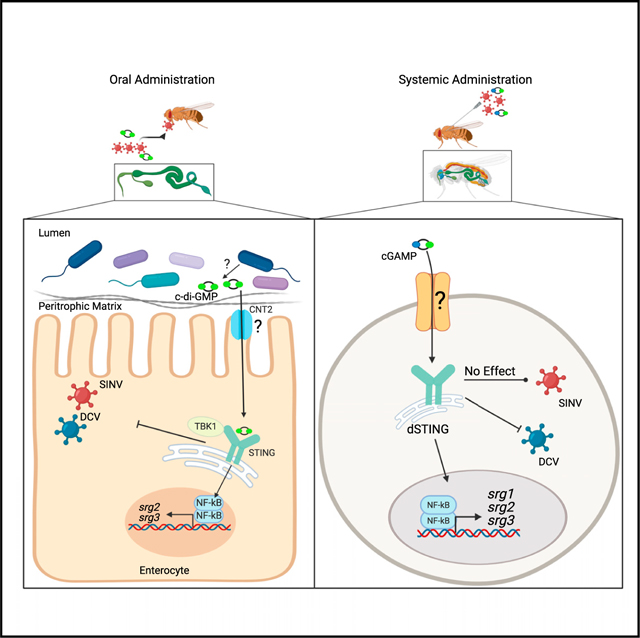
SUMMARY
Enteric pathogens overcome barrier immunity within the intestinal environment that includes the endogenous flora. The microbiota produces diverse ligands, and the full spectrum of microbial products that are sensed by the epithelium and prime protective immunity is unknown. Using Drosophila, we find that the gut presents a high barrier to infection, which is partially due to signals from the microbiota, as loss of the microbiota enhances oral viral infection. We report cyclic dinucleotide (CDN) feeding is sufficient to protect microbiotadeficient flies from enhanced oral infection, suggesting that bacterial-derived CDNs induce immunity. Mechanistically, we find CDN protection is dSTING- and dTBK1-dependent, leading to NF-kB-dependent gene expression. Furthermore, we identify the apical nucleoside transporter, CNT2, as required for oral CDN protection. Altogether, our studies define a role for bacterial products in priming immune defenses in the gut.
In brief
Segrist et al. find the cyclic dinucleotide, c-di-GMP, primes antiviral immunity in intestinal epithelial cells through the conserved apical purine transporter, CNT2, and the ancient dSTING-dTBK1 antiviral pathway.
Graphical abstract
INTRODUCTION
The gut houses an extensive microbiota which influences homeostasis, nutrient uptake, and immunity. The mutualistic interaction between host and microbiome relies on the ability of microbes to colonize the gut and the host’s ability to tolerate and control them. There is a vast spectrum of microbiota-derived ligands, and how they shape immunity is not fully characterized, though products like toll-like receptor (TLR) ligands and peptidoglycan prime enterocytes to promote immunity (Rakoff-Nahoum et al., 2004; Sansone et al., 2015). Commensals produce cyclic dinucleotides (CDNs) (Danilchanka and Mekalanos, 2013), but their impact on barrier immunity is unknown. Drosophila, with its high conservation, powerful genetic tools, and simplified biology, provides important insights into the complex interactions between host, microbiota, and enteric viral infection (Buchon et al., 2014; Xu et al., 2013).
The conserved protein STING binds endogenous CDNs produced by cGAS and exogenous bacterial CDNs and controls pathogen infections, from mammals to invertebrates (Ahn and Barber, 2019; Cai and Imler, 2021). CDNs are not cell permeable, therefore, extracellular CDNs are transported into cells. Recently, CDN transporters with tissue-specific activity were discovered (Lahey et al., 2020; Luteijn et al., 2019; Ritchie et al., 2019; Zhou et al., 2020a; Zhou et al., 2020b), and an intestinal epithelial transporter has not been identified.
In mammals, STING activation upon infection leads to interferon signaling, NF-kB activation, and autophagy (Ahn and Barber, 2019; Cai and Imler, 2021). Less is known regarding STING sensing of commensals. Indeed, STING is implicated in intestinal homeostasis, particularly in myeloid cells (Aden et al., 2018; Ahn et al., 2017; Shmuel-Galia et al., 2021). Additionally, Drosophila STING (dSTING) responds to systemic sources of CDNs (Cai et al., 2020; Martin et al., 2018). However, it is unclear whether STING and CDNs play a role in antiviral immunity in the intestine.
As the breadth of antiviral pathways in enterocytes is unknown, we determined if dSTING was antiviral in the gut using the arthropod-borne virus Sindbis virus (SINV) and a picornalike virus, Drosophila C virus (DCV) (Johnson and Christian, 1999; Sansone et al., 2015; Xu et al., 2013). We found that dSTING is broadly antiviral in the gut and specifically active in enterocytes. We uncovered a circuit whereby orally acquired CDNs activate a dSTING-, dTBK1- dependent antiviral response that requires the apical nucleoside transporter CNT2. Furthermore, a CDN-STING-NF-kB pathway leads to induction of non-canonical targets to control infection. We explored this pathway during systemic infection and found striking tissue- and virus-specific differences. Diverse CDNs, CNT2, and dSTING protect against oral infection of SINV and DCV, but c-di-GMP and CNT2 do not protect against systemic infection. Moreover, dSTING and 2’3’cGAMP protect flies against systemic DCV infection but not systemic SINV infection. Therefore, our studies demonstrate exogenous CDNs activate enteric antiviral immune defenses and have more complex roles during systemic infection.
RESULTS
dSTING controls enteric virus infection
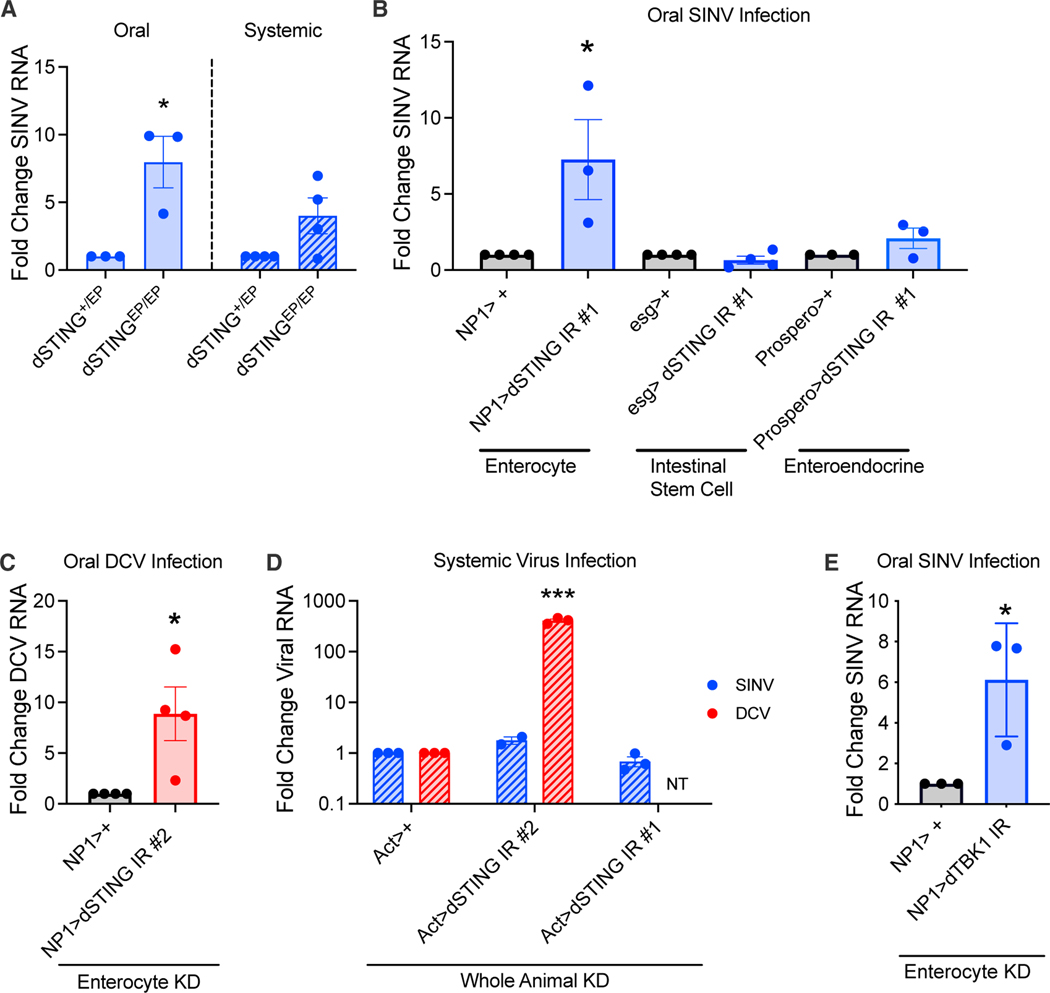
First, we determined if dSTING influences enteric immunity and observed increased virus infection in dSTING mutants (Figure 1A and S1A). Next, we explored which cell type (enterocytes, intestinal stem cells, or enteroendocrine cells) is required for dSTING’s antiviral activity. We used tissue-specific drivers and in vivo RNAi to deplete dSTING in each cell type and challenged these flies with SINV. dSTING was specifically required in enterocytes for antiviral defense against SINV (Figure 1B) and DCV (Figures 1C and S1D). >90% of intestinal cells are enterocytes, so we verified dSTING depletion with an enterocyte-specific or ubiquitous driver (Figures S1B and S1C).
