Abstract
Background
The introduction of primary HPV screening has doubled the number of colposcopy referrals because of the direct referral of HPV-positive women with a borderline or mild dyskaryosis (BMD) cytology (ASC-US/LSIL) triage test. Further risk-stratification is warranted to improve the efficiency of HPV-based screening.
Methods
This study evaluated the discriminative power of FAM19A4/miR124-2 methylation, HPV16/18 genotyping and HPV16/18/31/33/45 genotyping in HPV-positive women with BMD (n = 294) in two Dutch screening trials. Absolute CIN3+ risks and colposcopy referrals within one screening round were calculated.
Results
Methylation analysis discriminated well, yielding a CIN3+ risk of 33.1% after a positive result and a CIN3+ risk of 9.8% after a negative result. HPV16/18 and HPV16/18/31/33/45 genotyping resulted in a 27.6% and 24.6% CIN3+ risk after a positive result, and a 13.2% and 9.1% CIN3+ risk after a negative result. Colposcopy referral percentages were 41.2%, 43.2%, and 66.3% for FAM19A4/miR124-2 methylation, HPV16/18 and HPV16/18/31/33/45 genotyping, respectively. The CIN3+ risk after a negative result could be lowered to 2.8% by combining methylation and extended genotyping, at the expense of a higher referral percentage of 75.5%.
Conclusion
The use of FAM19A4/miR124-2 methylation and/or HPV genotyping in HPV-positive women with BMD can lead to a substantial reduction in the number of direct colposcopy referrals.
Subject terms: DNA methylation, Diagnostic markers, Cervical cancer, Molecular medicine
Background
Human papillomavirus (HPV)-based cervical screening has an increased sensitivity for the detection of high-grade cervical intraepithelial neoplasia (CIN) and cervical cancer compared with cytology-based screening, but has a suboptimal specificity [1, 2] and triage of HPV-positive women is essential to avoid unnecessary colposcopy referrals. However, first evaluations of primary HPV screening in Europe and Australia with triage by cytology and/or HPV16/18 genotyping showed a substantial increase in direct colposcopy referrals compared with cytology-based screening [3, 4].
The Netherlands was the first country that implemented primary HPV screening with cytology triage in the national cervical screening programme in 2017 [5]. HPV-positive women with borderline or mild dyskaryosis (BMD) cytology (comparable with atypical squamous cells of undetermined significance (ASC-US) and low-grade squamous intraepithelial neoplasia (LSIL) in the Bethesda classification) [6] or worse are directly referred for colposcopy. HPV-positive women with normal cytology are re-tested at 6 months and referred for colposcopy in case of BMD cytology or worse [7]. The replacement of the cytological screening programme by primary HPV-based screening with cytology triage has resulted in more clinically relevant findings [1, 2] at the cost of an approximately two-fold increase of colposcopy referrals and ≤CIN1 diagnosis [8, 9]. The increase in colposcopy referrals is mainly caused by the direct referral of women with BMD cytology [9], who often do not harbour high-grade CIN (CIN2/3) [10–12]. Further triage of HPV-positive women with BMD cytology with an objective molecular test that has a higher specificity for high-grade CIN might reduce the number of women referred for colposcopy, while maintaining clinical sensitivity.
HPV16/18 genotyping (partial genotyping) or HPV16/18/31/33/45 genotyping (extended genotyping) has been considered for triaging HPV-positive women with BMD cytology, because CIN3+ risks vary between individual high-risk HPV genotypes [13–15]. An alternative triage strategy is DNA methylation analysis of host-cell genes. Hypermethylation of promotor regions of certain tumour suppressor genes is a crucial step in cervical carcinogenesis [16, 17]. Methylation levels have shown to increase with duration of the HPV infection and with increasing CIN grade, reaching very high levels in cervical cancer [18–21]. FAM19A4/miR124-2 methylation analysis has shown a high sensitivity for cervical cancer and advanced high-grade CIN (i.e., CIN lesions associated with a HPV infection of at least 5 years) in HPV-positive women [22, 23].
In this study, we evaluate the performance of FAM19A4/miR124-2 methylation, HPV16/18 genotyping, HPV16/18/31/33/45 genotyping and combinations thereof in HPV-positive women with BMD cytology in two large Dutch population-based screening cohorts.
