Abstract
Ixodes scapularis is the primary vector of tick-borne pathogens in North America but notably does not transmit pathogenic Rickettsia species. This tick harbors the transovarially transmitted endosymbiont Rickettsia buchneri, which is widespread in I. scapularis populations, suggesting that it confers a selective advantage for tick survival such as providing essential nutrients. The R. buchneri genome includes genes with similarity to those involved in antibiotic synthesis. There are two gene clusters not found in other Rickettsiaceae, raising the possibility that these may be involved in excluding pathogenic bacteria from the tick. This study explored whether the R. buchneri antibiotic genes might exert antibiotic effects on pathogens associated with I. scapularis. Markedly reduced infectivity and replication of the tick-borne pathogens Anaplasma phagocytophilum, R. monacensis, and R. parkeri were observed in IRE11 tick cells hosting R. buchneri. Using a fluorescent plate reader assay to follow infection dynamics revealed that the presence of R. buchneri in tick cells, even at low infection rates, inhibited the growth of R. parkeri by 86–100% relative to R. buchneri-free cells. In contrast, presence of the low-pathogenic species R. amblyommatis or the endosymbiont R. peacockii only partially reduced the infection and replication of R. parkeri. Addition of host-cell free R. buchneri, cell lysate of R. buchneri-infected IRE11, or supernatant from R. buchneri-infected IRE11 cultures had no effect on R. parkeri infection and replication in IRE11, nor did these treatments show any antibiotic effect against non-obligate intracellular bacteria E. coli and S. aureus. However, lysate from R. buchneri-infected IRE11 challenged with R. parkeri showed some inhibitory effect on R. parkeri infection of treated IRE11, suggesting that challenge by pathogenic rickettsiae may induce the antibiotic effect of R. buchneri. This research suggests a potential role of the endosymbiont in preventing other rickettsiae from colonizing I. scapularis and/or being transmitted transovarially. The confirmation that the observed inhibition is linked to R. buchneri's antibiotic clusters requires further investigation but could have important implications for our understanding of rickettsial competition and vector competence of I. scapularis for rickettsiae.
Keywords: Ixodes scapularis, tick, Rickettsia, endosymbiont, antibiosis, interference, competition
Introduction
The blacklegged tick Ixodes scapularis Say (Acari: Ixodidae) is the primary vector of zoonotic tick-borne pathogens in North America. It transmits seven pathogens, including those causing Lyme borreliosis (Borrelia burgdorferi, Borrelia mayonii), human anaplasmosis (Anaplasma phagocytophilum), and human babesiosis (Babesia microti) (1). Interestingly, in contrast to other major human-biting species of Ixodes in different parts of the world, I. scapularis does not transmit pathogenic Rickettsia species. Ixodes ricinus and I. persulcatus, the tick vectors primarily responsible for the transmission of B. burgdorferi, A. phagocytophilum, tick-borne encephalitis virus, and Babesia spp. in Europe and Asia, respectively, are commonly infected with Rickettsia spp. linked to human disease. Both R. helvetica and R. monacensis are commonly detected in I. ricinus across Europe (2–7), and various other rickettsiae including R. raoultii and R. slovaca, which are primarily vectored by other tick species, have also been identified in I. ricinus (3, 8). Meanwhile, I. persulcatus is infected with a wider range of Rickettsia spp., with R. helvetica, R. raoultii, R. sibirica, R. heilongjiangensis, and “Candidatus R. tarasevichiae” often detected in this tick species (3, 4, 9–15). In eastern Australia, Ixodes holocyclus is a vector of R. australis, the causative agent of Queensland tick typhus (16, 17).
Instead, I. scapularis hosts a rickettsial endosymbiont, R. buchneri (18), which dominates the tick microbiome, particularly in females where it typically constitutes almost 100% of the microbiome (19–24). These bacteria, formerly known as “rickettsial endosymbiont of I. scapularis” (REIS), have been detected in I. scapularis populations throughout its range (Figure 1) and are often present at high prevalence (18–23, 25–53), suggesting an established relationship between the tick and its endosymbiont. Rickettsia buchneri reside primarily in the ovaries of adult female ticks (18), although there is also some evidence of their presence in salivary glands (22, 52). The endosymbiont is transovarially transmitted and can be found in all life stages (22, 26), yet it is still unclear where it resides within adult males and immature stages, or what roles it may play in tick biology. The existence of genes in the endosymbiont encoding complete biosynthetic pathways for biotin and folate (54, 55) suggests that it may aid the tick by supplying essential nutrients lacking in blood. Phylogenetic analyses imply that R. buchneri is ancestral to the spotted fever group (SFG) rickettsiae (54), which contains the majority of tick-transmitted Rickettsia species; R. buchneri is most closely related to R. monacensis from I. ricinus, and the rickettsial endosymbiont of Ixodes pacificus, “R. monacensis” strain Humboldt (18, 56). Like other Rickettsiales, the SFG rickettsiae are obligate intracellular Gram-negative bacteria (57).
Figure 1.
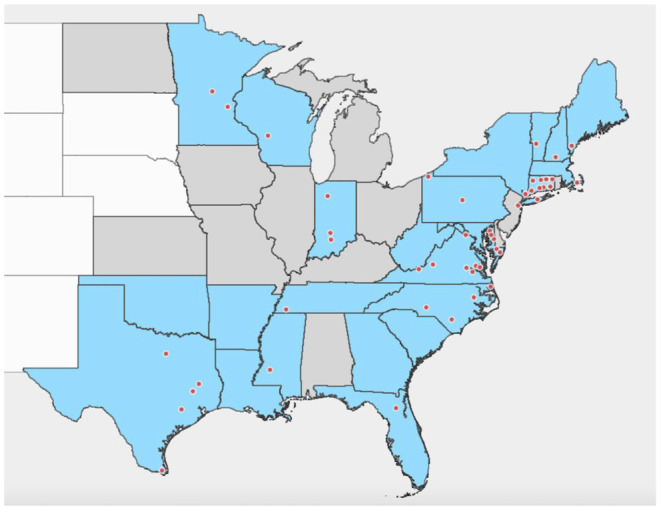
Distribution of detections of R. buchneri and “Rickettsial endosymbiont of Ixodes scapularis” in Ixodes scapularis in the United States of America. Shaded area indicates states with established populations of Ixodes scapularis in 2016 based on Eisen and Eisen (1). States shaded blue are those where R. buchneri/REIS has been detected in I. scapularis; records with county-level data are shown by dots. Based on data from references published 2007–2021 (18–23, 25–53).
Cases of SFG rickettsioses are on the rise in the United States (58). While human cases of severe illness due to infection with R. rickettsii (Rocky Mountain spotted fever) appear to be rare, there have been increases in cases of milder spotted fever, thought to be primarily due to infection with other less pathogenic Rickettsia species and the geographic expansion of the lone star tick Amblyomma americanum (59). This tick commonly bites humans and is a potential vector of both R. rickettsii (60, 61) and R. parkeri (62), which causes a relatively mild eschar-associated rickettsiosis (63). Additionally, R. amblyommatis, originally considered an endosymbiont of A. americanum and highly prevalent in this tick, has been linked to mild disease (64–66), so it may also be contributing. The main vector of R. parkeri, the Gulf Coast tick A. maculatum, is also expanding its distribution (67), and populations are increasingly being found in more northerly US states (68, 69). The distribution of I. scapularis overlaps in large parts of the country with those of tick species responsible for the transmission of pathogenic SFG Rickettsia spp., particularly A. americanum and Dermacentor variabilis. These ticks may share similar habitats and hosts, therefore making it possible for I. scapularis to come into contact with pathogenic Rickettsia species. However, field-collected I. scapularis are very rarely infected with Rickettsia species other than R. buchneri. This might suggest that the presence of R. buchneri plays a role in excluding other Rickettsia spp. from its tick host. Evidence of competition (or “interference”) between different Rickettsia species exists in other ticks; for example, the presence of the endosymbiont R. peacockii in the Rocky Mountain wood tick Dermacentor andersoni has been associated with reduced transovarial transmission of pathogenic R. rickettsii in the tick (70). In addition, infection of D. variabilis with either R. montanensis or R. rhipicephali prevented the transovarial transmission of the competing rickettsia in reciprocal challenge experiments (71), and A. americanum infected with R. amblyommatis were less likely to acquire R. parkeri than uninfected ticks (72). Similarly, while A. americanum larvae infected with R. amblyommatis were able to acquire R. rickettsii, its prevalence was significantly lower compared to that in R. amblyommatis-free larvae (73). Furthermore, milder symptoms were observed in guinea pigs infected with R. rickettsii by dually infected nymphs than those infected by R. amblyommatis-free nymphs (73), suggesting that R. rickettsii load was reduced by the presence of the additional Rickettsia species. In field studies, a high prevalence of “Candidatus Rickettsia andeanae” in A. maculatum populations was hypothesized to be linked to the exclusion of R. parkeri from these ticks (74). Mechanisms for the competition between Rickettsia species, or whether these might differ for endosymbiotic and pathogenic species, have not been elucidated.
Two genome sequences of R. buchneri are currently available. The REIS (Wikel) genome was extracted from the genome sequence of I. scapularis from the Wikel colony (54), and the R. buchneri ISO7T genome was sequenced from the R. buchneri-type strain isolated from the ovaries of a female I. scapularis removed from a dog in Minnesota (18). A gene cluster encoding aminoglycoside antibiotic biosynthesis machinery has been identified in R. buchneri (54), which is not present in other rickettsiae, and therefore antibiotic production might represent a mechanism by which R. buchneri is able to exclude pathogenic Rickettsia species from I. scapularis. Genes from the cluster were found to be highly similar to those of kanamycin and gentamicin synthesis gene clusters found in members of Actinobacteria and Firmicutes (54), yet to date no experimental studies have examined whether these genes are functional in R. buchneri. In this study, we report that an aminoglycoside biosynthesis gene cluster, almost identical to that described by Gillespie et al. (54), is also present in the R. buchneri ISO7T genome, along with a second gene cluster encoding genes similar to those for polyketide and non-ribosomal peptide antibiotic synthesis, which appears to be only partially present in the REIS (Wikel) genome. Additionally, this study shows that genes from these clusters are actively transcribed and examines whether competition exists between R. buchneri and rickettsial pathogens associated with ticks, using in vitro experiments to provide preliminary evidence that the presence of the endosymbiont in tick cells has an inhibitory effect on the infection and replication of other intracellular bacteria.
Materials and Methods
Bioinformatic Analysis of Rickettsia buchneri Antibiotic Gene Clusters
Annotation of the sequenced R. buchneri ISO7T genome [GenBank: JFKF01000000.1; (18)] identified the presence of two clusters of genes with similarity to bacterial genes for aminoglycoside, polyketide, and non-ribosomal peptide synthesis. This strain of R. buchneri was isolated from the ovaries of an I. scapularis female collected from a dog in Minnesota (18). To determine potential functions of proteins in the two putative antibiotic clusters, amino acid sequences from the R. buchneri ISO7T genome were searched against other available sequences using the NCBI protein–protein BLAST algorithm, performed with default parameters. Putative protein function was determined by examining data for each protein in the InterPro database as well as performing literature searches. The gene clusters were also compared to the R. buchneri genome derived from the I. scapularis genome sequence, REIS (Wikel) [GenBank: CM000770.1; (54)]. Protein sequences obtained from annotated genomes were aligned using ClustalW (75) and MUSCLE (76) in MacVector version 18.1.5.
Cell and Rickettsia Culture
Embryonic tick cell lines ISE6 (77), IRE11 (78), and AAE2 (79), derived from I. scapularis, I. ricinus, and A. americanum, respectively, were maintained at 34°C in L15C300 medium supplemented with 5% heat-inactivated fetal bovine serum (FBS), 5% tryptose phosphate broth (TPB), and 0.1% lipoprotein concentrate (LPC; MP Biomedicals, Irvine, CA, USA), adjusted to pH 7.2–7.5 with 1 M NaOH, as previously described (77). Rickettsia buchneri ISO7T [(18); hereafter referred to as Rb-WT] and R. buchneri expressing GFPuv from the plasmid pRAM18dRGA [(80); hereafter referred to as Rb-GFPuv] were maintained in IRE11 at 28°C in a modified L15C300 medium containing 10% FBS, 5% TPB, 0.06% NaHCO3, 6 mM HEPES, and 0.1% LPC; pH was not adjusted. Rickettsia parkeri Tate's Hell expressing mKate from plasmid pRAM18dSFA (Rp-mKate) (80, 81), R. peacockii-GFPuv [pRAM18dSGK; (80)], R. monacensis IrR/Munich with mKate (pRAM18dSFA), and A. phagocytophilum HGE1 expressing mCherry from an intergenic Himar1 transposon insertion (82) (A. phagocytophilum-mCherry) were grown in ISE6 cells, and R. amblyommatis Darkwater (kindly supplied by Chris Paddock, CDC) were grown in AAE2 cells. Infected tick cell cultures were maintained at 34°C in L15C300 with 10% FBS, 5% TPB, 0.1% LPC, 0.25% NaHCO3, and 25 mM HEPES, adjusted to pH 7.5 (83). The infection level of cell cultures was assessed by Giemsa staining. Vero cells (African green monkey kidney) were grown in Gibco RPMI 1640 (Thermo Fisher, Waltham, MA, USA) supplemented with 10% FBS and 2 mM L-glutamine at 34°C following established methods (84). All cultures were grown in 25-cm2 culture flasks (CELLSTAR, Greiner Bio-One, Monroe, NC, USA).
Host cell-free bacteria were prepared as previously described (83); heavily infected tick cells were added to tubes containing rock tumbler grit (60/90 coarse silicon carbide, Lortone, Mukilteo, WA, USA), vortexed for 30 s, then passed through a 2 μm filter to remove cellular debris. Cell-free bacteria were then collected by centrifugation at 13,200 × g for 5 min at 4°C.