Figure 1. The dSTING-dTBK1 pathway is antiviral in the intestine.
(A) Control (dSTINGEP/+) and dSTING mutant (dSTINGEP/EP) flies were orally or systemically infected with SINV. n = 3.
(B) Flies with STING depleted in enterocytes (NP1>dSTING IR #1), intestinal stem cells (esg>dSTING IR #1), and enteroendocrine cells (Prospero>dSTING IR #1) and their controls (NP1>+, esg>+, or Prospero>+) were orally infected with SINV. n = 3–4.
(C) Control (NP1>+) and dSTING-enterocyte-depleted (NP1>dSTING IR #2) flies were orally infected with DCV. n = 4.
(D) Control (Act>+) or dSTING-depleted (Act>dSTING IR #1, #2) flies were systemically infected with SINV or DCV. NT indicates not tested. n = 2–3.
(E) Control (NP1>+) or dTBK1-enterocyte-depleted (NP1>dTBK1 IR) flies were orally infected with SINV. n = 3.
(A–E) qRT-PCR was performed to quantify viral RNA levels. Each dot represents an independent replicate with bars indicating mean ± SEM. Statistical significance was determined by a Student’s t test; * denotes p < 0.05; *** denotes p < 0.001. See also Figures S1 and S2 and Tables S1 and S3.
dSTING controls systemic Zika virus and DCV infection (Goto et al., 2018; Liu et al., 2018). Given this, we evaluated the role of dSTING during systemic SINV infection. As expected, dSTING depletion leads to increased infection of DCV in whole animals (Figure 1D) (Goto et al., 2018; Liu et al., 2018) but does not control systemic SINV infection (Figures 1A and 1D).
Mammalian STING signals through TBK1 to activate antiviral signaling pathways (Abe and Barber, 2014; Ishikawa and Barber, 2008; Watson et al., 2012). The Drosophila TBK1 homolog (ik2, dTBK1) is dispensable during in vitro DCV infection (Goto et al., 2018; Kuranaga et al., 2006). However, dTBK1 polymorphisms are associated with variance in suppression of bacterial infection, suggesting that it plays a role in host defences (Lazzaroetal.,2006; Lazzaro et al., 2004). dTBK1 is an essential gene (Shapiro and Anderson, 2006), so we used tissue-specific approaches to explore the role of dTBK1 in enteric immunity and depleted dTBK1 in enterocytes (Figure S1E). Next, we orally challenged these flies with SINV and observed increased viral infection (Figure 1E).
To rule out the possibility that increased infection in dSTING- or dTKB1-enterocyte-depleted flies was due to increased gut permeability, we performed a SMURF assay (Rera et al., 2012). We also performed the assay in young or aged flies, as aging increases gut permeability (Table S1) (Dambroise et al., 2016; Rera et al., 2012). However, depletion of dSTING or dTBK1 did not affect gut permeability (Table S1). Since the microbiota can influence virus infection, we quantified microbiota levels and observed no change in bacterial counts in dSTING- or dTBK1-deficient flies (Table S1).
RNAi is dispensable during enteric SINV infection
The RNAi pathway is strongly antiviral during systemic viral infection, including DCV and SINV in Drosophila (Galiana-Arnoux et al., 2006; van Rij et al., 2006). However, RNAi is less active in certain tissues, including the brain, salivary gland, or intestine (Liu et al., 2018; Mondotte et al., 2018; Palmer et al., 2020). Indeed, Dcr2 and Ago2 mutants have increased systemic infection compared to controls (Figure S2A) (Liu et al., 2018). However, during oral DCV infection, Ago2 but not Dcr2 mutants had increased DCV RNA in the gut (Figure S2B) (Mondotte et al., 2018). In contrast, Ago2 and Dcr2 do not impact oral SINV infection (Figure S2B).
CDNs protect against virus infection and induce gene expression in the gut
Studies suggest that intestinal STING may sense the microbiota (Ahn et al., 2017; Shmuel-Galia et al., 2021). Therefore, we hypothesized enteric dSTING may be activated by CDNs produced by local commensals and thus investigated the role of orally acquired CDNs in enteric antiviral immunity. Nearly all bacteria, including human commensals, produce CDNs as essential second messengers (Corrigan and Gründling, 2013; Rö mling et al., 2013). Indeed, bioinformatics reveals 4 of the 7 major bacteria in the Drosophila microbiota encode proteins homologous to CDN synthases (Table S2) (Simm et al., 2004; Woodward et al., 2010).
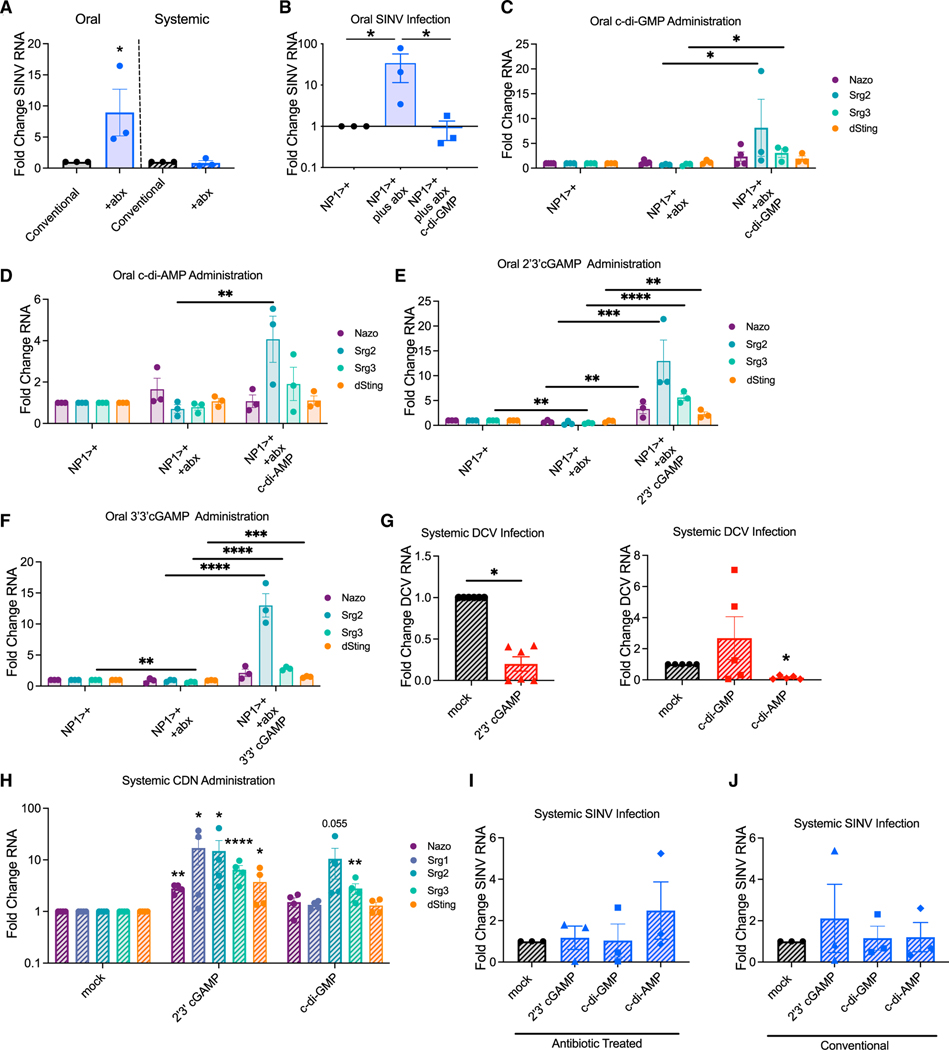
To assess the role of the microbiota during SINV infection, we orally infected conventional or antibiotical treated flies. Microbiota-depleted animals had increased SINV infection in the gut (Figure 2A). This led us to evaluate whether exogenous CDNs, potentially from the microbiota, activate dSTING-dependent antiviral immunity. Therefore, we explored whether feeding flies the bacterial CDN, c-di-GMP, protected them from infection. We fed c-di-GMP only to antibiotic-treated flies to prevent any confounding effects CDNs could have on the microbiota rather than the intestine. c-di-GMP feeding protects antibiotic-treated flies from the increase in infection caused by loss of the microbiota (Figure 2B).