Methods
Study population
This study is a post hoc analysis within the VUSA-Screen trial and POBASCAM trial. HPV-positive women with borderline or mild dyskaryosis (BMD) (VUSA-Screen n = 167 and POBASCAM n = 192) were selected. Fifty-two samples were excluded due to insufficient leftover material for methylation analysis (VUSA-Screen n = 41 and POBASCAM n = 11).
VUSA-Screen trial
The VUSA-Screen trial is a population-based cervical screening study within the Dutch screening programme with enrolment between October 2003 and August 2005. The VUSA-Screen trial was approved by the Ministry of Public Health (2002/02-WBO; ISBN-10: 90-5549-452-6) and registered in the trial register (NTR215, ISRCTN64621295). The study was designed to evaluate the effectiveness of combined HPV and cytology testing in the Dutch national screening programme. A detailed description of the VUSA-Screen trial has been published before [24]. Cervical scrapes were classified according to the CISOE-A classification, the standard classification system for cytology in the Netherlands, which can be translated into the Bethesda classification system [6]. HPV testing was performed by the HC2 high-risk HPV DNA test (QIAGEN, Hilden, Germany) on cervical scrapes [25]. HPV-positive women with BMD were directly referred for colposcopy. In this post hoc analysis, only HPV-positive women with BMD cytology were included.
POBASCAM trial
The POBASCAM trial is a population-based cervical screening study within the Dutch screening programme with enrolment between January 1999 and September 2002. The POBASCAM trial was approved by the Medical Ethics Committee of the VU University Medical Center (no 96/103A) and by the Ministry of Public Health (VWS no 328 650) and registered in the trial register (NTR218; ISRCTN20781131). The study was designed to evaluate the effectiveness for CIN2/3 detection of combined HPV and cytology testing in the intervention group compared with sole cytology testing and blinded HPV testing in the control group. A detailed description of the POBASCAM trial has been published before [26, 27]. Cervical scrapes were classified according to the CISOE-A classification. HPV testing was performed on cervical scrapes using the GP5+/6+ PCR-EIA [28]. Women with BMD in the control group were referred for colposcopy if repeat cytology at 6 or 18 months showed BMD or worse. In this post hoc analysis, only HPV-positive women with BMD cytology from the control group were included.
In both studies, histology results were classified as no dysplasia, CIN grade 1, 2, 3, or cervical cancer. Adenocarcinoma in situ was classified as CIN3. Histopathologic follow-up data were collected through the nationwide network and registry of histopathology and cytopathology (PALGA) [29].
HPV genotyping and FAM19A4/miR124-2 methylation analysis
HC2-positive samples from the VUSA-Screen trial were tested with GP5+/6+ PCR-EIA. All EIA-positive samples from both trials were typed by reverse line blot (RLB) assay for identification of 14 high-risk HPV types (i.e., 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) [28]. Samples that were either GP5+/6+ PCR-EIA positive and/or HC2 positive, but negative on RLB for HPV type 16, 18, 31, 33 and 45 were considered negative for HPV16/18/31/33/45.
FAM19A4/miR124-2 methylation analysis was performed as described previously [30, 31], blinded for cytology and histology outcomes, by quantitative methylation specific PCR (qMSP) on bisulphite converted DNA from cervical scrapes using the QIAsure Methylation Test® (Qiagen, Hilden, Germany) (VUSA-Screen cohort) or its prototype version (POBASCAM cohort). This prototype version was identical regarding to the primers and probes design, PCR conditions and PCR system. All samples had a ß-actin cycle threshold (Ct) value below 26.4 to assure successful bisulphite conversion and sample quality. Thirteen samples had an invalid FAM19A4/miR124-2 methylation result (VUSA-Screen n = 3 and POBASCAM n = 10) and were excluded from further analysis. Methylation status was labelled positive if the QIAsure Methylation Test® result exceeded the preset ΔΔCt value threshold for methylation positivity according to the manufacturer’s instructions.