Reverse Transcriptase PCR
Cell-free Rb-WT were prepared from infected IRE11 cultures as described above. Bacterial pellets were washed once in SPG buffer, then resuspended in 1 ml TRI Reagent (Sigma-Aldrich, St Louis, MO, USA), vortexed, and rested at room temperature for 10 min. Samples were centrifuged at 12,000 × g to remove particulates and the supernatant transferred to new tubes and mixed 1:2 with 100% ethanol, followed by vortexing. RNA was purified using a Direct-zol RNA Miniprep Kit (Zymo Research, Irvine, CA, USA). Contaminating DNA was removed by treating three times with Ambion TURBO DNA-free Kit (Thermo Fisher), followed by purification with an RNA Clean & Concentrator Kit (Zymo Research). Reverse transcriptase PCR (RT-PCR) was carried out using the Access RT-PCR system (Promega, Madison, WI, USA) in 25 μl reactions consisting of 5 × reaction buffer, 10 μM dNTPs, 10 μM of each primer, 0.5 μl Tf1 DNA polymerase, 14.5 μl nuclease-free water, and 1 μl sample. Nuclease-free water was used as a negative control, and no reverse transcriptase (no RT) controls included water instead of the DNA polymerase to confirm the absence of contaminating DNA. RT-PCR was performed in a Techne TC-312 Thermocycler with the following cycling conditions: 45 min at 45°C; 2 min at 94°C; 40 cycles of 30 s at 94°C, 1 min at 55°C or 58°C, and 2 min at 68°C; final extension 1 min at 68°C. Amplification of HTH, lagD, ppsE_1, kanC, and btrB was performed with an annealing temperature of 58°C, and amplification of glycogen synthase, homoserine kinase, and lgrB at 55°C. Primers used in this study are shown in Table 1. RT-PCR products were visualized on a 1.2% agarose gel stained with GelGreen (Biotium, Fremont, CA, USA).
Table 1.
Primers used for the amplification of Rickettsia buchneri antibiotic cluster genes.
| Product name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| HTH domain | AGC TGA TTT AGA AAG AAA GGC A | GAG GTA ACA TCA ATA CAG GGA AG |
| ppsE_1 | CCT GGA GGT ATA AGA TCT GCT AAT G | GCT CCT TGT CCT GGG AAT AAA |
| lgrB | CTA CCG GAC AAC CTA AAG GAA TAG | CGG AAA CCT CGA ACC TTA ACT |
| btrB (choline dehydrogenase) | CGG GTT AAA TCC TTT CCC TAC TC | AGT AAC GAC AAG TCC CAT GTA AG |
| kanC | GGA GGA ATC CCA GGA AAC ATA G | CAA TGA GCA TAC CTA ACC CTA CA |
| lagD | AGT TCG GGT ATT GCC ACA TAT T | TGG TAT GCC ATA GGT AAG GAT TTC |
| Homoserine kinase | GTT CTA GCG CAA TAC CCT CTT | CGC GCA ATG TCC CAA ATA C |
| Glycogen synthase | TCC TGG CTA CTC GGT ACA TTA | CTC TGG CAA TAC GAC CAA CA |
| ppsE_1 (qRT-PCR) | ACG TAC TCC TAT GAA GCT CCG | GCT CCT TGT CCT GGG AAT AAA |
| lgrE (qRT-PCR) | TTT TCC CTT TCG CAG GTG GG | ACC CCA GAT ATT TTC CAC GTC C |
| btrB (qRT-PCR) | CGG GTT AAA TCC TTT CCC TAC TC | GCA TTC ATG CCT GCA AAA ATA G |
| kanC (qRT-PCR) | TAC ATG TCC AAG AGT ATG GCC G | AGC AGA GGC GAT AAA GCT AGT |
In vitro Competition Assays
Competition assays were set up in 24-well plates to compare the infection and replication of R. monacensis-mKate, Rp-mKate, and A. phagocytophilum-mCherry in tick cells with and without Rb-GFPuv infection. For the R. monacensis and R. parkeri experiments, three 24-well plates were prepared; the first contained uninfected IRE11, the second contained IRE11 heavily infected with Rb-GFPuv, and the third contained a mixture of infected and uninfected cells to give an approximate level of Rb-GFPuv infection of 45%. Cells (0.4 ml) were applied to each well, giving 1 × 106 cells/well in the R. monacensis experiment or 3 × 105 cells/well in the R. parkeri experiment. Plates were incubated in a humidified candle jar at 28°C. After 24 h, plates were infected with 250 μl fresh medium containing host cell-free R. monacensis-mKate or Rp-mKate, in 10-fold serial dilutions. Each dilution was applied to each plate in triplicate, and the remaining wells were used as negative controls. The number of mKate-positive colonies in the lowest-dilution wells was determined by fluorescent microscopy and used to extrapolate the number of rickettsiae in each dilution. Wells were observed on a Nikon Diaphot fluorescent microscope, and adhesion/invasion and replication were determined for each dilution on each plate by visualization of red fluorescent rickettsiae over 14 days. Additionally, the ability of Rp-mKate to infect IRE11 was assessed daily over the 14 days by observing at least 500 cells per well and determining the percentage of cells containing replicating fluorescent R. parkeri.
For A. phagocytophilum experiments, five 24-well plates were prepared; two contained uninfected IRE11, two contained IRE11 heavily infected with Rb-GFPuv, and the fifth contained a mixture of infected and uninfected cells to give an approximate level of Rb-GFPuv infection of 25%. Cells (0.5 ml) were applied to each well to give 1 × 106 cells/well. One uninfected IRE11 plate and one infected IRE11 plate were incubated at 27°C in a humidified candle jar; the remaining three plates were incubated at 34°C with 4% CO2. After 24 h, plates were infected with 250 μl medium containing 10-fold serial dilutions of host cell-free A. phagocytophilum-mCherry, added to plates in triplicate. The number of Anaplasma in each dilution was estimated by testing each dilution in a sixth 24-well plate seeded with ISE6 cells and counting the number of mCherry-positive colonies in the lowest-dilution wells and then extrapolating to each dilution. Wells were observed on a Nikon Diaphot fluorescent microscope, and adhesion/invasion and replication were determined for each dilution on each plate by visualization of red fluorescent bacteria.
Fluorescent Plate Reader Assays
A fluorescent plate reader assay was used to measure the growth dynamics of fluorescent Rickettsia in IRE11 cells. Uninfected IRE11 and IRE11 heavily infected with Rb-GFPuv (>95% cells infected) were adjusted to 1 × 105 cells/ml in fresh 10% FBS medium. Heavily infected and uninfected cells were then mixed to create additional populations with 25%, 50%, and 75% cells infected (Figure 2). A volume of 200 μl of each cell population was added in triplicate to wells of a clear-bottomed black-sided 96-well plate (Falcon, Corning, NY, USA), with dH2O added between wells to prevent drying, and incubated at 28°C in a humidified candle jar for 24 h to allow cells to settle to the bottom of wells. Host cell-free Rp-mKate were resuspended in 1 ml fresh medium and enumerated on a Petroff–Hausser chamber. The cell-free bacteria were diluted to create 1,000:1, 100:1, and 10:1 multiplicity of infection (MOI) ratios and then added to IRE11 cells in 96-well plates in a volume of 10 μl. The plate was returned to the humidified candle jar and incubated at 28°C. Fluorescence readings were taken 24 h later and then every 24 h up to 14 days postinfection. Readings were taken at room temperature (~22–25°C) on a BioTek Synergy H1 microplate reader at excitation/emission 395/509 for GFPuv and 588/633 for mKate and adjusted to uninfected IRE11 to account for background fluorescence.
Figure 2.
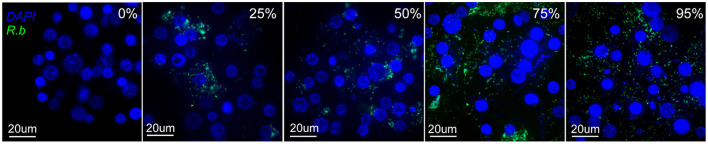
Images of IRE11 cells with different levels of R. buchneri-GFPuv infection. Live IRE11 cells were stained with NucBlue Live ReadyProbes Reagent (Hoechst 33342; Invitrogen) and spun onto slides with a Cytospin centrifuge, following Wang et al. (81). Images were captured on an Olympus BX61 DSU Confocal Microscope using a ×60 objective and a double-wavelength filter (DAPI; FITC). DNA shown in blue, GFPuv shown in green.
Plate reader experiments for R. amblyommatis used a similar protocol to the above, except that only uninfected and 25% and >95% infected cell populations were used, and only 1,000:1 and 10:1 Rp-mKate challenges were performed. Additionally, only mKate fluorescence was measured for these experiments. The R. peacockii experiments were carried out using the same protocol as for Rb-GFPuv. Prior to IRE11 plate experiments, cell-free R. amblyommatis and R. peacockii (grown in AAE2 and ISE6 cells, respectively) were transferred to IRE11 and cultured in modified L15C300 medium at 28°C for at least 2 weeks to adjust them to growing in these conditions. Both Rickettsia species were found to grow well in IRE11 cells.
To assess the effect of cell-free R. buchneri and lysate of R. buchneri on the replication of Rp-mKate, a 96-well plate was set up with 200 μl of uninfected IRE11 adjusted to 1 × 105 cells/ml in fresh medium, as above. Cells were treated with either 50 μl medium (negative control), 50 μl of cell-free Rb-WT prepared from 2.5 × 105 heavily infected IRE11, 50 μl of a 1:10 dilution of the cell-free Rb-WT, 50 μl cell lysate from 2.5 × 105 IRE11 heavily infected with Rb-WT, or 50 μl cell lysate from 2.5 × 105 uninfected IRE11. Lysates were prepared by sonicating cells on ice at full power for a total of 1 min (separated into 3 × 20-s bursts, with 20-s intervals resting on ice). After 2 h, cells were challenged with cell-free Rp-mKate at 1,000:1, 100:1, or 10:1, and the plate was incubated at 28°C in a humidified candle jar. mKate fluorescence was measured every 24 h for 14 days, as described above, and adjusted to uninfected, untreated IRE11.
To determine if R. parkeri could induce R. buchneri antibiosis activity, an additional plate reader assay was used to examine whether lysates from R. buchneri challenged with Rp-mKate for varying lengths of time exhibited inhibitory effects on the growth of Rp-mKate in IRE11 cells. Wells of a 6-well plate were seeded with 2 ml IRE11 75% infected with Rb-WT, at 1 × 105/ml. One well served as a no challenge control, while to the remaining wells cell-free Rp-mKate at a ratio of 100:1 was added. The plate was incubated at 28°C in a candle jar. After 24 h, cells from the control and one of the challenged wells were collected, and rickettsiae were isolated from IRE11 as above. Cell pellets were then frozen at −70°C. At 48, 72, 120, and 168 h after infection, this was repeated for cells from each of the remaining wells. Lysates were prepared from the pellets by four freeze thaw cycles of −70 to 37°C. The rickettsial lysates from each treatment were resuspended in 120 μl medium, and then 10 μl was added to 12 wells of a 96-well plate, each containing 200 μl IRE11 at 1 × 105/ml, prepared 24 h previously. Wells were then challenged with Rp-mKate at ratios of 1,000:1, 100:1, and 10:1. Medium without Rp-mKate was added to control wells for each treatment. The plate was incubated at 28°C in a humidified candle jar, and mKate fluorescence was measured every 24 h for 14 days, as described above.
Antibiotic Susceptibility Assays
To test the antibiotic activity of R. buchneri against extracellular bacteria, antibiotic susceptibility testing was performed against Escherichia coli D21 and Staphylococcus aureus MN8 using disk diffusion assays. IRE11 infected with Rb-WT and uninfected IRE11 was pelleted by centrifugation at 350 × g for 6 min at 4°C, and the pellets were frozen at −70°C. Cell-free Rb-WT was prepared by vortexing with rock tumbler grit and filtration through a 2-μm filter, as described above, then the bacteria were pelleted by centrifugation at 13,600 × g for 7 min at 4°C, and the cell pellet frozen at −70°C. Lysates of the three samples were prepared by freeze-thawing (four cycles of −70 to 37°C) and then centrifugation at 13,600 × g for 5 min at 4°C. Bacterial or cell lysates were then resuspended in 50 μl medium, of which 30 μl was added to 50 μl of absolute methanol. A volume of 20 μl of each sample was then added to separate filter paper disks. Spectinomycin at 10 and 100 μg was added to two additional filter paper disks as positive controls. Disks were air-dried in a biosafety cabinet for 20–30 min and then placed onto Mueller–Hinton agar plates streaked with E. coli D21 or S. aureus MN8. Plates were incubated for 18 h at 37°C.
The disk diffusion assays were repeated using pellets of live IRE11, Rb-WT-infected IRE11 (25%, 50%, and >95% infected), and cell-free Rb-WT, as well as supernatant from IRE11 cultures at various levels of infection with Rb-WT (25, 50, >95%) or uninfected. Cells were pelleted by centrifugation at 350 × g for 6 min at 12°C, supernatant was removed to separate tubes, and then pellets were resuspended in 100 μl medium. A volume of 20 μl of resuspended cell pellets or 20 μl supernatant was added to filter paper disks. Spectinomycin was added to additional disks at 100 μg for positive controls. Disks were air-dried for 20-30 min in a biosafety cabinet, then applied to Mueller–Hinton agar plates streaked with E. coli D21 or S. aureus MN8. Plates were incubated for 18 h at 37°C.