Figure 2. Exogenous CDNs are antiviral and induce gene expression changes.
(A) Conventional and antibiotics-treated control flies (w1118) were orally or systemically infected with SINV. n = 3.
(B) Conventional and antibioticas-treated control flies (NP1>+) were mock or c-di-GMP fed and infected orally with SINV. n = 3.
(C–F) Conventional and antibiotics-treated control flies (NP1>+) were mock or (C) c-di-GMP, (D) c-di-AMP, (E) 2’3’cGAMP, or (F) 3’3’cGAMP fed. n = 3–4.
‘(G) Control flies (w1118) were co-injected with DCV and mock, 2’3’cGAMP, c-di-GMP, or c-di-AMP.
(H) Control flies (w1118) were injected with mock, 2’3’ cGAMP, or c-di-GMP. n = 4.
(I and J) (I) Antibiotic-treated and (J) conventional control flies (w1118) were co-injected with SINV and mock, 2’3’cGAMP, c-di-GMP, or c-di-AMP. n = 3.
(A–J) qRT-PCR was performed to quantify viral RNA or RNA of indicated genes.
(A and G) A Student’s t test was performed to determine statistical significance.
(B–F and G –J) A one-way ANOVA with multiple comparisons was performed to determine statistical significance.
(A–J) Each dot represents an independent replicate with bars indicating mean ± SEM. * denotes p < 0.05; ** denotes p < 0.01; *** denotes p < 0.001; **** denotes p < 0.0001. See also Figure S3 and Tables S2 and S3.
Previous studies found CDN injection into the fly body cavity activates dSTING-dependent expression of genes, including srg1, srg2, and srg3 (Cai et al., 2020). Thus, we determined if CDN feeding induced similar gene expression changes in the gut. srg2 and srg3 expression is induced by c-di-GMP feeding (Figure 2C), but srg1 was undetectable in the gut by qPCR (data not shown) (Dutta et al., 2015; Hung et al., 2020). We also monitored nazo and dSTING, which are induced by dSTING-NF-kB antiviral signaling during systemic infection (Goto et al., 2018; Liu et al., 2018). c-di-GMP feeding did not change nazo or dSTING expression (Figure 2C).
We tested if the CDNs, c-di-AMP (bacteria) or 2’3’cGAMP (cGAS) and 3’3’cGAMP (cGAS/bacteria), induce expression of these genes. We confirmed antibiotic treatment ablated the microbiota (Table S3). c-di-AMP induced srg2 expression, 3’3’cGAMP induced dSTING, srg2, and srg3 expression, and 2’3’ cGAMP induced dSTING, nazo, srg2, and srg3 expression (Figures 2D–2F). In contrast, SINV did not induce expression of srg2 and srg3 (Figure S3A).
CDN injection protects animals against systemic DCV infection (Cai et al., 2020; Slavik et al., 2021). Similarly, co-injection of 2’3’ cGAMP or c-di-AMP but not c-di-GMP protected flies from systemic DCV infection (Figure 2G). As expected, systemic 2’3’ cGAMP induced nazo, srg2, and srg3 expression (Figure 2H) (Cai et al., 2020). Surprisingly, systemic c-di-GMP induced gene expression changes despite its lack of antiviral activity against DCV(Figure 2H). Since dSTING is not antiviral systemically against INV (Figure 1A), we hypothesized exogenous CDNs would not inhibit systemic SINV infection. Indeed, we found CDNs do not protect flies from systemic SINV infection (Figures 2I and 2J).
Canonical NF-kB signaling is not induced by oral infection or CDNs in the gut
Mammalian STING and dSTING activate NF-kB (Goto et al., 2018; Ishikawa and Barber, 2008; Ishikawa et al., 2009; Liu et al., 2018; Martin et al., 2018; Sun et al., 2009). Therefore, we examined whether feeding SINV or c-di-GMP induces canonical NF-kB signaling in the gut. Drosophila encode two NF-kB signaling pathways, TOLL and IMD, which control expression of specific AMPs (Cornwell and Kirkpatrick, 2001; Lemaitre et al., 1996). Toll AMP expression (Drosomycin) did not change, and IMD AMP expression (Diptericin, Cecropin A2 [CecA2], and Attacin A [AttaA]) decreased after antibiotic treatment but did not change after feeding SINV or c-di-GMP (Figures S3B and S3C). Similarly, systemic CDNs do not induce expression of these AMPs (Figure S3D). In addition, SINV infection did not alter the expression of dSTING, nazo, or dTBK1(Figure S3B).
We also determined whether dSTING or dTBK1 impacted basal TOLL or IMD signaling in the intestine. Diptericin or Drosomycin expression did not change in STING mutant, TBK1 mutant, or enterocyte-depleted intestines compared to controls (Figures S3E and S3F).
Systemic virus infection induces NF-kB (Relish)-dependent expression of dSTING (Goto et al., 2018; Liu et al., 2018). Therefore, we monitored dSTING expression in NF-kB (Relish) mutant flies and observed no change in dSTING expression in the intestine and no change in dSTING expression basally or during early SINV infection (Figure S3G).
dSTING is required for CDN-dependent antiviral activity
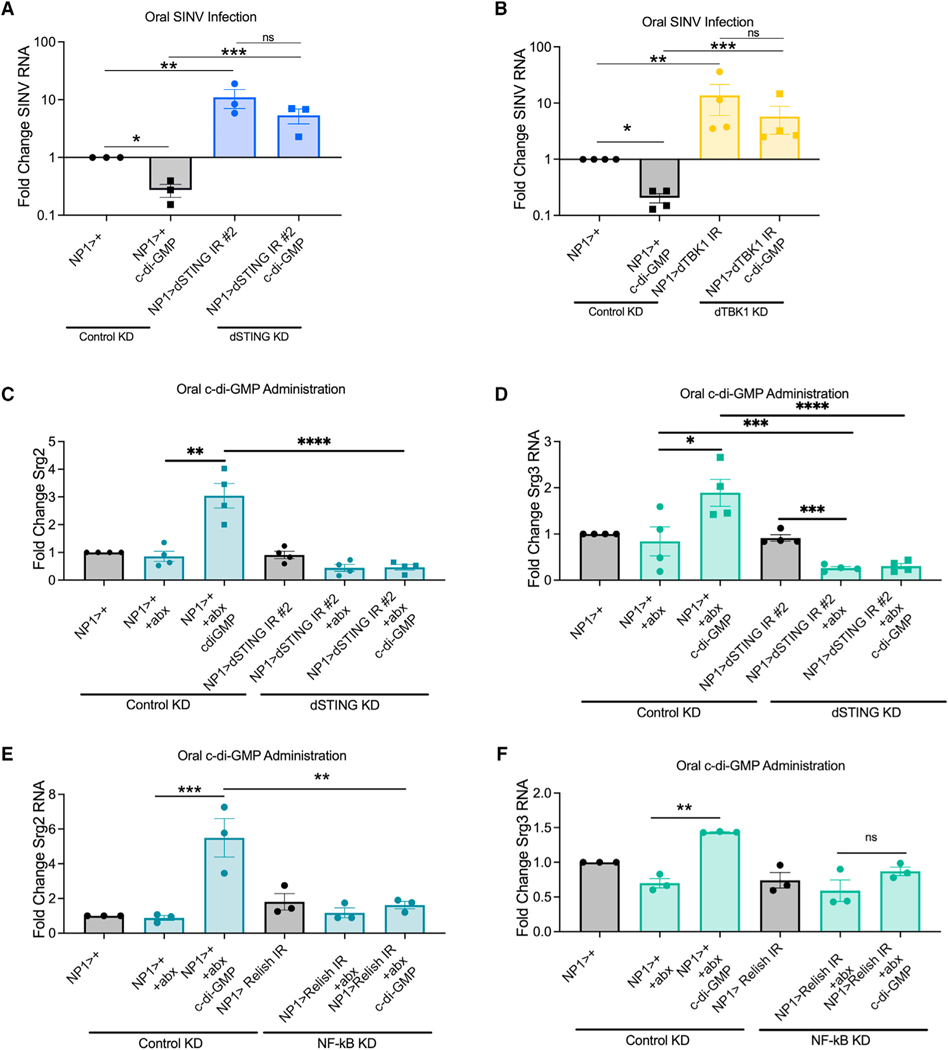
Next, we asked if the antiviral activity of orally acquired CDNs is dSTING- or dTBK1-dependent. Indeed, CDN-mediated protection from enteric SINV infection was lost in dSTING-enterocyte-depleted flies and in dTBK1-enterocyte-depleted flies compared to control flies (Figures 3A and 3B). We confirmed antibiotic treatment ablated the microbiota (Table S3).
