Abstract
In this study, we have investigated 201 gastric biopsy specimens obtained from dyspeptic patients for the presence of Helicobacter pylori. By means of fluorescent in situ hybridization (FISH) with rRNA-targeted fluorescence-labeled oligonucleotide probes specific for H. pylori, this pathogen was detected in 63 biopsy specimens. By using conventional culturing, H. pylori was isolated from 49 of these 63 gastric biopsy specimens. In contrast, FISH failed to identify H. pylori in four samples from which the pathogen was cultured. The lowest sensitivity was obtained by using the urease test. H. pylori was detected indirectly by this method in 43 of 67 biopsy specimens, which were positive for the pathogen as determined by FISH and/or culturing. All 49 H. pylori isolates that were detected by FISH and culturing underwent antimicrobial susceptibility testing for clarithromycin, a macrolide drug that is a key component in the therapy of peptic ulcer disease caused by this pathogen. Clarithromycin susceptibility testing of cultured isolates was carried out by the E-test, whereas FISH was used on biopsy specimens to detect clarithromycin-resistant mutant strains. No discrepancies were found between these two methods. Thirty-seven strains were clarithromycin sensitive, and eight H. pylori isolates were resistant to the macrolide. From another four biopsy specimens, a mixture of clarithromycin-sensitive and -resistant strains was identified by both methods. Thus, FISH is a reliable technique for determining the clarithromycin susceptibility of this pathogen. Taken together, FISH is a more sensitive and rapid technique than culturing for detection of H. pylori in gastric biopsy specimens. However, in the microbiology routine diagnostic laboratory, the combination of both FISH and conventional culturing significantly increases the sensitivity in detection of H. pylori.
Colonization of the human gastric mucosa with Helicobacter pylori induces chronic gastritis and peptic ulcer disease (2, 3) In addition, H. pylori plays a role in the etiology of gastric cancer and cancer of the mucosa-associated lymphoid tissue (4, 15). On one hand, the assessment of an H. pylori infection is based on noninvasive tests, such as the urea breath test or serological methods (12, 21). On the other hand, the “gold standard” for the diagnosis of an H. pylori infection is the direct detection and culturing of the pathogen, which require gastric biopsy specimens obtained from invasive gastroduodenoscopy (5).
In most European countries, one week of triple therapy is recommended to eradicate H. pylori and to cure the peptic ulcer disease caused by this pathogen (5). The triple therapy comprises a proton pump inhibitor in combination with two antibiotics, including amoxicillin, clarithromycin, or metronidazole (5). In many cases, the macrolide drug clarithromycin is the key component of these combination therapies, since macrolide resistance occurring in H. pylori is the most important cause of treatment failure (6, 13). Thus, antimicrobial susceptibility testing appears to be necessary and cost-effective prior to the first therapy and is mandatory after the first treatment failure.
Clarithromycin resistance in H. pylori is based on single base mutations within the peptidyltransferase-encoding region of the 23S rRNA gene (16, 18, 25). In our institute, we have established the fluorescent in situ hybridization (FISH) technique with rRNA-targeted fluorescence-labeled oligonucleotide probes specific for three described mutations, in which the adenine residues at positions 2143 and 2144 are replaced by guanine (A2143G and A2144G) or cytosine (A2143C) (20). Recently we have reported that the whole-cell hybridization of H. pylori rRNA is in principle a culturing-independent, reliable, and specific method for detection of the pathogen in gastric tissue sections and determination of clarithromycin susceptibility (20). However, the aim of this study was to directly compare the practicality and reliability of the FISH technique with the conventional culturing method in the routine diagnostic laboratory under daily working conditions. Therefore, over a period of 9 months, gastric biopsies obtained from 201 patients with dyspeptic symptoms were prepared and processed to carry out the comparison of these two methods for H. pylori detection. The results of this investigation reveal that the combination of the two methods significantly increases the sensitivity of H. pylori identification, whereas the single application of each method for clarithromycin susceptibility testing gave concordant results.