To further test the antibiotic activity of supernatant from Rb-WT-infected IRE11 cultures against R. parkeri grown in mammalian cells, Vero cell cultures were grown at 34°C in 2.5 ml RPMI medium supplemented with 2.5 ml supernatant from either uninfected IRE11 or IRE11 heavily infected with Rb-WT. Cell-free Rp-mKate was added to Vero cultures and flasks checked daily for evidence of infection, which was assessed by the appearance of plaques in the cell layer as well as the timing and size of plaques.
Expression of Antibiotic Genes in Response to R. parkeri Challenge
To determine whether the expression of antibiotic genes by Rb-WT was induced by challenge with R. parkeri, and to determine the time of maximal expression, a time course experiment was set up comparing unchallenged and Rp-mKate-challenged Rb-WT-infected IRE11. A 6-well plate was prepared with each well containing IRE11 75% infected with Rb-WT at 1 × 105/ml in 2 ml modified L15C300 medium. One well served as a no challenge control, while to the remaining wells cell-free Rp-mKate at a ratio of 100:1 in 10 μl was added. The plate was incubated at 28°C in a candle jar. After 24 h, cells from the control well were collected and centrifuged for 2 min at 500 × g, and the cell pellet was resuspended in 750 μl RNAlater solution (Qiagen, Hilden, Germany) and stored at−20°C. Cells from the challenge wells were collected at 24, 48, 72, 120, and 168 h after infection, resuspended in RNAlater solution, and stored at −20°C. RNA isolation was performed using TRIzol–chloroform extraction. cDNA was then prepared using Takara RT PrimeScript Kit (Takara Bio, Kusatsu, Japan), after a 30-min treatment with a gDNA eraser at room temperature to remove contaminating DNA. Quantitative PCR (qPCR) was performed on an Agilent Mx3005P RT-PCR system using the Agilent Brilliant II SYBR Green Master Mix, using 10-μM primers against various targets from both gene clusters (Table 1) and 1 μl cDNA preparation. Since both R. buchneri and R. parkeri were present in samples, I. scapularis GAPDH was used as a reference gene. Nuclease-free water and no reverse transcriptase controls were included on each plate. Each sample was run in triplicate. Cycling conditions were 10 min at 95°C, followed by 40 cycles of 30 s at 95°C, 1 min at 55°C (glycogen synthase, GAPDH), or 52°C (btrB, kanC, lgrE, ppsE_1), 1 min at 72°C. Results were compared to GAPDH expression to adjust for total cDNA per sample. To measure primer efficiency, standard curves consisting of cDNA from R. buchneri-infected IRE11 were also added to reaction plates. To account for differences in primer efficiency, relative quantification was calculated with a PCR efficiency correction (85).
Statistical Analysis
All statistical analyses were carried out using GraphPad Prism version 9.1.2. For plate reader experiments, the growth of Rp-mKate in IRE11 cultures infected with other Rickettsia or treated with lysates was analyzed using a two-way ANOVA with Dunnett's multiple-comparison test, using Rp-mKate growth in uninfected IRE11 wells as the control. Relative gene expression was analyzed with a two-way ANOVA with Dunnett's multiple-comparison test, comparing the expression in Rp-challenged groups to that in the unchallenged control group. Statistical significance was assigned when p < 0.05.
Results
Description and Expression of Antibiotic Gene Clusters in Rickettsia buchneri
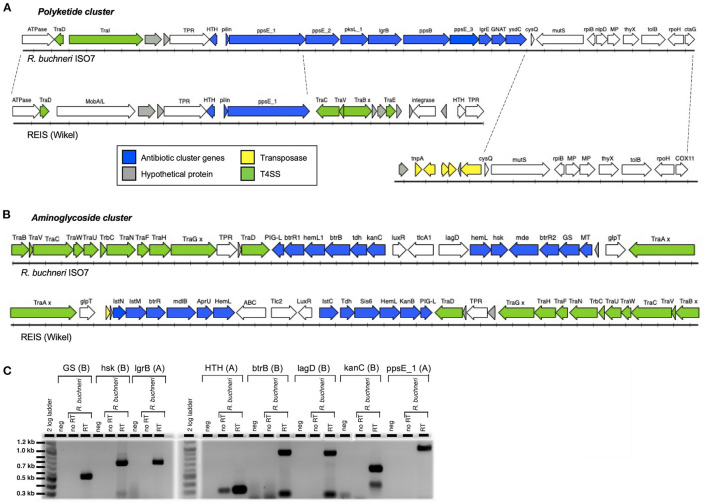
Analysis of the sequenced R. buchneri ISO7T genome revealed that it contains two gene clusters encoding proteins similar to those involved in antibiotic synthesis. Neither gene cluster is present in other members of Rickettsiaceae. The first cluster contains eleven genes including those for polyketide and other non-ribosomal peptide synthesis enzymes (Table 2; Figure 3A). Biosynthesis pathways for these compounds (which include the penicillins, cyclosporin A, vancomycin, and erythromycin) involve large multi-modular enzymes that act as assembly lines for the catalysis of chain elongation and addition of modifications (86). These are usually clustered with additional genes encoding tailoring enzymes that further modify the resulting compound, for example by methylation or cyclization, and/or by releasing it from the assembly line (87, 88). Proteins in the R. buchneri polyketide cluster show similarity to sequences from the Gammaproteobacteria Legionella israelensis, Erwinia amylovora, Pantoea ananatis, and Pectobacterium spp. as well as Cyanobacteria (Supplementary Figure S1; Supplementary Data S1). Interestingly, one of the hypothetical proteins in the cluster appears to be a type IV pilin, whose sequence obtained no blastp hits except those from the two R. buchneri genomes. Only the first three genes in the cluster are conserved in the REIS (Wikel) genome, while the remaining genes are not present (Figure 3A; Table 2; Supplementary Figure S1). Instead, the region of the REIS genome downstream of ppsE_1 encodes numerous tra genes (REIS_1815 = traC, REIS_1814 = traV, REIS_1813 = truncated traB, REIS_1810 = traE). The region upstream of the cluster is homologous in both genomes with a similar arrangement of genes (Figure 3A). Interestingly, the genes following ysdC in the Rb ISO7 genome are homologous to those in a different region of the REIS genome, with the next gene after ysdC, REISMN_1205, identical to REIS_1393 (Figure 3A). Notably the area upstream of REIS_1393 is heavily populated with transposase sequences, suggesting that a transposition event may have led to the loss of the remaining polyketide cluster genes in the REIS genome.
Table 2.
Genes in Rickettsia buchneri putative polyketide synthesis cluster.
| Label | Locus tag | Accession | Length (aa) | Annotationa |
|---|---|---|---|---|
| HTH domain* | REISMN_01150 (REIS_1819) | KDO03565.1 | 142 | Helix-turn-helix transcriptional regulator |
| Hypothetical | REISMN_01155 (REIS_1817) | KDO03566.1 | 62 | hypothetical protein; type IV pilin |
| ppsE_1* | REISMN_01160 (REIS_1816) | KDO03567.1 | 1448 | Beta-ketoacyl-acyl-carrier-protein synthase I; type I polyketide synthase; erythronolide synthase; acyltransferase domain-containing protein |
| ppsE_2 | REISMN_01165 | KDO03568.1 | 636 | Beta-ketoacyl-acyl-carrier-protein synthase I; type I polyketide synthase; SDR family NAD(P)-dependent oxidoreductase |
| pksL_1 | REISMN_01170 | KDO03569.1 | 522 | Polyketide synthase PksL; SDR family NAD(P)-dependent oxidoreductase; type I polyketide synthase |
| lgrB* | REISMN_01175 | KDO03570.1 | 630 | Linear gramicidin synthase subunit B; non-ribosomal peptide synthetase |
| ppsB | REISMN_01180 | KDO03571.1 | 878 | Plipastatin synthase subunit B; non-ribosomal peptide synthetase |
| ppsE_3 | REISMN_01185 | KDO03572.1 | 554 | Beta-ketoacyl-acyl-carrier-protein synthase I; polyketide synthase |
| lgrE | REISMN_01190 | KDO03573.1 | 239 | Linear gramicidin dehydrogenase LgrE; thioesterase |
| Hypothetical | REISMN_01195 | KDO03574.1 | 265 | hypothetical protein; GNAT family N-acetyltransferase |
| ysdC | REISMN_01200 | KDO03575.1 | 392 | Putative aminopeptidase ysdC; M42 family metallopeptidase |
Asterisks indicate genes with transcription confirmed by RT-PCR.
Locus tags for corresponding genes in the REIS (Wikel) genome are shown in brackets.
Bold type indicates annotation recorded in the genome; regular type indicates additional annotations gained through protein BLAST searches.
Figure 3.
Antibiotic gene clusters in Rickettsia buchneri. Diagrams showing the gene arrangement of R. buchneri ISO7 antibiotic gene clusters containing polyketide (A) and aminoglycoside (B) synthesis genes, and neighboring regions of the genome, and their comparison with those in the REIS (Wikel) genome. GS—glycogen synthase; hsk—homoserine kinase; MT—methyltransferase; MP—metallopeptidase; x denotes a mutation resulting in a truncation. (C) RT-PCR of R. buchneri ISO7 RNA showing the transcription of selected genes from antibiotic clusters. The gene product and antibiotic cluster (A = polyketide, B = aminoglycoside) are shown above the wells of the gel. See Tables 2, 3 for additional information.
Gillespie et al. (54) also identified an additional polyketide synthase with a putative frameshift mutation (REIS_0330). This prompted an examination of the Rb ISO7 genome for additional polyketide synthase genes. A cluster of three were identified, and these are annotated as pksL_2 (REISMN_07055), pksR (REISMN_07060), and pksN (REISMN_07065). Corresponding genes encoding identical proteins are annotated in the REIS (Wikel) RefSeq on GenBank, with corresponding locus tags REIS_RS15050 (hypothetical protein), REIS_RS10770 (methyltransferase), and REIS_RS01405 (KR domain-containing protein), respectively.
The second cluster contains fifteen genes, with eleven showing similarity to genes involved in aminoglycoside antibiotic synthesis, as well as genes coding for putative antibiotic exporters and an antibiotic resistance factor (Table 3; Figure 3B). This second antibiotic cluster was also identified in the REIS (Wikel) genome (54) and found to be associated with the Rickettsiales-amplified genetic element, RAGE-A. The aminoglycoside antibiotics include streptomycin, kanamycin, and gentamicin and are produced through complex biosynthetic pathways involving many enzymatic reactions; few of these pathways have been fully characterized (89–91). Proteins of the R. buchneri aminoglycoside cluster show similarity to those from antibiotic synthesis gene clusters from Actinobacteria and Firmicutes (54), while the putative multidrug exporter mde/mdlB, transcriptional regulator LuxR, and ABC transporter lagD show greater similarity to proteins from the Gammaproteobacteria (Supplementary Figure S2; Supplementary Data S2). Meanwhile, the nucleotide translocase tlcA1/tlc2 is related to those from other Rickettsia species (54). This cluster is conserved in both R. buchneri genomes (Figure 3B; Table 3), with the majority of sequences identical (Supplementary Figure S2).
Table 3.
Genes in Rickettsia buchneri putative aminoglycoside synthesis cluster.
| Label | Locus tag | Accession | Length (aa) | Annotationa |
|---|---|---|---|---|
| PIG-L (PIG-L) | REISMN_01820 (REIS_1505) | KDO03398.1 | 234 | GlcNAc-PI de-N-acetylase; PIG-L family deacetylase |
| btrR_1 (KanB) | REISMN_01825 (REIS_1504) | KDO03399.1 | 421 | L-Glutamine:2-deoxy-scyllo-inosose aminotransferase; DegT/DnrJ/EryC1/StrS family aminotransferase |
| hemL1 (HemL) | REISMN_01830 (REIS_1503) | KDO03400.1 | 420 | Glutamate-1-semialdehyde 2,1-aminomutase 1; aminotransferase class III-fold pyridoxal phosphate dependent enzyme |
| btrB* (Sis6) | REISMN_01835 (REIS_1502) | KDO03401.1 | 518 | Choline dehydrogenase; GMC family oxidoreductase |
| tdh (Tdh) | REISMN_01840 (REIS_1501) | KDO03402.1 | 342 | L-Threonine 3-dehydrogenase |
| kanC* (IstC) | REISMN_01845 (REIS_1500) | KDO03403.1 | 383 | 2-Deoxy-scyllo-inosose synthase |
| luxR (LuxR) | REISMN_01850 (REIS_1499) | KDO03404.1 | 275 | luxR family transcriptional regulator |
| tlcA1 (Tlc2) | REISMN_01855 (REIS_1498) | KDO03405.1 | 521 | ADP/ATP translocase 1 |
| lagD* (ABC) | REISMN_01860 (REIS_1497) | KDO03406.1 | 605 | Lactococcin-G-processing and transport ATP-binding protein LagD; ABC transporter ATP-binding protein/permease |
| hemL (HemL) | REISMN_01865 (REIS_1496) | KDO03407.1 | 435 | Glutamate-1-semialdehyde 2,1-aminomutase |
| Homoserine kinase* (AprU) | REISMN_01870 (REIS_1495) | KDO03408.1 | 342 | Homoserine kinase |
| mde protein (mdlB) | REISMN_01875 (REIS_1494) | KDO03409.1 | 594 | Putative multidrug export ATP-binding/permease protein |
| btrR_2 (btrR) | REISMN_01880 (REIS_1493) | KDO03410.1 | 392 | L-Glutamine:2-deoxy-scyllo-inosose aminotransferase; DegT/DnrJ/EryC1/StrS family aminotransferase |
| Glycogen synthase* (IstM) | REISMN_01885 (REIS_1492) | KDO03411.1 | 422 | Glycogen synthase |
| Methyl transferase (IstN) | REISMN_01890 (REIS_1491) | KDO03412 | 262 | Hypothetical protein; class I SAM-dependent methyltransferase |
Asterisks indicate genes with transcription confirmed by RT-PCR. Protein labels and locus tags for corresponding genes in the REIS (Wikel) genome are shown in brackets.