Figure 3. dSTING and Relish are required for CDN-induced gene expression or CDN-mediated protection against viral infection.
(A and B) (A) Control (NP1>+) and dSTING-enterocyte-depleted flies (NP1>dSTING IR #2) and (B) dTBK1-enterocyte-depleted flies (NP1>dTBK1 IR) were antibiotic treated, mock or c-di-GMP fed, and orally infected with SINV. qRT-PCR was performed to quantify viral RNA levels. n = 3–4.
(C–F) Conventional and antibiotic-treated control (NP1>+) and (C and D) dSTING-enterocyte-depleted flies (NP1>dSTING IR #2) and (E and F) NF-kB- (Relish) enterocyte-depleted flies (NP1>Relish IR) were mock or c-di-GMP fed. qRT-PCR was performed to quantify (C and E) srg2 and (D and F) srg3 RNA levels. n = 3–4.
(A–F) Each dot represents an independent replicate with bars indicating mean ± SEM. A one-way ANOVA with multiple comparisons was performed to determine statistical significance. * denotes p < 0.05; ** denotes p < 0.01; *** denotes p < 0.001; **** denotes p < 0.0001. See also Figure S3 and Table S3.
CDN-dependent gene expression is dSTING, dTBK1, and NF-kB dependent
Since CDNs induce expression of srg2 and srg3, we interrogated whether this was dSTING or dTBK1 dependent. Oral c-di-GMP no longer induced expression of srg2 and srg3 when STING was depleted in enterocytes (Figures 3C and 3D). Additionally, c-di-GMP-induced expression of srg3 was dTBK1 dependent (Figures S3H and S3I).
Systemic CDNs induce dSTING- and NF-kB-dependent induction of srg2 and srg3 expression (Cai et al., 2020). In addition, NF-kB controls enteric viral infection of DCV and SINV (Sansone et al., 2015). Therefore, we explored whether NF-kB is required for intestinal CDN-dependent induction of srg2 and srg3. c-diGMP-dependent induction of srg2 and srg3 was NF-kB dependent (Figures 3E and 3F). We confirmed antibiotic treatment ablated the microbiota (Table S3).
CNT2 is required for CDN-mediated protection from enteric infection
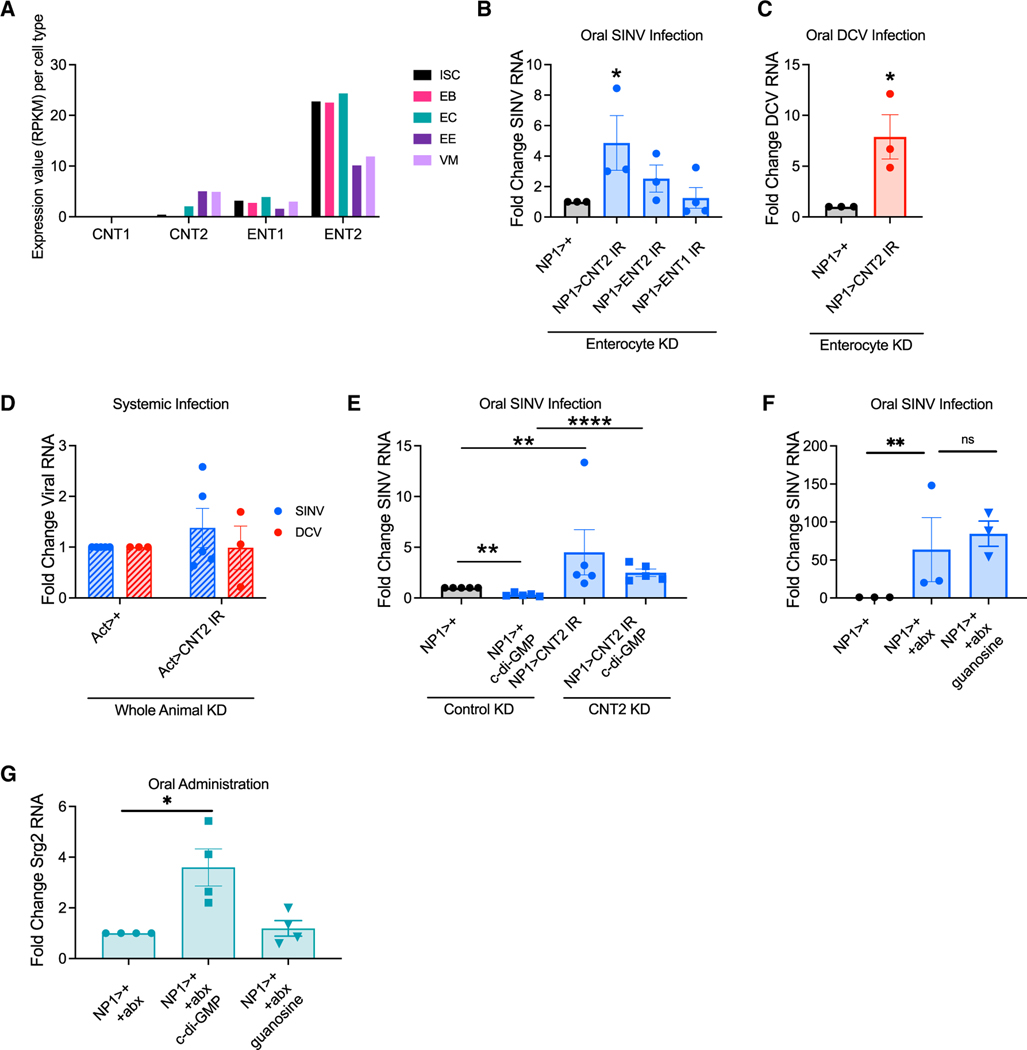
CDNs are charged molecules and not cell permeable (McWhirter et al., 2009). Therefore, CDNs use transporters to cross the membrane (Lahey et al., 2020; Luteijn et al., 2019; Ritchie et al., 2019; Zhou et al., 2020a; Zhou et al., 2020b). Epithelial transporters for CDNs have not been described; however, there are two conserved classes of nucleoside transporters expressed on enterocytes, namely, apical concentrative transporters (CNTs) and basolateral equilibrative transporters (ENTs) (Sankar et al., 2002). Single-cell RNA-seq studies of the Drosophila intestine revealed expression of cnt2, ent1, and ent2 but not cnt1 (Figures 4A and S4A) (Dutta et al., 2015; Hung et al., 2020). We hypothesized that the apical CNT2 would be more likely to interact with CDNs than the basolateral ENTs, as they are not exposed to intestinal contents. Nevertheless, we tested the role of the three conserved transporters expressed in enterocytes in antiviral immunity. We verified efficient depletion of the transporters in the gut (Figure S4B). ENT1 and ENT2 were not antiviral against SINV (Figure 4B). In contrast, CNT2 enterocyte depletion increased enteric SINV and DCV infection (Figures 4B and 4C). Importantly, loss of CNT2 in enterocytes did not impact gut permeability or microbiota loads (Table S1). Since CDNs are antiviral against DCV during systemic infection, and this is dSTING dependent (Cai et al., 2020), we tested whether CNT2 controls systemic DCV or SINV infection and found it did not (Figure 4D). This suggests that CNT2 plays an enterocyte-specific antiviral role.
