Abstract
In order to gain a better understanding of the molecular epidemiology of Mycobacterium bovis isolates in Cameroon, 75 isolates of M. bovis collected in three provinces of northern Cameroon were studied by spoligotyping. For 65 of these isolates, typing was also carried out by pulsed-field gel electrophoresis (PFGE) with DraI, and 18 of the isolates were also typed by restriction fragment length polymorphism (RFLP) analysis with probe IS6110-RHS. Molecular typing of the isolates by these techniques revealed a high degree of homogeneity, with 10 spoligotypes for 75 isolates, four PFGE profiles for 65 isolates, and three RFLP types for 18 isolates. Some types were present in the three different provinces, while some were confined to one or two areas. These results suggest that geographical mapping of M. bovis strains could be helpful for the control of bovine tuberculosis at the regional level. An interesting feature of all the spoligotypes was the absence of spacer 30, suggesting a common origin for all of the Cameroon isolates tested; an evolutionary scenario for the isolates is discussed. In addition, a comparison of the three techniques showed that for M. bovis strain differentiation in Cameroon and in surrounding countries, spoligotyping would be a more discriminating and practical tool for molecular typing than the other two techniques used in this study.
Bovine tuberculosis (TB) is endemic in many African countries, but economic constraints preclude the use of skin test and slaughter control strategies, which have proved effective in the developed world. In Cameroon, the majority of cattle herds are concentrated in the north (13), which is surrounded by Nigeria, Chad, and the Central African Republic. From visible lesion data obtained in the main slaughterhouses, it would appear that the prevalence of bovine TB in Cameroon is high (7). In addition, frequent cattle movement across the different areas of the country and across frontiers favors strain dissemination. In order to reduce the transmission of bovine TB, a bill from the Ministry of Livestock, Fisheries, and Animal Industries of Cameroon (no. 76/420) was introduced in 1976 to prevent the circulation of cattle between Adamaoua and the other two areas of northern Cameroon, i.e., Extreme North and North. This action resulted in the isolation of cattle within Adamaoua.
To date, few studies have been performed to determine the correct prevalence of Mycobacterium bovis infection at local and regional levels (3, 16, 17, 19), and there are no available data regarding the variability of M. bovis isolates within Cameroon. The aim of this study was to apply a number of molecular typing techniques to M. bovis isolates from different slaughterhouses located in three different provinces of northern Cameroon—North, Extreme North, and Adamaoua—in order gain a better understanding of the geographical distribution of M. bovis strains. The typing techniques used in this study were spoligotyping (11), pulsed-field gel electrophoresis (PFGE) (14) and, for some isolates, restriction fragment length polymorphism (RFLP) analysis with probe IS6110-RHS (10, 20). Whenever possible, more than one technique was applied to each isolate, since the ability of the various techniques to differentiate between isolates has been reported to vary according to the geographical localization of the isolates of M. bovis (1, 4, 18, 23).
MATERIALS AND METHODS
Mycobacterial strains. (i) M. bovis isolates.
Samples were collected in 1989–1990 and 1995–1996 from cattle in different slaughterhouses. These slaughterhouses were located in different provinces of northern Cameroon—North, Extreme North, and Adamaoua. This sampling regimen allowed the isolation of 123 isolates of M. bovis which had classical cultural and biochemical properties (6). A total of 75 isolates were available for DNA typing. All 75 were subjected to spoligotyping, 65 were subjected to PFGE, and only 18 were subjected to RFLP analysis with probe IS6110-RHS. The geographical distribution of the isolates according to the DNA typing techniques used is presented in Table 1.
TABLE 1.
Geographical distribution of M. bovis strains collected in northern Cameroon according to the DNA typing technique used
| Province | No. of strains tested by:
|
||
|---|---|---|---|
| Spoligotyping | PFGE | RFLP analysis | |
| North | 28 | 27 | 8 |
| Extreme North | 23 | 23 | 10 |
| Adamaoua | 24 | 15 | |
| Total | 75 | 65 | 18 |
(ii) Reference strains.
