Abstract
The accuracy of antimicrobial susceptibility data submitted by microbiology laboratories to national and international surveillance systems has been debated for a number of years. To assess the accuracy of data submitted to the World Health Organization by users of the WHONET software, the Centers for Disease Control and Prevention distributed six bacterial isolates representing key antimicrobial-resistance phenotypes to approximately 130 laboratories, all but one of which were outside of the United States, for antimicrobial susceptibility testing as part of the World Health Organization's External Quality Assurance System for Antimicrobial Susceptibility Testing. Each laboratory also was asked to submit 10 consecutive quality control values for several key organism-drug combinations. Most laboratories were able to detect methicillin (oxacillin) resistance in Staphylococcus aureus, high-level vancomycin resistance in Enterococcus faecium, and resistance to extended-spectrum cephalosporins in Klebsiella pneumoniae. Many laboratories, particularly those using disk diffusion tests, had difficulty in recognizing reduced susceptibility to penicillin in an isolate of Streptococcus pneumoniae. The most difficult phenotype for laboratories to detect was reduced susceptibility to vancomycin in an isolate of Staphylococcus epidermidis. The proficiency testing challenge also included a request for biochemical identification of a gram-negative bacillus, which most laboratories recognized as Enterobacter cloacae. Although only a small subset of laboratories have submitted their quality control data, it is clear that many of these laboratories generate disk diffusion results for oxacillin when testing S. aureus ATCC 25923 and S. pneumoniae ATCC 49619 that are outside of the acceptable quality control range. The narrow quality control range for vancomycin also proved to be a challenge for many of the laboratories submitting data; approximately 27% of results were out of range. Thus, it is important to establish the proficiency of laboratories submitting data to surveillance systems in which the organisms are tested locally, particularly for penicillin resistance in pneumococci and glycopeptide resistance in staphylococci.
Resistance to a variety of antimicrobial agents is emerging in bacterial pathogens throughout the world (8, 21, 50). Increases in the prevalence of penicillin resistance in Streptococcus pneumoniae (13, 36, 39, 47), methicillin resistance in Staphylococcus aureus (1, 3, 38), vancomycin resistance in enterococci (5, 9, 16, 28), extended-spectrum β-lactamase-production in enteric gram-negative bacilli (2, 18, 37), and fluoroquinolone resistance in Neisseria gonorrhoeae (17) are just a few examples of the rising problem of resistance documented by both national and international surveillance systems in the past few years. The antimicrobial susceptibility testing data collected by the various surveillance systems are generated in several different fashions. In some systems, bacterial isolates are sent to a central laboratory for testing using a standardized reference method (12, 43), and the data are compiled and maintained by the central laboratory. In other systems, the Etest method is used by participating laboratories to test isolates locally, and the data are forwarded to a central database (10, 35). A third approach to surveillance is to collect the antimicrobial susceptibility testing data directly from microbiology laboratories by either a direct computer link (40) or through diskettes sent to a central laboratory in which standardized computer database software such as the WHONET is used (49). These systems usually employ a series of data checks and quality control filters before the data are entered into the larger database. The final surveillance system consists of the microbiology laboratories throughout the world that perform routine antimicrobial susceptibility testing of bacterial isolates but that do not belong to one of the above systems (40). This vast network, which generates tens of thousands of susceptibility results daily, is a significant source of resistance data. It is often through this system of astute clinical microbiologists that new resistant organisms are detected (30).
The proliferation of surveillance systems internationally has raised the issue of the accuracy of the data collected, particularly for systems in which testing occurs outside of a central site. The need to obtain and review the quality control data from noncentralized laboratories or to assess the competence of laboratories both before data collection begins and during testing has resulted in the call for wider proficiency testing of laboratories.
In 1995, the World Health Organization (WHO) initiated a program of quality control and proficiency testing focused on antimicrobial susceptibility testing to assist laboratories in both developed and developing countries to assess the accuracy of their antimicrobial susceptibility testing data. The goals of the program were (i) to assist laboratories, particularly those that were WHONET software users, in evaluating the quality of their antimicrobial susceptibility testing data, (ii) to validate the susceptibility testing data submitted to WHO and the WHO Collaborating Center for Surveillance of Antimicrobial Resistance in Boston, (iii) to provide guidance for laboratories wishing to develop quality assurance programs, and (iv) to provide laboratories with organisms that manifest novel antimicrobial resistance so that they could confirm that their susceptibility testing methods were capable of detecting the emerging resistance patterns. The program, which began with 17 laboratories, all of which were located outside of the United States, has evolved into the WHO External Quality Assurance System for Antimicrobial Susceptibility Testing (WHO-EQAS), which now includes approximately 130 laboratories in 42 countries (including one laboratory in the United States). Several countries have set up national quality assurance systems further distributing the proficiency testing strains to an additional 200 laboratories. Herein, we describe the results reported by approximately 130 laboratories for the first six proficiency testing challenges.
MATERIALS AND METHODS
Organism challenges.
Three sets of two organisms each were sent to approximately 130 laboratories in 42 countries over a 3-year period (1996 through 1999). Most of the laboratories were WHONET software users or laboratories that had participated in WHO laboratory training programs. Some laboratories participated in only one or two of the challenges, and additional laboratories were continually added to the list of participants throughout the study period. New regulations disallowing the importation of infectious substances into various countries also limited the participation of several laboratories. Thus, the number of results and the antimicrobial agents tested for each organism in this report vary. The list of countries represented among the participating laboratories is shown in Table 1 by WHO region. The organisms were tested multiple times at the Centers for Disease Control and Prevention (CDC) by broth microdilution and disk diffusion using National Committee for Clinical Laboratory Standards (NCCLS) methods (32–34) to establish the reference MIC and disk diffusion values. In some cases (e.g., for methicillin-resistant S. aureus, vancomycin-resistant Enterococcus faecium, and erythromycin-resistant S. pneumoniae), PCR assays for mecA, vanA, and mefE, respectively, were used to confirm the resistance mechanisms (54). A data collection sheet was provided with each organism, requesting information on the antimicrobial susceptibility testing method used (MIC or disk diffusion), source of media and reagents, interpretive criteria used (e.g., NCCLS [32–34], the Comité de l'Antibiogramme de la Société de Française Microbiologie [CA-SFM] [22], or the British Society for Antimicrobial Chemotherapy [BSAC] [57]), and disk potency. The data sheets included a suggested set of antimicrobial agents to test and provided space to fill in additional drugs that were tested in the laboratory. Laboratories were asked to provide both quantitative results (MICs or zone diameters) and the qualitative interpretations (i.e., susceptible, intermediate, or resistant [S, I, or R, respectively]) for each antimicrobial agent tested. Data sheets were returned to CDC for analysis. The data were entered into a SAS data set (SAS, Cary, N.C.). Because NCCLS, CA-SFM, and BSAC often use conflicting interpretive criteria, only laboratories using NCCLS methods and interpretive criteria were considered for this manuscript, to make comparison of data feasible. If the laboratory provided an incorrect interpretation (i.e., an interpretive error) for an MIC or disk diffusion result, the correct interpretation (S, I, or R) was entered into Tables 3, 5, 6, 7, 8, or 9 (see below), and the error was noted in a footnote to these tables. The interpretive errors from laboratories using disk diffusion are included in Table 10 below, and interpretive errors from laboratories using MIC methods are listed in Table 11 below.