Figure 4. CNT2 is required for CDN-dependent enteric defense.
(A) Reanalysis of published single-cell RNA-seq expression data for cnt1, cnt2, ent1, and ent2 across cell types in the Drosophila intestine (Dutta et al., 2015).
(B) Control (NP1>+) or CNT2-, ENT2-, or ENT1-enterocyte-depleted (NP1>cnt2 IR, NP1>ent2 IR, NP1>ent1 IR) flies were orally infected with SINV. n = 3–4.
(C) Control (NP1>+) or CNT2-enterocyte-depleted (NP1>cnt2 IR) flies were orally infected with DCV. n = 3.
(D) Control (Act>+) and CNT2-depleted animals (Act>cnt2 IR) were systemically infected with SINV or DCV. n = 3–4.
(B–D) A Student’s t test was performed to determine statistical significance.
(E) Control (NP1>+) and CNT2-enterocyte-depleted (NP1>cnt2 IR) flies were antibiotic treated, mock or c-di-GMP fed, and orally infected with SINV. n = 5.
(F) Conventional or antibiotic-treated control (NP1>+) flies were mock or guanosine fed and orally infected with SINV. n = 3.
(B–F) qRT-PCR was performed to quantify viral RNA levels.
(G) Antibiotic-treated control (NP1>+) flies were fed vehicle, c-di-GMP, or guanosine. qRT-PCR was performed to quantify srg2 RNA levels. n = 4.
(E–G) A one-way ANOVA with multiple comparisons was performed to determine statistical significance.
(A–G) Each dot represents an independent experiment with bars indicating mean ± SEM. * denotes p < 0.05; ** denotes p < 0.01; **** denotes p < 0.0001. See also Figure S4 and Tables S1 and S3.
CNT2 preferentially transports purines (Ritzel et al., 1998), but it is unknown whether CNT2 transports purine containing CDNs. Therefore, we tested if CNT2 is required for CDN-dependent antiviral activity in the gut. We confirmed ablation of the microbiota (Table S3). Indeed, CDN-dependent antiviral activity was CNT2 dependent (Figure 4E).
To rule out the possibility that oral c-di-GMP is degraded to guanosine, a known CNT2 ligand, and is antiviral through a purine salvage pathway, we fed antibiotic-treated control flies guanosine and orally infected them with SINV. We confirmed ablation of the microbiota (Table S3). Unlike CDNs, guanosine feeding did not protect flies from SINV infection (Figure 4F) and does not induce expression of srg2 in the intestine (Figure 4G).
DISCUSSION
Altogether, we defined a circuit whereby exogenous CDNs and dSTING play a fundamental role in antiviral defense of enterocytes to diverse viruses. CDN-dependent antiviral activity requires the apical nucleoside transporter CNT2. Intracellularly, this CDN pathway requires both dSTING and dTBK1, which has not previously been implicated in immune defense in flies (Goto et al., 2018). Moreover, enteric CDNs induces gene expression changes in enterocytes, which are dSTING-, dTBK1-, and NF-kB-dependent. Canonical NF-kB genes are not induced by CDN stimulation, which indicates that CDNs induce a distinct, non-canonical gene program. Future studies will define the activities of these CDN-induced genes in viral infection.
There are striking differences in requirements for the CDN-STING pathway between enteric and systemic infection. For example, dSTING and dCNT2 are antiviral during oral SINV infection of the intestine but not during systemic infection. Also, c-diGMP protects the intestine from oral but not systemic virus infection. Moreover, the canonical insect antiviral pathway, RNAi, has minimal influence on enteric virus infection despite being essential during systemic infection. This work highlights the importance of CDN-STING signaling, the breadth of enteric antiviral pathways, and virus- and tissue-specific differences in antiviral signaling.
Also, distinct CDNs led to overlapping gene expression changes and distinct antiviral activity in the same tissue. This may be due to differences between CDN binding strength to dSTING and subsequent variation in strength of STING activation or suggests the presence of negative regulators of CDNs, or activation of STING in different tissues may be due to the expression of different transporters with distinct specificities for these CDNs. For example, known CDN transporters display tissue-specific activity and varying ability to transport different CDNs (Lahey et al., 2020; Luteijn et al., 2019; Ritchie et al., 2019; Zhou et al., 2020a; Zhou et al., 2020b).
Our discovery that exogenous CDNs impact immunity has parallels in mammalian systems and has broad implications in enteric biology (Aden et al., 2018; Ahn et al., 2017; Shmuel-Galia et al., 2021). First, CDN recognition may be deeply conserved, as it is likely commensals from flies to humans produce CDNs. Second, microbiota shifts and dysbiosis influence immunity and health. This may be partially due to changes in the presence, type, amount, and location of CDNs in the intestine, which would subsequently impact STING activity, consequently altering the threshold for inflammatory responses and innate immune protection. Future studies will undoubtedly explore how changes in levels of commensal-derived CDNs contribute to disease.
Limitations of the Study
We suggest that the Drosophila microbiota produces CDNs as nearly all bacteria synthesize CDNs and some species of the Drosophila microbiota have CDN synthase homologs, but further mediated protection from infection. This interaction is not due work must be performed to confirm these findings. Additionally, to c-di-GMP degradation into the CNT2 ligand guanosine, which CNT2 is antiviral in the intestine and is required for c-di-GMP- leads us to hypothesize that CNT2 acts as a c-di-GMP transporter on enterocytes. However, in vitro and biochemical work needs to verify if CNT2 is a bone fide CDN transporter.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be direct to and will be fulfilled by the lead contact, Sara Cherry (cherrys@pennmedicine.upenn.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resource table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Beta-Actin | Santa Cruz Technology | Cat#4778; RRID:AB 626632 |
| dmSTING | (Martin et al., 2018) | N/A |
| DCV capsid | (Cherry and Perrimon, 2004) | N/A |
| anti-chicken-Alexa 488 | Fisher Scientific | Cat# A-11039; RRID:AB_142924 |
| anti-mouse IgG-HRP | Fisher Scientific | Cat#31430; RRIS: AB_228307 |
| anti-Rabbit- IgG-HRP | GE Healthcare | Cat# NA934; RRID:AB_772206 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| SINV (HRsp) | Richard W. Hardy (Burnham et al., 2007) | N/A |
| DCV | Peter D. Christian (Johnson and Christian, 1999) | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| FD&C Blue Dye #1 | Spectrum Chemical | Cat#FD110–25GM; CAS# 3844–45-9 |
| c-di-GMP | Invivogen | Cat#tlrl-nacdg; CAS# 61093–23-0 |
| c-di-AMP | Invivogen | Cat#tlrl-nacda; CAS# 54447–84-6 |
| 3'3'cGAMP | Invivogen | Cat#tlrl-nacga; CAS#849214–04-6 |
| 2'3'cGAMP | Invivogen | Cat#tlrl-nacga23–1; CAS# 1441190–66-4 |
| guanosine | Millipore Sigma | Cat#G6264; CAS#118–00-3 |
|
| ||
| Critical commercial assays | ||
|
| ||
| SuperScript III cDNA synthesis kit | Invivogen | Cat#18080044 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#4367659 |
|
| ||
| Deposited data | ||
|
| ||
| Single cell RNA-seq data of Drosophila intestine | (Dutta et al., 2015) | GEO. GSE120537; https.//doi.org/10.1016/j.celrep.2015.06.009 |
| Single cell RNA-seq data of Drosophila intestine | (Hung et al., 2020) | GEO. GSE120537; https.//doi.org/10.1073/pnas.1916820117 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| D. melanogaster. DL1 | Norbert Perrimon (Cherry and Perrimon, 2004) | N/A |
| Aedes albopictus. C6/36 | ATCC | ATCC.CRL-1660; RRID. CVCL Z230 |
|
| ||
| Experimental models: Organisms and strains | ||
|
| ||