DNA from M. tuberculosis H37Rv was used in order to obtain probe IS6110-RHS for RFLP and was included as a control in RFLP analysis, spoligotyping, and PFGE. A reference strain of M. bovis BCG (BCG Pasteur P3) was also used as a control in spoligotyping.
Spoligotyping.
For amplification of the direct repeat (DR) locus, we used either genomic DNA extracted by the method of Wilson (22) or cell lysates obtained by heat treatment. Spoligotyping was performed according to the technique of Kamerbeek et al. (11), as described for M. bovis by Aranaz et al. (2).
PFGE.
Bacteria were grown in 40 ml of 7H9 broth (Difco, Detroit, Mich.) at 37°C to the early exponential phase of growth. The cells were harvested by centrifugation, and PFGE was carried out with DraI (Roche/Boehringer, Mannheim, Germany) by the method of Lévy-Frébault et al. (14). The molecular weight marker used as a reference was Lambda Ladder PFG Marker (Biolabs, Beverly, Mass.).
RFLP.
Cells were grown at 37°C in 7H9 broth and DNA extraction was performed as described previously (22).
Probe IS6110-RHS was obtained by amplification of a fragment of 244 bp situated at the right-hand side of the PvuII site using primers INS-1 and INS-2 (Oligo Express, Paris, France) as described by Hermans et al. (10). The PCR was carried out with a total volume of 50 μl/microtube containing 75 ng of each primers INS-1 and INS-2, 1× Taq polymerase buffer (Roche/Boehringer), 100 μM (each) deoxynucleoside triphosphate (Roche/Boehringer), 20 ng of M. tuberculosis H37Rv DNA, and 0.1 U of Taq polymerase (Roche/Boehringer). Fifty microliters of mineral oil (Sigma Aldrich, St. Louis, Mo.) was added. DNA amplification was performed using a Programmable Thermal Controller thermocycler (MJ Research, Inc). Two series of cycles were performed: fives cycles at 94°C for 1 min, 65°C for 1.5 min, and 72°C for 2 min and 35 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min. These cycles were followed by a final elongation for 10 min at 72°C. The amplified DNA was purified by extraction from a 1.2% agarose gel (Eurobio, les Ullis, France) in Tris-acetate-EDTA using a Geneclean II R kit (Bio 101 Inc., Vista, Calif.) according to supplier instructions. The purified IS6110-RHS fragment was end labeled with alkaline phosphatase by use of an AlkPhos Direct kit (Amersham Life Science, Buckinghamshire, United Kingdom) and the protocol recommended by the supplier.
DNAs digested with PvuII (Roche/Boehringer) were allowed to migrate at 45 V for 16 h with a 1-kb ladder marker (Gibco BRL, Paisley, Scotland) and M. tuberculosis H37Rv DNA as a positive control. DNA transfer and labeling were then performed. For all of these steps, the recommendations of van Emden et al. (20) were followed.
RESULTS
Spoligotyping.
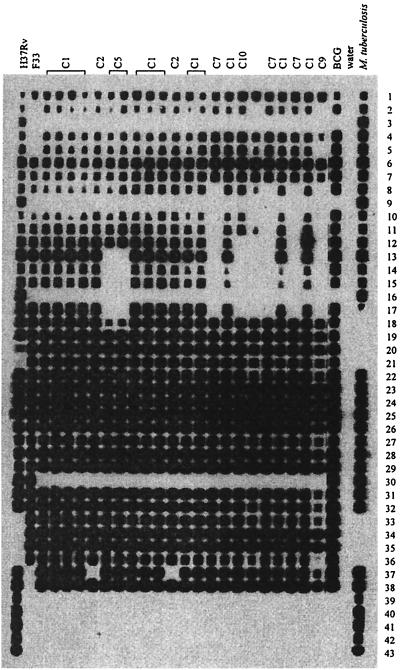
Ten different spoligotypes were obtained from the 75 isolates tested. These types were designated C1 to C10. The 10 spoligotypes are represented in Fig. 1, and their distribution in each province is shown in Table 2 and in Fig. 2. For all of these types, the absence of spacers 3, 9, 16, and 39 to 43, which is a characteristic of the majority of M. bovis strains, was noted. The unique feature of M. bovis strains from Cameroon compared to strains from other countries was the consistent absence of spacer 30 in the DR locus.
