Abstract
Background:
Decipher Biopsy is a commercially-available gene expression classifier used in risk-stratification of newly diagnosed prostate cancer (PCa). Currently, there are no prospective data evaluating its clinical utility. We seek to assess the clinical utility of Decipher Biopsy in localized PCa patients.
Methods:
A multi-institutional study of 855 men who underwent Decipher Biopsy testing between February 2015 – October 2019. All patients were tracked through the prospective Michigan Urological Surgery Improvement Collaborative and linked to the Decipher Genomics Resource Information Database (GRID®; NCT02609269). Patient matching was performed by an independent third-party (ArborMetrix Inc.) using two or more unique identifiers. Cumulative incidence curves for Time to Treatment (TTT) and Time to Failure (TTF) were constructed using Kaplan-Meier estimates. Multivariable Cox proportional hazard models were used to evaluate the independent association of high-risk Decipher scores with the conversion from AS to radical therapy and treatment failure (biochemical failure or receipt of salvage therapy).
Results and limitations:
855 patients underwent Decipher Biopsy testing during the study period. Of the 855 men, 264 proceeded to AS (31%), and 454 (53%) received radical therapy. In men electing AS, after adjusting for NCCN risk group, age, PSA, prostate-volume, body-mass-index, and percent positive cores, a high-risk Decipher score was independently associated with shorter TTT (HR 2.51, 95% CI 1.52–4.13 p <0.001). Similarly, in patients that underwent radical therapy, a high-risk Decipher score was independently associated with TTF (HR 2.98, 95%CI 1.22–7.29, p=0.01) on multivariable analysis. Follow-up time was a limitation.
Conclusion:
In a prospective statewide registry, high-risk Decipher Biopsy score was strongly and independently associated with conversion from AS to definitive treatment and treatment failure. These real-world data support the clinical utility of Decipher Biopsy. An ongoing phase 3 randomized trial (NCT04396808) will provide level 1 evidence of the clinical impact of Decipher biopsy testing.
Keywords: Prostate cancer, gene expression testing, Decipher, biopsy, active surveillance, biomarkers
Introduction:
Long-term data support the safety of active surveillance (AS) for men with low-risk, and well-selected favorable intermediate-risk prostate cancer.1–3 However, some men on AS experience disease progression and metastasis, and there remains variability in the use of AS.4 Much of this variability is potentially related to concerns of disease progression and lack of confidence in clinical and pathologic tools given the documented clinical heterogeneity within traditional National Comprehensive Cancer Network (NCCN) or Cancer of the Prostate Risk Assessment (CAPRA) risk groups.5,6 Strategies to improve risk stratification of these patients include the use of clinical nomograms, multiparametric MRI, and tissue-based gene expression classifiers.7,8
There are several commercially-available gene expression classifiers intended to provide additional prognostic information to men with newly-diagnosed prostate cancer. All of these tests measure expression of a specific set of genes using tissue from prostate needle cores.9–11 While each of these tests has robust analytical validity, the majority of the published data are retrospective studies using archival specimens. Additionally, in the case of Decipher, while there is a substantial body of data using prostatectomy specimens to predict metastasis, there are less prospective outcomes data evaluating the performance of Decipher Biopsy in men with newly diagnosed prostate cancer.12–16 Given the increasing use of genomic classifiers in men considering AS, there is a critical need to understand both the performance of a given assay and its potential impact on clinical care.17In this study, we performed a retrospective analysis using a large prospective statewide collaborative registry to better understand the real-world clinical utility of Decipher Biopsy in men initially managed with either AS or who receive definitive therapy with surgery or radiotherapy.
Methods:
Study Cohort
Michigan Urological Surgery Improvement Collaborative (MUSIC) is a statewide collaborative with prospective data collection funded by Blue Cross Blue Shield of Michigan. It is composed of 44 practices and includes over 90% of practicing Urologists in the state of Michigan.18 All practices have either an exemption or approval from the local internal review board to participate in the collaborative. Each MUSIC practice employs data abstractors with standardized training who prospectively collect patient information, including demographics, clinical variables, treatment, and additional variables.
Between February 2015 – October 2019, 855 patients from 25 different sites in the MUSIC collaborative with clinically-localized prostate cancer underwent testing with the Decipher Prostate Biopsy assay (Decipher Biosciences) during their routine clinical care. Decipher data was obtained through the Decipher prospective Genomics Resource Information Database (GRID®; NCT02609269), a prospective registry, which was cross-referenced with the MUSIC registry based on two or more identifiers (medical record number, patient name, date of birth, treatment date, and biopsy date) using an independent third-party (Arbormetrix Inc.). Patients were eligible for inclusion in the study if they underwent Decipher Biopsy testing for localized prostate cancer and could be matched with the Decipher GRID registry. Importantly, the biopsy used for Decipher Biopsy testing is termed the index biopsy, while the biopsy resulting in the diagnosis of prostate cancer is referred to as the diagnostic biopsy. For 750/855 (88%) of patients these were the same biopsy.
Patients included in the time on AS analysis were required to have met two criteria, as per established MUSIC protocols: 1) clinicians have explicitly stated in the medical record that AS is their primary management strategy, and 2) patients must not have received any definitive treatment within six months of diagnosis.19 Patients managed by watchful waiting (as specified by their treating physician) were excluded.