| D. melanogaster. w1118 | N. Perrimon (Harvard Medical School, Boston, MA) | N/A |
| D. melanogaster: 5905 | Bloomington Drosophila Stock Center | FBal0018186; RRID.BDSC_5905 |
| D. melanogaster. Myo1A-Gal4 | E. Baehrecke (University of Massachusetts Medical School, Worchester, MA) | FBtp0098092 |
| D. melanogaster. Act-Gal4 | Bloomington Drosophila Stock Center | FBst0004414; RRID.BDSC_4414 |
| D. melanogaster. Escargot-Gal4 | N. Perrimon (Harvard Medical School, Boston, MA) | N/A |
| D. melanogaster. Prospero-Gal4 | Bloomington Drosophila Stock Center | FBst080572. RRID.BDSC_80572 |
| D. melanogaster: UAS-ik2 IR | Bloomington Drosophila Stock Center | FBst0034709; RRID.BDSC_34709 |
| D. melanogaster. dSTING [EY0649] | Bloomington Drosophila Stock Center | BDSC.16729; RRID.BDSC_16729 Cell Reports 37, 110150, December 28, 2021 el |
| D. melanogaster: UAS-STING IR (TRiP. JFO1138) (STING #1) | Bloomington Drosophila Stock Center | BDSC:31565; RRID:BDSC_31565 |
| D. melanogaster. UAS-STING IR (GD1905) (STING #2) | Vienna Drosophila Resource Center | RRID:Flybase_FBst0463487; Cat#VDRC:4031 |
| D. melanogaster: UAS-CNT2 IR (KK100597) | Vienna Drosophila Resource Center | RRID:Flybase_FBst0477654; Cat# VDRC:105828 |
| D. melanogaster: UAS-ENT2 IR (KK104419) | Vienna Drosophila Resource Center | RRID:Flybase_FBst04272337; Cat# VDRC:100464 |
| D. melanogaster: UAS- ENT1 IR (GD783) | Vienna Drosophila Resource Center | RRID:Flybase_FBst0468399; Cat#VDRC :49328 |
| D. melanogaster: UAS-Relish IR (TRiP.HMS00070) | Bloomington Drosophila Stock Center | FBst0033661; RRID: BDSC_33661 |
| D. melanogaster: AGO2 414 | Bloomington Drosophila Stock Center | FBst0313641 |
| D. melanogaster: Dcr-2L811fsX | Bloomington Drosophila Stock Center | FBst0033053; RRID:BDSC_33053 |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S4 | N/A | |
|
| ||
| Software and Algorithms | ||
|
| ||
| Graphpad Prism 8 | Graphpad | https://www.graphpad.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster
All fly stocks used are Wolbachia free and listed in the key resource table. Flies were maintained on standard cornmeal food at room temperature. 4–7 day old female flies of the indicated genotype were used. Only female flies were used as we use flies to study mosquito intestinal biology, and only female mosquitoes take blood meals and are enterically infected with arboviruses. Flies used in this paper had never previously been experimented on. Siblings were randomly sorted into experimental groups.
Cell culture
DL1 cells (sex unknown) were cultured in Schneider’s medium (Invitrogen-GIBCO, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Sigma), 100 U/mL of penicillin, and 100 mg/mL streptomycin (Invitrogen-GIBCO, Carlsbad, CA) (Rose et al., 2011) at 25°C.
Virus generation
Sindbis virus (HRsp) was generated in BHK cells and propagated in C6/36 cells (Burnham et al., 2007; Rose et al., 2011) at 37C. DCV was propagated by infecting Drosophila cells maintained at 25°C and collecting cells and supernatant after 75% of cells had died (Cherry and Perrimon, 2004; Xu et al., 2012). Infected cells and supernatant were then subject to ultracentrifugation steps and resuspended in PBS.
METHOD DETAILS
Drosophila genetics and infection
For antibiotic experiments flies were fed an antibiotic cocktail (Doxycycline 640 μg/mL, Ampicillin 640 μg/mL, Kanamycin 1 mg/mL, and Rifampicin 200 μg/mL) for × days on standard cornmeal food.
Oral infections were performed as previously described (Sansone et al., 2015; Xu et al., 2013). In brief, flies were starved and infected on cornmeal food (SINV, 150ul, 2.1 × 109 pfu/mL) or whatman (DCV, 10 μl, 5.17 × 107 pfu/mL) supplemented with sucrose (4% final solution). Flies of the indicated genotype were infected with SINV and placed onto new food with virus × days later or were infected with DCV and flipped onto fresh, virus-free whatman × days later. At 6dpi, intestines (n = 15) were collected and pooled for RT-qPCR.
For systemic injections flies of the stated genotypes were inoculated with 50nL of DCV using an Eppendorf Femtojet (Cherry and Perrimon, 2004; Liu et al., 2018; Moy et al., 2014). For CDN systemic administration 50 nL of a 1 mg/mL CDN solution (PBS) or PBS only was injected intrathoracically into flies of the indicated genotype. The indicated number of independent experiments with pools of 5 flies were used for all RT-qPCR experiments.
For CDN and guanosine feeding experiments flies were antibiotic treated, starved, and transferred onto whatman with virus and c-di-GMP (200 μg/mL, endotoxin free, Invivogen #tlrl-nacdg) or guanosine (200 μg/mL, for use in cell culture, Milipore Sigma, #G6264) and infected as above. c-di-GMP or guanosine were replaced three days later with virus and intestines (n = 15) were collected at 6dpi. For gene expression studies after CDN or guanosine feeding, flies were mock or antibiotic treated for three days. Then starved for one h and transferred to whatman with PBS, mock (sterile water, sterile water and DMSO), c-di-GMP (sterile water), or guanosine (sterile water and DMSO), and sucrose (4%). At 6 h after feeding, 15 intestines were dissected for RT-qPCR and 5 intestines were dissected for plating. The indicated number of independent experiments were used for all RT-qPCR experiments.
TCID50
Flies were orally infected with DCV and 30 intestines were dissected and pooled in 100 μL Schneider’s media at 7dpi. Fly intestines were homogenized and centrifuged (13,000 rpm, 10 min) to separate tissue from supernatant. Supernatants were serially diluted onto DL1 cells (5 × 104) in a 96 well format. At 48 hpi, DL1 cells were fixed and stained with anti-DCV capsid antibody (1:1000, (Cherry and Perrimon, 2004)). The Reed Muench method was used to determine TCID50, samples were normalized by the gut numbers in each sample. The end point dilution method was used to determine the limit of detection as previously described (Chan et al., 2009). We normalized the log transformed TCID50/mL LOD value to TCID50/mL/gut to directly compare the LOD to experimental TCID50/mL/gut values. The LOD was 0.872 Log10TCID50/mL/gut.
RNA extraction and qRT-PCR
Total RNA was extracted from 15 fly intestines or 5 whole flies using TRIzol (Invitrogen), DNase 1 treatment, and the RNA Clean and Concentrator Kit (Zymo Research) according to the manufacturers’ protocol. The SuperScript III cDNA synthesis kit (Invitrogen) was used to generate cDNA. cDNA was analyzed in triplicate with gene specific primers and SYBR Green PCR Master Mix (Applied Biosystems) for at least three independent experiments. RT-qPCR was used to quantify viral RNA levels or RNA levels of the genes indicated. All data was normalized to rp49 by relative quantification using the ddCT method. Primers are listed in Table S2.
Western blotting
5 animals were homogenized in protein lysis buffer consisting of 1x radioimmunoprecipitation assay (RIPA) buffer, Protease Inhibitor cocktail (cOmplete Protease Inhibitor Cocktail, Sigma Aldrich), and Phenylmethylsulfonyl fluoride (Sigma Aldrich). Protein samples were mixed with SDS sample buffer (Sigma-Aldrich) containing 5% beta-mercaptoethanol and boiled for 10 min. Samples were chilled on ice for two minutes and ran on a 10% SDS-PAGE gel (Bio-rad) and transferred to a PVDF membrane (EMD Milipore). Anti-beta Actin (1:1000, Santa Cruz #4778) and anti-dmSTING (1:500,(Martin et al., 2018)) antibodies were incubated with the membrane overnight at 4°. The membranes were then incubated with anti-ms-HRP or anti-Rb-HRP antibody respectively (1:10000, Fisher Scientific (mouse), Sigma-Aldrich (rabbit) and visualized with ECL (Amersham).