FIG. 1.
Spoligotyping patterns of M. bovis isolates in northern Cameroon. C1 to C10 are spoligotypes of some isolates from Cameroon. Reference strains were H37Rv (M. tuberculosis H37Rv); BCG (M. bovis BCG P3), which had the same profile as the most frequent spoligotype in France. F33 is another example of a French spoligotype and M. tuberculosis (last lane) is an example of an M. tuberculosis isolate. Numbers 1 to 43 correspond to 43 spacer oligonucleotides of the DR locus, which were covalently linked to the membrane.
TABLE 2.
Distribution by province and by year of isolation of the 10 spoligotypes of M. bovis observed in northern Cameroon
| Spoligotype | No. (%) of spoligotypes in the following province at the indicated time:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| North
|
Extreme North
|
Adamaoua
|
Grand total | |||||||
| 1989–1990 | 1995–1996 | Total | 1989–1990 | 1995–1996 | Total | 1989–1990 | 1995–1996 | Total | ||
| C1 | 3 | 18 | 21 (75) | 17 | 17 (74) | 2 | 7 | 9 (38) | 47 (63) | |
| C2 | 1 | 1 | 1 | 1 | 2 (3) | |||||
| C3 | 1 | 1 | 1 | |||||||
| C4 | 1 | 2 | 3 (11) | 1 | 1 | 2 | 2 (8) | 6 (8) | ||
| C5 | 1 | 1 | 1 | 1 | 2 (3) | |||||
| C6 | 1 | 1 | 3 | 3 | 1 | 1 | 5 (7) | |||
| C7 | 8 | 8 (33) | 8 (11) | |||||||
| C8 | 1 | 1 | 1 | |||||||
| C9 | 1 | 1 | 1 | |||||||
| C10 | 2 | 2 (8) | 2 | |||||||
| Total | 4 | 24 | 28 | 0 | 23 | 23 | 14 | 10 | 24 | 75 |
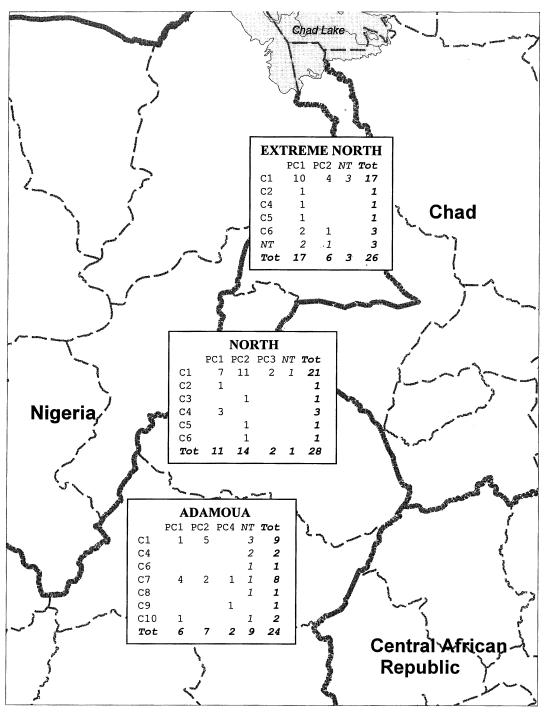
FIG. 2.
Geographical distribution of the spoligotypes and PFGE profiles of M. bovis strains collected in the three provinces of northern Cameroon (Extreme North, North, and Adamaoua). C1 to C10, spoligotypes; PC1 to PC4, PFGE profiles; NT, not tested; Tot, total.