Decipher Biopsy
Decipher Biopsy testing was performed as previously described.20 Briefly, Decipher testing utilizes a microarray platform to perform whole-transcriptome RNA expression analysis. Scores are derived from the expression of 22 genes. Decipher scores are a continuous variable reported on a scale of 0 to 1. These scores are then stratified to three risk-categories: low (<0.45), intermediate (≥ 0.45 – < 0.6) and high (≥ 0.6). In this study, Decipher scores were analyzed as an ordinal variable based on pre-defined risk categories (low, intermediate and high) and also as a continuous variable. Use of both methods allows evaluation of Decipher scores associations across risk categories, and for every 0.1 increase in score.
Endpoints
The primary endpoints for analysis were determined a priori. For men on AS, the primary endpoint was time to treatment (TTT), which was defined as time from Decipher Biopsy testing until radical therapy (i.e. radical prostatectomy or radiotherapy). For men that underwent radical therapy, the primary endpoint was time to treatment failure (TTF), defined from date of treatment completion until biochemical failure or receipt of salvage therapy. Treatment failure for patients undergoing prostatectomy was defined as PSA ≥0.2 ng/mL. For patients who received radiotherapy, treatment failure was defined in accordance with the Phoenix definition of biochemical failure (PSA rise of ≥ 2 ng/mL above the nadir).21 Salvage therapy included any form of oncologic therapy used for a patient with a detectable PSA after radical therapy, including salvage hormone therapy, radiotherapy, or other ablative therapies (e.g. cryotherapy) for prostate cancer.
Patients who received initial cryotherapy or androgen deprivation therapy alone were classified as “other” in our treatment stratification. Patients under this treatment class were included in our analysis to compare Decipher Biopsy scores to clinicopathologic variables; however, these patients were excluded from our time to event analyses. Patients who underwent definitive treatment without PSA follow-up data were also excluded from primary endpoint analyses.
Sensitivity Analysis
TTT for patients initially managed with AS was also analyzed where time zero was the date of initial diagnostic biopsy rather than at time of initial Decipher biopsy (index biopsy).
Statistical Analysis
Patient characteristics were compared between those with low/intermediate Decipher scores and those with high Decipher scores based on previous analyses.22 Median Decipher scores were compared across NCCN-risk groups, Cancer of the Prostate Risk Assessment (CAPRA) risk groups, and Grade Groups using Wilcoxon tests, and violin plots were constructed. One minus Kaplan-Meier method was utilized for time-to-event analysis to generate cumulative incidence curves for duration on AS (TTT) and time to treatment failure (TTF). Multivariable Cox proportional hazards regression was performed to evaluate the association of Decipher scores with time on AS and time to failure following radical therapy. Variables included in multivariable models were selected a priori based on published literature23 and included age, NCCN risk category, Log PSA, Log prostate volume, body mass index, percent positive cores, and Decipher score. The TTF model additionally included treatment modality (i.e. RP vs. radiotherapy). Statistical analyses were performed in R version 3.6.2 (R Foundation, Vienna, Austria).24 All statistical tests were two-sided using a 5% significance level.
Results:
Patient Characteristics
The 855 patients in this cohort had a median age of 66 years (IQR 60–72 years). Patient characteristics are given in Table 1, and a breakdown of patient characteristics by Decipher risk category is given in Supplementary Table 1. A total of 641 (75%) patients were self-reported white and 112 (13%) were self-reported black race. At the time of diagnosis, the median PSA level was 5.9 ng/ml (IQR 4.4–8.9 ng/ml), and 83% of patients had cT1 or T2a disease at diagnosis. Also, 60% of patients had either Grade Group 1 or 2 disease at diagnosis. Of the 105 patients (12%) who were diagnosed with prostate cancer prior to their index Decipher biopsy, the median time from diagnostic biopsy to their index Decipher biopsy was 15.7 months (IQR 11.7–32.4 months. An analysis of all men diagnosed with prostate cancer in MUSIC during our study period, stratified by receipt of Decipher Biopsy testing, demonstrates men undergoing Decipher Biopsy testing were more likely to have intermediate-risk disease. (p < 0.001; Supplementary Table 2).
Table 1.
Patient characteristics.