TABLE 1.
Number of participating laboratories by WHO region and country
| Region/country | No. of laboratoriesa |
|---|---|
| African Region | |
| Algeria | 1 |
| Kenya | 4 |
| South Africa | 1 |
| Sudan | 1 |
| Uganda | 1 |
| Eastern Mediterranean Region | |
| Jordan | 1 |
| Kuwait | 2 |
| Lebanon | 1 |
| Saudi Arabia | 15 |
| European Region | |
| Bulgaria | 2 |
| Croatia | 15 |
| Czech Republic | 2 |
| Finland | 19 |
| Greece | 1 |
| Hungary | 1 |
| Italy | 1 |
| Poland | 1 |
| Russian Federation | 1 |
| Slovakia | 1 |
| Turkey | 1 |
| Region of the Americas | |
| Argentina | 24 |
| Colombia | 1 |
| Guatemala | 1 |
| United States | 1 |
| Uruguay | 3 |
| Venezuela | 3 |
| Southeast Asian Region | |
| Hong Kong | 1 |
| India | 2 |
| Indonesia | 1 |
| Myanmar | 1 |
| Sri Lanka | 1 |
| Thailand | 2 |
| Western Pacific Region | |
| China | 11 |
| Japan | 59 |
| Korea | 1 |
| Malaysia | 1 |
| Mongolia | 1 |
| Philippines | 1 |
| Viet Nam | 2 |
Because some laboratories joined the system later than others and some completed testing of only the first four organisms, the number of participating laboratories listed exceeds 130.
TABLE 3.
Results of testing K. pneumoniae (WHO-1)
| Antimicrobial agent (no. of labs that tested)a | Reference result (interpretive category)
|
No. of labs reporting disk diffusion results (range of zone diameters in mm)
|
No. of labs reporting MIC results (range of MICs in μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MIC in μg/ml | Disk zone diameter in mm | S | I | R | S | I | R | |
| Aztreonam (9) | 32 (R) | 16 (I) | 3 (28–32) | 1 (20)b | 0 | 0 | 1 (16) | 4 (>16–>32) |
| Cefazolin (74) | >32 (R) | 7 (R) | 0 | 0 | 24 (6–14) | 0 | 0 | 50 (>16–>128) |
| Cefotaxime (92) | >64 (R) | 12 (R) | 6 (28–34) | 18 (15–22)bc | 32 (6–14) | 6 (4–8) | 8 (16–32)d | 22 (>32) |
| Ceftriaxone (16) | >64 (R) | 12 (R) | 3 (27–32) | 0 | 13 (6–13)e | 0 | 0 | 0 |
| Ceftazidime (105) | 128 (R) | 11 (R) | 9 (18–30)d | 4 (15–17)f | 57 (6–14) | 0 | 4 (16)d | 31 (>16–>32) |
| Cephalothin (45) | 6 (R) | 5 (18–22) | 2 (15–16) | 35 (6–14) | 0 | 0 | 3 (32–>256) | |
Labs returning results, 130.
One incorrectly called susceptible.
Seven incorrectly called resistant.
One incorrectly called resistant.
One incorrectly called intermediate.
Two incorrectly called resistant.
TABLE 5.
Results of testing for S. aureus WHO-2
| Antimicrobial agent (no. of labs that tested antimicrobial agent)a | Reference result (category)
|
No. of labs reporting disk diffusion results (range of zone diameters in mm)
|
No. of labs reporting MIC results (range of MICs in μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MIC in μg/ml | Disk zone diameters in mm | S | I | R | S | I | R | |
| Erythromycin (121) | >8 (R) | 6 (R) | 1 (25) | 3 (17–22)bc | 68 (6–13) | 0 | 0 | 49 (>4–>32) |
| Oxacillin (117) | 32 (R) | 6 (R) | 9 (13–23) | 1 (12) | 57 (6–10) | 0 | 0 | 50 (>4–>64) |
| Penicillin (99) | 128 (R) | 7 (R) | 1 (32) | 0 | 64 (6–28) | 0 | 0 | 34 (>8–>16)b |
| Trimethoprim-sulfamethoxazole (106) | ≤0.25 (S) | 21 (S) | 68 (16–35)de | 0 | 0 | 37 (0.12–<2) | 0 | 1 (5)b |
Labs returning results, 127.
One incorrectly called susceptible.
Two incorrectly called resistant.
One incorrectly called intermediate.
One incorrectly called resistant.
TABLE 6.
Results of testing for E. faecium WHO-3
| Antimicrobial agent (no. of labs that tested antimicrobial agents)a | Reference result (category)
|
No. of labs reporting disk diffusion results (range of zone diameters in mm)
|
No. of labs reporting MIC results (range of MICs in μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MIC in μg/ml | Disk zone diameter in mm | S | I | R | S | I | R | |
| Ampicillin (95) | 4 (S) | 19 (S) | 36 (17–36)b | 0 | 19 (6–16)c | 39 (1–8)d | 0 | 1 (>4) |
| Penicillin (71) | 32 (R) | 6 (R) | 4 (15–20)e | 0 | 32 (6–13) | 10 (8) | 0 | 25 (>1–>16) |
| Teicoplanin (26) | 256 (R) | 6 (R) | 1 (17) | 3 (11–12)e | 19 (6–10) | 1 (0.25) | 0 | 2 (>16–32) |
| Vancomycin (117) | 512 (R) | 6 (R) | 3 (21–30) | 0 | 63 (6–12) | 0 | 0 | 51 (>8–64)d |
| β-Lactamase (39) | Negative | 36 negative, 3 positive | ||||||
Labs returning results, 122.
Three incorrectly called intermediate.
One incorrectly called susceptible.
One incorrectly called intermediate.
One incorrectly called resistant.
TABLE 7.
Results of testing for S. pneumoniae WHO-4
| Antimicrobial agent (no. of labs that tested antimicrobial agent)a | Reference result (category)
|
No. of labs reporting disk diffusion results (range of zone diameters in mm)
|
No. of labs reporting MIC results (range of MICs in μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MIC in μg/ml | Disk zone diameter in mm | S | I | R | S | I | R | |
| Clindamycin (115) | 0.12 (S) | 22 (S) | 94 (19–40) | 1 (18)b | 0 | 19 (≤0.12–≤0.25) | 1 (>8) | |
| Erythromycin (128) | 4 (R) | 10 (R) | 9 (21–34)c | 6 (16–19) | 94 (6–15)bd | 4 (≤0.5) | 0 | 15 (1–16)e |
| Penicillin (76) | 0.06 (S) | naf | 61 (0.007–0.06) | 14 (0.12–1.0)g | 1 (≥2.0) | |||
| Oxacillin screen (89) | naf | 15 | 32 (20–28)h | 57 (6–19)i | ||||
Labs returning results, 128.
One incorrectly called susceptible.
One incorrectly called intermediate.
Two incorrectly called intermediate.
Three incorrectly called intermediate.
na, not applicable.
Three incorrectly called susceptible.
Results are for zone diameters of ≥20 mm.
Results are for zone diameters of ≤19 mm. A result of ≤19 mm is not necessarily resistant; NCCLS recommends that a penicillin MIC test be performed.
TABLE 8.