Bold type indicates annotation recorded in the genome; regular type indicates additional annotations gained through protein BLAST searches.
To determine whether these genes are actively transcribed by R. buchneri, eight genes from the two antibiotic clusters were selected for RT-PCR analysis (Tables 2, 3). Transcripts for all eight genes were detected, as indicated by bands of the expected sizes observed on gel electrophoresis (Figure 3C). Negative controls and no RT controls showed no products, except for HTH where a less robust band was seen, suggesting some DNA contamination, although there is clearly less amplification than in the RT well. Smaller bands observed in the btrB, lagD, and kanC reactions likely represent nonspecific products.
In vitro Antibiosis Experiments
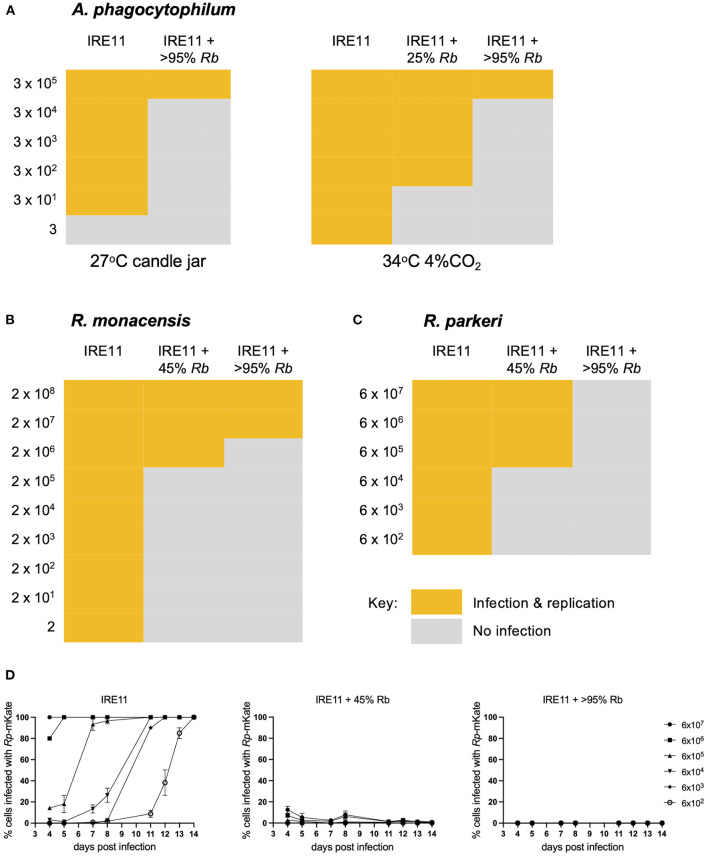
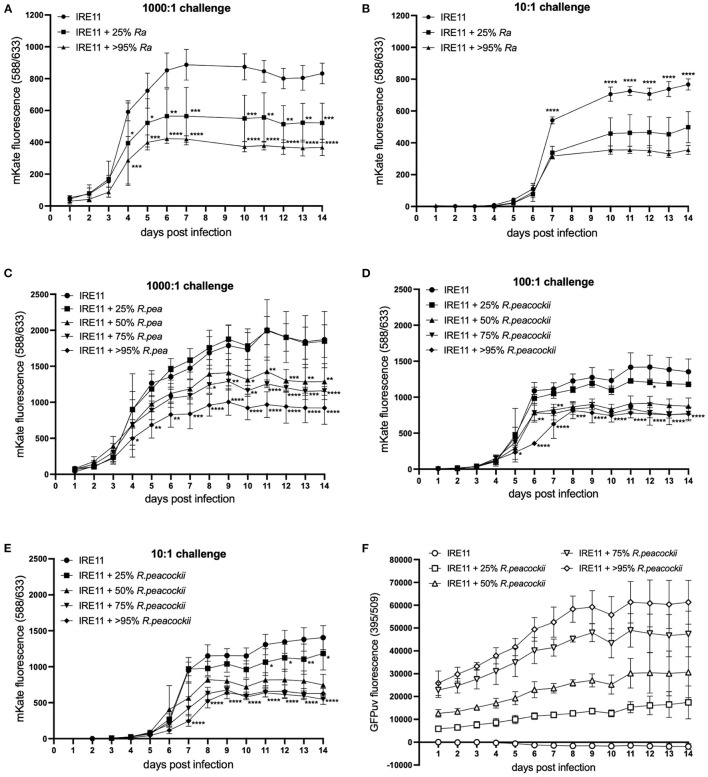
To determine whether R. buchneri might exhibit antibiosis against other bacteria infecting tick cells, cell-free red fluorescent A. phagocytophilum, R. monacensis, or R. parkeri was used to challenge tick cell cultures containing different levels of green fluorescent R. buchneri. Infectivity was measured 14 days after inoculating serial dilutions of cell-free rickettsiae/Anaplasma into replicated wells of a 24-well tissue culture plate seeded with R. buchneri-infected IRE11 cells. All three pathogens showed markedly reduced ability to infect and replicate in tick cell cultures infected with R. buchneri (Figure 4), and even a low level of infection with Rb-GFPuv led to a reduction in pathogen infection and replication. Fluorescent microscopy revealed that R. monacensis and A. phagocytophilum did not replicate in tick cells infected with R. buchneri (Supplementary Figure S3). Infectivity of R. monacensis, R. parkeri, or A. phagocytophilum in tick cell cultures infected with R. buchneri was reduced by 3–5 orders of magnitude compared to that in cells without R. buchneri (Figure 4), confirming that the presence of R. buchneri was inhibitory to the growth of other intracellular tick-borne bacteria. Furthermore, examining the percentage of infected cells by fluorescent microscopy during Rp-mKate infection showed that in IRE11 without R. buchneri, cells became completely infected at all dilutions over the 14-day period (Figure 4D). In contrast, Rp-mKate infection only reached low levels (<10%) in IRE11 cultures containing the endosymbiont in 45% cells, and almost no cells became infected with Rp-mKate in cultures with >95% cells harboring R. buchneri.
Figure 4.
Rickettsia buchneri inhibits infection and replication of other tick-borne bacteria in tick cell culture. IRE11 cells infected with different levels of R. buchneri-GFPuv were infected with serial dilutions of red fluorescent A. phagocytophilum (A), R. monacensis (B), or R. parkeri (C), and plates were monitored over 14 days for infection and replication, compared to uninfected IRE11 cells. (D) The percentage of IRE11 cells infected with R. parkeri-mKate was assessed by fluorescent microscopy. Measurements not taken on days 6, 9, and 10.
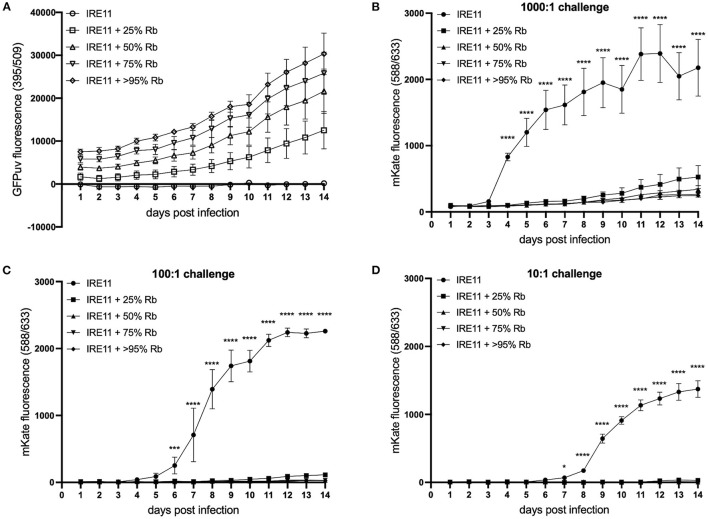
To further investigate the growth dynamics during coinfection, a fluorescent plate reader was used to measure the replication of Rp-mKate in IRE11 cells infected with Rb-GFPuv at levels of 25, 50, 75, and >95%, in comparison to uninfected IRE11. GFPuv measurement could clearly differentiate the various levels of infection with Rb-GFPuv and indicated a steady replication of the endosymbiont over the 14-day experiment (Figure 5A). No increases in GFPuv fluorescence were seen in uninfected IRE11. With a high (1,000:1) challenge, fluorescence from Rp-mKate growth indicated a rapid infection and replication of Rp-mKate in uninfected IRE11 from day 3 which then began to level off from day 6 onward (Figure 5B). In contrast, there were significantly lower rates of mKate fluorescence increase in IRE11 harboring Rb-GFPuv at all levels of infection from day 4 onward (p < 0.0001); compared to uninfected IRE11 at day 14, there was an 89% reduction in Rp-mKate in IRE11 with >95% Rb-GFPuv, 88% reduction in IRE11 with 75% Rb-GFPuv, 84% reduction in IRE11 with 50% Rb-GFPuv, and 76% reduction in IRE11 with 25% Rb-GFPuv. Similarly, 100:1 challenge of uninfected IRE11 with Rp-mKate resulted in a rapid increase in fluorescence from days 5 to 6, reaching a peak by day 12, indicating replication and spread of Rp-mKate in the cells (Figure 5C). However, in IRE11 with Rb-GFPuv, significant differences in mKate fluorescence were observed from day 6 (p < 0.001), and at day 14 the reduction was 99% in 50%, 75% and >95% infected cells, and 95% in IRE11 with 25% Rb-GFPuv. With a low challenge (10:1), mKate fluorescence increased from days 7 to 8 and reached its height at days 13–14 in uninfected IRE11, whereas in Rb-GFPuv-infected IRE11 mKate fluorescence was reduced by 99%-100% in 50%, 75%, and >95% infected cells, and by 98% in 25% infected cells (Figure 5D), with significant differences seen from day 8 (p < 0.0001) compared to IRE11 without Rb-GFPuv. No changes in mKate fluorescence were observed in control wells to which Rp-mKate was not added (data not shown). Together, these results suggest that the presence of R. buchneri in IRE11 has a significant inhibitory effect on the ability of R. parkeri to successfully infect and replicate in the culture.
Figure 5.
Rickettsia buchneri prevents infection and replication of Rickettsia parkeri in tick cell culture. Uninfected IRE11 and IRE11 infected with different levels of R. buchneri-GFPuv were challenged with different doses of R. parkeri-mKate. Rickettsial replication in IRE11 cells was monitored for 14 days by measuring GFPuv and mKate fluorescence on a microplate reader. (A) GFPuv fluorescence indicating replication of R. buchneri-GFPuv. (B–D) mKate fluorescence indicating growth of R. parkeri-mKate at challenge doses of 1,000:1 (B), 100:1 (C), and 10:1 (D); lines show mean and error bars standard deviation of three replicate wells. Means were compared to the uninfected control IRE11 using a two-way ANOVA with Dunnett's multiple-comparison test; statistically significant values are marked by asterisks *p < 0.05, ***p < 0.001, ****p < 0.0001. Data are representative of two independently performed experiments (see Supplementary Figure S4).
In order to begin to separate whether the observed inhibition of pathogen growth might be due to antibiosis by R. buchneri or competitive exclusion as observed previously (70–73) between various species of Rickettsia (that do not contain antibiotic synthesis gene clusters), additional plate reader experiments were performed using different Rickettsia species in place of R. buchneri as the resident bacteria. Firstly, this was assessed using the low-pathogenic species R. amblyommatis at infection levels of 25 and >95% in IRE11 cells, and uninfected control IRE11, which were then challenged with Rp-mKate at 1,000:1 (high) and 10:1 (low). In contrast to results obtained with Rb-GFPuv, the presence of R. amblyommatis in tick cells resulted in only partial inhibition of Rp-mKate replication. Growth of Rp-mKate in R. amblyommatis-infected IRE11 was inhibited in a manner that was relative to the level of R. amblyommatis infection (Figures 6A,B); i.e., at the low level of infection (25%), there was a lower inhibition of Rp-mKate (35% inhibition at 10:1 challenge, and 37% at 1,000:1, compared to IRE11 without R. amblyommatis at day 14), while at the high level of infection (>95%) there was a higher inhibition of Rp-mKate (54% inhibition at 10:1 challenge and 56% at 1,000:1).
Figure 6.
Rickettsia parkeri replication in the presence of R. amblyommatis or R. peacockii. Replication of R. parkeri-mKate in tick cells was monitored for 14 days by measuring mKate fluorescence on a microplate reader. (A,B) R. parkeri-mKate replication in IRE11 cells with or without R. amblyommatis at 28°C in a candle jar at challenge doses of 1,000:1 (A) and 10:1 (B); readings not taken on day 8 or 9. (C–E) R. parkeri-mKate replication in IRE11 cells with or without R. peacockii-GFPuv at 28°C in a candle jar at challenge doses of 1,000:1 (C), 100:1 (D), and 10:1 (E). (F) GFPuv fluorescence indicating replication of R. peacockii-GFPuv. Data show mean and error bars standard deviation of three replicate wells. Means were compared to the uninfected control IRE11 using a two-way ANOVA with Dunnett's multiple-comparison test; statistically significant values are marked by asterisks *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are representative of two independent experiments (see Supplementary Figure S4).