| All | |
|---|---|
| Age (time of diagnosis) | |
| Median (IQR) | 66 (60–72) |
| <55 | 78 (9.1%) |
| 55–64 | 277 (32.4%) |
| 65–74 | 500 (58.5%) |
| Race | |
| Black | 112 (13.1%) |
| Asian | 8 (0.9%) |
| Native American | 1 (0.1%) |
| White | 641 (75%) |
| Unknown/Other | 93 (10.9%) |
| BMI (kg/m2) | |
| Median (IQR) | 28.6 (25.8–31.7) |
| ≤ 25 | 161 (19.1%) |
| 25.1 – 30 | 386 (46%) |
| > 30 | 293 (34.9%) |
| Charlson Comorbidity index | |
| 0 | 20 (2.3%) |
| 1 | 121(14.2%) |
| ≥2 | 714 (83.5%) |
| Index Decipher Biopsy | |
| Grade Group | |
| 1 | 184 (21.9%) |
| 2 | 302 (36%) |
| 3 | 194 (23.1%) |
| 4–5 | 159 (19%) |
| PSA | |
| Median (IQR) | 6.1 (4.4–9.2) |
| 0–4 ng/ml | 163 (19%) |
| 4.1–10 ng/ml | 507 (59.1%) |
| 10.1–20 ng/ml | 111 (12.9%) |
| >20 | 77 (9%) |
| Clinical T-stage | |
| T1 | 607 (71.6%) |
| T2 | 224 (26.4%) |
| T3/4 | 17 (2%) |
| Number of positive cores | |
| 1–2 | 274 (32.6%) |
| 3–4 | 222 (26.4%) |
| >4 | 344 (41%) |
| NCCN Risk Group | |
| Low | 159 (19.1%) |
| Favorable-Intermediate | 258 (30.9%) |
| Unfavorable-Intermediate | 331 (39.7%) |
| High | 86 (10.3%) |
| Treatment | |
| AS | 264 (30.9%) |
| EBRT | 218 (25.5%) |
| RP | 236 (27.6%) |
| Other | 137 (16%) |
Abbreviations: AS, active surveillance; BMI, body mass index; EBRT, external beam radiation therapy; IQR, interquartile range; NCCN, National Comprehensive Cancer Network; PSA, prostate-specific antigen; RP, radical prostatectomy
Initial management was AS in 264 individuals (30.9%), RP in 236 (27.6%), and RT in 218 (25.5%). There were also 55 patients treated with primary ADT alone, 25 patients who underwent primary cryotherapy, and 66 patients classified as “other” who had no treatment recorded. For the patients initially managed on AS, 116 (44%) underwent subsequent primary treatment at a median of 15.2 months (IQR 8.0–22.5). For patients who stayed on AS, the median follow-up was 25.2 months (IQR 12.7–34.1). For patients who underwent RP (inclusive of those who were managed by AS prior to RP), pathologic characteristics are given in Supplementary Table 3. A total of 58 patients (18%) experienced treatment failure following RP, and the median time to failure was 6.4 months (IQR 4.3–9.6) after RP. Additionally, 10 (3.7%) patients experienced treatment failure following RT, and the median time to failure was 13.3 (IQR 4.0–22.0) months. Patients who underwent definitive treatment with no subsequent BCR had a median follow-up of 15.8 months (IQR 8.4–22.7).
Correlation of Decipher with Clinical Variables