Data and statistical analysis
Data of the VUSA-Screen trial and POBASCAM trial were pooled and absolute CIN3+ risks were calculated. All CIN3+ detected within one screening round (i.e., up to 4 years) were included. Separate estimates were retrieved for single and combined triage strategies using FAM19A4/miR124-2 methylation, HPV16/18 genotyping and HPV16/18/31/33/45 genotyping. The performance of five triage strategies was evaluated: (I) FAM19A4/miR124-2 methylation, (II) HPV16/18 genotyping, (III) HPV16/18/31/33/45 genotyping, (IV) HPV16/18 genotyping combined with FAM19A4/miR124-2 methylation and (V) HPV16/18/31/33/45 genotyping combined with FAM19A4/miR124-2 methylation. Strategy (I) was labelled positive if the methylation result was positive. Strategy (II) was labelled positive if HPV 16 and/or 18 was present. Strategy (III) was labelled positive if HPV 16, 18, 31, 33, and/or 45 was present. Strategy (IV) was labelled positive if HPV 16 and/or 18 was present or if the methylation result was positive. Strategy (V) was labelled positive if HPV 16, 18, 31, 33, and/or 45 was present or if the methylation result was positive. Sensitivity and specificity were estimated with Wald 95% confidence intervals (95% CI). Direct referral percentage was calculated as the percentage of positives from each screening strategy and the number of referrals needed to detect one CIN3+ was calculated by dividing the number of screen positives by the number of true positives. We constructed 95% confidence intervals (95% CI) for the risk differences in STATA (version 14.1, StataCorp, College Station, Texas, USA). If the 95% CI did not contain the value 0, the difference was considered significant. All other statistical analysis were performed with SPSS Statistics (version 26, IBM Corp, Armonk, NY, USA).
Results
The final study population consisted of 294 HPV-positive women with BMD and valid results for HPV genotyping and FAM19A4/miR124-2 methylation analysis (VUSA-Screen n = 123, POBASCAM n = 171), in whom 56 CIN3 and 1 cervical carcinoma were detected. The median age of women included was 35.0 years (range: 29–60 years) and median time to CIN3+ detection was 0.7 years (IQR: 0.2–1.3 years). In total, 121/294 women (41.2%) tested methylation positive, 127/294 women (43.2%) tested HPV16/18 positive and 195/294 women (66.3%) tested HPV16/18/31/33/45 positive.
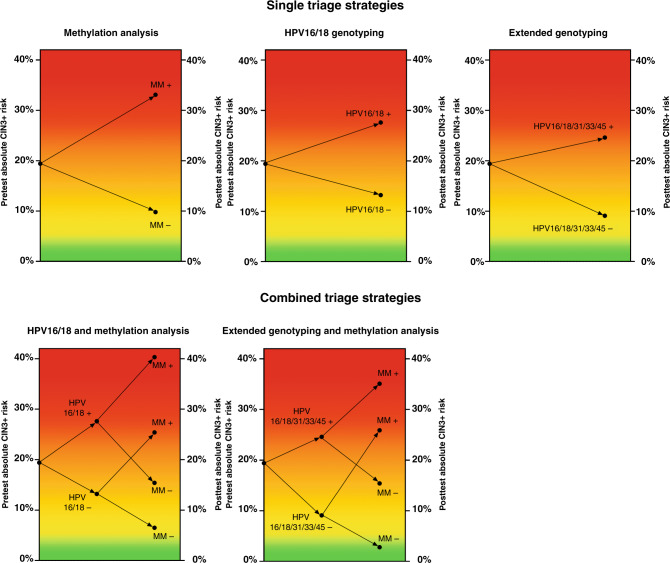
Table 1 shows the absolute CIN3+ risks after triaging HPV-positive women with BMD cytology by FAM19A4/miR-124 methylation, HPV16/18 genotyping, HPV16/18/31/33/45 genotyping or combinations thereof. Fig. 1 visualises the pre- and post-test CIN3+ risks after application of the single or combined triage strategies. A positive methylation test resulted in the highest absolute CIN3+ risk (33.1%; 95% CI: 24.7–41.4%) and methylation analysis showed the largest risk difference between test-positives and test-negatives (23.2%; 95% CI 13.7–32.7%). The risk difference between HPV16/18-positive and HPV16/18-negative women was 14.4% (95% CI: 5.1–23.7%) and the risk difference between HPV16/18/31/33/45-positive and HPV16/18/31/33/45-negative women was 15.5% (95% CI: 7.2–23.8%). Women who tested negative for FAM19A4/miR124-2 methylation, HPV16/18 or HPV16/18/31/33/45 had a CIN3+ risk of >9%. Combinations of HPV genotyping and methylation lowered the CIN3+ risk among test-negatives with a CIN3+ risk of 6.5% (95% CI: 1.8–11.1%) among women who were both HPV16/18-negative and FAM19A4/miR124-2 methylation-negative and a CIN3+ risk of 2.8% (95% CI: 0–6.6%) among women who were both HPV16/18/31/33/45-negative and FAM19A4/miR124-2 methylation-negative. Supplementary Table 1 reports the absolute CIN3+ risks within the VUSA-Screen trial and POBASCAM trial separately.
