Abstract
In a previous study, we showed that Acinetobacter genomic DNA group 3 was the most common species among blood culture isolates and was commonly found on superficial carriage sites of the healthy and the sick, which are different findings from those reported in Europe and North America. We used amplified ribosomal DNA restriction analysis and pulsed-field gel electrophoresis to study further the molecular epidemiology of acinetobacters in our region. Over a study period of 6 weeks with 136 consecutive routine clinical isolates (1.33% of all specimens), genomic DNA groups 2 (Acinetobacter baumannii), 3, and 13TU were obtained from 59 of 69 positive patients. There is a significant difference in the specimen sources of the three genomic DNA groups, with group 13TU being significantly associated with the respiratory tract (chi-square exact test, P = 0.0064). Settle plates showed a significantly heavier environmental load from the intensive care unit (ICU) than from the four surgical wards examined (22 of 70 versus 76 of 120 plates with <5 colonies; chi-square test, P < 0.0001). Genomic group 3 accounted for 6 of 12 clusters of possibly related strains among patients, between patients and the ICU environment, and in the ICU environment. Genomic groups 2 and 3 accounted for 21% of the 132 genomically identified isolates recovered from 21 of 41 local vegetables, 53 of 74 fish and meat samples, and 22 of 60 soil samples. Group 13TU was present only in patients' immediate surroundings. The role played by the environment and by human carriage should be evaluated in order to devise a cost-effective infection control program pertinent to our situation of acinetobacter endemicity.
Acinetobacter spp. are important nosocomial pathogens associated with a growing number of hospital-acquired infections worldwide (2, 14). In hot, humid areas, such as Hong Kong, Acinetobacter infection is endemic, with higher incidences of nosocomial infection, including bacteremia and pneumonia, than those reported elsewhere (2, 14, 28, 34, 35). The clinically important species, such as Acinetobacter baumannii (genomic DNA group 2), are intrinsically resistant to the first-line antimicrobial agents, e.g., ampicillin and cefuroxime. Acinetobacter spp. have a propensity to readily develop resistance to second- and third-line agents such as cefotaxime, ciprofloxacin, and imipenem, giving rise to therapeutic problems (2, 30, 32). Outbreaks of Acinetobacter infections, often caused by multiresistant strains, have been widely reported, commonly in intensive care units (ICUs) in North America and Europe. Epidemiological features and risk factors of these outbreaks have also been well described (2, 5, 8, 10, 14, 16, 17, 18, 20, 24, 36, 43). In contrast, there is a paucity of information in regions of Acinetobacter endemicity, such as Hong Kong (28, 34, 35). It has recently been shown that there is a significant difference between Hong Kong and Europe in the genomic DNA groups of isolates obtained from blood cultures and various superficial carriage sites (4, 7, 33). Species other than A. baumannii appear to be of greater epidemiological significance than was previously appreciated (7). This raises the question of whether identified risk factors of infection and control measures that are promulgated and practiced in areas where the infection is not endemic are wholly applicable to our region (8, 9, 24, 36, 42). We report here the molecular epidemiology of isolates obtained from clinical specimens and compare it with that in the environment both inside and outside the hospital. We discuss the infection control issues in light of our findings.
MATERIALS AND METHODS
The setting.
Prince of Wales Hospital, Shatin, Hong Kong, People's Republic of China, is a 1,400-bed general hospital built in 1984. There are 10 surgical and 10 medical wards, in addition to wards of other specialties, with one to two single rooms in most of these 26- to 34-bed wards. There were about 136,000 deaths and discharges in 1998. There is a 22-bed ICU for both medical and surgical cases. There is an infection control committee and an infection control team, with two infection control nurses and one infection control doctor. Infection control practices are largely based on those practiced in the United Kingdom. Based on laboratory results, particularly of organisms of infection control interest, or admission diagnoses, the infection control nurses inform and liaise with the ward staff to ensure that the right level of isolation and infection control measures are carried out. Universal precautions are practiced by all health care workers. Infection control problems are also discussed in clinical meetings.
Isolates from routine clinical specimens.
All acinetobacters isolated from clinical specimens by the routine laboratory were stored in nutrient agar slants until examination.
Isolates from hospital environments.
Modified Leeds Acinetobacter medium (MLAM) containing vancomycin (3 instead of 10 μg/ml) was used as the selective medium for settle plates to sample the environment of the ICU and four surgical wards (7). Petri dishes (85-mm diameter) containing MLAM were exposed to the air at different locations for 6 h continuously in the ICU and for 8 h in the four surgical wards on the day of sampling. The duration of exposure was determined by previous experiments so as to achieve discrete colonies with minimal fungal contamination. Repeat sampling was carried out from the same locations in the wards, at the same hour and on the same day of the week. The plates were left on the level surfaces near patients (locker surfaces, windowsills) or on equipment, e.g., respirators, in such a position that they could not be disturbed. Representative isolates were stored for genomic DNA group identification (hereafter referred to as genomic identification) and studies of clonal relatedness.
Isolates from vegetables, meat, and fish.
Samples of vegetables, pork, beef, and freshwater fish were purchased from local markets situated in different districts. Vegetables were aseptically cut into 1-in. pieces, weighed (5 to 10 g), and placed in 90 ml of saline in sterile plastic bottles. The meat and fish samples were weighed and placed in saline (1 g/ml). All samples were then vigorously shaken for 15 min. Serial dilutions (1:10) were made with the suspension, and inocula (0.5 ml) in duplicates of these dilutions were then spread with glass rods onto MLAM plates.