Results of testing for E. cloacae WHO-5
| Antimicrobial agent (no. of labs that tested antimicrobial agent)a | Reference result (category)
|
No. of labs reporting disk diffusion results (range of zone diameters in mm)
|
No. of labs reporting MIC results (range of MICs in μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MIC in μg/ml | Disk zone diameter in mm | S | I | R | S | I | R | |
| Ampicillin (124) | >64 (R) | 6 (R) | 1 (22) | 0 | 100 (5–8) | 1 (4) | 0 | 22 (>16–>128) |
| Cefotaxime (81) | 64 (R) | 8 (R) | 0 | 1 (17) | 64 (6–13) | 0 | 2 (32) | 14 (>32–128) |
| Ceftazidime (111) | >128 (R) | 6 (R) | 0 | 0 | 98 (6–13) | 0 | 0 | 13 (>16–>128) |
| Ceftriaxone (45) | >64 (R) | 8 (R) | 1 (22) | 0 | 42 (6–12) | 0 | 0 | 2 (64) |
| Ciprofloxacin (109) | ≤0.06 (S) | 30 (S) | 100 (22–37)b | 0 | 1 (6) | 8 (0.15–<1) | 0 | 0 |
| Gentamicin (115) | ≤0.25 (S) | 20 (S) | 93 (15–28)bc | 0 | 0 | 22 (0.12–<1) | 0 | 0 |
| Imipenem (121) | 1–2 (S) | 22 (S) | 96 (17–29)d | 0 | 1 (6) | 24 (0.5–<4)e | 0 | 0 |
| Tobramycin (77) | 0.5 (S) | 18 (S) | 66 (16–28) | 1 (13) | 1 (11) | 9 (0.12–<4) | 0 | 0 |
| Trimethoprim-sulfamethoxazole (118) | ≤0.12 (S) | 24 (S) | 97 (16–32)bc | 0 | 2 (6) | 19 (0.001–<4) | 0 | 0 |
| Identification (126) | E. cloacae | 114 correct, 4 Enterobacter species, 8 incorrect | ||||||
Labs returning results, 130.
One incorrectly called intermediate.
Two incorrectly called resistant.
Two incorrectly called intermediate.
One incorrectly called resistant.
TABLE 9.
Results of testing for S. epidermidis WHO-6
| Antimicrobial agent (no. of labs that tested antimicrobial agent)a | Reference result (category)
|
No. of labs reporting disk diffusion results (range of zone diameters in mm)
|
No. of labs reporting MIC results (range of MICs in μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MIC in μg/ml | Disk zone diameter in mm | S | I | R | S | I | R | |
| Ciprofloxacin (87) | >8 (R) | 6 (R) | 2 (26–30) | 1 (20)b | 82 (6–10) | 1 (1) | 0 | 1 (64) |
| Clindamycin (117) | 0.25 (S) | 26 (S) | 90 (22–42) | 2 (19)c | 0 | 25 (<0.12–0.5) | 0 | 0 |
| Erythromycin (124) | 0.5 (S) | 28 (S) | 92 (24–42) | 7 (19–22)d | 0 | 22 (≤0.2–0.5) | 3 (1) | 0 |
| Gentamicin (116) | 16 (R) | 10 (R) | 9 (15–30)b | 4 (13–14)e | 77 (6–12)f | 0 | 0 | 26 (>8–32) |
| Oxacillin (127) | >16 (R) | 6 (R) | 14 (18–30) | 0 | 86 (6–16)gh | 0 | 0 | 27 (>4–>256) |
| Penicillin (121) | >2 (R) | 10 (R) | 0 | 0 | 93 (6–25) | 0 | 0 | 28 (8–64) |
| Rifampin (90) | ≤0.12 (S) | 40 (S) | 77 (23–48) | 0 | 3 (6–10) | 10 (<0.008–<1) | ||
| Teicoplanin (49) | 16 (I) | 14 (S) | 30 (14–24)f | 10 (11–13)e | 1 (6) | 1 (4) | 3 (8–16) | 4 (>16–32) |
| Trimethoprim-sulfamethoxazole (121) | 76/4 (R) | 6 (R) | 2 (30–32) | 5 (12–15)b | 88 (6–10) | 1 (2) | 0 | 25 (>2–160) |
| Vancomycin (127) | 8 (I) | 18 (S) | 92 (15–34) | 0 | 4 (6–14)c | 5 (1–4) | 26 (8–16) | 0 |
Labs returning results, 130.
One incorrectly called resistant.
One incorrectly called susceptible.
Three incorrectly called susceptible.
Two incorrectly called resistant.
One incorrectly called intermediate.
Seven incorrectly called susceptible.
Three incorrectly called intermediate.
TABLE 10.
Interpretation errors by disk diffusion users
| Antimicrobial agent | No. and category of errors for bacterial strain:
|
|||||
|---|---|---|---|---|---|---|
| K. pneumoniae WHO-1 | S. aureus WHO-2 | E. faecium WHO-3 | S. pneumoniae WHO-4 | E. cloacae WHO-5 | S. epidermidis WHO-6 | |
| Ampicillin | 1 very major | |||||
| 3 minor | ||||||
| Aztreonam | 1 minor | |||||
| Cefotaxime | 8 minor | |||||
| Ceftazidime | 1 major | |||||
| 2 minor | ||||||
| Ceftriaxone | 1 minor | |||||
| Ciprofloxacin | 1 minor | 1 minor | ||||
| Clindamycin | 1 minor | 1 minor | ||||
| Erythromycin | 3 minor | 1 very major | 3 minor | |||
| 3 minor | ||||||
| Gentamicin | 2 major | 1 major | ||||
| 1 minor | 3 minor | |||||
| Imipenem | 2 minor | |||||
| Oxacillin | 7 major | |||||
| 3 minor | ||||||
| Penicillin | 1 major | |||||
| Teicoplanin | 1 minor | 3 minor | ||||
| Trimethoprim-sulfamethoxazole | 1 major | 1 major | 1 minor | |||
| 1 minor | 1 minor | |||||
| Vancomycin | 1 very major | |||||
TABLE 11.
Interpretation errors by MIC users
| Antimicrobial agent | No. and category of errors for bacterial strain:
|
|||||
|---|---|---|---|---|---|---|
| K. pneumoniae WHO-1 | S. aureus WHO-2 | E. faecium WHO-3 | S. pneumoniae WHO-4 | E. cloacae WHO-5 | S. epidermidis WHO-6 | |
| Ampicillin | 1 minor | |||||
| Aztreonam | ||||||
| Cefotaxime | 1 minor | |||||
| Ceftazidime | 1 minor | |||||
| Ceftriaxone | ||||||
| Ciprofloxacin | ||||||
| Clindamycin | ||||||
| Erythromycin | 3 minor | |||||
| Gentamicin | ||||||
| Imipenem | 1 major | |||||
| Oxacillin | ||||||
| Penicillin | 1 very major | 3 minor | ||||
| Teicoplanin | ||||||
| Trimethoprim-sulfamethoxazole | 1 very major | |||||
| Vancomycin | 1 minor | |||||
RESULTS
Organism challenges.
The antimicrobial susceptibility testing methods used by participating laboratories for each of the challenge organisms are shown in Table 2.
TABLE 2.