Bacterial plating and quantification
5 intestines were dissected in sterile PBS in a sterile 6-well plate and homogenized in 100 μL of a 50% glycerol-water solution. Serial dilutions were plated onto MRS plates (10μl) and were allowed to grow at 29°C for two days. CFU/mL/gut or CFU/mL/Fly was calculated.
Smurf assay
The smurf assay was performed as previously described (Dambroise et al., 2016; Rera et al., 2012). In summary, flies were fed FD&C Blue Dye #1 on whatman and scored as Smurf+ or Smurf-. The proportion of Smurf+ flies was calculated by dividing the number of Smurf+ flies by the total number of flies scored for the indicated genotype. For the aging experiment, 63–67 day old flies were used.
QUANTIFICATION AND STATISTICAL ANALYSIS
P values for RT-qPCR experiments were obtained by performing a two-tailed t test or one-way ANOVA with multiple comparisons and correction for multiple tests on ddCT values from at least three independent experiments. Visualization of data was performed in Prism 8 (Graphpad). The statistical parameters for experiments can be found in the figure legends, n indicates independent experiments. The number of animals used per experiment can be found in the method details section. Significance was defined as p < 0.05.
Supplementary Material
Highlights.
Cyclic dinucleotides (CDNs) are sensed by enterocytes to induce antiviral protection
CDNs signal through dSTING and NF-kB to induce gene expression
Antiviral activity of CDNs is dependent on the apical nucleoside transporter CNT2
Antiviral activities of CDNs and dSTING are tissue specific
ACKNOWLEDGMENTS
We would like to thank Bloomington, VDRC, E. Baehrecke, N. Silverman, N. Perrimon, and Excelixis for fly stocks. We would like to thank R. Hardy for SINV (hrSp) and A. Goodman for dmSTING antibody. We would like to thank the lab of S.C. for helpful discussions and advice. Biorender was used for the graphical abstract. This work was supported by NIH T32-AI-007324 and NIH F31 AI147415–01 to E.S. and R01AI152362, R01AI122749, R01AI150246, and R01AI140539 to S.C. S.C. is a recipient of the Burroughs Welcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110150.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abe T, and Barber GN (2014). Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kB activation through TBK1. J. Virol. 88, 5328–5341. 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Häsler R, et al. (2018). ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J. Exp. Med. 215, 2868–2886. 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, and Barber GN (2019). STING signaling and host defense against microbial infection. Exp. Mol. Med. 51, 1–10. 10.1038/s12276019-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Son S, Oliveira SC, and Barber GN (2017). STING-Dependent Signaling Underlies IL-10 Controlled Inflammatory Colitis. Cell Rep. 21, 3873–3884. 10.1016/j.celrep.2017.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Silverman N, and Cherry S. (2014). Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham AJ, Gong L, and Hardy RW (2007). Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 367, 212–221. 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Cai H, and Imler JL (2021). cGAS-STING: insight on the evolution of a primordial antiviral signaling cassette. Fac Rev 10, 54. 10.12703/r/10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Holleufer A, Simonsen B, Schneider J, Lemoine A, Gad HH, Huang J, Huang J, Chen D, Peng T, et al. (2020). 2’3’-cGAMP triggers a STING- and NF-kB-dependent broad antiviral response in Drosophila. Sci. Signal. 13, eabc4537. 10.1126/scisignal.abc4537. [DOI] [PubMed] [Google Scholar]
- Chan KH, Lai ST, Poon LL, Guan Y, Yuen KY, and Peiris JS (2009). Analytical sensitivity of rapid influenza antigen detection tests for swine-origin influenza virus (H1N1). J. Clin. Virol. 45, 205–207. 10.1016/j.jcv.2009.05.034. [DOI] [PubMed] [Google Scholar]
- Cherry S, and Perrimon N. (2004). Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat. Immunol. 5, 81–87. 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell WD, and Kirkpatrick RB (2001). Cactus-independent nuclear translocation of Drosophila RELISH. J. Cell. Biochem. 82, 22–37. 10.1002/jcb.1144. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, and Gründling,€ A. (2013). Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol. 11, 513–524. 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- Dambroise E, Monnier L, Ruisheng L, Aguilaniu H, Joly JS, Tricoire H, and Rera M. (2016). Two phases of aging separated by the Smurf transition as a public path to death. Sci. Rep. 6, 23523. 10.1038/srep23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, and Mekalanos JJ (2013). Cyclic dinucleotides and the innate immune response. Cell 154, 962–970. 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, and Buchon N. (2015). Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell Rep. 12, 346–358. 10.1016/j.celrep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, and Imler JL (2006). Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7, 590–597. 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Goto A, Okado K, Martins N, Cai H, Barbier V, Lamiable O, Troxler L, Santiago E, Kuhn L, Paik D, et al. (2018). The Kinase IKKb Regulates a STING- and NF-kB-Dependent Antiviral Response Pathway in Drosophila. Immunity 49, 225–234.e4. 10.1016/j.immuni.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Hu Y, Kirchner R, Liu Y, Xu C, Comjean A, Tattikota SG, Li F, Song W, Ho Sui S, and Perrimon N. (2020). A cell atlas of the adult Drosophila midgut. Proc. Natl. Acad. Sci. USA 117, 1514–1523. 10.1073/pnas.1916820117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, and Barber GN (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, and Barber GN (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792. 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KN, and Christian PD (1999). Molecular characterization of Drosophila C virus isolates. J. Invertebr. Pathol. 73, 248–254. 10.1006/jipa.1998.4830. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, and Miura M. (2006). Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 126, 583–596. 10.1016/j.cell.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Lahey LJ, Mardjuki RE, Wen X, Hess GT, Ritchie C, Carozza JA, Böhnert V, Maduke M, Bassik MC, and Li L. (2020). LRRC8A:C/E Heteromeric Channels Are Ubiquitous Transporters of cGAMP. Mol. Cell 80, 578–591.e5. 10.1016/j.molcel.2020.10.021. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, and Clark AG (2004). Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303, 1873–1876. 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sackton TB, and Clark AG (2006). Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics 174, 1539–1554. 10.1534/genetics.105.054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, and Hoffmann JA (1996). The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983. 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Gordesky-Gold B, Leney-Greene M, Weinbren NL, Tudor M, and Cherry S. (2018). Inflammation-Induced, STING-Dependent Autophagy Restricts Zika Virus Infection in the Drosophila Brain. Cell Host Microbe 24, 57–68.e3. 10.1016/j.chom.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn RD, Zaver SA, Gowen BG, Wyman SK, Garelis NE, Onia L, McWhirter SM, Katibah GE, Corn JE, Woodward JJ, and Raulet DH (2019). SLC19A1 transports immunoreactive cyclic dinucleotides. Nature 573, 434–438. 10.1038/s41586-019-1553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Hiroyasu A, Guzman RM, Roberts SA, and Goodman AG (2018). Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep. 23, 3537–3550.e6. 10.1016/j.celrep.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, et al. (2009). A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206, 1899–1911. 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondotte JA, Gausson V, Frangeul L, Blanc H, Lambrechts L, and Saleh MC (2018). Immune priming and clearance of orally acquired RNA viruses in Drosophila. Nat. Microbiol. 3, 1394–1403. 10.1038/s41564-018-0265-9. [DOI] [PubMed] [Google Scholar]