MATERIALS AND METHODS
Human gastric biopsy specimens.
Two hundred one dyspeptic adults and children underwent gastroduodenoscopy at different medical departments of the Ludwig Maximilians-University Munich, Munich, Germany. During each endoscopic procedure, two antral mucosal biopsy specimens were obtained by the use of sterile biopsy forceps. Biopsy specimens were placed directly into transport media (Portagerm pylori; Biomerieux, Marcy l'Etoile, France) and immediately transported at room temperature to the microbiology laboratory. One to three hours after endoscopy, biopsy specimens were processed for (i) culturing H. pylori in a microaerophilic environment, (ii) the urease test to detect H. pylori in tissue samples indirectly, and (iii) fluorescent in situ hybridization of H. pylori.
Culturing of H. pylori.
Biopsy specimens were cut into small pieces and homogenized in a petri dish with a sterile scalpel. Equivalent portions of each biopsy specimens were used either for preparing cultures or for setting up the urease test. To culture H. pylori, biopsy specimens were smeared on the surface of Columbia agar plates supplemented with 5% sheep erythrocytes (Becton Dickinson, Heidelberg, Germany) and Schaedler agar plates supplemented with 5% sheep erythrocytes and vitamin K1 (Becton Dickinson). Inoculated vented plates were placed in an anaerobic jar together with a GENbox microaer paper sachet (Biomerieux, Marcy l'Etoile, France) to generate a microaerophilic environment (oxygen concentration, 7 to 10%; CO2 concentration, 20%) and incubated for 5 to 10 days. H. pylori microorganisms were identified on the basis of characteristic colony morphology, typical appearance on Gram staining, and positive urease, oxidase, and catalase tests.
Urease test.
The urease test (Jatrox-H. p.-Test; C. H. R. Hein Arzneimittel GmbH, Darmstadt, Germany) was carried out to detect H. pylori in tissue samples indirectly. Briefly, homogenized biopsy specimens were introduced into the test medium containing urea. Pink or red coloration of the test medium after 30 min to 3 h of incubation indicated a positive reaction and the presence of urease produced by H. pylori in the antrum tissue sample.
Fluorescent oligonucleotide probes.
All oligonucleotide probes used in this study have been previously described and evaluated (20). Briefly, probe Hpy-1 (5′-CACACCTGACTGACTATCCCG-3′) targeted to a 16S rRNA position was used to specifically identify H. pylori in gastric tissue sections, whereas probes ClaR1 (A2143G) (5′-CGGGGTCTTCCCGTCTT-3′), ClaR2 (A2144G) (5′-CGGGGTCTCTCCGTCTT-3′), and ClaR3 (A2143C) (5′-CGGGGTCTTGCCGTCTT-3′) were designed to detect 23S rRNA point mutations responsible for clarithromycin resistance of the pathogen. In contrast, probe ClaWT (5′-CGGGGTCTTTCCGTCTT-3′) was used as an internal control (data not shown) to identify clarithromycin-sensitive H. pylori strains which had not been detected by the mixture of probes ClaR1, ClaR2, and ClaR3 (20).
FISH.