Antimicrobial susceptibility testing methods used by participating laboratories by organisma
| Strain | No. of results | No. of participating laboratories that used method
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Disk diffusion | Broth microdilutiona | MIC 2000a | MicroScana | Sceptora | Sensititrea | Viteka | MIC method not identified | ||
| WHO-1 | 130 | 83 | 2 | 7 | 23 | 9 | 2 | 3 | 1 |
| WHO-2 | 127 | 75 | 3 | 7 | 25 | 9 | 2 | 5 | 1 |
| WHO-3 | 122 | 72 | 4 | 7 | 23 | 8 | 2 | 5 | 1 |
| WHO-4 | 128b | 109 | 2 | 4 | 1 | 5 | 1 | 0 | 6 |
| WHO-5 | 130 | 106 | 2 | 3 | 12 | 1 | 2 | 3 | 1 |
| WHO-6 | 130 | 105 | 2 | 0 | 19 | 1 | 1 | 2 | 0 |
Manufacturers: Broth microdilution, no manufacturer or method specified; MIC 2000, Eiken Ltd., Tokyo, Japan; MicroScan, Dade Behring, West Sacramento, Calif.; Sceptor, Becton Dickinson Microbiology Systems, Cockeysville, Md.; Sensititre, Trek Diagnostics, Westlake, Ohio; Vitek, bioMérieux, Hazelwood, Mo.
Forty laboratories performed the oxacillin screen test and determined the penicillin MIC of the organism using the Etest method.
Challenge WHO-1.
The first challenge strain (WHO-1) was an isolate of Klebsiella pneumoniae that produced a TEM-3 extended-spectrum β-lactamase (ESBL) (48) (Table 3). One hundred and thirty laboratories reported results; 63.9% (83 of 130) used a disk diffusion method, and 36.1% (47 of 130) used an MIC method (Table 2). The extended-spectrum cephalosporins and monobactams tested most frequently by participating laboratories are shown in Table 4. Six laboratories (four disk diffusion users and two MIC users) failed to test any extended-spectrum cephalosporins or aztreonam. Overall, 5.4% (7 of 130) laboratories (all disk diffusion users) reported WHO-1 to be susceptible to all extended-spectrum cephalosporins. For individual drugs, 91.4% of 105 laboratories reported an intermediate or resistant result for ceftazidime, 86.9% of 92 laboratories reported an intermediate or resistant result for cefotaxime, and 81.2% of 16 reported an intermediate or resistant result for ceftriaxone. On the other hand, 11.1% (5 of 45) of laboratories testing cephalothin, a narrow-spectrum cephalosporin, reported the organism incorrectly as susceptible to this agent. Of the nine laboratories reporting ceftazidime-susceptible results, six also reported susceptible results for cefotaxime or ceftriaxone. Only 2 of the 130 laboratories specifically reported the isolate as an “ESBL-producing strain.” None changed the results for extended-spectrum cephalosporins or aztreonam from susceptible to resistant based on detection of resistance to another extended-spectrum cephalosporin, as suggested by NCCLS (34). Failure to test extended-spectrum cephalosporins (n = 6) and reporting of susceptible results for all extended-spectrum cephalosporins (n = 7) (this includes the five laboratories that reported susceptible results for cephalothin) were considered unacceptable results.
TABLE 4.
Extended-spectrum cephalosporins and monobactams tested against WHO-1 by 130 laboratories
| Antimicrobial agent(s) tested | No. of laboratories | No. of laboratories reporting all extended-spectrum cephalosporins as susceptible |
|---|---|---|
| Ceftazidime, cefotaxime | 68 | 4 |
| Cefotaxime only | 18 | |
| Ceftazidime, ceftriaxone | 14 | 1 |
| Ceftazidime only | 14 | |
| No extended-spectrum cephalosporins nor monobactams | 6 | |
| Aztreonam, ceftazidime | 4 | |
| Aztreonam, ceftazidime, cefotaxime | 4 | |
| Aztreonam, cefotaxime | 1 | |
| Ceftazidime, cefotaxime, ceftriaxone | 1 | 1 |
| Ceftriaxone only | 1 | 1 |
Challenge WHO-2.
The second challenge strain (WHO-2) was a methicillin (oxacillin)-resistant, mecA-positive S. aureus (MRSA) (Table 5). One hundred twenty-seven laboratories reported results; 59.1% (75 of 127) reported disk diffusion results, and 40.9% reported MIC results. The MRSA strain demonstrated high-level resistance to oxacillin, erythromycin, clindamycin, and tetracycline. Approximately 91% of laboratories recognized the strain as an MRSA. Only one laboratory reported both penicillin and oxacillin as susceptible results, but eight other laboratories reported the strain as oxacillin or methicillin susceptible. These were considered unacceptable results. Three additional challenge strains of S. aureus were sent to the nine laboratories that reported the strain as oxacillin susceptible. Of the five laboratories returning results, four were successful in differentiating the two MRSA strains from the susceptible S. aureus strain.
Challenge WHO-3.
The third challenge (WHO-3) was a vancomycin-resistant strain of E. faecium (VRE; previously referred to as NJ-1 [55]) that was also borderline resistant to penicillin but (using NCCLS breakpoints) susceptible to ampicillin. Of the 122 laboratories that tested this organism, 59.0% used disk diffusion and 41.0% used an MIC method. Only 2.6% (3 of 117) of laboratories testing vancomycin failed to recognize this strain, which contained the vanA resistance gene, as highly resistant to vancomycin (Table 6). While 80.3% of laboratories correctly reported the strain as penicillin resistant, 21.0% incorrectly reported it as ampicillin resistant using NCCLS interpretive criteria. Three of 39 (7.7%) laboratories reported the isolate incorrectly as beta-lactamase positive. As the beta-lactam results were on the borderline and represented an unusual phenotype, for this challenge, reporting vancomycin-susceptible results (n = 3) was considered unacceptable performance.
Challenge WHO-4.
The fourth challenge (WHO-4) was an erythromycin-resistant (mefE-positive) strain of S. pneumoniae with reduced susceptibility to penicillin. The penicillin MIC varied from 0.06 μg/ml (susceptible) to 0.12 μg/ml (intermediate), but it invariably produced a zone diameter of 14 to 17 mm around a 1-μg oxacillin disk, which, according to NCCLS guidelines, indicates that an MIC test should be performed to determine if the isolate is susceptible or resistant to penicillin. Of the 89 laboratories that performed an oxacillin screen test, 35.9% reported a value of ≥20 mm, which indicates susceptibility to β-lactam agents (Table 7). Only 59.4% (76 of 128) of laboratories reported an MIC result for penicillin. Although NCCLS does not recommend penicillin or cephalosporin disk diffusion tests for pneumococci, two laboratories reported results for a 1-U penicillin disk, and six reported results for a 10-U penicillin disk. In addition, 21 laboratories reported values for 30-μg cefotaxime disks, and 15 reported values for 30-μg ceftriaxone disks. Thirteen laboratories (10.2%) incorrectly reported the organism as susceptible to erythromycin, and 2 reported it incorrectly as intermediate or resistant to clindamycin. Reporting oxacillin zone diameters of ≥20 mm, a penicillin MIC of ≥2 μg/ml or reporting erythromycin-susceptible results was considered unacceptable performance for this organism (n = 40).
Challenge WHO-5.