Decipher scores were assessed within biopsy Grade Group strata, and there was substantial variability of Decipher scores within each Grade Group (Figure 1A). Overall, increasing Decipher score was correlated with increasing Grade Group (p<0.001). This variability within standard clinical risk stratification categories was also observed when comparing Decipher scores according to NCCN risk group (p<0.001, Figure 1B) and CAPRA risk groups. (p<0.001, Figure 1C).
Figure 1.
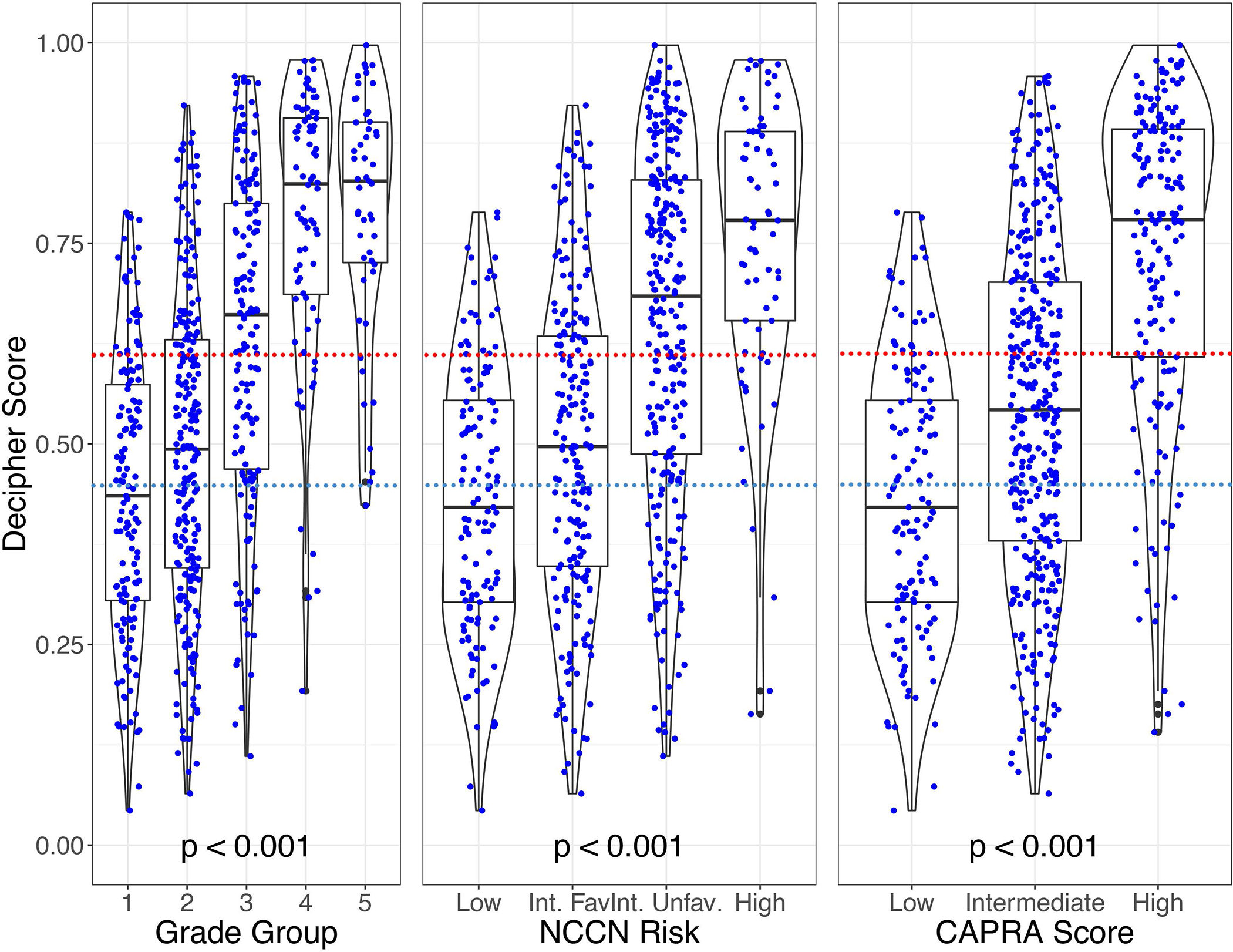
Violin plots showing the median and distribution of Decipher Biopsy scores for each pathologic grade group (Fig. 1A), NCCN risk strata (Fig. 1B), and CAPRA risk strata (Fig. 1C)(red dash line notates high-risk Decipher score cutpoint; blue dash line notates intermediate-risk Decipher score cutpoint)
Abbreviations:CAPRA, Cancer of the Prostate Risk Assessment; NCCN, National Comprehensive Cancer Network; Int. Fav, Favorable-intermediate risk; Int.Unfav.; Unfavorable-intermediate risk
Clinical Utility of Decipher in Active Surveillance
To assess the impact of Decipher score on length of time on AS, patients initially managed on AS were stratified according to Decipher risk category. A total of 241 patients were included in this analysis, with 23 patients being excluded due to incomplete follow-up data. In the cumulative incidence analysis, men with high-risk Decipher scores spent significantly less time on AS (median 13.6 months, IQR 3.5–24.9) than men with Decipher low/intermediate risk scores (median 33.0 months; p<0.001) (Figure 2). A repeat analysis assessing the differences between individual Decipher Biopsy risk groups (low, intermediate, and high) yields similar results (Supplemental Figure 1). At the time of data collection greater than 25% of patients with Decipher low/intermediate scores remained on AS, making the IQR unable to be reported. To evaluate patients initially managed on AS, we assessed grade group reclassification on subsequent biopsies for patients with NCCN low and favorable intermediate risk prostate cancer stratified by Decipher Biopsy risk (Supplementary Table 4). Among NCCN low risk patients, reclassification was observed in 20/58 (34%) of patients with lower Decipher scores compared to 3/8 (38%) with high Decipher scores. Among NCCN favorable intermediate risk patients, reclassification was observed in 11/43 (26%) of patients with lower Decipher scores compared to 2/7 (29%) with high Decipher scores.
Figure 2.
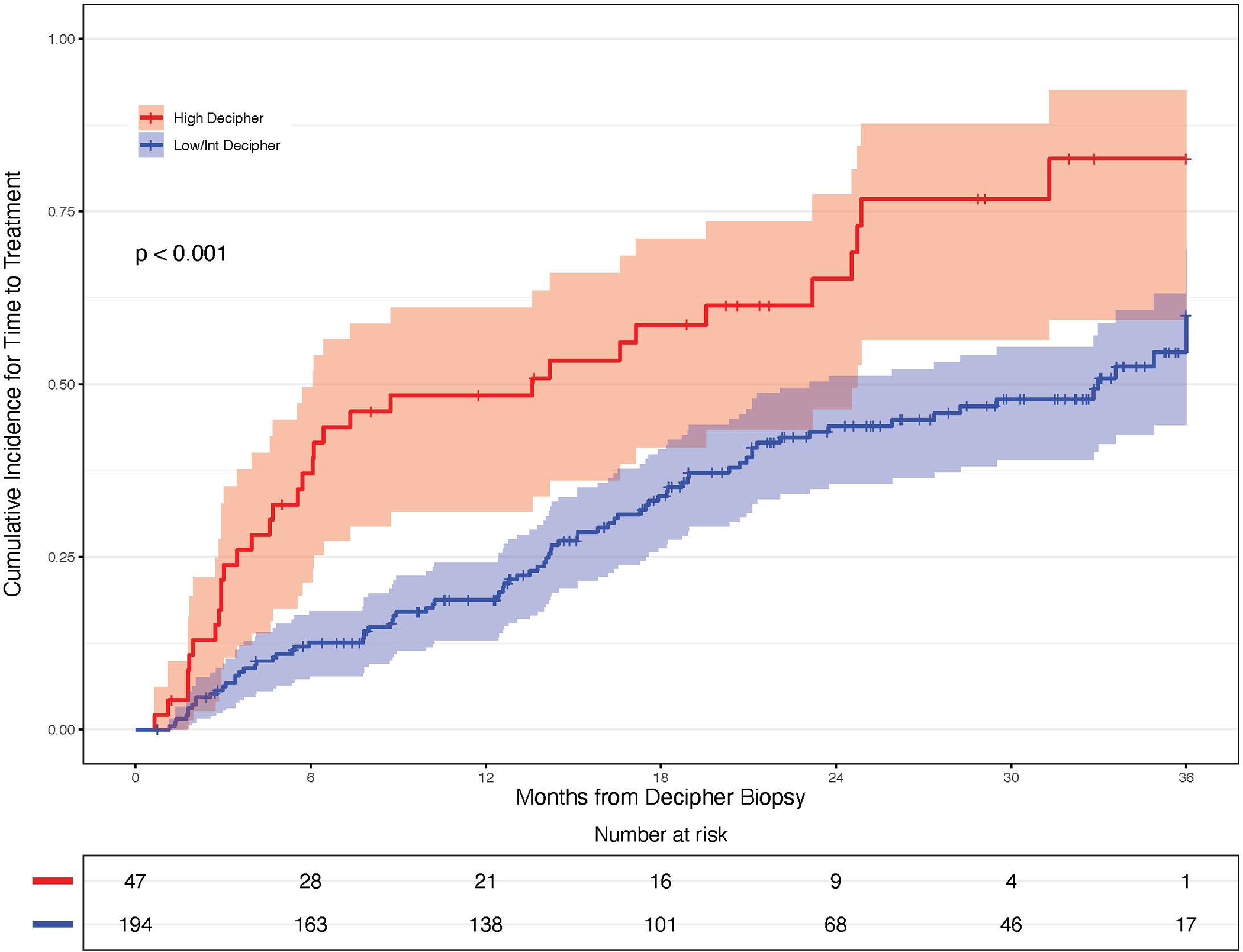