Table 1.
Absolute CIN3+ risks for single and combined triage tests.
| Test result | N | CIN3+ (n) | Absolute CIN3+ risk | 95% CI |
|---|---|---|---|---|
| Single triage strategies | ||||
| Methylation + | 121 | 40 | 33.1% | (24.7–41.4%) |
| Methylation − | 173 | 17 | 9.8% | (5.4–14.3%) |
| HPV16/18 + | 127 | 35 | 27.6% | (19.8–35.3%) |
| HPV16/18 − | 167 | 22 | 13.2% | (8.0–18.3%) |
| HPV16/18/31/33/45 + | 195 | 48 | 24.6% | (18.6–30.7%) |
| HPV16/18/31/33/45 − | 99 | 9 | 9.1% | (3.4–14.8%) |
| Combined triage strategies | ||||
| HPV16/18 + and methylation + | 62 | 25 | 40.3% | (28.1–52.5%) |
| HPV16/18 + and methylation − | 65 | 10 | 15.4% | (6.6–24.2%) |
| HPV16/18 − and methylation + | 59 | 15 | 25.4% | (14.3–36.5%) |
| HPV16/18 − and methylation − | 108 | 7 | 6.5% | (1.8–11.1%) |
| HPV16/18/31/33/45 + and methylation + | 94 | 33 | 35.1% | (25.5–44.8%) |
| HPV16/18/31/33/45 + and methylation − | 101 | 15 | 14.9% | (7.9–21.8%) |
| HPV16/18/31/33/45 − and methylation + | 27 | 7 | 25.9% | (9.4–42.5%) |
| HPV16/18/31/33/45 − and methylation − | 72 | 2 | 2.8% | (0–6.6%) |
N, group total; n, number of CIN3+ detected; CIN3+, cervical intraepithelial neoplasia grade 3 or worse; 95% CI, 95% confidence interval; +, positive; −, negative.
Fig. 1. Pre- and post-test CIN3+ risk plots for single and combined triage strategies.
Colour legend: green, low CIN3+ risk; orange, intermediate CIN3+ risk; red, high CIN3+ risk; MM+, methylation marker-positive; MM−, methylation marker-negative.
Table 2 shows the clinical sensitivity, specificity, colposcopy referral percentages and referrals needed to detect one CIN3+ of five triage strategies. Methylation analysis (strategy I) showed a sensitivity of 70.2% (95% CI: 58.3–82.1%) at a specificity of 65.8% (95% CI: 59.8–71.9%). This strategy resulted in the lowest referral percentage of 41.2% and the lowest number of referrals needed to detect one CIN3. Combined triage with HPV16/18/31/33/45 genotyping and methylation analysis (strategy V) leads to an increased sensitivity of 96.5% (95% CI: 91.7–100%), but specificity decreased to 29.5% (95% CI: 23.7–35.3%) and referral percentage increased to 75.5%.
Table 2.
Sensitivity, specificity, colposcopy referral percentages and referrals needed to detect one CIN3+ of five triage strategies for HPV-positive women with BMD cytology.
| Triage strategy | Sensitivity (95% CI) | Specificity (95% CI) | Colposcopy referral (%) | Referrals needed to detect one CIN3+ | |||
|---|---|---|---|---|---|---|---|
| I | Methylation analysis | 70.2% | (58.3–82.1%) | 65.8% | (59.8–71.9%) | 41.2% | 3.0 |
| II | HPV16/18 genotyping | 61.4% | (48.8–74.0%) | 61.2% | (55.0–67.4%) | 43.2% | 3.6 |
| III | HPV16/18/31/33/45 genotyping | 84.2% | (74.7–93.7%) | 38.0% | (31.8–44.2%) | 66.3% | 4.1 |
| IV | HPV16/18 genotyping and/or methylation analysis | 87.7% | (79.2–96.2%) | 42.6% | (36.3–48.9%) | 63.3% | 3.7 |
| V | HPV16/18/31/33/45 genotyping and/or methylation analysis | 96.5% | (91.7–100%) | 29.5% | (23.7–35.3%) | 75.5% | 4.0 |
HPV, human papillomavirus; 95% CI, 95% confidence interval.