The fifth challenge (WHO-5) was an Enterobacter cloacae strain that was sent for identification and antimicrobial susceptibility testing. It was resistant to all penicillins, cephamycins, and cephalosporins, including extended-spectrum cephalosporins, but was susceptible to the aminoglycosides, trimethoprim-sulfamethoxazole, ciprofloxacin, imipenem and meropenem (4, 42). Few laboratories had problems in recognizing the β-lactam resistance in this strain (Table 8), although 6.1% of laboratories (8 of 130) failed to identify this organism as an Enterobacter species. Susceptible results for ampicillin or extended-spectrum cephalosporins were considered to be unacceptable performance (n = 3). Because the focus of the survey was on susceptibility testing results and not identification, misidentifications were not considered as unacceptable performance.
Challenge WHO-6.
The sixth challenge (WHO-6) was a glycopeptide-intermediate strain of Staphylococcus epidermidis (20). The reference MIC of vancomycin for this organism was 8 μg/ml, and the MIC of teicoplanin was 16 μg/ml. Of the 130 laboratories testing this organism, 105 (80.8%) used disk diffusion, and 19.2% used an MIC method. Ninety-seven (74.6%) of 130 laboratories testing vancomycin reported this isolate as vancomycin susceptible (Table 9), which was considered unacceptable performance. This included a laboratory that reported a teicoplanin zone diameter of 6 mm, which also would be considered incorrect (since it is an unusually small zone diameter). Four laboratories that tested vancomycin by disk diffusion reported zone diameters of 6 to 14 mm, which are unusually small for this strain (52). The two reports of vancomycin zone diameters of 6 mm were also considered unacceptable performance.
Overall performance and interpretation and reporting errors.
Of the 74 laboratories that sent results on at least five of the six challenge organisms, 17 had fully acceptable results, 33 reported unacceptable results for a single challenge, 20 reported unacceptable results for 2 challenges, and 4 reported unacceptable results for 3 challenges. None reported unacceptable results for four or more of the challenges.
In addition to the unacceptable results reported above, which primarily represent very major testing errors (where the reference result was reported as resistant, and the proficiency test result was reported as susceptible), 61 of the S, I, or R disk diffusion categorical interpretations (Table 10) and 13 of the MIC categorical interpretations (Table 11) provided by the participating laboratories for the six challenge organisms were incorrect. These represent 5 very major errors, 15 major errors, and 54 minor errors. The majority of errors were reported for the S. epidermidis isolate (n = 24). Beta-lactam testing of the K. pneumoniae isolate also resulted in a large number of reporting errors. Some of the errors with oxacillin testing of the S. epidermidis isolate probably reflect recent changes in the NCCLS breakpoint for this organism-drug combination (34).
Quality control data.
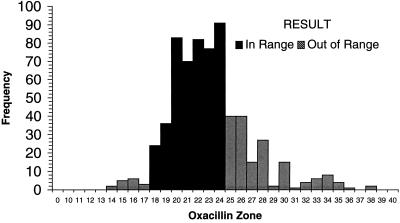
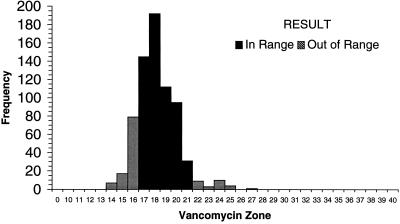
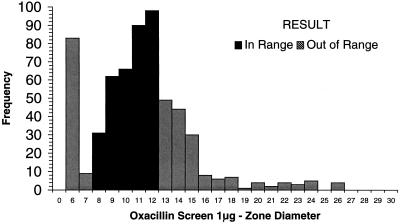
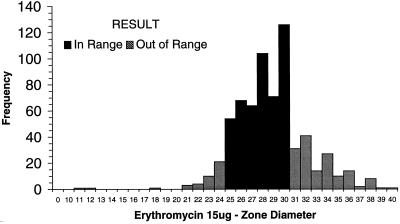
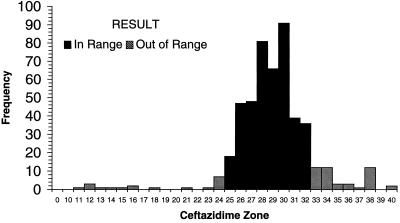
Each of the laboratories participating in the WHO-EQAS system was asked to submit 10 consecutive quality control results for Escherichia coli ATCC 25922, S. aureus ATCC 25923, and S. pneumoniae ATCC 49619 for analysis. Data were available from approximately 35 laboratories using NCCLS quality control ranges. Figure 1 demonstrates that many of the quality control results from oxacillin disk diffusion tests for S. aureus ATCC 25923 fell outside the specified control range. Of the 654 data points submitted by 35 laboratories, 16 values (2.5%) (from 3 different laboratories) were below the range and 162 values (24.8%) (from 22 laboratories) were above the range. By comparison, 130 of 705 disk diffusion results (18.4%) reported by 38 laboratories were out of range when testing vancomycin against the S. aureus control (Fig. 2). Twenty-eight laboratories reported values for vancomycin that were below the NCCLS quality control range, while 11 laboratories reported values beyond the upper end of the range. For oxacillin testing of S. pneumoniae ATCC 49619, 30 laboratories reported 167 of 606 of their results (27.6%) above the designated range, and 15 laboratories reported 92 results (15.2%) below the range (Fig. 3). For erythromycin testing of S. pneumoniae ATCC 49619, 41 of 677 values (6.1%) from 16 laboratories were below the range, and 149 values (22.0%) were above the range (Fig. 4). For E. coli ATCC 25922, ceftazidime results were more often in control. Of 500 reported values, only 19 (3.8%) were below the designated range, and 45 (9.0%) were above the range (Fig. 5).
FIG. 1.
Histogram showing quality control results for disk diffusion testing of S. aureus ATCC 25923 against oxacillin. The NCCLS quality control range against which results were compared was 18 to 24 mm. Data are all results received as of 30 August 1999 and tested by NCCLS methods.
FIG. 2.
Histogram showing quality control results for disk diffusion testing of S. aureus ATCC 25923 against vancomycin. The NCCLS quality control range against which results were compared was 17 to 21 mm. Data are all results received as of 25 August 1999 and tested by NCCLS methods.
FIG. 3.
Histogram showing quality control results for disk diffusion testing of S. pneumoniae ATCC 49619 against oxacillin. The NCCLS quality control range against which results were compared was 8 to 12 mm. Data are all results received as of 24 August 1999 and tested by NCCLS methods.
FIG. 4.
Histogram showing quality control results for disk diffusion testing of S. pneumoniae ATCC 49619 against erythromycin. The NCCLS quality control range against which results were compared was 25 to 30 mm. Data are all results received as of 24 August 1999 and tested by NCCLS methods.
FIG. 5.
Histogram showing quality control results for disk diffusion testing of E. coli ATCC 25922 against ceftazidime. The NCCLS quality control range against which results were compared was 25 to 32 mm. Data are all results received as of 30 August 1999 and tested by NCCLS methods.
DISCUSSION
The WHO-EQAS was designed to enhance the ability of laboratories to detect organisms with emerging antimicrobial resistance patterns and to ensure that the laboratories' reporting strategies for antimicrobial resistance were accurate. Overall, approximately 20% of laboratories reported fully acceptable results, while the number reporting unacceptable results for three or more of the challenges was low (<5%). Most laboratories had problems with the S. epidermidis and S. pneumoniae isolates (see below).