In this study, spoligotype C1 was identified as dominant, as 47 strains (63%) were of this genotype. This spoligotype was present in all three provinces of northern Cameroon. C4, present in six isolates (8%), and C6, present in five isolates (7%), were also found in the three areas tested. C7, observed in eight isolates (11%), was restricted to one province only, Adamaoua. In this area, 33% of the strains (8 of 24) belonged to this type. Three other spoligotypes (C8, C9, and C10) were also limited to this province but were present in very small numbers (one or two isolates).
PFGE.
The distribution of the four PFGE profiles according to the provinces of northern Cameroon is represented in Fig. 2. For the 65 isolates typed by PFGE, four PFGE profiles were obtained (data not shown); these were designated PC1 to PC4. PC1 and PC2 were clearly dominant, as they comprised 35 isolates (54%) and 26 isolates (40%), respectively. These PFGE profiles were present in all three provinces of northern Cameroon tested. It is interesting to note that the only difference observed between PC1 and PC2 corresponds to a fragment of 295 kb present in the PC2 profile. PC3 and PC4 were detected in only two isolates each (3%). In both cases, isolates with the same PFGE profile were from the same province of northern Cameroon.
RFLP analysis with probe IS6110-RHS.
RFLP results were obtained for 18 isolates collected from two of the three provinces, i.e. North (8 strains) and Extreme North (10 isolates). Three profiles were obtained (data not shown); these were designated RC1, RC2, and RC3. The majority of the isolates (83%) were of type RC1, which has only one copy of IS6110, located on a PvuII restriction fragment of 1.9 kb. This is the most prevalent M. bovis RFLP type observed globally (4). RC1 was present in six isolates (75%) in North and in nine isolates (90%) in Extreme North. RC2 (with bands of 1.9 and 4.07 kb) was observed in only two isolates collected in both areas, and RC3 (with bands of 1.9, 4.07, and 6.5 kb) was observed in only one isolate.
Comparison of the results obtained by the different molecular techniques.
Sixty-two isolates of M. bovis were typed by spoligotyping and by PFGE with DraI. The results are summarized in Fig. 2. Comparison of the different spoligotypes and DraI PFGE profiles revealed that some isolates with identical spoligotypes could be differentiated by PFGE and vice versa. For example, 40 C1 spoligotypes could be further differentiated into 3 PFGE types, whereas 32 PC1 PFGE types could be further differentiated into 7 spoligotypes and 26 PC2 PFGE types could be further differentiated into 5 spoligotypes. Thus, a combination of the two techniques provided greater discrimination than any single technique used alone. However, spoligotyping appeared more discriminative than PFGE with DraI since for the 62 strains typed by both techniques, nine spoligotypes and only four DraI PFGE profiles were identified. In addition, a combination of C1 and C7 (the two most frequent spoligotypes) encompassed 75.8% of the strains (47 of 62), whereas PC1-PC2 (the two most frequent DraI PFGE profiles) represented 93.5% of the strains (58 of 62). Although fewer isolates were typed in this study with IS6110 RHS, this technique appeared to have the lowest level of discrimination, since 14 of 16 isolates (87.5%) belonged to the dominant type, RC1.
DISCUSSION
For the 75 M. bovis isolates collected in three provinces of northern Cameroon, 10 different spoligotypes (C1 to C10), four DraI PFGE profiles, and three RFLP types were identified. These results indicate a high degree of homogeneity among M. bovis isolates in northern Cameroon compared with the results obtained in other countries, such as France (N. Haddad, A. Ostyn, B. Durand, C. Karoui, J. Inwald, S. Hughes, M. F. Thorel, and G. Hewinson, Abstr. 30th Int. Union Against Tuberc. Lung Dis. [IUATLD] World Conf., abstr. 199-PD, 1999). It is particularly interesting to note that Cameroon seems to belong to a group of countries in which there is a low level of heterogeneity among M. bovis isolates. This group includes Australia (4) and Tanzania for M. bovis (12) and China (21) and Tunisia (see reference 21 and references therein) for M. tuberculosis.