Isolates from soil samples.
Soil samples were obtained from different areas in Hong Kong. Weighed samples (1 g) were put into sterile containers with 10 ml of sterile distilled water in each, stirred to make a suspension, and left to settle for 30 min. The supernatant (2.5 ml) was then added to 10 ml of enrichment broth (Oxoid) and incubated at 30°C for 24 h with vigorous shaking. Serial dilutions (1:10) were made in enrichment broth, and inocula (0.1 ml) of each dilution in duplicates were then spread with glass rods onto MLAM plates.
Genus identification.
All MLAM plates were incubated at 30°C and examined daily for up to 3 days. Typical colonies were enumerated, picked, and examined further. Acinetobacter was identified by Gram staining, cell and colony morphology, activity in the oxidation/fermentation test, absence of motility, and negative oxidase and positive catalase reactions. The transformation assay of Juni was used to confirm the genus (22).
Method of genomic identification.
Genomic identification was carried out by amplified ribosomal DNA restriction analysis (ARDRA) as previously described (12), using two primers (5′-TGG CTC AGA TTG AAC GCT and 5′-TAC CTG TTA CGA CTT CA) and restriction endonucleases (CfoI, AluI, MboI, and MspI RsaI [Pharmacia, Uppsala, Sweden] and BfaI and BsmaI [New England Biolabs, Beverly, Mass.]). Restriction patterns were obtained by agarose (2%) gel electrophoresis and compared after ethidium bromide staining.
Relatedness of strains of the same genomic groups.
Random amplification of polymorphic DNA of groups of strains belonging to one genomic DNA group was performed with Ready-to-Go random amplification of polymorphic DNA beads and Primer 2 (both Pharmacia) as recommended by the manufacturer. Products were analyzed by agarose (2%) gel electrophoresis.
Enterobacterial repetitive intergenic consensus (ERIC) PCR typing was performed with the primer ERIC2 (5′-AAG TAA GTG ACT GGG GTG AGC G). Each reaction (25 μl) contained 50 pmol of the primer, 0.2 mM deoxynucleoside triphosphates, 1× reaction buffer, 0.25 U of Taq DNA polymerase (Pharmacia), and 10 to 50 ng of template DNA. Forty-five cycles of 95°C for 60 s, 36°C for 60 s, and 72°C for 120 s were performed with the reactions before analysis by 2% agarose gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) fingerprints were generated using a contour-clamped homogeneous electric field electrophoresis apparatus (Bio-Rad, Richmond, Calif.). The restriction endonuclease ApaI was used for the in situ digestion of intact Acinetobacter genomic DNA embedded in 1% agarose gel blocks prepared according to previously described methods (23). Samples were loaded into 1% certified PFGE-grade agarose gels and electrophoresed with 0.5× Tris-borate-EDTA buffer (TBE) (50 mM Tris, 40 mM borate, and 0.5 mM EDTA) with an electric field of 6 V/cm, an included angle of 120°C, and a pulse time of 5 to 35 s over 32 h at 14°C. Images of ethidium bromide solution (1 mg/liter)-stained gels were digitized using a gel documentation system (ImageMaster VDS; Pharmacia) and analyzed using the computer software GelCompar (Applied Maths, Kortrijk, Belgium). Clusters of possibly related isolates were identified using the Dice coefficient of similarity and unweighted pair group method using arithmetic averages at 70%, which indicates four- to six-fragment differences in gels with an average of 20 bands (37).
Statistical analyses.
Statistical analyses were carried out using the chi-square test (Epi Info, version 5.01b) and the chi-square exact test (StatXact, version 2.05). Unclassifiable strains were grouped together for analysis.
RESULTS
Acinetobacters from raw food and soil samples.
Table 1 shows the sources and the number of samples examined and the distribution of genomic DNA groups of isolates. Acinetobacters were cultured from 27 of 36 meat samples, 21 of 41 vegetable samples, and 26 of 38 fish samples. These food samples were tested at regular intervals throughout a period of 12 months. Soil samples were obtained from 10 sites, with and without human habitation. This was repeated three times in September 1998 and three times in January 1999. There is a significant difference in the distribution of genomic DNA groups between the two seasons (chi-square exact test, P = 0.003). Genomic DNA group 3 accounted for 30% of the isolates from the hot season but was not found during the winter. A significant difference is also seen in comparing the distribution of genomic DNA groups from vegetables and soil (winter plus summer) isolates (chi-square exact test, P = 0.0031). Group 2 (A. baumannii) was found in all environmental sources outside the hospital: vegetables, fish and meat, and soil, but a large proportion of isolates from these sources were not classifiable by ARDRA (35 to 51%). From all environmental sources (both inside and outside the hospital), genomic DNA group 3 was the most common.
TABLE 1.