To investigate whether endosymbiotic rickettsiae may have greater exclusionary effect on pathogenic bacteria than other pathogenic Rickettsia species, as well as to further investigate the contribution of the involvement of potential antibiotic production by R. buchneri, additional plate reader competition assays were conducted using R. peacockii (an endosymbiont of D. andersoni) in place of Rb-GFPuv. Wells contained either uninfected IRE11 or IRE11 infected with R. peacockii-GFPuv at levels of 25, 50, 75, and >95%. The wells were challenged with Rp-mKate at 1,000:1, 100:1, and 10:1. In the high-challenge (1,000:1) wells, mKate fluorescence was significantly lower than that in IRE11 without R. peacockii-GFPuv at day 4 onward in cells infected with >95% R. peacockii-GFPuv (Figure 6C). mKate fluorescence in IRE11 with 75% or 50% R. peacockii-GFPuv was significantly lower from day 8 and day 10 onward, respectively, while growth of Rp-mKate in IRE11 with 25% R. peacockii-GFPuv was not significantly different from IRE11 without the endosymbiont (Figure 6C). At day 14, compared to that in IRE11 without R. peacockii-GFPuv, there was a 31% reduction in mKate fluorescence in IRE11 with 50% R. peacockii-GFPuv, a 38% reduction in IRE11 with 75% R. peacockii-GFPuv, and a 51% reduction in IRE11 with >95% R. peacockii-GFPuv. Similarly, in the 100:1 challenge wells, mKate fluorescence in 50, 75, and >95% R. peacockii-GFPuv-infected IRE11 was significantly different from that in IRE11 without R. peacockii-GFPuv, from day 5 (>95%) or day 6 (50 and 75%) onward, while mKate fluorescence in 25% R. peacockii-GFPuv-infected IRE11 was similar to the control (Figure 6D). At day 14, compared to that in IRE11 without R. peacockii-GFPuv, there were reductions of 35%, 43%, and 43% in mKate fluorescence measured in IRE11 with 50, 75, and >95% R. peacockii-GFPuv, respectively. In the 10:1 challenge experiment, mKate fluorescence was significantly reduced in IRE11 50, 75, and >95% infected with R. peacockii-GFPuv relative to IRE11 without R. peacockii-GFPuv from day 7 onward (Figure 6E). In IRE11 25% infected with R. peacockii-GFPuv, mKate fluorescence was significantly lower than the control from days 11 to 14 (Figure 6E). At day 14, in comparison to that in IRE11 without R. peacockii-GFPuv, mKate fluorescence was reduced by 16, 47, 61, and 55% in IRE11 with 25, 50, 75, and >95% R. peacockii-GFPuv, respectively. Measurement of GFPuv fluorescence could differentiate the different populations of IRE11 infected with 25, 50, 75, and >95% R. peacockii-GFPuv and implied a steady replication of R. peacockii-GFPuv over time (Figure 6F). No increase in GFPuv fluorescence was observed in uninfected IRE11; rather, there was a decline, likely due to lysis of IRE11 by Rp-mKate in infected cells (Figure 6F). Together, these results suggest that neither R. amblyommatis nor R. peacockii possess the ability to inhibit growth of competing R. parkeri to the same extent as R. buchneri.
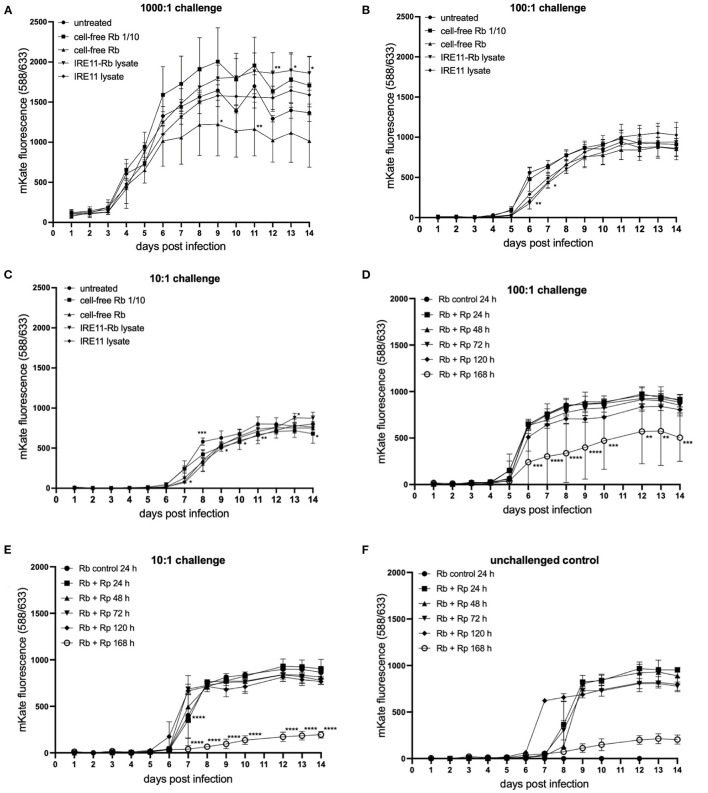
Further experiments were performed to examine whether the inhibitory effect of R. buchneri on Rp-mKate infection was due to the secretion of antibiotic products. Uninfected IRE11 was added to a 96-well plate then treated with either cell-free Rb-WT, a 1:10 dilution of cell-free Rb-WT, lysate from IRE11 heavily infected with Rb-WT, or lysate from uninfected IRE11. After 2 h, cells were challenged with Rp-mKate. Although there were some significant differences in the growth of Rp-mKate under some of the treatments, these were inconsistent across the three different challenge doses, suggesting that rather they were due to experimental variations, for example in challenge dose or cell density (Figures 7A–C). Taking these data together, there seems to be no obvious effect of any of the treatments on the growth of R. parkeri in comparison to untreated control cells. Furthermore, these results suggest that R. buchneri need to be intracellular to inhibit R. parkeri growth and that there is also no antibiotic effect of extracellular addition of Rb-WT lysate.
Figure 7.
Rickettsia parkeri replication in IRE11 treated with cell-free R. buchneri or R. buchneri lysate. Replication of R. parkeri-mKate in tick cells was monitored for 14 days by measuring mKate fluorescence on a microplate reader. (A–C) R. parkeri-mKate replication in IRE11 at challenge doses of 1,000:1 (A), 100:1 (B), and 10:1 (C) in the presence of cell-free R. buchneri, lysate from R. buchneri-infected IRE11, or lysate from uninfected IRE11. (D–F) R. parkeri-mKate replication in IRE11 treated with lysates from R. buchneri challenged with Rp-mKate for 24, 48, 72, 120, or 168 h. Treated cells were challenged with 100:1 (D) or 10:1 (E) Rp-mKate. Unchallenged control indicates that viable Rp-mKate are present in cell lysate (F). Data show mean and error bars standard deviation of three replicate wells. Means were compared to the uninfected control IRE11 using a two-way ANOVA with Dunnett's multiple-comparison test; statistically significant values are marked by asterisks *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
An additional experiment was performed to determine whether challenging R. buchneri-infected IRE11 with Rp-mKate would increase the inhibitory activity of lysates against Rp-mKate. Lysates were prepared from cell-free Rb-WT isolated from IRE11 challenged with Rp-mKate for 1–7 days and then used to treat IRE11 cells in a 96-well plate. The wells were then challenged as previously with Rp-mKate at challenge doses of 1,000:1, 100:1, and 10:1, and mKate fluorescence was measured over 14 days and compared to mKate fluorescence in cells treated with lysate from unchallenged Rb-WT-infected IRE11. At 1,000:1 challenge, there was no difference in mKate fluorescence between the control and any of the treatments (Supplementary Figure S5), showing a growth curve similar to that in other plate reader experiments. However, at challenge doses of 100:1 and 10:1, there were significant differences in mKate fluorescence in the wells treated with lysates from Rb-WT challenged with Rp-mKate for 7 days (168 h) in comparison to the control, with 43 and 77% reductions at day 14, respectively (Figures 7D,E). The mKate fluorescence in the other treatment groups was similar to that in the control wells. Unchallenged control wells were also incorporated into the experiment to check the viability of any rickettsiae in the lysate; mKate fluorescence was observed increasing on days 6–8, suggesting that viable Rp-mKate were present in the lysate (Figure 7F). However, their growth in wells treated with 168-h-challenged lysate was also much lower than in other treatment wells. The fact that the mKate fluorescence in the unchallenged control wells peaked later than in challenged wells suggests that the low level of viable Rp-mKate contributed little to the results seen in the Rp-mKate-challenged wells. Overall, these results indicate that after 7 days in the presence of Rp-mKate, lysates from Rb-WT were able to inhibit the replication of Rp-mKate in IRE11 cells at lower doses (100:1 and 10:1), which could be due to increased antibiotic activity at this time point.
Expression of Antibiotic Genes in Response to R. parkeri Infection
To investigate whether the expression of genes from the putative antibiotic clusters of R. buchneri was upregulated in response to the presence of potentially competing bacteria, a time-course experiment was set up and qRT-PCR was used to examine the relative expression of selected genes from each antibiotic cluster in the presence and absence of Rp-mKate during infection. Over a 7-day time course, several of the examined genes appeared to be upregulated in response to multiple days' challenge with Rp-mKate (Figure 8); however, there was wide variation between replicate experiments and the differences were not statistically significant.
Figure 8.
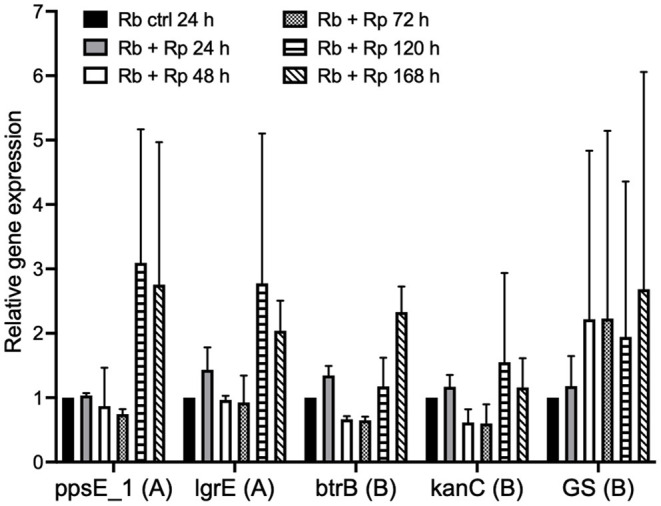
Expression of R. buchneri antibiotic cluster genes in response to Rickettsia parkeri challenge. Relative expression of R. buchneri antibiotic cluster genes in response to R. parkeri-mKate challenge over 7 days. Bars show the expression of each gene relative to the control (unchallenged R. buchneri) as the mean and standard deviation of two independent experiments for each qRT-PCR assay. Ct values were normalized to GAPDH expression in infected IRE11 cells. Genes from the polyketide and aminoglycoside clusters are denoted by (A) and (B), respectively. Means were compared to the unchallenged control using a two-way ANOVA with Dunnett's multiple-comparison test.
Investigation of R. buchneri Antibiotic Activity
The antibiotic activity of R. buchneri against extracellular bacteria was investigated using antibiotic susceptibility assays. Filter paper disks were treated with cell-free Rb-WT, Rb-WT-infected IRE11, and supernatant from IRE11 cultures infected with various levels of Rb-WT (25%, 50%, and >95%). Spectinomycin (10 and 100 μg)-treated disks were used as positive controls, and uninfected IRE11 cells and IRE11 culture supernatant were used as negative controls. Disks were placed onto Mueller–Hinton agar plates streaked with E. coli strain D21 or S. aureus strain MN8. None of the IRE11- or Rb-WT-derived treatments resulted in any inhibition of E. coli or S. aureus growth, whereas spectinomycin-treated disks prevented bacterial growth (Table 4).
Table 4.
Results from antibiotic susceptibility tests (disk diffusion assays) against E. coli and S. aureus.
| E. coli D21 | S. aureus MN8 | |
|---|---|---|
| Experiment 1—cell lysates | ||
| Cell-free Rb | - | - |
| Rb-infected IRE11 | - | - |
| Uninfected IRE11 | - | - |
| Spectinomycin 10 μg | + | + |
| Spectinomycin 100 μg | + | + |
| Experiment 2—live cells and supernatant | ||
| Cell-free Rb | - | - |
| IRE11 + 25% Rb | - | - |
| IRE11 + 50% Rb | - | - |
| IRE11 + >95% Rb | - | - |
| Uninfected IRE11 | - | - |
| IRE11 + 25% Rb supernatant | - | - |
| IRE11 + 50% Rb supernatant | - | - |
| IRE11 + >95% Rb supernatant | - | - |
| IRE11 supernatant | - | - |
| Spectinomycin 100 μg | + | + |
+ inhibition of bacterial growth; - no inhibition of bacterial growth.
Supernatant from IRE11 heavily infected with Rb-WT was also tested for activity against Rp-mKate growing in Vero cells. Addition of the supernatant to Vero cultures prior to infection with Rp-mKate did not inhibit infection; there were no differences observed in the progression of plaque formation and size of plaques in comparison to a duplicate experiment using supernatant from an uninfected IRE11 culture (Supplementary Figure S6).
Discussion
There is growing evidence that competition between endosymbiotic and pathogenic Rickettsia species in tick vectors may play an important role in the persistence and transmission of rickettsial pathogens (70–73). This study describes the existence of two putative antibiotic synthesis gene clusters in the genome of the I. scapularis endosymbiont R. buchneri isolated in Minnesota. Furthermore, in vitro experiments show that R. buchneri exerts an inhibitory effect on the growth of pathogenic rickettsiae in tick cell culture. Inhibition of R. parkeri growth by R. buchneri was greater than that exhibited by the other rickettsiae examined in this study, the low pathogenic R. amblyommatis, and the endosymbiont R. peacockii, which may suggest that this is due to the presence of antibiotic synthesis genes in R. buchneri that are lacking in other rickettsiae. Even at low infection rates, the presence of R. buchneri resulted in significant perturbation of R. parkeri growth, while the presence of either R. amblyommatis or R. peacockii showed a more direct competition where higher infection rates lead to a greater reduction in the growth of R. parkeri. These results correlate with what is known from current field and laboratory data, which suggest reduced horizontal and vertical transmission of R. parkeri or R. rickettsii by ticks in the presence of coinfecting R. amblyommatis or R. peacockii (70, 72, 73) and an almost complete absence of any coinfecting Rickettsia species in R. buchneri-infected I. scapularis. While a link between this inhibition and antibiotic synthesis is yet to be proven, this work raises the possibility that antibiotic production by R. buchneri may be a mechanism for exclusion of competing intracellular bacteria from its host tick.