- Moy RH, Gold B, Molleston JM, Schad V, Yanger K, Salzano MV, Yagi Y, Fitzgerald KA, Stanger BZ, Soldan SS, and Cherry S. (2014). Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity 40, 51–65. 10.1016/j.immuni.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer WH, Dittmar M, Gordesky-Gold B, Hofmann J, and Cherry S. (2020). Drosophila melanogaster as a model for arbovirus infection of adult salivary glands. Virology 543, 1–6. 10.1016/j.virol.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, and Medzhitov R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rera M, Clark RI, and Walker DW (2012). Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl. Acad. Sci. USA 109, 21528–21533. 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie C, Cordova AF, Hess GT, Bassik MC, and Li L. (2019). SLC19A1 Is an Importer of the Immunotransmitter cGAMP. Mol. Cell 75, 372–381.e5. 10.1016/j.molcel.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzel MW, Yao SY, Ng AM, Mackey JR, Cass CE, and Young JD (1998). Molecular cloning, functional expression and chromosomal localization of a cDNA encoding a human Na+/nucleoside cotransporter (hCNT2) selective for purine nucleosides and uridine. Mol. Membr. Biol. 15, 203–211. [DOI] [PubMed] [Google Scholar]
- Rö mling U, Galperin MY, and Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, and Cherry S. (2011). Natural resistance-associated macrophage protein is a cellular receptor for sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10, 97–104. 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar N, Machado J, Abdulla P, Hilliker AJ, and Coe IR (2002). Comparative genomic analysis of equilibrative nucleoside transporters suggests conserved protein structure despite limited sequence identity. Nucleic Acids Res. 30, 4339–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone CL, Cohen J, Yasunaga A, Xu J, Osborn G, Subramanian H, Gold B, Buchon N, and Cherry S. (2015). Microbiota-Dependent Priming of Antiviral Intestinal Immunity in Drosophila. Cell Host Microbe 18, 571–581. 10.1016/j.chom.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, and Anderson KV (2006). Drosophila Ik2, a member of the I kappa B kinase family, is required for mRNA localization during oogenesis. Development 133, 1467–1475. 10.1242/dev.02318. [DOI] [PubMed] [Google Scholar]
- Shmuel-Galia L, Humphries F, Lei X, Ceglia S, Wilson R, Jiang Z, Ketelut-Carneiro N, Foley SE, Pechhold S, Houghton J, et al. (2021). Dysbiosis exacerbates colitis by promoting ubiquitination and accumulation of the innate immune adaptor STING in myeloid cells. Immunity 54, 1137–1153.e8. 10.1016/j.immuni.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, and Rö mling U. (2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134. 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Slavik KM, Morehouse BR, Ragucci AE, Zhou W, Ai X, Chen Y, Li L, Wei Z, Bähre H, Kö nig M, et al. (2021). cGAS-like receptors sense RNA and control 3’2’-cGAMP signalling in Drosophila. Nature 597, 109–113. 10.1038/s41586-021-03743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, and Jiang Z. (2009). ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA 106, 8653–8658. 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, and Andino R. (2006). The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995. 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, and Cox JS (2012). Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815. 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, and Portnoy DA (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Grant G, Sabin LR, Gordesky-Gold B, Yasunaga A, Tudor M, and Cherry S. (2012). Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe 12, 531–543. 10.1016/j.chom.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hopkins K, Sabin L, Yasunaga A, Subramanian H, Lamborn I, Gordesky-Gold B, and Cherry S. (2013). ERK signaling couples nutrient status to antiviral defense in the insect gut. Proc. Natl. Acad. Sci. USA 110, 15025–15030. 10.1073/pnas.1303193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Chen X, Planells-Cases R, Chu J, Wang L, Cao L, Li Z, Ló pezCayuqueo KI, Xie Y, Ye S, et al. (2020a). Transfer of cGAMP into Bystander Cells via LRRC8 Volume-Regulated Anion Channels Augments STINGMediated Interferon Responses and Anti-viral Immunity. Immunity 52, 767–781.e6. 10.1016/j.immuni.2020.03.016. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fei M, Zhang G, Liang WC, Lin W, Wu Y, Piskol R, Ridgway J, McNamara E, Huang H, et al. (2020b). Blockade of the Phagocytic Receptor MerTK on Tumor-Associated Macrophages Enhances P2X7R-Dependent STING Activation by Tumor-Derived cGAMP. Immunity 52, 357–373.e9. 10.1016/j.immuni.2020.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resource table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Beta-Actin | Santa Cruz Technology | Cat#4778; RRID:AB 626632 |
| dmSTING | (Martin et al., 2018) | N/A |
| DCV capsid | (Cherry and Perrimon, 2004) | N/A |
| anti-chicken-Alexa 488 | Fisher Scientific | Cat# A-11039; RRID:AB_142924 |
| anti-mouse IgG-HRP | Fisher Scientific | Cat#31430; RRIS: AB_228307 |
| anti-Rabbit- IgG-HRP | GE Healthcare | Cat# NA934; RRID:AB_772206 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| SINV (HRsp) | Richard W. Hardy (Burnham et al., 2007) | N/A |
| DCV | Peter D. Christian (Johnson and Christian, 1999) | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| FD&C Blue Dye #1 | Spectrum Chemical | Cat#FD110–25GM; CAS# 3844–45-9 |
| c-di-GMP | Invivogen | Cat#tlrl-nacdg; CAS# 61093–23-0 |
| c-di-AMP | Invivogen | Cat#tlrl-nacda; CAS# 54447–84-6 |
| 3'3'cGAMP | Invivogen | Cat#tlrl-nacga; CAS#849214–04-6 |
| 2'3'cGAMP | Invivogen | Cat#tlrl-nacga23–1; CAS# 1441190–66-4 |
| guanosine | Millipore Sigma | Cat#G6264; CAS#118–00-3 |
|
| ||
| Critical commercial assays | ||
|
| ||
| SuperScript III cDNA synthesis kit | Invivogen | Cat#18080044 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat#4367659 |
|
| ||
| Deposited data | ||
|
| ||
| Single cell RNA-seq data of Drosophila intestine | (Dutta et al., 2015) | GEO. GSE120537; https.//doi.org/10.1016/j.celrep.2015.06.009 |
| Single cell RNA-seq data of Drosophila intestine | (Hung et al., 2020) | GEO. GSE120537; https.//doi.org/10.1073/pnas.1916820117 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| D. melanogaster. DL1 | Norbert Perrimon (Cherry and Perrimon, 2004) | N/A |
| Aedes albopictus. C6/36 | ATCC | ATCC.CRL-1660; RRID. CVCL Z230 |
|
| ||
| Experimental models: Organisms and strains | ||
|
| ||
| D. melanogaster. w1118 | N. Perrimon (Harvard Medical School, Boston, MA) | N/A |
| D. melanogaster: 5905 | Bloomington Drosophila Stock Center | FBal0018186; RRID.BDSC_5905 |
| D. melanogaster. Myo1A-Gal4 | E. Baehrecke (University of Massachusetts Medical School, Worchester, MA) | FBtp0098092 |
| D. melanogaster. Act-Gal4 | Bloomington Drosophila Stock Center | FBst0004414; RRID.BDSC_4414 |
| D. melanogaster. Escargot-Gal4 | N. Perrimon (Harvard Medical School, Boston, MA) | N/A |
| D. melanogaster. Prospero-Gal4 | Bloomington Drosophila Stock Center | FBst080572. RRID.BDSC_80572 |
| D. melanogaster: UAS-ik2 IR | Bloomington Drosophila Stock Center | FBst0034709; RRID.BDSC_34709 |
| D. melanogaster. dSTING [EY0649] | Bloomington Drosophila Stock Center | BDSC.16729; RRID.BDSC_16729 Cell Reports 37, 110150, December 28, 2021 el |
| D. melanogaster: UAS-STING IR (TRiP. JFO1138) (STING #1) | Bloomington Drosophila Stock Center | BDSC:31565; RRID:BDSC_31565 |
| D. melanogaster. UAS-STING IR (GD1905) (STING #2) | Vienna Drosophila Resource Center | RRID:Flybase_FBst0463487; Cat#VDRC:4031 |
| D. melanogaster: UAS-CNT2 IR (KK100597) | Vienna Drosophila Resource Center | RRID:Flybase_FBst0477654; Cat# VDRC:105828 |
| D. melanogaster: UAS-ENT2 IR (KK104419) | Vienna Drosophila Resource Center | RRID:Flybase_FBst04272337; Cat# VDRC:100464 |
| D. melanogaster: UAS- ENT1 IR (GD783) | Vienna Drosophila Resource Center | RRID:Flybase_FBst0468399; Cat#VDRC :49328 |
| D. melanogaster: UAS-Relish IR (TRiP.HMS00070) | Bloomington Drosophila Stock Center | FBst0033661; RRID: BDSC_33661 |
| D. melanogaster: AGO2 414 | Bloomington Drosophila Stock Center | FBst0313641 |
| D. melanogaster: Dcr-2L811fsX | Bloomington Drosophila Stock Center | FBst0033053; RRID:BDSC_33053 |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S4 | N/A | |
|
| ||
| Software and Algorithms | ||
|
| ||
| Graphpad Prism 8 | Graphpad | https://www.graphpad.com |