Genomic DNA groups of isolates obtained from clinical specimens and different environments in the hospital and the community
| Source | Samples (no. positive/total) (no. of patients) | Isolates (no. identified to genomic DNA group/ total) (no. of patients) | No. of isolates (no. of patients) from genomic DNA group:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 13TU | 10 | 16 | 17 | Other | Ua | |||
| Clinical specimens | 136/10,253 (69) | 127/136 (68) | 39 (30) | 36 (24) | 35 (22) | 1 (1) | 16 (11) | ||||
| Settle plates | |||||||||||
| ICU | 67/70 | 64/105 | 2 | 5 | 33 | 3 | 1 | 1 | 5 | 4 | 10 |
| Surgical wards | 35/120 | 46/107 | 3 | 9 | 16 | 6 | 2 | 3 | 7 | ||
| Local vegetables | 21/41 | 45/46 | 1 | 15 | 1 | 4 | 4 | 20 | |||
| Fish and meat | 53/74 | 53/53 | 1 | 25 | 27 | ||||||
| Soil | |||||||||||
| September | 15/30 | 20/20 | 2 | 6 | 1 | 4 | 7 | ||||
| January | 7/30 | 14/14 | 1 | 3 | 1 | 4 | 5 | ||||
U, unclassifiable by ARDRA.
Acinetobacters from clinical specimens between 26 October and 6 December 1998.
During the period from 26 October to 6 December 1998, in the routine microbiology laboratory at the Prince of Wales Hospital, Shatin, Hong Kong, People's Republic of China, of the 2,221 sets of blood cultures received, 11 cultures from seven patients yielded acinetobacters (0.5%). Acinetobacters were also reported in 7 (7 patients) (≥105 CFU/ml) of the 4,794 (0.15%) samples of urine, 76 (55 patients) of the 1,772 (4.3%) respiratory tract specimens, and 42 (29 patients) of the 1,466 (2.9%) miscellaneous specimens (bile samples, wound swabs, catheter tips, and tissues). A total of 97 isolates were collected from 35 patients in the ICU, and 39 isolates from 34 patients were collected from patients in the rest of the hospital. Nine isolates (one obtained from a catheter tip and eight from the respiratory tract) from nine ICU patients were not genomically identified, because one isolate was lost and eight isolates were from patients who had multiple positive specimens from the same sources within 3 days. All 39 clinical isolates from the wards and 88 of the 97 ICU isolates were genomically identified (Table 1). Of the patients examined, clinical isolates from 59 (87%) of them belonged to genomic DNA groups 2, 3, and 13TU.
Table 2 shows the distribution of genomic DNA groups obtained from different specimen sources. Although the number of positive patients is similar for each of the genomic DNA groups 2, 3, and 13TU, there is a significant difference in the distribution of sites from which these genomic DNA groups were obtained (chi-square exact test, P = 0.0064). Group 13TU was significantly associated with the respiratory tract, being isolated from 19 of 37 respiratory sites but only 5 of 46 other sites (chi-square test, P < 0.005; odds ratio, 4.98; 95% confidence limits, 1.53 to 17.24). Among patients with wounds or intravenous catheters, A. baumannii was found in 60% (9 of 15) and 45% (5 of 11), respectively.
TABLE 2.
Distribution of the main genomic DNA groups in clinical specimens from different sources
| Genomic DNA group | No. of patients (no. of specimens) with specimens from:
|
|||||
|---|---|---|---|---|---|---|
| Blood cultures | Central intravenous catheters | Miscellaneousa | Wound swabs | Respiratory tracts | Total | |
| 2 | 1 (1) | 5 (5) | 5 (5) | 9 (14) | 12 (14) | 30 (39) |
| 3 | 4 (8) | 4 (4) | 7 (7) | 2 (2) | 12 (15) | 24 (36) |
| 13TU | 1 (1) | 1 (1) | 3 (4) | 19 (29) | 22 (35) | |
| Otherb | 2 (2) | 1 (3) | 2 (2) | 8 (10) | 12 (17) | |
| Totalc | 7 (11) | 11 (13) | 13 (13) | 15 (22) | 37 (68) | 68 (127) |
Miscellaneous sources were urine, ears, eyes, bile, and continuous ambulatory peritoneal dialysis fluid.
Includes 16 isolates that showed 10 patterns that were not classifiable by ARDRA.
Some patients had >1 isolate from the same site belonging to different genomic groups.
Acinetobacters from the ICU and four surgical wards.
Ten settle plates were placed in a standardized manner as described above for 6 h on seven weekly occasions in the ICU. Of the 70 plates used, 67 (96%) yielded acinetobacters, with a range of 1 to 42 colonies per plate. The mean colony counts per location over the 7 weeks ranged from 4 to 19 per plate. All together, 32% of the plates contained less than 5 colonies, 53% had 5 to 15 colonies, and 15% had more than 15 colonies. Of the 105 representative isolates saved, 64 from different parts of the ICU were identified to the genomic level (Table 1).
Forty settle plates were placed in four surgical wards (with a total of 120 beds) at the same times and locations for 8 h on three occasions (22 September and 7 and 20 October 1998). Acinetobacters (range, 1 to 36 colonies) were found on 107 (89%) of the 120 plates used, with 63% of the plates yielding less than 5 colonies, 32% yielding 5 to 15 colonies, and 5% yielding more than 15 colonies. There was a significantly smaller number of plates containing <5 colonies from the ICU than from the wards (22 of 70 versus 76 of 120) (chi-square test, P < 0.0001; odds ratio, 3.77; 95% confidence limits, 1.93 to 7.42). All 46 isolates obtained on 20 October from the four wards were genomically identified (Table 1).