Cumulative incidence curve comparing patients with high-risk Decipher scores to those with low/intermediate Decipher scores to analyze time to treatment, from time of index biopsy
Abbreviation: Int, intermediate
When analyzing Decipher scores as an ordinal variable on multivariable analysis, high-risk Decipher scores were associated with a significantly greater likelihood of undergoing definitive treatment (HR 2.51, 95% CI 1.52–4.13; p <0.001, Table 2). Similar findings were observed when analyzing Decipher scores as a continuous variable (HR 1.20 per 0.1 unit score increase, 95% CI 1.07–1.35; p= 0.002, Supplemental Table 5).
Table 2.
Decipher scores analyzed as ordinal variable
| Risk of Progression to Definitive Therapy Decipher as an ordinal variable | |||
| Variable | HR | 95% Confidence Interval | p-value |
| High-risk Decipher | 2.51 | 1.52, 4.13 | <0.001 |
| Age | 0.98 | 0.95, 1.01 | 0.29 |
| Ordinal NCCN risk strata | 1.67 | 1.27, 2.20 | <0.001 |
| Log PSA | 0.8 | 0.64, 1.0 | 0.05 |
| Log Prostate Volume (cc) | 0.9 | 0.56, 1.42 | 0.65 |
| BMI (kg/m2) | 0.98 | 0.94, 1.02 | 0.36 |
| Percent Positive Cores (10% increase) | 1.24 | 1.06, 1.45 | 0.008 |
| Risk of Treatment Failure after Radical Therapy Decipher as an ordinal variable | |||
| Variable | HR | 95% Confidence Interval | p-value |
| High-risk Decipher | 2.98 | 1.22, 7.29 | 0.02 |
| Age | 0.95 | 0.91, 1.0 | 0.03 |
| Ordinal NCCN risk strata | 1.02 | 0.64, 1.62 | 0.92 |
| Log PSA | 0.98 | 0.74, 1.29 | 0.86 |
| Log Prostate Volume (cc) | 1.62 | 0.68, 3.89 | 0.27 |
| BMI (kg/m2) | 0.98 | 0.91, 1.05 | 0.55 |
| Percent Positive Cores (10% increase) | 1.16 | 1.01, 1.33 | 0.03 |
Abbreviations: BMI, Body Mass Index; HR, hazard ratio; NCCN, National Comprehensive Cancer Network; Ordinal NCCN risk strata: low, favorable intermediate, unfavorable intermediate, high; PSA, Prostate-specific antigen
Association of Decipher Scores and Time to Treatment Failure after Radical Therapy
To assess the performance of Decipher scores in predicting treatment failure, we analyzed a total of 479 evaluable patients who underwent definitive treatment either initially or after a period of AS and had complete follow-up data. These patients were stratified into two categories: Decipher low/intermediate-risk and Decipher high-risk using the locked Decipher cut-points. Patients with high-risk Decipher scores had significantly shorter TTF than lower Decipher score patients (p = 0.007, Figure 3). Similar results for TTF were demonstrated on repeat analysis assessing differences between low, intermediate, and high-risk Decipher Biopsy groups (Supplemental Figure 2).
Figure 3.