Although genes from the clusters are actively transcribed by R. buchneri, no evidence of antibiotic activity was found in lysates or supernatants from R. buchneri-infected cultures, against either R. parkeri or the extracellular bacteria E. coli and S. aureus. However, when grown in the presence of R. parkeri for 7 days, lysates from R. buchneri-infected cells showed some inhibitory activity against lower challenge doses (100:1 and 10:1) of Rp-mKate, suggesting that R. parkeri challenge could be a trigger that might induce antibiotic activity of R. buchneri. However, results from qRT-PCR examining the expression of certain genes in the clusters in the presence of R. parkeri were highly variable and therefore inconclusive. Further analyses to obtain additional information about the regulation of the antibiotic synthesis clusters are required to understand how they respond to R. parkeri challenge. For example, it is unknown whether the clusters are transcribed as operons or in what ratios the different components of the clusters are required for antibiotic production. The antibiotic compounds produced by R. buchneri might only act intracellularly to prevent cells inhabited by the endosymbiont being invaded by other bacteria which could compete for resources. However, the fact that even when only a quarter of cells are occupied by R. buchneri results in inhibition of R. parkeri growth suggests that there must be some effect of R. buchneri on neighboring cells, which could potentially be mediated through the delivery of compounds to adjacent cells that are either antimicrobial or make cells refractory to infection. Further in-depth studies are required to elucidate the mechanisms of anti-rickettsial activity as well as how the putative compounds may be transported.
The proteins encoded by the aminoglycoside gene cluster show similarity to those from Actinobacteria, particularly Streptomyces spp. and Firmicutes (54), while the polyketide gene cluster encodes proteins with similarity to those of Gammaproteobacteria, particularly Erwinia amylovora, Pantoea ananatis, and Legionella spp. This suggests these gene clusters were likely obtained from environmental bacteria; indeed, members of these phyla have been identified in the I. scapularis microbiome (21, 22, 24, 32, 92). The R. buchneri genome is known to be highly plastic and contains multiple mobile genetic elements and sections of genetic material from other microorganisms (54). As ticks spend a large portion of their life in the environment in close association with soil, leaf litter, and vegetation, and also come into contact with the skin and blood of animals during host-seeking and feeding, it is likely that they encounter numerous environmental bacteria from which these gene clusters could have been transferred. Further examination of the gene clusters and similar pathways in the bacterial taxa from which these most likely originated may give us a better understanding of the likely antimicrobial compounds that could be synthesized by R. buchneri, and this could lead to future isolation and characterization of these products and determination of their antimicrobial activity. For example, Legionella spp. are known to contain multiple gene clusters for synthesis of polyketides/non-ribosomal peptides (93), and polyketide synthases were identified in Pantoea and Erwinia species that showed antagonistic activity (attributed to antibiosis) against the rice pathogen Xanthomonas oryzae (94). Similarly, strains of P. ananatis and P. agglomerans contain antibiotic biosynthesis clusters that allow them to compete with E. amylovora (95, 96). Interestingly, the Rb ISO7 polyketide cluster includes a putative type IV pilin; these proteins have diverse functions including adherence, motility, biofilm formation, host cell manipulation, DNA transfer, and protein secretion (97). It will be interesting to further investigate the role of this protein in R. buchneri, as many functions of this class of pilins could be related to the antibiotic activity of the cluster. Most genes in the polyketide cluster are absent from the REIS (Wikel) genome, and there may have been some recombination of this region resulting in excision of a large portion of the cluster that is found in Rb ISO7. This might suggest that this gene cluster is not essential for the endosymbiont, and the lack of antimicrobial products it synthesizes is compensated for by the presence of the aminoglycoside cluster. The REIS (Wikel) genome is derived from that of a lab colony of I. scapularis, whereas the Rb ISO7 genome originates from a field-collected tick, so an alternative explanation for the loss of these genes in REIS (Wikel) is that they are not essential for survival in the lab but could be necessary under natural conditions for protection against challenge from environmental microbes, for example. Further experiments to compare R. buchneri with REIS (Wikel) might determine whether absence of the polyketide cluster has any effects on survival or competition with other bacteria.
Given the inhibitory effect of R. buchneri on the in vitro infection and replication of the intracellular pathogens A. phagocytophilum, R. monacensis, and R. parkeri, one role of the endosymbiont may be the exclusion of pathogens from the tick. As R. buchneri is primarily restricted to the ovaries of female ticks, in nature it might be involved in preventing the colonization of this organ and subsequent transovarial transmission of intracellular pathogens, such as other Rickettsia species. While inhibition of A. phagocytophilum was seen in tick cell culture, it is unclear whether R. buchneri has any effect on A. phagocytophilum infection within the tick vector. Sakamoto et al. found that A. phagocytophilum levels were higher in male ticks, which have significantly lower titers of R. buchneri (19), while Steiner et al. found no correlation between R. buchneri and A. phagocytophilum infection prevalence (48), yet few other studies have examined interactions between these two bacteria in tick populations. Similarly, there is little data on whether R. buchneri has any effect on I. scapularis infection with the spirochete B. burgdorferi. Steiner and colleagues found that B. burgdorferi infection rates were significantly higher when R. buchneri was not detected, but only for male I. scapularis (48). One microbiome study found that Rickettsia reads were significantly less abundant in B. burgdorferi-positive ticks (98), while another found no differences in bacterial composition between B. burgdorferi-positive and negative ticks, for either males or females (20). Neither of these pathogens are transovarially transmitted, with A. phagocytophilum localizing to the salivary glands (99) and B. burgdorferi residing in the midgut (100). That both pathogens are highly prevalent in I. scapularis populations supports the hypothesis that any antibiotic activity R. buchneri may have is likely restricted to the ovaries. In this study, R. buchneri inhibition of tick-borne pathogens was observed in a tick cell culture system that is likely not directly comparable to life inside the tick. Further studies examining the effect of R. buchneri on infection by these pathogens using ex vivo ovaries (101) and/or live I. scapularis should provide additional insights into the dynamics of this competition. With the ability to remove R. buchneri from ticks with the use of ciprofloxacin (26), in vivo studies to further examine the consequences for ticks lacking the endosymbiont and whether they are susceptible to infection with pathogenic rickettsiae can be performed in future.
Ixodes pacificus, the main tick vector of B. burgdorferi and A. phagocytophilum in the western US, also harbors a highly prevalent rickettsial endosymbiont, “R. monacensis” strain Humboldt (56, 102, 103), and is not known to vector pathogenic rickettsiae. However, application of genome similarity sequence-threshold criteria indicates that this endosymbiont is a new distinct species closely related to R. buchneri and R. monacensis (104). In addition, I. pacificus is often coinfected with Rickettsia phylotype G022 (102, 103). While little is currently known about phylotype G022, it is more closely related to pathogenic SFG rickettsiae than to R. buchneri and Rickettsia strain Humboldt (103). Cheng et al. suggested that it is likely to be the “Tillamook agent” previously isolated from I. pacificus and shown to be mildly pathogenic in guinea pigs (105, 106). However, this agent has recently been characterized and found to be a separate species (R. tillamookensis sp. nov.) related to the transitional group of Rickettsia (107), meaning that I. pacificus is associated with two potentially pathogenic rickettsiae in addition to its endosymbiont. The Rickettsia strain Humboldt genome (NZ_LAOP01000001.1) does not appear to contain antibiotic gene clusters similar to those found in R. buchneri, which might be one reason that I. pacificus can be coinfected with both its endosymbiont and potentially pathogenic species. Interestingly, field-collected I. scapularis have occasionally been found containing R. amblyommatis, R. montanensis, or R. parkeri (25, 31, 36, 41, 42, 44, 53, 108), which could potentially occur through “spillover” from host feeding alongside infected A. americanum or D. variabilis; however, these infected I. scapularis appear to be individuals lacking R. buchneri since the endosymbiont was not detected in these ticks. Only one coinfection of I. scapularis with R. buchneri and R. parkeri has been reported (44), and this was in a blood-fed tick collected from Louisiana black bears (Ursus americanus luteolus) also being fed on by R. parkeri-infected A. maculatum, making it likely that the pathogen was present in the infected blood meal.
Interference between rickettsiae has been little studied since it was proposed 40 years ago, but existing research shows that infection with a first Rickettsia species may reduce transovarial transmission of a second Rickettsia (70, 71, 73), reduce acquisition of the second Rickettsia from infected hosts, and/or reduce its replication in the tick (72, 73), all of which could potentially lead to reduced transmission of pathogenic rickettsiae in enzootic cycles. In this study, the presence of either R. amblyommatis or R. peacockii led to a reduction in the ability of R. parkeri to infect and replicate in tick cells, reflecting what has been observed in in vivo studies in ticks (70–73). However, the mechanisms by which this interference occurs remain unexplored. One potential mechanism that has been suggested is immune priming (26), in which extracellular Rickettsia could stimulate the tick innate immune response, making the vector less susceptible to infection with a second Rickettsia. Symbionts have been shown to be important for immune development and protection from pathogens in other arthropods (109–111). It is also possible that occupation of tick cells or tissues by one Rickettsia species could physically prevent them being infected by a second Rickettsia, reducing their ability to effectively spread and replicate in the tick.
In summary, this research provides evidence that the endosymbiont of I. scapularis, R. buchneri, exerts an inhibitory effect on the growth of pathogenic tick-borne bacteria in cell culture and possesses two gene clusters encoding putative antibiotic biosynthesis machinery. This might suggest that besides being a potential nutritional endosymbiont, R. buchneri could also provide the service of preventing pathogenic Rickettsia species from occupying the ovaries, which could be detrimental to the tick's biology as has been shown to be the case for R. rickettsii in D. andersoni and D. variabilis (112, 113). While a correlation between the presence of antibiotic clusters and the ability to inhibit the growth of pathogenic Rickettsiae was found in this study, confirmation that the observed inhibition is directly linked to R. buchneri's antibiotic clusters requires further investigation. Supportive evidence from in vivo studies could have important implications for our understanding of rickettsial interference and the vector competence of I. scapularis for SFG rickettsiae.
Data Availability Statement
The datasets generated for this study can be found in the Data Repository for the University of Minnesota, https://doi.org/10.13020/ZQXG-JF78.
Author Contributions
BC, JO, TK, and UM conceived and designed the experiments. BC, NB, TK, and X-RW performed the experiments. BC and TK analyzed the data. CT developed the fluorescent plate reader assay. BC drafted the manuscript and prepared the figures. All authors contributed to the manuscript revision and approved the submitted version.