Relatedness of clinical isolates from the same patients.
Among the 69 patients, there were 9 patients who had positive specimens from at least two sites, and in all of them, isolates from different sites belonged to different genomic DNA groups. Of the 69 patients, there were 21 sites (19 patients) from which multiple specimens (≥2) from the same sites were positive: 16 respiratory, 2 central intravenous catheter, 2 blood culture, and 1 wound swab sample. The results of ARDRA showed that isolates from the same sites belonged to different genomic DNA groups for 12 of the 19 sites. For another four sites, the multiple isolates were shown to belong to the same genomic DNA groups but were different from one another by ERIC and/or PFGE. In another three sites, the multiple isolates were shown to be possibly related by PFGE (Dice coefficient of similarity of ≥70%) (37). Isolates from the remaining two sites were not examined. Seven patients had ≥3 positive respiratory specimens, and in four of them, the same strain was found on at least two occasions.
Relatedness of clinical and environmental isolates obtained from the ICU.
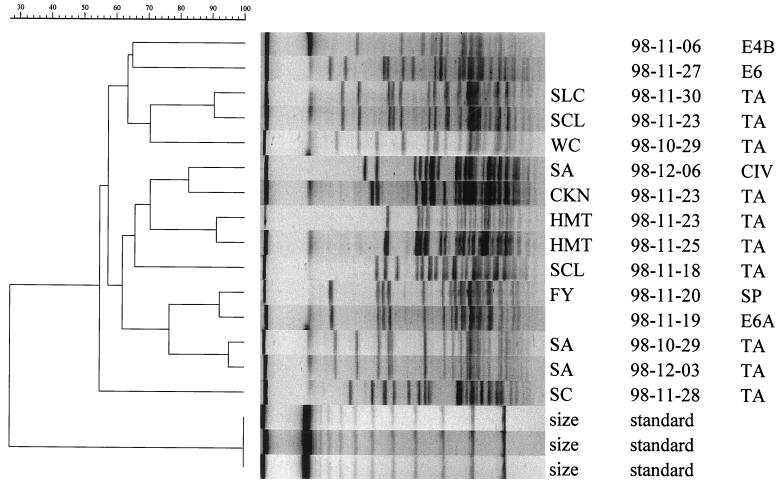
From the environmental collection, 44 of the 64 genomically identified isolates belonging to genomic DNA groups 2, 3, 13TU, and an unclassifiable group (U19) were examined by ERIC PCR and PFGE. The results were compared with those of 68 clinical isolates of the same genomic DNA groups from 29 patients who had multiple specimens or a prolonged stay in the ICU. The results by PFGE are tabulated in Table 3. This table shows strains from clinical and environmental sources which shared a Dice coefficient of similarity of ≥70% and therefore could possibly be related (37). There were six clusters of clinical isolates from different patients, four clusters involving both environmental and clinical isolates, and two environmental clusters, each involving two ICU locations. Genomic DNA group 3 was responsible for 6 of the 12 clusters (Table 3). A. baumannii was involved in the cluster covering the longest duration (34 days). Figure 1 shows the PFGE dendrogram of isolates belonging to group 13TU, demonstrating two clusters, each involving two patients and one ICU location.
TABLE 3.
ICU clusters found in 76 clinical and environmental isolates showing Dice coefficients of similarity of ≥70% by PFGE for genomic DNA groups 2, 3, and 13TU
| Genomic DNA group | Total no. examined
|
Clinical clusters
|
Clinical and environmental clusters
|
Environmental clusters
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinical isolates | Patients | Environmental isolates | No. of patients | Duration (days)a | No. of patients | No. of locations | Duration (days) | No. of locations | Duration (days) | |
| 2 | 9 | 6 | 5 | 5 | 34 | |||||
| 2 | 1 | |||||||||
| 3 | 28 | 17 | 14 | 2 | 1 | 1 | 1 | 11 | 2 | 1 |
| 2 | 20 | 2 | 1 | 6 | 2 | 7 | ||||
| 13TU | 12 | 8 | 3 | 2 | 7 | 2 | 1 | 35 | ||
| 2 | 14 | |||||||||
| U19b | 3 | 1 | 2 | 1 | 1 | 10 | ||||
The number of days between the earliest and the latest isolates within the cluster.
An ARDRA pattern that was not recognized by the current identification scheme.
FIG. 1.
Dendrogram and fingerprints of 15 isolates of genomic DNA group 13TU obtained by PFGE, showing two clusters of clinical isolates (SLC, SCL, and WC; SA and CKN) and one cluster of two clinical isolates (FY and SA) with one environmental isolate (E6A), with a Dice coefficient of similarity of ≥70%. First column, isolate identification; second column, date of isolation (year-month-day); third column, source of isolate. E4B, E6, E6A, environmental isolates; TA, tracheal aspirate; CIV, central intravenous catheter; SP, sputum.