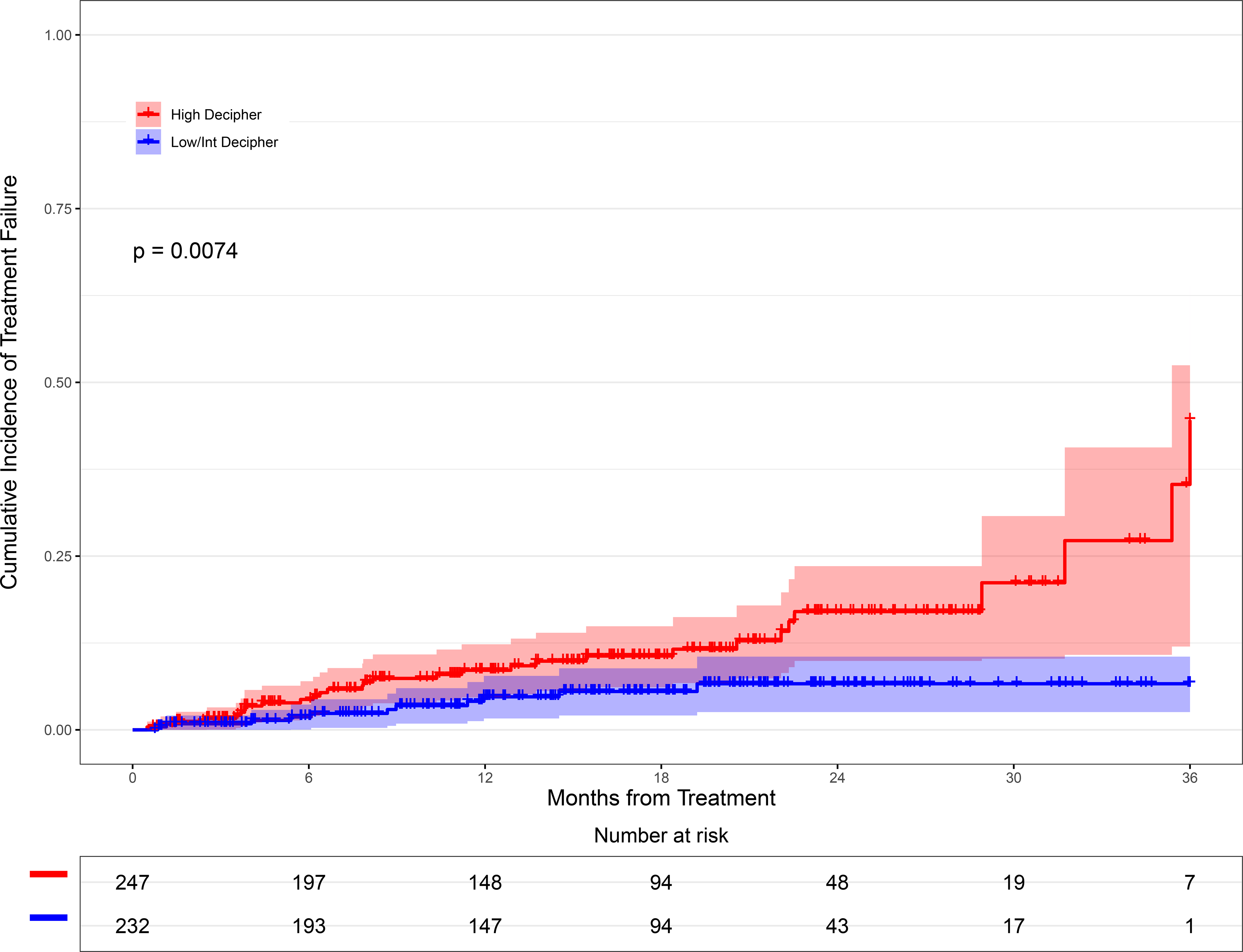
Cumulative incidence curve comparing patients with high-risk Decipher scores to those with low/intermediate Decipher scores to analyze time to failure from the index biopsy
Abbreviation: Int, intermediate
This finding held true on multivariable analysis, when Decipher scores were analyzed as an ordinal variable with a HR of 2.98 (95%CI 1.22–7.29, p=0.01, Table 2). Similar results were observed when analyzing Decipher scores as a continuous variable, HR 1.26 (95%CI 1.02–1.544, p = 0.026, Supplemental Table 5).
Discussion:
Decipher initially received Molecular Diagnostic Services (MolDx) approval for Centers for Medicare and Medicaid Services (CMS) coverage for use as a post-RP test to help with post-operative decision making. More recently, the Decipher Biopsy pre-treatment test has been evaluated in multiple studies.16,25,26 These studies range from showing disease reclassification, change in management, and improved prognostication and discrimination in men with low-, intermediate-, and high-risk disease. Due to the accumulation of data across these risk strata, Decipher Biopsy recently received MolDx approval for localized prostate cancer in cases where the test may impact clinical decision-making. Despite prospective data supporting clinical utility in the post-RP setting, there are limited data describing utility in the biopsy setting. To date, there have been no prospective studies evaluating the use of Decipher Biopsy on common oncologic endpoints for early-stage disease. These intermediate endpoints (e.g., TTT and TTF), although unlikely to be formal statistical surrogate endpoints for overall survival, are clinically relevant as they trigger subsequent therapy.
The present study addresses a current knowledge gap in the field of gene expression biomarkers. To date, there are multiple retrospective studies demonstrating that Decipher, Oncotype Dx, and Prolaris are prognostic in men with favorable-risk disease pre-treatment. However, this is the first study performed on purely prospectively collected data analyzing Decipher in men on AS and following radical therapy, studying the performance in the intended use population. Our data reveal that men with high Decipher scores are more likely to receive radical therapy sooner by more than 1.5 years, and that men with high Decipher scores are over two-fold more likely to experience treatment failure after radical therapy. These data are consistent with prior retrospective studies supporting the prognostic performance of Decipher in newly diagnosed prostate cancer. Cooperberg and colleagues retrospectively analyzed 408 patients with low risk prostate cancer using the Decipher microarray platform, which allows simultaneous assessment of a large number of molecular signatures.6 They demonstrated that the only independent predictor of biochemical recurrence after adjusting for CAPRA and PSA density was the average genomic risk score.