Discussion
We evaluated FAM19A4/miR124-2 methylation, HPV16/18 genotyping (partial genotyping) and HPV16/18/31/33/45 genotyping (extended genotyping) for additional risk-stratification of HPV-positive women with BMD cytology to reduce direct colposcopy referral rates, while maintaining high sensitivity for CIN3+ detection. The choice of additional triage tests depends on the CIN3+ risk, the number of repeat tests and the number of colposcopy referrals that are deemed acceptable.
Of the single triage strategies, FAM19A4/miR124-2 methylation resulted in the highest CIN3+ risk of 33.1% and the largest risk-difference between test-positives and test-negatives (23.2%). Recently Bonde et al. also showed in a large multicentre cohort that FAM19A4/miR124-2 methylation detected the very large majority of CIN3 in HPV-positive women with ASC-US/LSIL [32]. In our study, triage with FAM19A4/miR124-2 methylation led to a referral percentage of 41.2% and a sensitivity of 70.2%. Still, the CIN3+ risk after a negative test for any of the single strategies was >9%. Combined strategies lowered the CIN3+ risk among test-negatives substantially with the lowest CIN3+ risk of 2.8% among women who were both HPV16/18/31/33/45-negative and FAM19A4/miR124-2 methylation-negative. Despite a high referral percentage of 75.5%, a benefit of this combined strategy is that the CIN3+ risk among triage-negative women seems sufficiently low to recommend return to the regular cervical screening programme.
FAM19A4/miR124-2 methylation has shown a very high sensitivity for cervical cancer [23] and advanced CIN2/3 lesions (i.e., CIN lesions with a preceding HPV infection of at least 5 years) [22]. Hence, it is assumed that methylation analysis identifies CIN2/3 lesions at highest risk of progression to cervical cancer. This is supported by a recent Finnish study which showed that methylation analysis could serve as a prognostic biomarker to differentiate between regressive and progressive CIN2 lesions [33]. Furthermore, the results of a prospective cohort of women with CIN2/3 with 2 years of follow-up showed that a negative FAM19A4/miR124-2 methylation test is associated with high spontaneous regression [34] (Kremer, Dick et al., manuscript in preparation). FAM19A4/miR124-2 methylation has also prognostic value for development of cervical cancer in the long term. De Strooper et al. found a significantly lower 14-year cervical cancer risk among HPV-positive, FAM19A4/miR124-2 methylation-negative women compared with HPV-positive, cytology-negative women. Collectively, these data indicate that FAM19A4/miR124-2 methylation-negative CIN2/3 have high regression rates and low cervical cancer progression risks. As a consequence, methylation analysis may be considered when deciding whether women can be returned to routine screening.
In the new HPV-based screening programme in the Netherlands, the proportion of HPV-positive women who were directly referred for colposcopy because of BMD cytology was 70.3% in year 2019 [35]. The cumulative CIN3+ risk of these women after 2.5 years was 6% [9] and considerably lower than the cumulative CIN3+ risk of 19.4% in our studies after four years of follow-up. This decrease in CIN3+ risk may be related to changes in HPV and cytology test accuracy. A change in the performance of the HPV test is not improbable because the proportion of HPV-positive women was nearly twice as high in the national screening programme as compared with the cohort studies. A change in the performance of cytology can neither be ruled out because cytology in the national programme was conducted with knowledge of HPV status, whereas cytology in the cohort studies was conducted without knowledge of HPV status. Despite these potential screening test generalisability issues, we project that the impact of FAM19A4/miR124-2 methylation on the number of referrals and detection of CIN3+ will be considerable. Surely, the high colposcopy referral rates and low CIN3+ risk in the national screening programme makes the need for additional triaging of HPV-positive women with BMD even larger than anticipated based on our studies. For the implementation of methylation analysis, applicability and reproducibility of the test among different sample types and DNA extraction methods is essential. Floore et al. evaluated the intra- and inter-laboratory agreement of the FAM19A4/miR124-2 methylation test with the use of several cervical scrape collection media and several DNA extraction methods and showed a good to excellent intra- and inter-laboratory agreement on the assay and the full workflow [36].