DISCUSSION
Acinetobacter spp. are important nosocomial pathogens in our region, with a prevalence of infection higher than reported elsewhere (28, 34, 35). There are many studies in the literature showing that genomic DNA group 2 (A. baumannii), together with genomic DNA groups 1, 3, and 13TU (known as the Acinetobacter calcoaceticus-A. baumannii complex [Acb complex]), is predominantly involved in infection (2, 5, 8, 9, 13, 17, 20, 24, 28, 36, 39). Many of these studies were, however, based on outbreak strains or did not use reliable methods for genomic identification (18, 40). Our study is the first report using modern genomic identification and nomenclature to study clinical isolates from a situation where they are endemic. We did not study the clinical significance of positive samples. Others have shown infection rates of 30 to 55% among patients with positive specimens (13, 39). Ng et al. reported that 72% of Acinetobacter blood cultures were of clinical significance (28). We used patient-based data to compare the distribution of genomic DNA groups according to sites and found a significant difference among the three genomic DNA groups. Genomic DNA group 13TU was significantly associated with specimens from the respiratory tract. Genomic DNA groups 2 (A. baumannii) and 3 appeared to be able to infect, colonize, or contaminate different sites readily. Genomic DNA group 3 was shown to be the most common species among blood culture isolates (7). It was the most common species found on superficial carriage sites among healthy volunteers (7) and, as shown here, was the most common one in the environment, both inside and outside the hospital. For hospitalized patients, genomic DNA group 13TU was the most common species carried in the throat (31% [reference 7 and our unpublished data]) and, as shown here, was also significantly related to specimens from the respiratory tract. Genomic DNA group 2 was the most common species in all other carriage sites sampled from the hospitalized patients—hairlines, fourth toe webs, groins, and noses (7). Colonization of hospitalized patients occurs rapidly, during the first week of stay (13). Our data show that two or more genomic DNA groups may be isolated from the same clinical sites on different occasions in some patients. A prospective clinical study is required to determine whether certain genomic DNA groups are significantly more likely to be associated with infection than others at certain sites. If this were the case, it would be helpful for clinical management to genomically identify acinetobacters from clinical specimens. To this end, a method of identification which is rapid and easy to perform is required. The recent use of monoclonal antibodies to identify group 13TU is a promising development (29).
Heterogeneity among isolates of the same genomic DNA group from the same site was shown to be a common feature in previous and present studies (7). Seven patients had multiple respiratory samples yielding acinetobacters of the same genomic DNA group on three or more occasions. For three of these patients, PFGE showed that different strains were involved. Limitations in the number of colonies picked from the primary culture for identification might have exaggerated the picture of heterogeneity. One or more strains may contaminate, colonize, or infect the sampled site at any one time. The continuous presence of isolates belonging to two or more genomic DNA groups or two or more clones of the same genomic DNA group cannot be excluded. There were other patients whose specimens from wounds or tracheal aspirates consistently yielded isolates showing similar PFGE and/or ERIC patterns. Repeat isolation of the same strain(s) may represent a greater likelihood of infection or colonization than of transient contamination. In outbreaks of infection caused by an epidemic strain, typing is carried out for containment measures. For endemic infections, there may be an urgent need to study virulence markers of strains associated with nosocomial infections. Their identification would allow the judicious use of antibiotics, an important measure in the prevention of such infections in the ICU. To be of practical use, a fast, reliable, and cost-effective method for typing is required.
There appear to be geographical differences in the distribution of genomic DNA groups. In Europe and North America, A. baumannii (genomic DNA group 2) is the most common species found in clinical specimens, and genomic DNA groups 8 and 9 are the most common ones found in carriage studies. Because of the low isolation rates of carriage studies, the natural habitat of A. baumannii remains uncertain. A recent study in London reported that 17% of vegetables (countries of origin not stated) bought from local supermarkets yielded Acinetobacter spp., with genomic DNA groups 2 and 11 being the most common, each with a frequency of 27% (3). It is therefore suggested that vegetables may be a habitat for A. baumannii and may provide a route by which these bacteria are introduced into hospitals. We showed that A. baumannii and genomic DNA group 3—the former in small numbers only—can be readily recovered from all environmental sources: locally grown vegetables, raw food, soil, and the hospital environment. Raw foods may be contaminated by a variety of sources and may serve as vehicles of transmission. We cannot exclude the possibility that some of the vegetable isolates could have resulted from contamination by handlers, although we used only the inside leaves for sampling. It is curious that soil samples taken during the winter from the same locations that were sampled in the summer did not yield any isolates of genomic DNA group 3. Genomic DNA group 13TU appears to have a restricted habitat, being isolated only from human carriage sites (7), clinical specimens, and patients' immediate environments. Resistance to desiccation has been well described for A. baumannii, but information on other species of the Acb complex is scarce (19, 21, 26). Dry vectors could be secondary reservoirs during an outbreak and during sporadic cases (1, 5).