Because of ongoing changes in the interpretive criteria for antimicrobial susceptibility testing (e.g., NCCLS, BSAC, or CA-SFM), it is possible to perform a test correctly but report inaccurate results, particularly if old guidelines or criteria are used. There were 74 instances in this study in which the results of the susceptibility tests were interpreted incorrectly, which, in a clinical setting, would have resulted in inaccurate information being transmitted to the patient's chart. The goal of quality assurance is to detect and correct problems such as these in a continuous fashion to improve the accuracy of testing, record keeping, and reporting (41, 44). The WHO-EQAS proficiency testing program was designed specifically to help laboratories determine whether all aspects of their current antimicrobial susceptibility testing methods are accurate. To this end, feedback letters detailing the reporting errors were sent to each participating laboratory as part of the quality assurance aspect of the study.
For the proficiency testing portion of the WHO-EQAS, it was gratifying to observe that most laboratories were able to detect methicillin resistance in S. aureus and high-level vancomycin resistance in E. faecium. Only a few laboratories had problems detecting resistance to the extended-spectrum cephalosporins mediated by the chromosomal AmpC β-lactamase in the E. cloacae isolate; additional laboratories also had problems identifying this isolate biochemically, but the two groups of laboratories did not completely overlap. Assessing the accuracy of bacterial identification was not a major goal of this program, but since NCCLS recommends testing for ESBL production among E. coli, K. pneumoniae, and Klebsiella oxytoca isolates, we wanted to determine if most laboratories could differentiate E. cloacae from K. pneumoniae.
The ESBL-producing K. pneumoniae was one of the first isolates sent to most participating laboratories and preceded publication of the NCCLS guidelines for ESBL detection (34). While approximately 88% of laboratories reported that the K. pneumoniae isolate was resistant to at least one extended-spectrum cephalosporin, only 1.5% reported that it was specifically an extended-spectrum β-lactamase producer. None of the laboratories modified the interpretations of the other cephalosporins to “resistant” as currently suggested by NCCLS (34). Based on feedback from participating laboratories, the issue of ESBL reporting is a source of confusion for many laboratories and may take additional educational efforts before improvement will be seen (11, 53).
The key problem areas identified in our surveys were detection of reduced susceptibility to penicillin in pneumococci and detection of reduced susceptibility to glycopeptides in staphylococci. Approximately 36% of the laboratories reported oxacillin zone diameters of ≥20 mm for the S. pneumoniae strain WHO-4, indicating a failure to detect reduced susceptibility to penicillin. Of those laboratories that did report a zone diameter of ≤19 mm, only 68.2% reported an MIC result. Previous reports have noted that the oxacillin screening test for pneumococci is very sensitive and rarely fails to detect penicillin-resistant pneumococci. However, the test lacks specificity, often yielding zone diameters of ≤19 mm for strains of pneumococci for which the penicillin MICs are 0.03 to 0.06 μg/ml (15, 27). For this reason, the NCCLS has recommended that all pneumococci yielding oxacillin zone diameters of ≤19 mm should be tested for penicillin resistance using an MIC method (32). The range of penicillin MICs reported for this isolate was also remarkably wide, from 0.007 to 2.0 μg/ml. The modal penicillin MIC reported at CDC was 0.06 μg/ml; thus, several laboratories significantly overcalled the resistance level of this organism. Of the 76 laboratories reporting a penicillin MIC result, 40 used Etest and the other 36 used a variety of methods (Table 2). Proficiency testing results from pneumococcal challenges conducted in United States laboratories as collected in recent surveys by the College of American Pathologists (CAP) showed similar testing problems with β-lactam drugs, indicating that testing of pneumococci for susceptibility to penicillin and cephalosporins in the United States is far from optimal (14). One of the key problems recognized in both the WHO-EQAS and CAP studies is the continued use of penicillin, cefotaxime, and ceftriaxone disks for pneumococcal testing. Although NCCLS has not approved interpretive criteria for these disks, laboratories continue to use them. The zone diameters and interpretations sent to the WHO-EQAS project indicate that a combination of gram-negative enteric, staphylococcal, and occasionally enterococcal breakpoints is being used to interpret results, which frequently leads to under reporting of resistance. Clearly, a portion of the problem relates to the expense of performing MIC versus disk diffusion tests in many developing countries. While disk diffusion methods for β-lactam drugs were proposed many years ago (26), they have not been embraced by NCCLS because of their poor predictive values in multicenter studies. More recent attempts to optimize disk testing using multidisk systems have been encouraging (25), but the accuracy of the method has yet to be independently confirmed. Given the wide intermediate range for penicillin (0.1 to 1.0 μg/ml), it is very difficult to devise a disk diffusion test using a 2-U or 10-U penicillin disk that accurately differentiates susceptible and resistant isolates. Because the breakpoint of 0.1 μg/ml is so important for optimal therapy of pneumococcal meningitis (19, 24, 31, 56) and because high-level ceftriaxone- and cefotaxime-resistant pneumococci have emerged (45), the importance of using accurate MIC methods for penicillin and extended-spectrum cephalosporins cannot be overstated. Although the Etest method has been shown to be an accurate predictor of penicillin and cephalosporin resistance (29), it is often too expensive for widespread use in developing countries.
The emergence of reduced susceptibility to vancomycin in S. aureus is a serious public health issue (23, 46). CDC and its advisory committees have published recommendations on preventing the spread of vancomycin-resistant organisms (7) and separate guidelines on preventing the development and spread of staphylococci with reduced susceptibility to vancomycin (6). In the WHO-EQAS survey, few laboratories were successful in detecting reduced susceptibility to vancomycin in the S. epidermidis isolate. This is primarily because such organisms cannot readily be detected by disk diffusion testing (52). Testing of staphylococci, particularly for vancomycin resistance, in laboratories where disk diffusion is used will require the use of an alternate method, such as brain heart infusion agar plates containing 6 μg of vancomycin per ml (developed for enterococci), which have been shown to detect most of the glycopeptide-intermediate staphylococci to date (52). This test, which is inexpensive to prepare, should allow laboratories to detect strains of staphylococci with emerging glycopeptide resistance. Many laboratories also had problems with oxacillin testing of this isolate. Recently, NCCLS changed the oxacillin breakpoints for coagulase-negative staphylococci (34, 51). Based on the results submitted to WHO-EQAS, it appears that many laboratories are unaware of this change.
Quality control testing ensures that the reagents and media and the technologist conducting the test are performing in a predictable and reliable fashion. Unfortunately, adherence to a quality control program is often lax in both developed and developing countries, where it is considered to be either too time consuming or too expensive to perform or both. This is probably why only 30 to 40 laboratories have sent quality control data to WHO-EQAS to date. The quality control data received from the participating laboratories showed many failures. However, it is encouraging that the laboratories continue to generate and use such data to optimize their testing methods. The data from the WHO-EQAS studies have been reviewed by NCCLS to determine whether some of the narrow ranges, such as that for oxacillin and S. pneumoniae ATCC 49619, require revision. Quality control testing is at the heart of good surveillance data and should be reviewed and verified prior to accepting data from sentinel laboratories participating in surveillance systems, particularly if such data are used to guide changes in empiric antimicrobial chemotherapy.