A strength of our study is that we used data of two large population-based HPV DNA screening trials within the Dutch screening programme. As shown in Supplementary Table 1, data of both studies were consistent with comparable CIN3+ risks and therefore results were pooled. Histology endpoints were retrieved from the nationwide network and registry of histo- and cytopathology in the Netherlands (PALGA) [29], which covers all pathology labs in the Netherlands. A limitation is that gynaecological reports were not collected which means that we do not know how many women complied with colposcopy and could potentially lead to an underestimation of CIN3+ risks.
To conclude, the implementation of primary HPV screening with cytology triage in the Netherlands has led to a two-fold increase in direct colposcopy referrals and ≤CIN1 diagnosis, mainly because of the direct referral of women with BMD. Additional risk-stratification of HPV-positive women with BMD by FAM19A4/miR124-2 methylation could reduce direct colposcopy referral rate with 60%, while retaining high CIN3+ sensitivity. The combination of FAM19A4/miR124-2 methylation with HPV16/18/31/33/45 genotyping would reduce colposcopy referrals with only 25%, but the low residual CIN3+ risk would obviate the need of short-term follow-up testing.
Supplementary information
Acknowledgements
We thank Lise De Strooper for the data collection.
Author contributions
Study design: CJLMM and JB. Data management: SD and FJV. Statistical analysis: SD, FJV and BLW. Data interpretation: SD, FJV, CJLMM, JB and DAMH. Writing the first draft of the manuscript: SD, FJV, CLJMM, DAMH and JB. All authors were involved in writing the paper and had final approval of the submitted and published version.
Funding information
This project was partially supported by the EU Horizon 2020 program (project ID 666800), the CoheaHr research consortium (EC FP7 Health 2013 Innovation 1 CoheaHr), RISCC Network (grant number 847845) and the Dutch Cancer Society (grant number KWF VU 2014-7238).
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The VUSA-Screen trial was approved by the Ministry of Public Health (2002/02-WBO; ISBN-10: 90-5549-452-6) and registered in the trial register (NTR215, ISRCTN64621295). The POBASCAM trial was approved by the Medical Ethics Committee of the VU University Medical Center (no 96/103A) and by the Ministry of Public Health (VWS no 328 650) and registered in the trial register (NTR218; ISRCTN20781131). All participants provided written informed consent. The work in this study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Competing interests
CJLMM and DAMH are minority stockholders of Self-screen BV, a spin-off company of VUmc, which develops, manufactures and licences the high-risk HPV assay and methylation marker assays for cervical cancer screening and holds patents on these tests. CJLMM is part-time director of Self-screen BV. He has been on the speakers bureau and served occasionally on the scientific advisory board (expert meeting) of GSK, QIAGEN, SPMSD/Merck. He has been co-investigator on a SPMSD sponsored trial, of which his institute received research funding. CJLMM has a very small number of shares of MDxHealth and previously had shares of QIAGEN. DAMH has been on the speaker’s bureau of QIAGEN and serves occasionally on the scientific advisory boards of Pfizer and Bristol-Myers Squibb. SD, FJV, BLW and JB have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Stèfanie Dick, Frederique J. Vink
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01614-4.
References
- 1.Arbyn M, Ronco G, Anttila A, Meijer CJ, Poljak M, Ogilvie G, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30:F88–99. doi: 10.1016/j.vaccine.2012.06.095. [DOI] [PubMed] [Google Scholar]
- 2.Ronco G, Dillner J, Elfstrom KM, Tunesi S, Snijders PJ, Arbyn M, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383:524–32.. doi: 10.1016/S0140-6736(13)62218-7. [DOI] [PubMed] [Google Scholar]
- 3.Maver PJ, Poljak M. Primary HPV-based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clin Microbiol Infect. 2020;26:579–83.. doi: 10.1016/j.cmi.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Machalek DA, Roberts JM, Garland SM, Thurloe J, Richards A, Chambers I, et al. Routine cervical screening by primary HPV testing: early findings in the renewed National Cervical Screening Program. Med J Aust. 2019;211:113–9. doi: 10.5694/mja2.50223. [DOI] [PubMed] [Google Scholar]
- 5.Polman NJ, Snijders PJF, Kenter GG, Berkhof J, Meijer C. HPV-based cervical screening: Rationale, expectations and future perspectives of the new Dutch screening programme. Prev Med. 2019;119:108–17.. doi: 10.1016/j.ypmed.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Bulk S, Van Kemenade FJ, Rozendaal L, Meijer CJ. The Dutch CISOE-A framework for cytology reporting increases efficacy of screening upon standardisation since 1996. J Clin Pathol. 2004;57:388–93.. doi: 10.1136/jcp.2003.011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Veen N Framework for the Execution of Cervical Cancer Population Screening 2017 Available from: https://www.rivm.nl/documenten/framework-for-execution-of-cervical-cancer-population-screening.