Funding
This research was supported by generous funding from the National Institutes of Health grants R01 AI49424 and R01 AI081690 to UM (http://www.grants.nih.gov/grants/oer.htm) and funds from the University of Minnesota Agricultural Experiment Station. The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Rod Felsheim (formerly Department of Entomology, University of Minnesota) for the preliminary analysis of the antibiotic gene clusters in the Rickettsia buchneri ISO7 genome.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.748427/full#supplementary-material
References
- 1.Eisen RJ, Eisen L. The blacklegged tick, Ixodes scapularis: an increasing public health concern. Trends Parasitol. (2018) 34:295–309. 10.1016/j.pt.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oteo JA, Portillo A. Tick-borne rickettsioses in Europe. Ticks Tick Borne Dis. (2012) 3:271–8. 10.1016/j.ttbdis.2012.10.035 [DOI] [PubMed] [Google Scholar]
- 3.Capligina V, Seleznova M, Akopjana S, Freimane L, Lazovska M, Krumins R, et al. Large-scale countrywide screening for tick-borne pathogens in field-collected ticks in Latvia during 2017-2019. Parasit Vectors. (2020) 13:351. 10.1186/s13071-020-04219-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sormunen JJ, Andersson T, Aspi J, Bäck J, Cederberg T, Haavisto N, et al. Monitoring of ticks and tick-borne pathogens through a nationwide research station network in Finland. Ticks Tick Borne Dis. (2020) 11:101449. 10.1016/j.ttbdis.2020.101449 [DOI] [PubMed] [Google Scholar]
- 5.Klitgaard K, Kjær LJ, Isbrand A, Hansen MF, Bødker R. Multiple infections in questing nymphs and adult female Ixodes ricinus ticks collected in a recreational forest in Denmark. Ticks Tick Borne Dis. (2019) 10:1060–5. 10.1016/j.ttbdis.2019.05.016 [DOI] [PubMed] [Google Scholar]
- 6.Knoll S, Springer A, Hauck D, Schunack B, Pachnicke S, Strube C. Regional, seasonal, biennial and landscape-associated distribution of Anaplasma phagocytophilum and Rickettsia spp. infections in Ixodes ticks in northern Germany and implications for risk assessment at larger spatial scales. Ticks Tick Borne Dis. (2021) 12:101657. 10.1016/j.ttbdis.2021.101657 [DOI] [PubMed] [Google Scholar]
- 7.Kjær LJ, Klitgaard K, Soleng A, Edgar KS, Lindstedt HEH, Paulsen KM, et al. Spatial patterns of pathogen prevalence in questing Ixodes ricinus nymphs in southern Scandinavia, 2016. Sci Rep. (2020) 10:1–14. 10.1038/s41598-020-76334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remesar S, Díaz P, Portillo A, Santibáñez S, Prieto A, Díaz-Cao JM, et al. Prevalence and molecular characterization of Rickettsia spp. in questing ticks from north-western Spain. Exp Appl Acarol. (2019) 79:267–78. 10.1007/s10493-019-00426-9 [DOI] [PubMed] [Google Scholar]
- 9.Shao JW, Zhang XL, Li WJ, Huang HL, Yan J. Distribution and molecular characterization of rickettsiae in ticks in Harbin area of Northeastern China. PLoS Negl Trop Dis. (2020) 14:e0008342. 10.1371/journal.pntd.0008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Pan YS, Jiang BG, Ye RZ, Chang QC, Shao HZ, et al. Prevalence of multiple tick-borne pathogens in various tick vectors in Northeastern China. Vector-Borne Zoonotic Dis. (2021) 21:162–71. 10.1089/vbz.2020.2712 [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Liang X, Wang H, Sun X, Bai X, Hu B, et al. Molecular evidence of the spotted fever group Rickettsiae in ticks from Yunnan Province, Southwest China. Exp Appl Acarol. (2020) 80:339–48. 10.1007/s10493-020-00467-5 [DOI] [PubMed] [Google Scholar]
- 12.Rar V, Livanova N, Tkachev S, Kaverina G, Tikunov A, Sabitova Y, et al. Detection and genetic characterization of a wide range of infectious agents in Ixodes pavlovskyi ticks in Western Siberia, Russia. Parasit Vectors. (2017) 10:258. 10.1186/s13071-017-2186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katargina O, Geller J, Ivanova A, Värv K, Tefanova V, Vene S, et al. Detection and identification of Rickettsia species in Ixodes tick populations from Estonia. Ticks Tick Borne Dis. (2015) 6:689–94. 10.1016/j.ttbdis.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 14.Igolkina Y, Bondarenko E, Rar V, Epikhina T, Vysochina N, Pukhovskaya N, et al. Genetic variability of Rickettsia spp. in Ixodes persulcatus ticks from continental and island areas of the Russian Far East. Ticks Tick Borne Dis. (2016) 7:1284–9. 10.1016/j.ttbdis.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 15.Cheng C, Fu W, Ju W, Yang L, Xu N, Wang Y, et al. Diversity of spotted fever group Rickettsia infection in hard ticks from Suifenhe, Chinese–Russian border. Ticks Tick Borne Dis. (2016) 7:715–9. 10.1016/j.ttbdis.2016.02.023 [DOI] [PubMed] [Google Scholar]
- 16.Graves SR, Stenos J. Tick-borne infectious diseases in Australia. Med J Aust. (2017) 206:320–4. 10.5694/mja17.00090 [DOI] [PubMed] [Google Scholar]
- 17.Graves SR, Jackson C, Hussain-Yusuf H, Vincent G, Nguyen C, Stenos J, et al. Ixodes holocyclus tick-transmitted human pathogens in North-Eastern New South Wales, Australia. Trop Med Infect Dis. (2016) 1:4. 10.3390/tropicalmed1010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtti TJ, Felsheim RF, Burkhardt NY, Oliver JD, Heu CC, Munderloh UG. Rickettsia buchneri sp. nov, a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int J Syst Evol Microbiol. (2015) 65:965–70. 10.1099/ijs.0.000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto J, Diaz GES, Wagner EA. Bacterial communities of Ixodes scapularis from Central Pennsylvania, USA. Insects. (2020) 11:718. 10.3390/insects11100718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thapa S, Zhang Y, Allen MS. Bacterial microbiomes of Ixodes scapularis ticks collected from Massachusetts and Texas, USA. BMC Microbiol. (2019) 19:138. 10.1186/s12866-019-1514-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ, et al. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl Environ Microbiol. (2015) 81:6200–9. 10.1128/AEM.01562-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zolnik CP, Prill RJ, Falco RC, Daniels TJ, Kolokotronis SO. Microbiome changes through ontogeny of a tick pathogen vector. Mol Ecol. (2016) 25:4963–77. 10.1111/mec.13832 [DOI] [PubMed] [Google Scholar]
- 23.Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol. (2015) 24:2566–79. 10.1111/mec.13187 [DOI] [PubMed] [Google Scholar]
- 24.Thapa S, Zhang Y, Allen MS. Effects of temperature on bacterial microbiome composition in Ixodes scapularis ticks. Microbiologyopen. (2019) 8:e00719. 10.1002/mbo3.719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinkerhoff RJ, Clark C, Ocasio K, Gauthier DT, Hynes WL. Factors affecting the microbiome of Ixodes scapularis and Amblyomma americanum. PLoS ONE. (2020) 15:e0232398. 10.1371/journal.pone.0232398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olafson PU, Buckmeier BG, May MA, Thomas DB. Molecular screening for rickettsial bacteria and piroplasms in ixodid ticks surveyed from white-tailed deer (Odocoileus virginianus) and nilgai antelope (Boselaphus tragocamelus) in southern Texas. Int J Parasitol Parasites Wildl. (2020) 13:252–60. 10.1016/j.ijppaw.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Khafaji AM, Armstrong SD, Varotto Boccazzi I, Gaiarsa S, Sinha A, Li Z, et al. Rickettsia buchneri, symbiont of the deer tick Ixodes scapularis, can colonise the salivary glands of its host. Ticks Tick Borne Dis. (2020) 11:101299. 10.1016/j.ttbdis.2019.101299 [DOI] [PubMed] [Google Scholar]
- 28.Livengood J, Hutchinson ML, Thirumalapura N, Tewari D. Detection of Babesia, Borrelia, Anaplasma, Rickettsia spp. in adult black-legged ticks (Ixodes scapularis) from Pennsylvania, United States, with a Luminex multiplex bead assay. Vector-Borne Zoonotic Dis. (2020) 20:406–11. 10.1089/vbz.2019.2551 [DOI] [PubMed] [Google Scholar]
- 29.Hodo CL, Forgacs D, Auckland LD, Bass K, Lindsay C, Bingaman M, et al. Presence of diverse Rickettsia spp. and absence of Borrelia burgdorferi sensu lato in ticks in an East Texas forest with reduced tick density associated with controlled burns. Ticks Tick Borne Dis. (2020) 11:101310. 10.1016/j.ttbdis.2019.101310 [DOI] [PubMed] [Google Scholar]
- 30.Oliver JD, Price LD, Burkhardt NY, Heu CC, Khoo BS, Thorpe CJ, et al. Growth dynamics and antibiotic elimination of symbiotic Rickettsia buchneri in the tick Ixodes scapularis (Acari: Ixodidae). Appl Environ Microbiol. (2020) 87:e01672–20. 10.1128/AEM.01672-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pokutnaya D, Molaei G, Weinberger DM, Vossbrinck CR, Diaz AJ. Prevalence of infection and co-infection and presence of Rickettsial endosymbionts in Ixodes scapularis (Acari: Ixodidae) in Connecticut, USA. J Parasitol. (2020) 106:30–7. 10.1645/19-116 [DOI] [PubMed] [Google Scholar]
- 32.Molaei G, Little EAH, Stafford KC, Gaff H. A. seven-legged tick: Report of a morphological anomaly in Ixodes scapularis (Acari: Ixodidae) biting a human host from the Northeastern United States. Ticks Tick Borne Dis. (2020) 11:101304. 10.1016/j.ttbdis.2019.101304 [DOI] [PubMed] [Google Scholar]
- 33.Gleim ER, Conner LM, Zemtsova GE, Levin ML, Wong P, Pfaff MA, et al. Rickettsiales in ticks removed from outdoor workers, southwest Georgia and northwest Florida, USA. Emerg Infect Dis. (2019) 25:1019–21. 10.3201/eid2505.180438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokarz R, Tagliafierro T, Sameroff S, Cucura DM, Oleynik A, Che X, et al. Microbiome analysis of Ixodes scapularis ticks from New York and Connecticut. Ticks Tick Borne Dis. (2019) 10:894–900. 10.1016/j.ttbdis.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 35.Gleim ER, Zemtsova GE, Berghaus RD, Levin ML, Conner M, Yabsley MJ. Frequent prescribed fires can reduce risk of tick-borne diseases. Sci Rep. (2019) 9:9974. 10.1038/s41598-019-46377-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landesman WJ, Mulder K, Fredericks LP, Allan BF. Cross-kingdom analysis of nymphal-stage Ixodes scapularis microbial communities in relation to Borrelia burgdorferi infection and load. FEMS Microbiol Ecol. (2019) 95: fiz167. 10.1093/femsec/fiz167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-Vicente S, Tagliafierro T, Coleman JL, Benach JL, Tokarz R. Polymicrobial nature of tick-borne diseases. MBio. (2019) 10:e02055–19. 10.1128/mBio.02055-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendell NL, Reynolds ES, Blanton LS, Hermance ME, Londoño AF, Hart CE, et al. Detection of Rickettsiae, Borreliae, and Ehrlichiae in ticks collected from Walker County, Texas, 2017-2018. Insects. (2019) 10:315. 10.3390/insects10100315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagen R, Verhoeve VI, Gillespie JJ, Driscoll TP. Conjugative transposons and their cargo genes vary across natural populations of Rickettsia buchneri infecting the tick Ixodes scapularis. Genome Biol Evol. (2018) 10:3218–29. 10.1093/gbe/evy247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubie TR, Turner J, Noden BH. Questing behavior and analysis of tick-borne bacteria in Ixodes scapularis (Acari: Ixodidae) in Oklahoma. J Med Entomol. (2018) 55:1569–74. 10.1093/jme/tjy133 [DOI] [PubMed] [Google Scholar]
- 41.Castellanos AA, Medeiros MCI, Hamer GL, Morrow ME, Eubanks MD, Teel PD, et al. Decreased small mammal and on-host tick abundance in association with invasive red imported fire ants (Solenopsis invicta). Biol Lett. (2016) 12:20160463. 10.1098/rsbl.2016.0463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sayler K, Rowland J, Boyce C, Weeks E. Borrelia burgdorferi DNA absent, multiple Rickettsia spp. DNA present in ticks collected from a teaching forest in North Central Florida. Ticks Tick Borne Dis. (2017) 8:53–9. 10.1016/j.ttbdis.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 43.Gleim ER, Garrison LE, Vello MS, Savage MY, Lopez G, Berghaus RD, et al. Factors associated with tick bites and pathogen prevalence in ticks parasitizing humans in Georgia, USA. Parasit Vectors. (2016) 9:125. 10.1186/s13071-016-1408-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell EA, Williamson PC, Billingsley PM, Seals JP, Ferguson EE, Allen MS. Frequency and distribution of Rickettsiae, Borreliae, and Ehrlichiae detected in human-parasitizing ticks, Texas, USA. Emerg Infect Dis. (2016) 22:312–5. 10.3201/eid2202.150469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trout Fryxell RT, Steelman CD, Szalanski AL, Billingsley PM, Williamson PC. Molecular detection of Rickettsia species within ticks (Acari: Ixodidae) collected from Arkansas United States. J Med Entomol. (2015) 52:500–8. 10.1093/jme/tjv027 [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Kakumanu ML, Ponnusamy L, Vaughn M, Funkhouser S, Thornton H, et al. Prevalence of Rickettsiales in ticks removed from the skin of outdoor workers in North Carolina. Parasit Vectors. (2014) 7:607. 10.1186/s13071-014-0607-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mays SE, Hendricks BM, Paulsen DJ, Houston AE, Trout Fryxell RT. Prevalence of five tick-borne bacterial genera in adult Ixodes scapularis removed from white-tailed deer in western Tennessee. Parasit Vectors. (2014) 7:473. 10.1186/s13071-014-0473-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leydet BF, Liang FT. Detection of human bacterial pathogens in ticks collected from Louisiana black bears (Ursus americanus luteolus). Ticks Tick Borne Dis. (2013) 4:191–6. 10.1016/j.ttbdis.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williamson PC, Billingsley PM, Teltow GJ, Seals JP, Turnbough MA, Atkinson SF. Borrelia, Ehrlichia, Rickettsia spp. in ticks removed from persons, Texas, USA. Emerg Infect Dis. (2010) 16:441–6. 10.3201/eid1603.091333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mead DG, Huang J, Fritzen CM, Dunn JR, Cohen SB, Dunlap BG, et al. Absence of Rickettsia rickettsii and occurrence of other spotted fever group Rickettsiae in ticks from Tennessee. Am J Trop Med Hyg. (2010) 83:653–7. 10.4269/ajtmh.2010.09-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yabsley MJ, Nims TN, Savage MY, Durden LA. Ticks and tick-borne pathogens and putative symbionts of Black Bears (Ursus americanus floridanus) from Georgia and Florida. J Parasitol. (2009) 95:1125–8. 10.1645/GE-2111.1 [DOI] [PubMed] [Google Scholar]
- 52.Steiner FE, Pinger RR, Vann CN, Grindle N, Civitello D, Clay K., et al. Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp, Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol. (2009) 45:289–97. 10.1093/jmedent/45.2.289 [DOI] [PubMed] [Google Scholar]
- 53.Swanson KI, Norris DE. Co-circulating microorganisms in questing Ixodes scapularis nymphs in Maryland. J Vector Ecol. (2007) 32:243. 10.3376/1081-1710(2007)32[243:CMIQIS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, et al. A rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. (2012) 194:376–94. 10.1128/JB.06244-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, et al. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS ONE. (2015) 10:e0144552. 10.1371/journal.pone.0144552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alowaysi M, Chen J, Stark S, Teague K, LaCourse M, Proctor J, et al. Isolation and characterization of a Rickettsia from the ovary of a Western black-legged tick, Ixodes pacificus. Ticks Tick Borne Dis. (2019) 10:918–23. 10.1016/j.ttbdis.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salje J. Cells within cells: Rickettsiales and the obligate intracellular bacterial lifestyle. Nat Rev Microbiol. (2021) 19:375–90. 10.1038/s41579-020-00507-2 [DOI] [PubMed] [Google Scholar]
- 58.Centers for Disease Control and Prevention . Epidemiology and Statistics - Rocky Mountain Spotted Fever. (2018). Available online at: https://www.cdc.gov/rmsf/stats/index.html (accessed July 13, 2021).