Overall, the results of the WHO-EQAS challenges have been encouraging. However, there is a clear need for educational programs that emphasize proper laboratory testing methods, the importance of quality control, and the basic concepts of quality assurance. The majority of participants in the WHO-EQAS used disk diffusion as their testing method. While this technique is suitable for routine surveillance for resistance in most nonfastidious pathogens, it does have its drawbacks, particularly for testing beta-lactam agents against pneumococci and glycopeptides against staphylococci. Through the use of alternate screening methods and a strict program of quality control to ensure the quality of the media, the emergence of resistance to these classes of drugs in these organisms should be detected. In conclusion, surveillance for antimicrobial resistance is critical, but the data must be validated prior to use.
ACKNOWLEDGMENTS
We thank Jana Swenson, Christine Steward, Bertha Hill, and Meredith Radney at CDC for technical assistance with testing and shipping and for helpful discussions, Philippe Munger at WHO in Geneva for assistance with shipping the organisms, and all WHO-EQAS participants for providing results.
REFERENCES
- 1.Aires de Sousa M, Sanches I S, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlet G, Sanson-le Pors M J, Rouveau M, Fournier G, Marie O, Schlemmer B, Philippon A. Outbreak of nosocomial infections due to Klebsiella pneumoniae producing SHV-4 beta-lactamase. Eur J Clin Microbiol Infect Dis. 1990;9:797–803. doi: 10.1007/BF01967377. [DOI] [PubMed] [Google Scholar]
- 3.Ayliffe G A J. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24(Suppl. 1):S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morb Mortal Wkly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Recommendations for preventing the spread of vancomycin resistance: recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC) Morb Mortal Wkly Rep. 1995;44(RR-12):1–13. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Interim guideline for prevention and control of staphylococcal infection associated with reduced susceptibility to vancomycin. Morb Mortal Wkly Rep. 1997;46:626–628. , 635–636. [PubMed] [Google Scholar]
- 8.Chen D K, McGeer A, de Azavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 9.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones R N, Salazar J C, Pfaller M A, Doern G V Columbian Antimicrobial Resistance Study Group. Multicenter evaluation of antimicrobial resistance to six broad-spectrum β-lactams in Columbia using the Etest method. Diagn Microbiol Infect Dis. 1997;29:265–272. doi: 10.1016/s0732-8893(97)00157-0. [DOI] [PubMed] [Google Scholar]
- 11.Coudron P E, Moland E S, Sanders C C. Occurrence and detection of extended spectrum β-lactamases in members of the family Enterobacteriaceae at a veterans medical center: seek and you may find. J Clin Microbiol. 1997;35:2593–2597. doi: 10.1128/jcm.35.10.2593-2597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diekema D J, Pfaller M A, Jones R N, Doern G V, Winokur P L, Gales A C, Sader H S, Kugler K, Beach M. Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY antimicrobial surveillance program, 1997. Clin Infect Dis. 1999;29:595–607. doi: 10.1086/598640. [DOI] [PubMed] [Google Scholar]
- 13.Doern G V, Brueggemann A, Holley H P, Jr, Rauch A M. Antimicrobial resistance of Streptococcus pneumoniae from outpatients in the United States during the winter months of 1994 to 1995: results of a 30-center national surveillance study. Antimicrob Agents Chemother. 1996;40:1208–1213. doi: 10.1128/aac.40.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doern G V, Brueggemann A B, Pfaller M A, Jones R N. Assessment of laboratory performance with Streptococcus pneumoniae antimicrobial susceptibility testing in the United States: a report from the College of American Pathologists Microbiology Proficiency Survey Program. Arch Pathol Lab Med. 1999;123:285–289. doi: 10.5858/1999-123-0285-AOLPWS. [DOI] [PubMed] [Google Scholar]
- 15.Doern G V, Brueggemann A B, Pierce G. Assessment of the oxacillin disk screening test for determining penicillin resistance in Streptococcus pneumoniae. Eur J Clin Microbiol Infect Dis. 1997;16:311–314. doi: 10.1007/BF01695637. [DOI] [PubMed] [Google Scholar]
- 16.Endtz H P, van den Braak N, van Belkum A, Kluytmans J A J W, Koeleman J G M, Spanjaard L, Voss A, Weersink A J L, Vanderbroucke-Grauls C M J E, Buiting A G M, van Duin A, Verbrugh H A. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox K K, Knapp J S, Holmes K K, Hook III E W, Judson F N, Thompson S E, Washington J A, Whittington W L. Antimicrobial resistance in Neisseria gonorrhoeae in the United States, 1988–1994: the emergence of decreased susceptibility to the fluoroquinolones. J Infect Dis. 1997;175:1396–1403. doi: 10.1086/516472. [DOI] [PubMed] [Google Scholar]
- 18.French G L, Shannon K P, Simmons N. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam-β-lactamase inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J Clin Microbiol. 1996;34:358–363. doi: 10.1128/jcm.34.2.358-363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedland I R, Klugman K P. Failure of chloramphenicol therapy in penicillin-resistant pneumococcal meningitis. Lancet. 1992;339:405–408. doi: 10.1016/0140-6736(92)90087-j. [DOI] [PubMed] [Google Scholar]
- 20.Garrett D O, Jochimsen E, Murfitt K, Hill B, McAllister S, Nelson P, Spera R V, Sall R K, Tenover F C, Johnston J, Zimmer B, Jarvis W R. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect Control Hosp Epidemiol. 1999;20:167–170. doi: 10.1086/501605. [DOI] [PubMed] [Google Scholar]
- 21.Gold H S, Moellering R C., Jr Antimicrobial-drug resistance. N Engl J Med. 1996;335:1445–1453. doi: 10.1056/NEJM199611073351907. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein F, Soussy C-J, Thabaut A. Report of the Comité de l'Antibiogramme de la Société Française de Microbiologie: definition of the clinical antibacterial spectrum of activity. Clin Microbiol Infect. 1996;2:S40–S49. doi: 10.1111/j.1469-0691.1996.tb00874.x. [DOI] [PubMed] [Google Scholar]
- 23.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 24.Iyer P V, Kahler J H, Jacobs N M. Penicillin-resistant pneumococcal meningitis. Pediatrics. 1978;61:157–158. [PubMed] [Google Scholar]
- 25.Jacobs M R, Bajaksouzian S, Palavecino-Fasola E L, Holoszyc H M, Appelbaum P C. Determination of penicillin MICs for Streptococcus pneumoniae by using a two- or three-disk diffusion procedure. J Clin Microbiol. 1998;36:179–183. doi: 10.1128/jcm.36.1.179-183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs M R, Gaspar M N, Robins-Browne R M, Koornof H J. Antimicrobial susceptibility testing of pneumococci. J Antimicrob Chemother. 1980;6:53–64. doi: 10.1093/jac/6.1.53. [DOI] [PubMed] [Google Scholar]