- 8.Aitken CA, van Agt HME, Siebers AG, van Kemenade FJ, Niesters HGM, Melchers WJG, et al. Introduction of primary screening using high-risk HPV DNA detection in the Dutch cervical cancer screening programme: a population-based cohort study. BMC Med. 2019;17:228. doi: 10.1186/s12916-019-1460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loopik DL, Koenjer LM, Siebers AG, Melchers WJG, Bekkers RLM. Benefit and burden in the Dutch cytology-based vs high-risk human papillomavirus-based cervical cancer screening program. Am J Obstet Gynecol. 2021;224:200 e1–e9. doi: 10.1016/j.ajog.2020.08.026. [DOI] [PubMed] [Google Scholar]
- 10.Kocken M, Berkhof J, van Kemenade FJ, Louwers JA, Zaal A, Nobbenhuis MA, et al. Long-term CIN3+ risk in women with abnormal cytology; role of hrHPV testing. Br J Cancer. 2012;106:817–25.. doi: 10.1038/bjc.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katki HA, Schiffman M, Castle PE, Fetterman B, Poitras NE, Lorey T, et al. Five-year risks of CIN 2+ and CIN 3+ among women with HPV-positive and HPV-negative LSIL Pap results. J Low Genit Trac Dis. 2013;17:S43–9. doi: 10.1097/LGT.0b013e3182854269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katki HA, Schiffman M, Castle PE, Fetterman B, Poitras NE, Lorey T, et al. Five-year risks of CIN 3+ and cervical cancer among women with HPV testing of ASC-US Pap results. J Low Genit Trac Dis. 2013;17:S36–42. doi: 10.1097/LGT.0b013e3182854253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright TC, Jr, Stoler MH, Parvu V, Yanson K, Cooper C, Andrews J. Risk detection for high-grade cervical disease using Onclarity HPV extended genotyping in women, >/=21years of age, with ASC-US or LSIL cytology. Gynecol Oncol. 2019;154:360–7. doi: 10.1016/j.ygyno.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Gage JC, Schiffman M, Solomon D, Wheeler CM, Gravitt PE, Castle PE, et al. Risk of precancer determined by HPV genotype combinations in women with minor cytologic abnormalities. Cancer Epidemiol Biomark Prev. 2013;22:1095–101.. doi: 10.1158/1055-9965.EPI-12-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiffman M, Vaughan LM, Raine-Bennett TR, Castle PE, Katki HA, Gage JC, et al. A study of HPV typing for the management of HPV-positive ASC-US cervical cytologic results. Gynecol Oncol. 2015;138:573–8. doi: 10.1016/j.ygyno.2015.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405. doi: 10.1038/nrc3728. [DOI] [PubMed] [Google Scholar]
- 17.Durzynska J, Lesniewicz K, Poreba E. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res. 2017;772:36–50. doi: 10.1016/j.mrrev.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Bierkens M, Hesselink AT, Meijer CJ, Heideman DA, Wisman GB, van der Zee AG, et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013;133:1293–9. doi: 10.1002/ijc.28138. [DOI] [PubMed] [Google Scholar]
- 19.De Strooper LM, van Zummeren M, Steenbergen RD, Bleeker MC, Hesselink AT, Wisman GB, et al. CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol. 2014;67:1067–71.. doi: 10.1136/jclinpath-2014-202616. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz M, Eichelkraut K, Schmidt D, Zeiser I, Hilal Z, Tettenborn Z, et al. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer. 2018;18:1197. doi: 10.1186/s12885-018-5125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook DA, Krajden M, Brentnall AR, Gondara L, Chan T, Law JH, et al. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int J Cancer. 2019;144:2587–95.. doi: 10.1002/ijc.31976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Strooper LM, Meijer CJ, Berkhof J, Hesselink AT, Snijders PJ, Steenbergen RD, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Philos) 2014;7:1251–7. doi: 10.1158/1940-6207.CAPR-14-0237. [DOI] [PubMed] [Google Scholar]