In their analysis of the molecular typing profiles of M. bovis isolates from Tanzania, Daborn et al. (5) postulated the existence of two categories of M. bovis strains: autochthonous strains (with atypical cultural properties) and strains imported from Europe (with classical cultural properties). The isolates obtained from cattle lesions in northern Cameroon in this study possessed classical cultural properties, suggesting a European origin of infection. The first documented data concerning the introduction of cattle from Europe refer to the import in 1913 of Charolaise cattle from Saône-et-Loire, France, and France continued to be the major source of cattle exported to Cameroon, especially for the provinces of North Cameroon and for Adamaoua in particular (unpublished data). As far as we can determine, there is only one documented case of cattle importation from Switzerland (unpublished data). This information is consistent with the relative homogeneity observed for the spoligotypes identified in northern Cameroon. Moreover, the 75 strains which were typed shared one common characteristic, namely, the absence of spacer 30. In addition, the dominant spoligotype pattern, C1, is extremely similar to the most frequently observed spoligotype in France, which is identical to that of BCG (BCG-like) (Haddad et al,. 30th IUATLD World Conf.), differing only in the absence of spacer 30 in C1. It is therefore possible that M. bovis was introduced into Cameroon during the period of colonization by France from 1917 on, via the introduction of French breeds of cattle, which became commonly used from this time on (unpublished data). The appearance of C1 and its maintenance, in relation to the loss of spacer 30, could be related to the selection of an adaptive genetic factor in the natural environment of Cameroon. This hypothesis is supported by two observations. The first is the absence in Cameroon of the spoligotype most commonly observed in France BCG like (which differs from C1 by the absence of spacer 30 in C1). The second observation is that the loss of spacer 30 in M. bovis isolates in France is a very rare event. To date, the loss of spacer 30 has been observed in only 3% of the 1,000 French isolates which have been spoligotyped. Moreover, of these isolates, only one (0.1%) has been identified as having spoligotype C1, and no isolates have been observed with spoligotypes C2 to C10.
Recently, Fang et al. (8) noted that changes in the DR locus tend to be toward the loss of repeat spacers and their following spacer sequence, termed direct variable repeat, and postulated that the common ancestor of the M. tuberculosis complex had a DR structure similar to that in M. bovis BCG. The C1 spoligotype pattern is consistent with this hypothesis in that it could have evolved from a French (BCG-like) strain by the loss of spacer 30.
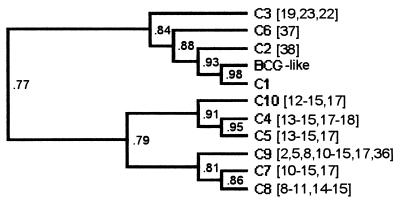
Moreover, the fact that all of the presently spoligotyped isolates from Cameroon lack spacer 30 suggests that all strains may have evolved from a C1 strain by direct or clonal expansion. The analysis of the dendrogram (Fig. 3) obtained with Diversity Database Software, version 2 (Bio-Rad, Hercules, Calif.), based on the similarities of the DR loci in different strains (Table 3), shows that such a link between C1 and all the other spoligotypes could exist, with a progressive loss of spacers. In this hypothesis, types C1 to C10 could have evolved as a result of selection or coselection of an adaptive factor beneficial for the maintenance of strains lacking spacer 30 in Cameroon. The relatively recent introduction of M. bovis into Cameroon would also explain the dominance of C1 and the small degree of divergence between spoligotypes. In this context, it would be interesting to investigate whether the absence of spacer 30 is shared by M. bovis isolates from neighboring countries or if this genotype is restricted to isolates in northern Cameroon.
FIG. 3.