- 59.Dahlgren SF, Paddock CD, Springer YP, Eisen RJ, Behravesh CB. Expanding range of Amblyomma americanum and simultaneous changes in the epidemiology of spotted fever group rickettsiosis in the United States. Am J Trop Med Hyg. (2016) 94:35–42. 10.4269/ajtmh.15-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levin ML, Zemtsova GE, Killmaster LF, Snellgrove A, Schumacher LBM. Vector competence of Amblyomma americanum (Acari: Ixodidae) for Rickettsia rickettsii. Ticks Tick Borne Dis. (2017) 8:615–22. 10.1016/j.ttbdis.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breitschwerdt EB, Hegarty BC, Maggi RG, Lantos PM, Aslett DM, Bradley JM. Rickettsia rickettsii transmission by a lone star tick, North Carolina. Emerg Infect Dis. (2011) 17:873–5. 10.3201/eid1705.101530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goddard J. Experimental infection of Lone star ticks. Amblyomma americanum (L), with Rickettsia parkeri and exposure of Guinea pigs to the agent. J Med Entomol. (2003) 40:686–9. 10.1603/0022-2585-40.5.686 [DOI] [PubMed] [Google Scholar]
- 63.Paddock CD, Sumner JW, Comer JA, Zaki SR, Goldsmith CS, Goddard J, et al. Rickettsia parkeri: A newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis. (2004) 38:805–11. 10.1086/381894 [DOI] [PubMed] [Google Scholar]
- 64.Yen W-Y, Stern K, Mishra S, Helminiak L, Sanchez-Vicente S, Kim HK. Virulence potential of Rickettsia amblyommatis for spotted fever pathogenesis in mice. Pathog Dis. (2021) 79:ftab024. 10.1093/femspd/ftab024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snellgrove AN, Krapiunaya I, Scott P, Levin ML. Assessment of the pathogenicity of Rickettsia amblyommatis, Rickettsia bellii, and Rickettsia montanensis in a Guinea pig model. Vector-Borne Zoonotic Dis. (2021) 21:232–41. 10.1089/vbz.2020.2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Billeter SA, Blanton HL, Little SE, Levy MG, Breitschwerdt EB. Detection of “Rickettsia amblyommii” in association with a tick bite rash. Vector-Borne Zoonotic Dis. (2007) 7:607–10. 10.1089/vbz.2007.0121 [DOI] [PubMed] [Google Scholar]
- 67.Paddock CD, Goddard J. The evolving medical and veterinary importance of the Gulf coast tick (Acari: Ixodidae). J Med Entomol. (2015) 52:230–52. 10.1093/jme/tju022 [DOI] [PubMed] [Google Scholar]
- 68.Molaei G, Little EAH, Khalil N, Ayres BN, Nicholson WL, Paddock CD. Established population of the Gulf Coast tick, Amblyomma maculatum (Acari: Ixodidae), infected with Rickettsia parkeri (Rickettsiales: Rickettsiaceae), in Connecticut. J Med Entomol. (2021) 58:1459–62. 10.1093/jme/tjaa299 [DOI] [PubMed] [Google Scholar]
- 69.Phillips VC, Zieman EA, Kim CH, Stone CM, Tuten HC, Jiménez FA. Documentation of the expansion of the Gulf Coast tick (Amblyomma maculatum) and Rickettsia parkeri: First report in Illinois. J Parasitol. (2020) 106:9–13. 10.1645/19-118 [DOI] [PubMed] [Google Scholar]
- 70.Burgdorfer W, Hayes S, Mavros A. Nonpathogenic rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsia. In: Burgdorfer W, Anacker R. editor. Rickettsiae and Rickettsial Diseases. New York: Academic Press. (1981) p. 585–594. [Google Scholar]
- 71.Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J Med Entomol. (2002) 39:809–13. 10.1603/0022-2585-39.6.809 [DOI] [PubMed] [Google Scholar]
- 72.Wright CL, Sonenshine DE, Gaff HD, Hynes WL. Rickettsia parkeri transmission to Amblyomma americanum by cofeeding with Amblyomma maculatum (Acari: Ixodidae) and potential for spillover. J Med Entomol. (2015) 52:1090–5. 10.1093/jme/tjv086 [DOI] [PubMed] [Google Scholar]
- 73.Levin ML, Schumacher LBM, Snellgrove A. Effects of Rickettsia amblyommatis infection on the vector competence of Amblyomma americanum ticks for Rickettsia rickettsii. Vector-Borne Zoonotic Dis. (2018) 18:579–87. 10.1089/vbz.2018.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paddock CD, Denison AM, Dryden MW, Noden BH, Lash RR, Abdelghani SS, et al. High prevalence of “Candidatus Rickettsia andeanae” and apparent exclusion of Rickettsia parkeri in adult Amblyomma maculatum (Acari: Ixodidae) from Kansas and Oklahoma. Ticks Tick Borne Dis. (2015) 6:297–302. 10.1016/j.ttbdis.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H., et al. Clustal W and Clustal X version 20. Bioinformatics. (2007) 23:2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 76.Edgar RC. MUSCLE, multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, et al. Invasion and intracellular development of the Human Granulocytic Ehrlichiosis agent in tick cell culture. J Clin Microbiol. (1999) 37:2518–24. 10.1128/JCM.37.8.2518-2524.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simser JA, Palmer AT, Fingerle V, Wilske B, Kurtti TJ, Munderloh UG. Rickettsia monacensis sp. nov, a Spotted Fever Group Rickettsia, from ticks (Ixodes ricinus) collected in a European city park. Appl Environ Microbiol. (2002) 68:4559–66. 10.1128/AEM.68.9.4559-4566.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurtti TJ, Simser JA, Baldridge GD, Palmer AT, Munderloh UG. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J Invertebr Pathol. (2005) 90:177–86. 10.1016/j.jip.2005.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurtti TJ, Burkhardt NY, Heu CC, Munderloh UG. Fluorescent protein expressing Rickettsia buchneri and Rickettsia peacockii for tracking symbiont-tick cell interactions. Vet Sci. (2016) 3:34. 10.3390/vetsci3040034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X-R, Burkhardt NY, Kurtti TJ, Oliver JD, Price LD, Cull B, et al. Mitochondrion-dependent apoptosis is essential for Rickettsia parkeri infection and replication in vector cells. mSystems. (2021) 6. 10.1128/mSystems.01209-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, et al. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. (2006) 6:42. 10.1186/1472-6750-6-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oliver JD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl Environ Microbiol. (2014) 80:1170–6. 10.1128/AEM.03352-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ammerman NC, Beier-Sexton M, Azad AF. Growth and maintenance of Vero cell lines. Curr Protoc Microbiol. (2008) 11:Appendix 4E. 10.1002/9780471729259.mca04es11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pfaffl MW. Quantification strategies in real-time PCR. In Bustin S. editor. A-Z of quantitative PCR. La Jolla, CA: International University Line. (2004) p. 87–112. [Google Scholar]
- 86.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. (2001) 18:380–416. 10.1039/a909079g [DOI] [PubMed] [Google Scholar]
- 87.Walsh CT. Polyketide and nonribosomal peptide antibiotics: Modularity and versatility. Science. (2004) 303:1805–10. 10.1126/science.1094318 [DOI] [PubMed] [Google Scholar]
- 88.Keating TA, Ehmann DE, Kohli RM, Marshall CG, Trauger JW, Walsh CT. Chain termination steps in nonribosomal peptide synthetase assembly lines: Directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. ChemBioChem. (2001) 2:99–107. [DOI] [PubMed] [Google Scholar]
- 89.Kudo F, Eguchi T. Biosynthetic genes for aminoglycoside antibiotics. J Antibiot. (2009) 62:471–81. 10.1038/ja.2009.76 [DOI] [PubMed] [Google Scholar]
- 90.Park JW, Park SR, Nepal KK, Han AR, Ban YH, Yoo YJ, et al. Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation. Nat Chem Biol. (2011) 7:843–52. 10.1038/nchembio.671 [DOI] [PubMed] [Google Scholar]
- 91.Ban YH, Song MC, Hwang J, Shin H, Kim HJ, Hong SK, et al. Complete reconstitution of the diverse pathways of gentamicin B biosynthesis. Nat Chem Biol. (2019) 15:295–303. 10.1038/s41589-018-0203-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. (2014) 15:58–71. 10.1016/j.chom.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnston CW, Plumb J, Li X, Grinstein S, Magarvey NA. Informatic analysis reveals Legionella as a source of novel natural products. Synth Syst Biotechnol. (2016) 1:130–6. 10.1016/j.synbio.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dewi RK. Suranto, Susilowati A, Wahyudi AT. Molecular identification of 16S rDNA and polyketide synthase genes of antagonist bacteria against Xanthomonas oryzae pathovar oryzae from rice phyllosphere. Asian J Plant Pathol. (2015) 9:148–57. 10.3923/ajppaj.2015.148.157 [DOI] [Google Scholar]
- 95.Walterson AM, Smith DDN, Stavrinides J. Identification of a Pantoea biosynthetic cluster that directs the synthesis of an antimicrobial natural product. PLoS ONE. (2014) 9:e96208. 10.1371/journal.pone.0096208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jin M, Fischbach MA, Clardy J. A. biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J Am Chem Soc. (2006) 128:10660–1. 10.1021/ja063194c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. (2012) 76:740–72. 10.1128/MMBR.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chauhan G, McClure J, Hekman J, Marsh PW, Bailey JA, Daniels RF, et al. Combining citizen science and genomics to investigate tick, pathogen, and commensal microbiome at single-tick resolution. Front Genet. (2020) 10:1322. 10.3389/fgene.2019.01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Telford SR, Dawson JE, Katavolos P, Warner CK, Kolbert CP, Persing DH. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci. (1996) 93:6209–14. 10.1073/pnas.93.12.6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Silva AM, Fikrig E. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg. (1995) 53:397–404. 10.4269/ajtmh.1995.53.397 [DOI] [PubMed] [Google Scholar]
- 101.Grabowski JM, Kissinger R. Ixodid tick dissection and tick ex vivo organ cultures for tick-borne virus research. Curr Protoc Microbiol. (2020) 59:e118. 10.1002/cpmc.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis. (2013) 4:280–7. 10.1016/j.ttbdis.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Phan JN, Lu CR, Bender WG, Smoak RM, Zhong J. Molecular detection and identification of Rickettsia species in Ixodes pacificus in California. Vector-Borne Zoonotic Dis. (2011) 11:957–61. 10.1089/vbz.2010.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diop A, El Karkouri K, Raoult D, Fournier PE. Genome sequence-based criteria for demarcation and definition of species in the genus Rickettsia. Int J Syst Evol Microbiol. (2020) 70:1738–50. 10.1099/ijsem.0.003963 [DOI] [PubMed] [Google Scholar]
- 105.Lane RS, Philip RN, Casper EA. Ecology of tick-borne agents in California. II. Further observations on rickettsiae. In: Burgdorfer W, Anacker R. editor. Rickettsiae and Rickettsial Disease. New York: Academic Press. (1981) p. 575–584. [Google Scholar]
- 106.Hughes LE, Clifford CM, Gresbrink R, Thomas LA, Keirans JE. Isolation of a spotted fever group Rickettsia from the Pacific Coast tick, Ixodes pacificus, in Oregon. Am J Trop Med Hyg. (1976) 25:513–6. 10.4269/ajtmh.1976.25.513 [DOI] [PubMed] [Google Scholar]
- 107.Gauthier DT, Karpathy SE, Grizzard SL, Batra D, Rowe LA, Paddock CD. Characterization of a novel transitional group Rickettsia species (Rickettsia tillamookensis sp. nov) from the western black-legged tick, Ixodes pacificus. Int J Syst Evol Microbiol. (2021) 71:004880. 10.1099/ijsem.0.004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Santanello C, Barwari R, Troyo A. Spotted fever group Rickettsiae in ticks from Missouri. Ticks Tick Borne Dis. (2018) 9:1395–9. 10.1016/j.ttbdis.2018.06.008 [DOI] [PubMed] [Google Scholar]
- 109.Hamilton PT, Perlman SJ. Host defense via symbiosis in Drosophila. PLoS Pathog. (2013) 9:e1003808. 10.1371/journal.ppat.1003808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol. (2012) 188:3395–403. 10.4049/jimmunol.1103691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oliver KM, Moran NA, Hunter MS. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci. (2005) 102:12795–800. 10.1073/pnas.0506131102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niebylski ML, Peacock MG, Schwan TG. Lethal effect of Rickettsia rickettsii on its tick vector (Dermacentor andersoni). Appl Environ Microbiol. (1999) 65:773–8. 10.1128/AEM.65.2.773-778.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schumacher L, Snellgrove A, Levin ML. Effect of Rickettsia rickettsii (Rickettsiales: Rickettsiaceae) infection on the biological parameters and survival of its tick vector - Dermacentor variabilis (Acari: Ixodidae). J Med Entomol. (2016) 53:172–6. 10.1093/jme/tjv166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the Data Repository for the University of Minnesota, https://doi.org/10.13020/ZQXG-JF78.