In situ hybridization of bacteria on glass slides was performed as previously described (1, 11, 19, 20) with the following modifications. For each hybridization reaction, the patient's biopsy specimen was shock frozen in Tissue- Tek freezing medium (Sakura Finetek, Torrance, Calif.). Thereafter, biopsy specimens were cut into 4-μm-thick sections using a cryomicrotome (Leica, Wetzlar, Germany) and placed on glass slides. Oligonucleotide probes used for this study were synthesized (Metabion, Munich, Germany) and 5′ labeled with the fluorochromes Cy3 (ClaR1, ClaR2, ClaR3, and ClaWT; red signal) or fluorescein isothiocyanate (Hpy-1; green signal). After air drying of the specimens, three tissue sections of each biopsy specimen were overlaid with 50 μl of hybridization buffer (0.9 M NaCl, 0.02 mM Tris-HCl [pH 8.0], 0.01% sodium dodecyl sulfate) containing 30% formamide and an oligonucleotide mixture (5 ng/μl) consisting of the probes ClaR1, ClaR2, ClaR3, and Hpy-1; ClaR1, ClaR2, and ClaR3; or Hpy-1. Hybridization was carried out at 46°C for 90 min in a humid chamber, and stringent washing was done at 48°C in a buffer containing 0.112 M NaCl, 20 mM Tris-HCl (pH 8.0), and 0.01% sodium dodecyl sulfate. Subsequently, samples were stained with DAPI (4′, 6′-diamidino-2-phenylindole), which detects the DNA of bacteria, fungi, and host cells, as described previously (1, 11, 19, 20). Besides specific labeling with fluorescent probes, staining with DAPI was used to identify the characteristic microscopic shape of H. pylori. Citifluor (Citifluor Ltd., London, United Kingdom) was used as a mounting medium on hybridized slides. Finally, the slides were analyzed with an epifluorescence microscope, Leica DMRBE (Leica, Heerburg, Switzerland), equipped with a standard filter set. Two different fluorochromes could be detected simultaneously. Microscopy was carried out blinded by two independent investigators.
Clarithromycin susceptibility testing.
For clarithromycin susceptibility testing of H. pylori by the E-test, colonies from Columbia or Schaedler agar plates were suspended in broth and homogenized carefully to minimize aeration. The inoculum suspension was prepared to a McFarland 3 turbidity standard. After a sterile swab was dipped into the inoculum, the entire surface of Mueller-Hinton agar plates supplemented with 5% sheep erythrocytes was swabbed in three directions. Before E-test strips (AB Biodisk, Solna, Sweden) were applied on the agar surface with sterile forceps, the moisture was allowed to be absorbed for 5 min. Inoculated plates were incubated in a microaerophilic environment at 37°C for 2 days, and the MIC for each strain was determined. Each clarithromycin susceptibility testing was repeated twice. In biopsies where a mixture of clarithromycin-sensitive and -resistant H. pylori strains was detected by FISH, at least 50 single colonies were subcultured for subsequent clarithromycin susceptibility testing.
Statistics.
In gastric biopsy specimens from which H. pylori was cultured (Table 1, groups I, II, and V), the number of CFU was determined. Values are means ± standard deviations of CFU obtained from all biopsies per group. The significance of the differences among groups I, II, and V was determined by the Student t test. P values of <0.05 were considered statistically significant.
TABLE 1.
Examination of 201 gastric biopsy specimens by FISH, conventional culturing, and the urease test for detection of H. pyloria
| Group no. | Results by:
|
No. of specimens | No. of CFU | ||
|---|---|---|---|---|---|
| FISH | Culture | Urease test | |||
| 0 | Negative | Negative | Negative | 134 | |
| I | Positive | Positive | Positive | 36 | 311.2 ± 110.2 |
| II | Positive | Positive | Negative | 13 | 41.2 ± 8.8 |
| III | Positive | Negative | Positive | 7 | |
| IV | Positive | Negative | Negative | 7 | |
| V | Negative | Positive | Negative | 4 | 4.0 ± 1.4 |
In gastric biopsy specimens from which H. pylori was cultured (groups I, II, and V), the number of CFU was determined. Values are means ± standard deviations of CFU obtained from all biopsies per group. Values of each group differ significantly from each other (P < 0.05).
RESULTS
Detection of H. pylori in gastric biopsy specimens.
