we found that stable knockdown of FTO resulted in upregulation of E-cadherin and down-regulation of vimentin in multiple GC-cell lines. It is suggested that FTO may be involved in the stabilisation and overexpression of EMT related genes. Among the previous evidence, Wang, et al, investigated the oncological importance of m6A regulators including FTO using The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) datasets, and revealed the oncogenic potential of FTO in GC patients [41]. Furthermore, these authors predicted the demethylated target gene of FTO was integrin B1 (ITGB1) and strengthened this hypothesis by performing in-vitro analysis. However, ITGB1 was not identified by m6A sequencing, but by relevant module and gene prediction using bioinformatic approaches. In order to resolve the remaining poorly understood issues in our current study, we are currently working on elucidating demethylated targets genes by using sophisticated methodologies such as m6A sequencing [46–50] as well as RNA-seq.
We acknowledge a couple of potential limitations to this present study. First, our clinical validation was performed using retrospectively collected GC specimens from a single institution, and therefore prospective validation using independent datasets is must to translate these findings to clinical care. We also did not further investigate the direct FTO target genes in our FTO knock-down and overexpression systems as well as pre-post MA2 treatment. We are currently working on elucidating targets genes, which will provide exciting mechanistic insights between FTO and EMT alteration in gastric cancer.
In conclusion, here we demonstrate that m6A regulators may serve as promising prognostic and recurrence biomarkers in GC patients. Our functional studies revealed that FTO is an important oncogene and may serve as a promising therapeutic target associating with the EMT alterations in gastric cancer.
Supplementary information
Acknowledgements
We would like to acknowledge Preethi Ravindranathan, Aki Sakatani, Michael Hsieh, and Kinnari Pankaj Modi for experimental advice, and acknowledge Divya Pasham, Ashley Cao, Maddie Brown, and Anna Wakita for experimental support.
Author contributions
Concept and design: TS, RK and AG; Acquisition, analysis, or interpretation of data: TS, RK; Drafting of the manuscript: TS, RK, YO, MO, YT and AG; Statistical analysis: TS, and RK; Administrative, technical, or material support: TS, YO, MO, YT and AG; Supervision: AG
Funding
This work was supported by CA184792, CA187956, CA227602, CA072851 and CA202797, grants from the National Cancer Institute, National Institutes of Health, and a pilot grant from the Stupid Strong Foundation to A Goel.
Data availability
All data are available within the article.
Ethics approval and consent to participate
All study-related procedures were performed as per the Declarations of Helsinki, wherein a written informed consent was obtained from each patient, and the institutional review boards of all participating institutions involved approved the study.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tadanobu Shimura, Raju Kandimalla.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01581-w.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–55. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–86. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 6.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–47. [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 8.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 10.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–21. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao BS, He C. Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biol. 2015;16:43. doi: 10.1186/s13059-015-0609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma Stem cells. Cell Rep. 2017;18:2622–34. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606 e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–42. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–43. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 24.Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–96 e11. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandimalla R, Gao F, Li Y, Huang H, Ke J, Deng X, et al. RNAMethyPro: a biologically conserved signature of N6-methyladenosine regulators for predicting survival at pan-cancer level. NPJ Precis Oncol. 2019;3:13. doi: 10.1038/s41698-019-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartzatt R. Anti-inflammatory drugs and prediction of new structures by comparative analysis. Antiinflamm Antiallergy Agents Med Chem. 2012;11:151–60. doi: 10.2174/187152312803305768. [DOI] [PubMed] [Google Scholar]
- 28.Zheng G, Cox T, Tribbey L, Wang GZ, Iacoban P, Booher ME, et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem Neurosci. 2014;5:658–65. doi: 10.1021/cn500042t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–84. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Li Z, Kong B, Song C, Cong J, Hou J, et al. Reduced m(6)A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget. 2017;8:98918–30. doi: 10.18632/oncotarget.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–42. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Zhang S, Huang S. FoxM1: a potential drug target for glioma. Future Oncol. 2012;8:223–6. doi: 10.2217/fon.12.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38:2285–92. doi: 10.3892/or.2017.5904. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Digestive Dis Sci. 2019;64:1503–13. doi: 10.1007/s10620-018-5452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8:4766–81. doi: 10.1002/cam4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Y, Huang J, Hu J. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gastric cancer. Front Oncol. 2019;9:1038. doi: 10.3389/fonc.2019.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan K, Liu X, Li J, Ding Y, Li J, Cui G, et al. Expression status and prognostic value of M6A-associated Genes in gastric cancer. J Cancer. 2020;11:3027–40. doi: 10.7150/jca.40866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Piao HY, Wang Y, Meng XY, Yang D, Zhao Y, et al. To develop and validate the combination of RNA methylation regulators for the prognosis of patients with gastric cancer. OncoTargets Ther. 2020;13:10785–95. doi: 10.2147/OTT.S276239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Zhou E, Zheng J, Zhang C, Zou Y, Lin J, et al. Prognostic and predictive value of m6A “Eraser” related gene signature in gastric cancer. Front Oncol. 2021;11:631803. doi: 10.3389/fonc.2021.631803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing JJ, Zhao X, Li H, Sun LP, Yuan Y. Expression profiles and prognostic roles of m6A writers, erasers and readers in gastric cancer. Future Oncol. 2021;17:2605–20. doi: 10.2217/fon-2020-0630. [DOI] [PubMed] [Google Scholar]
- 41.Wang D, Qu X, Lu W, Wang Y, Jin Y, Hou K, et al. N(6)-Methyladenosine RNA demethylase FTO promotes gastric cancer metastasis by down-regulating the m6A methylation of ITGB1. Front Oncol. 2021;11:681280. doi: 10.3389/fonc.2021.681280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 44.Roche, J, Gemmill, RM & Drabkin, HA. Epigenetic regulation of the epithelial to mesenchymal transition in lung cancer. Cancers (Basel). 2017:9:72. [DOI] [PMC free article] [PubMed]
- 45.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai Q, Fong R, Saikia M, Stephenson D, Yu YT, Pan T, et al. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 2007;35:6322–9. doi: 10.1093/nar/gkm657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chi KR. The RNA code comes into focus. Nature. 2017;542:503–6. doi: 10.1038/542503a. [DOI] [PubMed] [Google Scholar]
- 49.Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18:275–91. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 50.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–19. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within the article.