Recently, Lonergan et al reported a retrospective analysis of men with early stage prostate cancer managed by active surveillance who had undergone either Decipher, Prolaris, or Oncotype DX testing. They pooled all patients together regardless of test used and dichotomized outcomes according to the high-risk vs. lower-risk threshold provided on the test report of each assay. On multivariable analysis, they demonstrated that high genomic risk score was independently associated with biopsy reclassification from GG1 to GG2 or higher at initial surveillance biopsy (HR 2.81, 95%CI 1.21–6.52) and 1–3 years after the initial biopsy (HR 2.02, 95%CI 1.16–3.54). Furthermore, a high genomic risk score was independently associated with reclassification at first surveillance biopsy to GG3 or higher cancer (HR 6.77, 95%CI 1.58–29.08). It is unclear whether grouping of various test thresholds provides a consistent risk estimate across these platforms.27 Notably, multiparametric MRI did not significantly predict for any of these endpoints.22
Another key study assessing the performance of molecular classifiers in men with newly diagnosed favorable risk prostate cancer is the post-hoc analysis from the Prostate Cancer Active Surveillance Study (PASS).28 This prospective observational cohort retrospectively evaluated tissue from initial prostate biopsy specimens and determined Oncotype Dx risk scores to measure its association with adverse surgical pathology. While this study included only 101 men, it demonstrated that the Oncotype Dx score was independently associated with time to development of adverse pathology when accounting only for GG. However, when also adjusting for PSA density the classifier did not reach statistical significance (HR 1.17 (95%CI 1.00–1.43), p=0.06). This is in contrast to the current study, that included nearly every prognostic variable for time on AS reported recently by Cooperberg et al, where Decipher score remained independently prognostic.23
Our group previously demonstrated that the use of gene expression classifiers are associated with patient and provider decision-making.27 The current data provides evidence of an association between baseline molecular classifier risk and duration of AS. Combined with prior studies, these data suggest that men with early stage prostate cancer with a high Decipher score are more likely to rapidly transition off of AS and are more likely to recur after radical therapy. With growing understanding that early reclassification on AS is predominantly due to undersampling on initial diagnostic biopsy, these findings merit thoughtful assessment of risk prior to proceeding with AS in Decipher high risk patients and further support the importance of early confirmatory testing.1
This study has limitations. The length of follow-up is relevant for the endpoints analyzed, but longer follow-up is needed to assess the impact of Decipher score on endpoints, such as time to metastasis. In addition, the data relating Decipher score to the duration on AS are insufficient to demonstrate a causal relationship, given the nature of this study. Also, we can not account for the influence receipt of ADT has on lower rates of TTF in patients who received radiation for definitive treatment. Collectively, however, this does not explain the independent association of Decipher scores and TTF. There is variability in how molecular classifiers are used among providers, and data on the cause of withdrawal from AS (e.g., due to progression vs. patient anxiety) are not collected in MUSIC. Despite these limitations, this prospectively tracked, multi-institutional cohort allows for an improved understanding of Decipher testing in everyday clinical practice.
Conclusion:
In men with newly diagnosed prostate cancer, Decipher Biopsy score is independently associated with time on AS and time to treatment failure after radical therapy. The ongoing G-MAJOR clinical trial (NCT04396808) is assessing the use of molecular classifier testing in a prospective randomized trial in favorable risk prostate cancer to further establish the clinical utility of these tests.
Supplementary Material
Acknowledgments
Support:
Support for this project is provided by Blue Cross Blue Shield of Michigan; the Department of Defense Physician Research Training Award No. W81XWH-14-1-0287 (T.M.M.); Alfred A. Taubman Institute; Prostate Cancer Foundation (T.M.M. and D.E.S.); and National Cancer Institute Grant No. R01CA240991-01(T.M.M. and D.E.S.).
Disclosures/COI:
• Spratt: Personal Fees from Janssen, AstraZeneca, and Blue Earth. Funding from Janssen.
• Morgan: Research funding: Decipher Biosciences and Myriad Genetics. Advisory Board: Blue Earth
• Tosoian: Co-founder with equity interest, consulting fees: LynxDx, Inc.