Dendrogram obtained with isolates from Cameroon (UPGMA method). C1 to C10 are spoligotypes of some isolates from Cameroon; BCG-like is the most frequently observed spoligotype in France. Numbers in brackets indicate spacers lacking (in addition to spacers 3, 9, 16, 30, and 39 to 43, which are absent in spoligotypes C1 to C10).
TABLE 3.
Similarity matrix of our population of spoligotypes (UPGMA method; Dice coefficient method)a
| Spoligotype | % Similarity to the following spoligotype:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BCG-like | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | |
| BCG-like | 100.0 | 97.7 | 95.3 | 90.7 | 86.0 | 88.4 | 95.3 | 79.1 | 86.0 | 72.1 | 86.0 |
| C1 | 97.7 | 100.0 | 97.7 | 93.0 | 88.4 | 90.7 | 97.7 | 81.4 | 88.4 | 74.4 | 88.4 |
| C2 | 95.3 | 97.7 | 100.0 | 90.7 | 86.0 | 88.4 | 95.3 | 79.1 | 86.0 | 72.1 | 86.0 |
| C3 | 90.7 | 93.0 | 90.7 | 100.0 | 81.4 | 83.7 | 90.7 | 74.4 | 81.4 | 67.4 | 81.4 |
| C4 | 86.0 | 88.4 | 86.0 | 81.4 | 100.0 | 97.7 | 86.0 | 88.4 | 86.0 | 81.4 | 95.3 |
| C5 | 88.4 | 90.7 | 88.4 | 83.7 | 97.7 | 100.0 | 88.4 | 90.7 | 88.4 | 83.7 | 97.7 |
| C6 | 95.3 | 97.7 | 95.3 | 90.7 | 86.0 | 88.4 | 100.0 | 79.1 | 86.0 | 72.1 | 86.0 |
| C7 | 79.1 | 81.4 | 79.1 | 74.4 | 88.4 | 90.7 | 79.1 | 100.0 | 93.0 | 93.0 | 93.0 |
| C8 | 86.0 | 88.4 | 86.0 | 81.4 | 86.0 | 88.4 | 86.0 | 93.0 | 100.0 | 86.0 | 86.0 |
| C9 | 72.1 | 74.4 | 72.1 | 67.4 | 81.4 | 83.7 | 72.1 | 93.0 | 86.0 | 100.0 | 86.0 |
| C10 | 86.0 | 88.4 | 86.0 | 81.4 | 95.3 | 97.7 | 86.0 | 93.0 | 86.0 | 86.0 | 100.0 |
C1 to C10, spoligotypes detected in Cameroon; BCG-like, the most frequently observed spoligotype in France. UPGMA, unweighted pair group method with arithmetic mean.
The presence of spoligotypes C1, C4, and C6 in all three provinces may be explained by bovine transhumance, which occurs in more than 60% of cattle in Cameroon (12) and could have resulted in wide dissemination of certain spoligotypes. Conversely, from the limited data obtained in this study, some spoligotypes appear to be unique to a single province. For example, Adamaoua is the only province where spoligotypes C7 (with eight isolates), C8, C9, and C10 were detected. This result may be explained by the introduction, in 1976, of measures to prevent the circulation of cattle between Adamaoua and the other two provinces of northern Cameroon. However, sample bias cannot be excluded from our study, since all the isolates with these particular spoligotypes were collected in Adamaoua between 1989 and 1990, whereas these spoligotypes were not observed in isolates collected from this province in 1995–1996 or the other provinces in 1989–1990 (North) or 1995–1996 (North Province and Extreme North). Nevertheless, from the dendrogram shown in Fig. 3, it can be seen that types C7, C8, and C10 appear to be closely related in terms of their DR structure, consistent with their restricted location in Adamaoua. We can add C9, which seems more distant in the dendrogram, but which is in fact very close to C7, both quantitatively (94% identity) and qualitatively (nature of the deleted spacers). This result suggests that in vivo there may be a relatively rapid molecular clock for changes in the DR region, with a maximum time for change of 16 years, compared with the minimum time for change of 60 years observed in vitro for M. tuberculosis H37Rv (8). In our study, two isolates were identified with two genetic features unique to Adamaoua (spoligotype C7-PFGE profile PC4 and spoligotype C9-PFGE profile PC4), further evidence for regional differences in strain distribution. However, further work is required to gain a better understanding of strain distribution within northern Cameroon. It would be of interest to compare the spoligotypes of M. bovis isolates from countries which share a frontier with Adamaoua (Central African Republic and Nigeria) and from those which do not (e.g., Chad).