Over the study period for clinical specimens (26 October to 6 December 1998), acinetobacters were isolated from only 3 of the 1,272 patients admitted to the four surgical wards in which environmental sampling was undertaken and from 35 of 140 patients in the ICU. The environmental load in the ICU was significantly higher than in the four surgical wards, although there was no significant difference in the distribution of genomic DNA groups. Risk factors, such as antibiotic therapy, mechanical ventilation, and others, are well described for Acinetobacter carriage and infections among ICU patients (2, 8, 10, 14, 17, 18, 20, 24, 36, 42). The presence of a large number of patients with risk factors and a high volume of clinical activities, together with other factors in the ICU, could have contributed to the heavy load of acinetobacters in the environment. Our results of genomic DNA group identification indicated that genomic DNA group 3 was the most common species, accounting for 51.6% (33 of 64) of acinetobacters settling on surfaces in patients' immediate environments (i.e., environmental contamination). Comparing our results to those obtained by Gerner-Smidt in a Danish ICU in a situation where acinetobacters were endemic (1994 to 1995) (15), assuming all other factors were similar, our settle plates showed a two- to ninefold-higher count. Gerner-Smidt found 61.5 acinetobacter-carrying particles settling on 1 m2 per h, whereas our mean counts ranged from 117.4 to 557.5 m2 per h (4 to 19 colonies on the plate). In another ICU study in the United Kingdom regarding surveillance during non-outbreak situations, air sampling was found to be positive on 4 of 15 occasions (41), whereas we obtained positive samples on all 7 occasions when we sampled the environment. Thus, when compared with ICUs in Europe, our unit appears to have a heavier environmental load of acinetobacters, of which genomic DNA group 3 is the most common.
There were four clusters of possibly related clinical and environmental isolates, indicating that the environment is a reservoir (Table 3). Other clusters (six) contained possibly related isolates from different patients, indicating cross-transmission among patients. Transmission of gram-negative rods among ICU patients in a setting where these rods are endemic was regarded as low, accounting for only 6 to 10% of clinical isolates (6, 11). Yet we identified eight clusters (including two with isolates from 2 patients and one location) of possible cross-transmission among 68 clinical isolates from 29 patients over a period of 6 weeks (Tables 2 and 3). Contrary to nosocomial infections caused by other gram-negative rods, airborne sources have been suggested to be important in outbreaks of Acinetobacter infection (1). Samples from environmental sources often did not yield any acinetobacters, although their elimination when detected has been regarded as an important step toward the control of the outbreak (1, 5, 10, 18, 20, 39). Acinetobacter-carrying particles may be skin scales, droplets from respiratory secretions, or dust (15, 27). The digestive tract has also been shown to be a reservoir (38). It is not certain whether there is a critical level of contamination before the environment can be regarded as a significant reservoir for endemic infections. Given the ready occurrence of acinetobacters in carriage sites, raw food, and air in our region, it is unrealistic to aim for the eradication of the organism from the environment in the ICU. Means to interrupt the transmission among patients may theoretically be cost effective. Our ICU is a modern unit, and staff members practice hand washing and barrier precautions for contact isolation, yet our rates of infection and colonization remain high. It is uncertain whether regular cleaning can reduce the environmental load significantly. Other measures may include the isolation of infected or colonized patients. Rello suggested that standard preventive measures were inadequate to prevent Acinetobacter infections in intubated patients and therefore advocated the conversion of the ICU from open to individual isolation rooms (31). Mulin et al. reported a significant reduction in the bronchopulmonary colonization and infection rates associated with the use of private isolation rooms among ventilated patients in a surgical ICU (25). The role of the environment as a source of acinetobacters in cross-transmission among patients in a situation where acinetobacters are endemic, such as in our ICU, requires clarification before appropriate measures can be devised.
In summary, our findings indicate that genomic DNA groups of the Acb complex commonly isolated from clinical specimens can be readily found in the environment both inside and outside the hospital. In clinical specimens, these genomic DNA groups show a preference for certain sites, and genomic DNA group 3, the most common group in the environment, was as important as A. baumannii. Studies to determine the clinical significance of genomic DNA groups at different sites and virulence markers of clinically important strains are required so as to allow the judicious use of antibiotics in a situation of acinetobacter endemicity. There was evidence of ready cross-transmission of strains among patients and between patients and the environment. The role played by the environment in areas where acinetobacters are endemic should be evaluated in order to devise a cost-effective program for the control of nosocomial infections, for example in ICUs.
ACKNOWLEDGMENT
This study was supported by a grant (4233/97M) from the Research Grants Council, Hong Kong Special Administrative Region, Hong Kong, People's Republic of China.