- 23.Vink FJ, Meijer C, Clifford GM, Poljak M, Ostrbenk A, Petry KU, et al. FAM19A4/miR124-2 methylation in invasive cervical cancer: a retrospective cross-sectional worldwide study. Int J Cancer. 2020;147:1215–21. [DOI] [PMC free article] [PubMed]
- 24.Rijkaart DC, Berkhof J, van Kemenade FJ, Coupe VM, Rozendaal L, Heideman DA, et al. HPV DNA testing in population-based cervical screening (VUSA-Screen study): results and implications. Br J Cancer. 2012;106:975–81.. doi: 10.1038/bjc.2011.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vince A, Kutela N, Iscic-Bes J, Harni V, Ivanisevic M, Sonicki Z, et al. Clinical utility of molecular detection of human papillomavirus in cervical samples by hybrid capture technology. J Clin Virol. 2002;25:S109–12. doi: 10.1016/S1386-6532(02)00184-1. [DOI] [PubMed] [Google Scholar]
- 26.Bulkmans NW, Rozendaal L, Snijders PJ, Voorhorst FJ, Boeke AJ, Zandwijken GR, et al. POBASCAM, a population-based randomized controlled trial for implementation of high-risk HPV testing in cervical screening: design, methods and baseline data of 44,102 women. Int J Cancer. 2004;110:94–101. doi: 10.1002/ijc.20076. [DOI] [PubMed] [Google Scholar]
- 27.Rijkaart DC, Berkhof J, Rozendaal L, van Kemenade FJ, Bulkmans NW, Heideman DA, et al. Human papillomavirus testing for the detection of high-grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2012;13:78–88. doi: 10.1016/S1470-2045(11)70296-0. [DOI] [PubMed] [Google Scholar]
- 28.van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J Clin Microbiol. 2002;40:779–87.. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casparie M, Tiebosch AT, Burger G, Blauwgeers H, van de Pol A, van Krieken JH, et al. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007;29:19–24. doi: 10.1155/2007/971816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vink FJ, Lissenberg-Witte BI, Meijer C, Berkhof J, van Kemenade FJ, Siebers AG, et al. FAM19A4/miR124-2 methylation analysis as a triage test for HPV-positive women: cross-sectional and longitudinal data from a Dutch screening cohort. Clin Microbiol Infect. 2021;27:125.e1–e6. doi: 10.1016/j.cmi.2020.03.018. [DOI] [PubMed] [Google Scholar]
- 31.De Strooper LMA, Berkhof J, Steenbergen RDM, Lissenberg-Witte BI, Snijders PJF, Meijer C, et al. Cervical cancer risk in HPV-positive women after a negative FAM19A4/mir124-2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow-up. Int J Cancer. 2018;143:1541–8. doi: 10.1002/ijc.31539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonde J, Floore A, Ejegod D, Vink FJ, Hesselink A, van de Ven PM, et al. Methylation markers FAM19A4 and miR124-2 as triage strategy for primary human papillomavirus screen positive women: a large European multicenter study. Int J Cancer. 2021;148:396–405. doi: 10.1002/ijc.33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louvanto K, Aro K, Nedjai B, Butzow R, Jakobsson M, Kalliala I, et al. Methylation in predicting progression of untreated high-grade cervical intraepithelial neoplasia. Clin Infect Dis. 2020;70:2582–90.. doi: 10.1093/cid/ciz677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer WW, Berkhof J, Bleeker MC, Heideman DA, van Trommel NE, van Baal MW, et al. Role of FAM19A4/miR124-2 methylation analysis in predicting regression or non-regression of CIN2/3 lesions: a protocol of an observational longitudinal cohort study. BMJ Open. 2019;9:e029017. doi: 10.1136/bmjopen-2019-029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.RIVM. Monitor Bevolkingsonderzoek Baarmoederhalskanker 2019: Rijksinstituut voor Volksgezondheid en Milieu, Integraal Kankercentrum Nederland; Available from: https://www.rivm.nl/documenten/monitor-bevolkingsonderzoek-baarmoederhalskanker-2019.
- 36.Floore A, Hesselink A, Ostrbenk A, Alcaniz E, Rothe B, Pedersen H, et al. Intra- and inter-laboratory agreement of the FAM19A4/mir124-2 methylation test: Results from an international study. J Clin Lab Anal. 20193;3:e22854. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.