The hypothesis that the M. bovis isolates isolated from northern Cameroon have evolved from a common source which was introduced relatively recently into the cattle population is borne out by the results obtained by DraI PFGE typing. In this study, 94% of isolates were of one of two types which were distributed across all three provinces and which differed by only one band. These results are in contrast to those obtained for M. bovis isolates by Marois in France in 1997, who identified 22 types for 104 isolates (15), and by Feizebadi et al. in Australia in 1996, who reported 27 types for 69 isolates (9). Results similar to those observed in France and Australia have been reported for isolates from Canada, with 7 types for 28 isolates, from Ireland, with 10 types for 13 isolates, and from Iran, with 5 types for 6 isolates (9).
RFLP IS6110-RHS typing resulted in poor discrimination between the isolates tested. IS6110 typing is generally considered a poor marker of diversity for M. bovis strains of bovine origin (4) and, as in many other studies, the majority of strains were characterized by the presence of only one PvuII restriction band, of 1.9 kb. Thus, it is unlikely that we would have obtained a significant increase in strain diversity by typing all of the strains by RFLP with the IS6110-RHS probe. Recently, it has been reported that greater discrimination of M. bovis isolates can be achieved by RFLP typing with the polymorphic GC-rich sequence (PGRS) probe. This method can result in discrimination of isolates comparable to (18) or higher than (1, 4, 23) that of spoligotyping.
At the level of this study, spoligotyping seems promising for high-throughput molecular typing of M. bovis isolates in Cameroon. By using PFGE and RFLP with IS6110-RHS in combination with spoligotyping, we have already been able, for the 13 isolates belonging to spoligotype C1 and tested by the other two techniques, to further subdivide this group into four subgroups (PC1-RC1, PC1-RC2, PC1-RC3, and PC2-RC1). This result confirms that the use of more than one technique can be very helpful for further analysis of isolates. The techniques used in this study could be combined with PGRS-based RFLP for epidemiological studies, where further discrimination is required. Together, these techniques could be very useful on a regional scale in order to understand the consequences of transhumance across the provinces of Cameroon and across national frontiers, since the absence of systems for individual cattle identification precludes such investigations by more classical epidemiological methods. An improved understanding of the mechanisms and extent of strain dissemination would be very helpful for regional control programs.
ACKNOWLEDGMENTS
The work carried out at AFSSA Alfort was funded by the Ministry of Agriculture, Fisheries and Food, Paris, France. The work performed at VLA Weybridge was funded by the Ministry of Agriculture, Fisheries and Food (grant SE0129), London, Great Britain.