REFERENCES
- 1.Allen K D, Green H T. Hospital outbreak of multiresistant Acinetobacter anitratus: an airborne mode of spread? J Hosp Infect. 1987;9:110–119. doi: 10.1016/0195-6701(87)90048-x. [DOI] [PubMed] [Google Scholar]
- 2.Bergogne-Bérézin E, Towner K J. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlau J, Aucken H M, Houang E T S, Pitt T L. Isolation of Acinetobacter spp. including A. baumannii from vegetables: implication for hospital-acquired infections. J Hosp Infect. 1999;42:201–204. doi: 10.1053/jhin.1999.0602. [DOI] [PubMed] [Google Scholar]
- 4.Berlau J, Aucken H M, Malnick H, Pitt T L. Distribution of Acinetobacter spp. on skin of healthy humans. Eur J Clin Microbiol Infect Dis. 1999;18:179–183. doi: 10.1007/s100960050254. [DOI] [PubMed] [Google Scholar]
- 5.Catalano M, Quelle L S, Jeric P E, Di Martino A, Maimone S M. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J Hosp Infect. 1999;42:27–35. doi: 10.1053/jhin.1998.0535. [DOI] [PubMed] [Google Scholar]
- 6.Chetchotisakd P, Phelps C L, Hartstein A L. Assessment of bacterial cross-transmission as a cause of infections: patients in intensive care units. Clin Infect Dis. 1994;18:929–937. doi: 10.1093/clinids/18.6.929. [DOI] [PubMed] [Google Scholar]
- 7.Chu Y W, Leung C M, Houang E T S, Ng K C, Leung C B, Leung H Y, Cheng A F B. Skin carriage of acinetobacters in Hong Kong. J Clin Microbiol. 1999;37:2962–2967. doi: 10.1128/jcm.37.9.2962-2967.1999. . (Erratum, 37:4206.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cisneros J M, Reyes M J, Pachón J, Becerril B, Caballero F J, García-Garmendía J L, Ortiz C, Cobacho A R. Bacteremia due to Acinetobacter baumannii: epidemiology, clinical findings, and prognostic features. Clin Infect Dis. 1996;22:1026–1032. doi: 10.1093/clinids/22.6.1026. [DOI] [PubMed] [Google Scholar]
- 9.Corbella N, Pujol M, Ayats J, Sendra M, Ardanuy C, Domínguez M A, Liñares J, Ariza J, Gudiol F. Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii. Clin Infect Dis. 1996;23:329–334. doi: 10.1093/clinids/23.2.329. [DOI] [PubMed] [Google Scholar]
- 10.Crowe M, Towner K J, Humphreys H. Clinical and epidemiological features of an outbreak of Acinetobacter infection in an intensive therapy unit. J Med Microbiol. 1995;43:55–62. doi: 10.1099/00222615-43-1-55. [DOI] [PubMed] [Google Scholar]
- 11.D'agata E, Venkataraman L, de Girolami P, Samore M. Molecular epidemiology of acquisition of ceftazidime-resistant gram-negative bacilli in a nonoutbreak setting. J Clin Microbiol. 1997;35:2602–2605. doi: 10.1128/jcm.35.10.2602-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkshoorn L, van Harsselaar B, Tjernberg I, Bouvet P J M, Vaneechoutte M. Evaluation of amplified ribosomal DNA restriction analysis for identification of Acinetobacter genomic species. Syst Appl Microbiol. 1998;21:33–39. doi: 10.1016/S0723-2020(98)80006-4. [DOI] [PubMed] [Google Scholar]
- 13.Dy M E, Nord J A, LaBombardi V J, Kislak J W. The emergence of resistant strains of Acinetobacter baumannii: clinical and infection control implications. Infect Control Hosp Epidemiol. 1999;20:565–567. doi: 10.1086/501673. [DOI] [PubMed] [Google Scholar]
- 14.Forster D H, Daschner F D. Acinetobacter species as nosocomial pathogens. Eur J Clin Microbiol Infect Dis. 1999;17:73–77. doi: 10.1007/BF01682159. [DOI] [PubMed] [Google Scholar]
- 15.Gerner-Smidt P. Endemic occurrence of Acinetobacter calcoaceticus biovar anitratus in an intensive care unit. J Hosp Infect. 1987;10:265–272. doi: 10.1016/0195-6701(87)90008-9. [DOI] [PubMed] [Google Scholar]
- 16.Go E S, Urban C, Burns J, Kreiswirth B, Eisner W, Mariano N, Mosinka-Snipas K, Rahal J J. Clinical and molecular epidemiology of Acinetobacter infections sensitive only to polymyxin B and sulbactam. Lancet. 1994;344:1329–1332. doi: 10.1016/s0140-6736(94)90694-7. [DOI] [PubMed] [Google Scholar]
- 17.Gomez J, Simarro E, Banos V, Requena L, Ruiz J, Garcia F, Canteras M, Valdes M. Six-year prospective study of risk and prognostic factors in patients with nosocomial sepsis caused by Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 1999;18:358–361. doi: 10.1007/pl00015019. [DOI] [PubMed] [Google Scholar]
- 18.Horrevorts A, Bergman K, Kollee L, Breuker I, Tjernberg I, Dijkshoorn L. Clinical and epidemiological investigations of Acinetobacter genospecies 3 in a neonatal intensive care unit. J Clin Microbiol. 1995;33:1567–1572. doi: 10.1128/jcm.33.6.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houang E T S, Sormunen R T, Lai L, Chan C Y, Leong A S Y. Effect of desiccation on the ultrastructural appearance of Acinetobacter baumannii and Acinetobacter lwoffii. J Clin Pathol. 1998;51:786–788. doi: 10.1136/jcp.51.10.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husni R N, Goldstein L S, Arroliga A C, Hall G S, Fatica C, Stoller J K G, Steven M. Risk factors for an outbreak of multi-drug-resistant Acinetobacter nosocomial pneumonia among intubated patients. Chest. 1999;115:1378–1382. doi: 10.1378/chest.115.5.1378. [DOI] [PubMed] [Google Scholar]