- 27.Jetté L P, Sinave C. Use of an oxacillin disk screening test for detection of penicillin- and ceftriaxone-resistant pneumococci. J Clin Microbiol. 1999;37:1178–1181. doi: 10.1128/jcm.37.4.1178-1181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones R N, Sader H S, Erwin M E, Andersen S C the Enterococcus Study Group. Emerging multiply resistant enterococci among clinical isolates. I. Prevalence data from a 97 medical center surveillance study in the United States. Diagn Microbiol Infect Dis. 1995;21:85–93. doi: 10.1016/0732-8893(94)00147-o. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen J H, Ferraro M J, McElmell M L, Spargo J, Swenson J M, Tenover F C. Detection of penicillin and extended-spectrum cephalosporin resistance among Streptococcus pneumoniae clinical isolates by use of the Etest. J Clin Microbiol. 1994;32:159–163. doi: 10.1128/jcm.32.1.159-163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis K, Saubolle M A, Tenover F C, Rudinsky M F, Barbour S D, Cherry J D. Pertussis caused by an erythromycin-resistant strain of Bordetella pertussis. Pediatr Infect Dis J. 1995;14:388–391. doi: 10.1097/00006454-199505000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Naraqi S, Kirkpatrick G P, Kabins S. Relapsing pneumococcal meningitis: isolation of an organism with decreased susceptibility to penicillin. J Pediatr. 1974;85:671. doi: 10.1016/s0022-3476(74)80513-5. [DOI] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2–A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: eighth informational supplement M100–S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 35.Pfaller M A, Jones R N, Doern G V. Multicenter evaluation of the antimicrobial activity for six broad-spectrum beta-lactams in Venezuela using the Etest method: the Venezuelan Antimicrobial Resistance Study Group. Diagn Microbiol Infect Dis. 1998;30:45–52. doi: 10.1016/s0732-8893(97)00158-2. [DOI] [PubMed] [Google Scholar]
- 36.Pontani D, Washton H, Bouchillon S, Johnson J. Susceptibility of European respiratory tract isolates to trovafloxacin, ciprofloxacin, clarithromycin, azithromycin, and ampicillin. Eur J Clin Microbiol Infect Dis. 1998;17:413–419. doi: 10.1007/BF01691574. [DOI] [PubMed] [Google Scholar]
- 37.Prodinger W M, Fille M, Bauerfeind A, Stemplinger I, Amann S, Pfausler B, Lass-Flörl C, Dierich M P. Molecular epidemiology of Klebsiella pneumoniae producing SHV-5 β-lactamase: parallel outbreaks due to multiple plasmid transfer. J Clin Microbiol. 1996;34:564–568. doi: 10.1128/jcm.34.3.564-568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts R B, Tennenberg A M, Eisner W, Hargrave J, Drusin L M, Yurt R, Kreiswirth B N. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb Drug Resist. 1998;4:175–183. doi: 10.1089/mdr.1998.4.175. [DOI] [PubMed] [Google Scholar]
- 39.Rossi A, Ruvinsky R, Regueira M, Corso A, Pace J, Gentile A, Di Fabio J L. Distribution of capsular types and penicillin-resistance of strains of Streptococcus pneumoniae causing systemic infections in Argentinian children under 5 years of age: Streptococcus pneumoniae Working Group. Microb Drug Resist. 1997;3:135–140. doi: 10.1089/mdr.1997.3.135. [DOI] [PubMed] [Google Scholar]
- 40.Sahm D F, Tenover F C. Surveillance for the emergence and dissemination of antimicrobial resistance in bacteria. Infect Dis Clin N Am. 1997;11:767–783. doi: 10.1016/s0891-5520(05)70389-5. [DOI] [PubMed] [Google Scholar]
- 41.Salkin I F, Limberger R J, Stasik D. Commentary on the objectives and efficacy of proficiency testing in microbiology. J Clin Microbiol. 1997;35:1921–1923. doi: 10.1128/jcm.35.8.1921-1923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders C C. Chromosomal cephalosporinases responsible for multiple resistance to newer β-lactam antibiotics. Annu Rev Microbiol. 1987;41:573–593. doi: 10.1146/annurev.mi.41.100187.003041. [DOI] [PubMed] [Google Scholar]
- 43.Schito G C, Mannelli S, Pesce A. Trends in the activity of macrolide and beta-lactam antibiotics and resistance development: Alexander Project Group. J Chemother. 1997;9(Suppl. 3):18–28. [PubMed] [Google Scholar]
- 44.Shahangian S. Proficiency testing in laboratory medicine: uses and limitations. Arch Pathol Lab Med. 1998;122:15–30. [PubMed] [Google Scholar]
- 45.Sloas M M, Barrett F F, Chesney P J, English B K, Hill B C, Tenover F C, Leggiadro R J. Cephalosporin treatment failure in penicillin- and cephalosporin-resistant Streptococcus pneumoniae meningitis. Pediatr Infect Dis J. 1992;11:662–666. [PubMed] [Google Scholar]
- 46.Smith T, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Arduino M J, Zervos M J, Miller J M, Band J D, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus: epidemiology and clinical significance. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 47.Song J H, Lee N Y, Ichiyama S, Yoshida R, Hirakata Y, Fu W, Chongthaleong A, Aswapokee N, Chiu C H, Lalitha M K, Thomas K, Perera J, Yee T T, Jamal F, Warsa U C, Vinh B X, Jacobs M R, Appelbaum P C, Pai C H. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian network for surveillance of resistant pathogens (ANSORP) study. Clin Infect Dis. 1999;28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]
- 48.Sougakoff W, Goussard S, Gerbaud G, Courvalin P. Plasmid-mediated resistance to third generation cephalosporins caused by point mutations in TEM-type penicillinase genes. Rev Infect Dis. 1988;10:879–884. doi: 10.1093/clinids/10.4.879. [DOI] [PubMed] [Google Scholar]
- 49.Stelling J M, O'Brien T F. Surveillance of antimicrobial resistance: the WHONET program. Clin Infect Dis. 1997;24(Suppl. 1):S157–S168. doi: 10.1093/clinids/24.supplement_1.s157. [DOI] [PubMed] [Google Scholar]
- 50.Tenover F C, Hughes J M. The challenges of emerging infectious diseases: development and spread of multiply resistant bacterial pathogens. JAMA. 1996;275:300–304. [PubMed] [Google Scholar]
- 51.Tenover F C, Jones R N, Swenson J M, Zimmer B, McAllister S, Jorgensen J H for the NCCLS Staphylococcus Working Group. Methods for improved detection of oxacillin-resistance in coagulase negative staphylococci: results of a multicenter study. J Clin Microbiol. 1999;37:4051–4058. doi: 10.1128/jcm.37.12.4051-4058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, McAllister S A, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tenover F C, Mohammed J M, Gorton T S, Dembeck Z F. Detection and reporting of organisms producing extended-spectrum β-lactamases: a survey of laboratories in Connecticut. J Clin Microbiol. 1999;37:4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenover F C, Rasheed J K. Genetic methods for detecting antibacterial and antiviral resistance genes. In: Murray P, Baron E S, Pfaller M, Tenover F C, Yolken R, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 1578–1592. [Google Scholar]
- 55.Tenover F C, Tokars J, Swenson J M, Paul S, Spitalny K, Jarvis W. Ability of clinical laboratories to detect antimicrobial resistant enterococci. J Clin Microbiol. 1993;31:1695–1699. doi: 10.1128/jcm.31.7.1695-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viladrich P F, Guidol F, Linares J, Rufi G, Ariza J, Pallares R. Characteristics and antibiotic therapy of adult meningitis due to penicillin-resistant pneumococci. Am J Med. 1988;84:839–846. doi: 10.1016/0002-9343(88)90061-7. [DOI] [PubMed] [Google Scholar]
- 57.Working Party of the British Society for Antimicrobial Chemotherapy. Breakpoints in in-vitro antibiotic susceptibility testing. J Antimicrob Chemother. 1988;21:701–710. [PubMed] [Google Scholar]