REFERENCES
- 1.Aranaz A, Liebana E, Mateos A, Dominguez L, Cousins D. Restriction fragment length polymorphism and spacer oligonucleotide typing: a comparative analysis of fingerprinting strategies for Mycobacterium bovis. Vet Microbiol. 1998;61:311–324. doi: 10.1016/s0378-1135(98)00192-8. [DOI] [PubMed] [Google Scholar]
- 2.Aranaz A, Liebana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzales O, Rodrigues-Ferri F, Bunshoten A, van Embden J, Cousins D. Spoligotyping of Mycobacterium bovis strains from cattle and other animals: a tool for epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc R. Tuberculose et brucellose sur le bétail de la subdivision de Meïganga. MINEPIA report. Yaoundé, Cameroon: Cameroon Ministry of Agriculture; 1957. [Google Scholar]
- 4.Cousins D, Williams S, Liebana E, Aranaz A, Bunschoten A, van Embden J, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daborn C J, Kazwala R R, Kambarage D M. Bovine tuberculosis research programme in Tanzania: interim results. In: Berrada J, Bouchriti N, Bouslikhane M, editors. Animal tuberculosis in Africa and Middle East. Rabat, Morocco: Actes Editions; 1997. pp. 151–198. [Google Scholar]
- 6.David H, Lévy-Frébault V, Thorel M F. Méthodes de laboratoire pour mycobactériologie clinique. Paris, France: Commission des Laboratoires de Référence et d'Expertise de l' Institut Pasteur; 1989. [Google Scholar]
- 7.Doufissa A. l'Élevage bovin dans le M'béré. MINEPIA report. Yaoundé, Cameroon: Cameroon Ministry of Agriculture; 1993. [Google Scholar]
- 8.Fang Z, Morrison N, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;8:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feizebadi M M, Robertson I D, Cousins D V, Hampson D J. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:1136–1142. doi: 10.1128/jcm.34.5.1136-1142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermans PWM, van Soolingen D, Dale J W, Schuitema A R J, McAdam R A, Catty D, van Emden J D A. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunshoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differenciation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazwala R R, Sinclair K, Challans J, Kambarage D M, Sharp J M, van Embden J D A, Daborn C J, Nyange J. Zoonotic importance of Mycobacterium bovis complex organisms in Tanzania: a molecular biology approach. In: Berrada J, Bouchriti N, Bouslikhane M, editors. Animal tuberculosis in Africa and Middle East. Rabat, Morocco: Actes Editions; 1997. pp. 199–204. [Google Scholar]
- 13.Letenneur L. Etude du secteur élevage au Cameroun. Rapport provisoire. CIRAD/EMVT report. Montpellier, France: Centre de Coopération Internationale en Recherche Agronomique pour le Développement; 1995. [Google Scholar]
- 14.Lévy-Frébault V V, Thorel M F, Varnerot A, Gicquel B. DNA polymorphism in Mycobacterium paratuberculosis, “wood pigeon mycobacteria,” and related mycobacteria analyzed by field inversion gel electrophoresis. J Clin Microbiol. 1989;27:2823–2826. doi: 10.1128/jcm.27.12.2823-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marois C. Etude de la biodiversité des souches de Mycobacterium bovis par électrophorèse en champ pulsé. Rapport de stage de maîtrise MST GSE 18. Paris, France: Université Paris XII; 1997. [Google Scholar]
- 16.Martrenchar A, Njanpop B M, Yaya A, Njoya A, Tulasne J J. Problems associated with tuberculosis and brucellosis skin-test methods in northern Cameroon. Prevent Vet Med. 1993;15:221–229. [Google Scholar]
- 17.Merlin P, Tsangueu P. Incidence de la tuberculose bovine dans le nord-ouest du Cameroun. Rev Sci Technol Ser Sci Zootechnol. 1985;1:89–94. [Google Scholar]
- 18.Roring S, Brittain D, Bunschoten A E, Hughes M S, Skuce R A, van Embden J D, Neill S D. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet Microbiol. 1998;61:111–120. doi: 10.1016/s0378-1135(98)00178-3. [DOI] [PubMed] [Google Scholar]
- 19.Tanya V N. Screening for bovine tuberculosis at Wakwa. Rev Sci Technol Ser Sci Zootechnol. 1985;1:65–68. [Google Scholar]
- 20.van Emden JDA, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 3rd ed. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. pp. 2.4.1–2.4.5. [Google Scholar]
- 23.Zumarraga M J, Martin C, Samper S, Alito A, Latini O, Bigi F, Roxo E, Cicuta M E, Errico F, Ramos M C, Cataldi A, van Soolingen D, Romano M I. Usefulness of spoligotyping in molecular epidemiology of Mycobacterium bovis-related infections in South America. J Clin Microbiol. 1999;37:296–303. doi: 10.1128/jcm.37.2.296-303.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]