- 21.Jawad A, Seifert H, Snelling A M, Heritage J, Hawkey P M. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998;36:1938–1941. doi: 10.1128/jcm.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juni E. Interspecies transformation of Acinetobacter. Genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann M E, Pitt T L. Pulsed-field gel electrophoresis of bacterial DNA. In: Chart Henrik., editor. Methods in practical laboratory bacteriology. Boca Raton, Fla: CRC Press; 1994. pp. 83–92. [Google Scholar]
- 24.Lortholary O, Fagon J, Hoi A B, Slama M A, Pierre J, Giral P, Rosenzweig R, Gutmann L, Safar M, Acar J. Nosocomial acquisition of multiresistant Acinetobacter baumannii: risk factors and prognosis. Clin Infect Dis. 1995;20:790–796. doi: 10.1093/clinids/20.4.790. [DOI] [PubMed] [Google Scholar]
- 25.Mulin B, Bouget C, Clement C H, Bailly P, Julliot M, Viel J F, Thouverez M, Vielle I, Barale F, Talon D. Association of private isolation rooms with ventilator-associated Acinetobacter baumannii pneumonia in a surgical intensive care unit. Infect Control Hosp Epidemiol. 1997;18:499–503. doi: 10.1086/647655. [DOI] [PubMed] [Google Scholar]
- 26.Musa E K, Desai N, Casewell M K. The survival of Acinetobacter calcoaceticus inoculated on fingertips and on formica. J Hosp Infect. 1990;15:219–227. doi: 10.1016/0195-6701(90)90029-n. [DOI] [PubMed] [Google Scholar]
- 27.Ng K S, Kumarasinghe G, Inglis T J. Dissemination of respiratory secretions during tracheal tube suctioning in an intensive care unit. Ann Acad Med Singapore. 1999;28:178–182. [PubMed] [Google Scholar]
- 28.Ng T K C, Ling J M, Cheng A F B, Norrby S R. A retrospective study of clinical characteristics of Acinetobacter bacteremia. Scand J Infect Dis. 1996;101(Suppl.):26–32. [PubMed] [Google Scholar]
- 29.Pantophlet R, Brade L, Brade H. Use of a murine O-antigen-specific monoclonal antibody to identify Acinetobacter strains of unnamed genomic species 13 Sensu Tjernberg and Ursing. J Clin Microbiol. 1999;37:1693–1698. doi: 10.1128/jcm.37.6.1693-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinn J P. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin Infect Dis. 1998;27(Suppl. 1):S117–S124. doi: 10.1086/514912. [DOI] [PubMed] [Google Scholar]
- 31.Rello J. Acinetobacter baumannii infections in the ICU: customization is the key. Chest. 1999;115:1226–1229. doi: 10.1378/chest.115.5.1226. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz J, Nunez M L, Perez J, Simarro E, Martinez-Campos L, Gomez J. Evolution of resistance among clinical isolates of Acinetobacter over a 6-year period. Eur J Clin Microbiol Infect Dis. 1999;18:292–295. doi: 10.1007/s100960050280. [DOI] [PubMed] [Google Scholar]
- 33.Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, Vaneechoutte M. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol. 1997;35:2819–2825. doi: 10.1128/jcm.35.11.2819-2825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siau H, Yuen K Y, Ho P L, Wong S S, Woo P C. Acinetobacter bacteraemia in Hong Kong: prospective study and review. Clin Infect Dis. 1999;28:26–30. doi: 10.1086/515068. [DOI] [PubMed] [Google Scholar]
- 35.Siau H, Yuen K Y, Wong S S Y, Ho P L, Luk W K. The epidemiology of Acinetobacter infections in Hong Kong. J Med Microbiol. 1996;44:340–347. doi: 10.1099/00222615-44-5-340. [DOI] [PubMed] [Google Scholar]
- 36.Struelens M J, Carler E, Maes N, Serruys E, Quint W G V, van Belkum A. Nosocomial colonization and infection with multiresistant Acinetobacter baumannii: outbreak delineation using DNA macrorestriction analysis and PCR-fingerprinting. J Hosp Infect. 1993;25:15–32. doi: 10.1016/0195-6701(93)90005-k. [DOI] [PubMed] [Google Scholar]
- 37.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timsit J F, Garrait V, Misset B, Goldstein F W, Renaud B, Carlet J. The digestive tract is a major site for Acinetobacter baumannii colonization in intensive care unit patients. J Infect Dis. 1993;168:1336–1337. doi: 10.1093/infdis/168.5.1336. [DOI] [PubMed] [Google Scholar]
- 39.Villari P, Iacuzio L, Vozzella E A, Bosco U. Unusual genetic heterogeneity of Acinetobacter baumannii isolates in a university hospital in Italy. Am J Infect Control. 1999;27:247–253. doi: 10.1053/ic.1999.v27.a96961. [DOI] [PubMed] [Google Scholar]
- 40.Weaver R E, Actis L A. Identification of Acinetobacter species. J Clin Microbiol. 1994;32:1833. doi: 10.1128/jcm.32.7.1833-.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster C A, Crowe M, Humphreys H, Towner K J. Surveillance of an adult intensive care unit for long-term persistence of a multi-resistant strain of Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 1998;17:171–176. doi: 10.1007/BF01691113. [DOI] [PubMed] [Google Scholar]
- 42.Weinstein A A. Epidemiology and control of nosocomial infections in adult intensive care units. Am J Med. 1991;91:S179–S184. doi: 10.1016/0002-9343(91)90366-6. [DOI] [PubMed] [Google Scholar]
- 43.Wisplinghoff H, Perbix W, Seifert H. Risk factors for nosocomial bloodstream infections due to Acinetobacter baumannii: a case-control study of adult burn patients. Clin Infect Dis. 1999;28:59–66. doi: 10.1086/515067. [DOI] [PubMed] [Google Scholar]