References:
- 1.Tosoian JJM,M: Epstein JI: Landis P: Macura KJ: Simopoulos DN: Carter HB: Gorin MA Active Surveillance of Grade Group 1 Prostate Cancer: Long-term Outcomes from a Large Prospective Cohort. Eur Urol 2020;77(6):675–682. [DOI] [PubMed] [Google Scholar]
- 2.Klotz L, Vesprini D, Sethukavalan P, Vibhuti J, Liying Z, Suneil J, et al. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. J Clin Oncol 2015;33(3):272–277. [DOI] [PubMed] [Google Scholar]
- 3.Bul M, Zhu X, Valdagni R, Pickles T, Kakehi Y, Rannikko A, et al. Active surveillance for low-risk prostate cancer worldwide: the PRIAS study. Eur Urol 2013;63(4):597–603. [DOI] [PubMed] [Google Scholar]
- 4.Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med 2016;375(15):1415–1424. [DOI] [PubMed] [Google Scholar]
- 5.Spratt DE, Zhang J, Santiago-Jimenez M, Dess RT, Davis JW, Den RB, et al. Development and Validation of a Novel Integrated Clinical-Genomic Risk Group Classification for Localized Prostate Cancer. J Clin Oncol 2018;36(6):581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooperberg MR, Erho N, Chan JM, Feng FY, Fishbane N, Shuang G, et al. The Diverse Genomic Landscape of Clinically Low-risk Prostate Cancer. Eur Urol 2018;74(4):444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felker ER, Margolis DJ, Nassiri N, Marks LS. Prostate cancer risk stratification with magnetic resonance imaging. Urol Oncol 2016;34(7):311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooperberg MR, Broering JM, Carroll PR. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst 2009;101(12):878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JR, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12(3):245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark KM, Clark-Langone KC, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spratt DE, Yousefi K, Deheshi S, Ross AE, Den RB, Schaeffer EM, et al. Individual Patient-Level Meta-Analysis of the Performance of the Decipher Genomic Classifier in High-Risk Men After Prostatectomy to Predict Development of Metastatic Disease. J Clin Oncol 2017;35(18):1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, Karnes RJ. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol 2015;67(2):326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berlin A, Murgic J, Hosni A, Jenkins RB, Ghadessi M, Karnes RJ, et al. Genomic Classifier for Guiding Treatment of Intermediate-Risk Prostate Cancers to Dose-Escalated Image Guided Radiation Therapy Without Hormone Therapy. Int J Radiat Oncol Biol Phys 2019;103(1):84–91. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen PL, Martin NE, Choeurng V, Palmer-Aronsten B, Kolisnik T, Beard CJ, et al. Utilization of biopsy-based genomic classifier to predict distant metastasis after definitive radiation and short-course ADT for intermediate and high-risk prostate cancer. Prostate Cancer Prostatic Dis 2017;20(2):186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jairath NK, Dal Pra A, Vince R Jr., Dess RT, Jackson WC, Tosoian JJ, et al. A Systematic Review of the Evidence for the Decipher Genomic Classifier in Prostate Cancer. Eur Urol 2020. [DOI] [PubMed] [Google Scholar]
- 17.Kaye DR, Qi J, Morgan TM, Linsell S, Lane BR, Montie JE, et al. Association Between Early Confirmatory Testing and the Adoption of Active Surveillance for Men With Favorable-risk Prostate Cancer. Urology. 2018;118:127–133. [DOI] [PubMed] [Google Scholar]
- 18.Singhal U, Tosoian JJ, Qi J, Miller DC, Linsell SM, Cher M, et al. Overtreatment and Underutilization of Watchful Waiting in Men With Limited Life Expectancy: An Analysis of the Michigan Urological Surgery Improvement Collaborative Registry. Urology. 2020;145:190–196. [DOI] [PubMed] [Google Scholar]
- 19.Ginsburg KB, Cher ML, Montie JE. Defining Quality Metrics for Active Surveillance: The Michigan Urological Surgery Improvement Collaborative Experience. J Urol 2020;204(6):1119–1121. [DOI] [PubMed] [Google Scholar]
- 20.Klein EA, Haddad Z, Yousefi K, Lam LLC, Wang O, Choeurng V, et al. Decipher Genomic Classifier Measured on Prostate Biopsy Predicts Metastasis Risk. Urology. 2016;90:148–152. [DOI] [PubMed] [Google Scholar]
- 21.Roach M 3rd, Hanks G, Thames H Jr., Schellhammer P, Shipley WU, Sokol GH, et al. Defining biochemical failure following radiotherapy with or without hormonal therapy in men with clinically localized prostate cancer: recommendations of the RTOG-ASTRO Phoenix Consensus Conference. Int J Radiat Oncol Biol Phys 2006;65(4):965–974. [DOI] [PubMed] [Google Scholar]
- 22.Lonergan PE, Washington SL 3rd, Cowan JE, Zhao S, Nguyen HG, Shinohara K, et al. Risk Factors for Biopsy Reclassification over Time in Men on Active Surveillance for Early Stage Prostate Cancer. J Urol 2020;204(6):1216–1221. [DOI] [PubMed] [Google Scholar]
- 23.Cooperberg MR, Zheng Y, Faino AV, Newcomb LF, Zhu K, Cowan JE, et al. Tailoring Intensity of Active Surveillance for Low-Risk Prostate Cancer Based on Individualized Prediction of Risk Stability. JAMA Oncol 2020;6(10):e203187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan BKC. Data Analysis Using R Programming. Adv Exp Med Biol 2018;1082:47–122. [DOI] [PubMed] [Google Scholar]
- 25.Herlemann A, Huang HC, Alam R, Tosoian JJ, Kim HL, Klein EA, et al. Decipher identifies men with otherwise clinically favorable-intermediate risk disease who may not be good candidates for active surveillance. Prostate Cancer Prostatic Dis 2020;23(1):136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HL, Li P, Huang HC, Deheshi S, Marti T, Knudsen B, et al. Validation of the Decipher Test for predicting adverse pathology in candidates for prostate cancer active surveillance. Prostate Cancer Prostatic Dis 2019;22(3):399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu JC, Tosoian JJ, Qi J, Kaye D, Johnson A, Linsell S, et al. Clinical Utility of Gene Expression Classifiers in Men With Newly Diagnosed Prostate Cancer. JCO Precis Oncol 2018;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin DW, Zheng Y, McKenney JK, Brown MD, Lu R, Crager M, et al. 17-Gene Genomic Prostate Score Test Results in the Canary Prostate Active Surveillance Study (PASS) Cohort. J Clin Oncol 2020;38(14):1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