The results of the examinations of gastric biopsy specimens from 201 patients with dyspeptic symptoms for the presence of H. pylori using FISH, conventional culturing, and the urease test are shown in Table 1. In 134 of the patients' biopsy specimens (group 0), no H. pylori was detected. In 49 gastric tissue specimens, the pathogen was identified by both methods, FISH and culturing (groups I and II). In contrast, the urease test was positive for 36 (group I) and false negative for 13 (group II) of these 49 investigated samples. In another 14 gastric biopsy specimens (groups III and IV), H. pylori was detected by FISH but not by the culturing method (Fig. 1a). From this set of biopsies, seven were positive (group III) and seven were negative (group IV) as determined by the urease test. Finally, group V comprises four biopsy specimens from which Helicobacter was cultured, whereas the application of FISH and the urease test led to false-negative results.
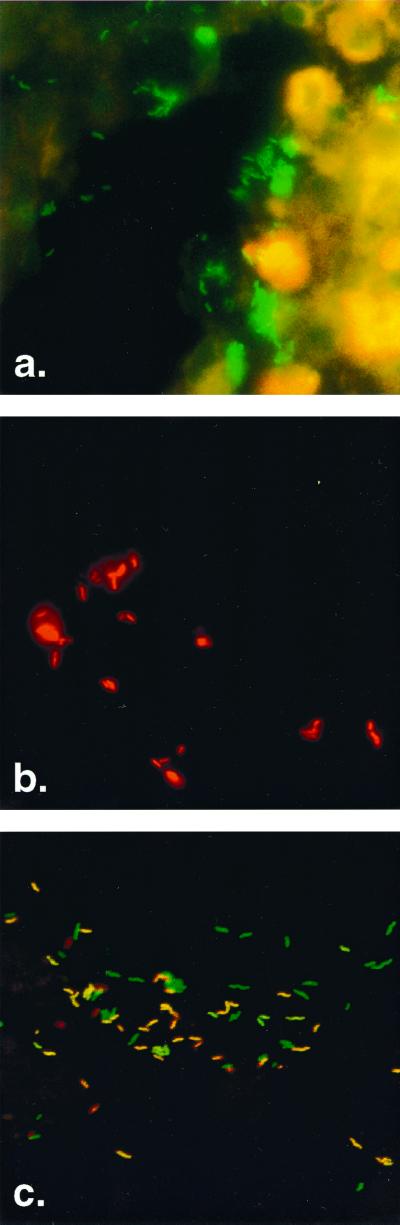
FIG. 1.
Detection of H. pylori within gastric antrum biopsy sections from patients with dyspeptic symptoms by whole-cell hybridization with fluorescence-labeled oligonucleotides. (a) Detection of H. pylori with green fluorescent probe Hpy-1–FITC. (b) Detection of clarithromycin-resistant H. pylori with a mixture of red fluorescent probes: ClaR1-Cy3, ClaR2-Cy3, and ClaR3-Cy3. (c) Detection of clarithromycin-sensitive and -resistant H. pylori in the same biopsy specimen of a patient by simultaneous application of probe Hpy-1–FITC and the mixture of probes ClaR1-Cy3, ClaR2-Cy3, and ClaR3-Cy3. Clarithromycin-resistant H. pylori bacteria are visible in yellow (mixed color of green and red), whereas clarithromycin-sensitive bacteria are green. The ratio of sensitive to resistant H. pylori strains in this gastric biopsy section is approximately 1:1.
Taken together, by using three independent microbiological methods, 67 of 201 gastric biopsy specimens investigated were positive for H. pylori. To calculate the sensitivity of all three methods, we suppose the ideal case of lacking false-negative results in this study. Thus, the urease test was the method with the lowest sensitivity (73.7%) for detecting Helicobacter indirectly. In our hands, FISH was the most sensitive technique (94.4%) for detecting the pathogen in gastric tissues, directly followed by the conventional culturing method, with a sensitivity of 82.7%. However, the combination of both FISH and culturing led to the identification of all 67 H. pylori-positive gastric biopsy specimens (100% sensitivity).
Determination of the bacterial load of H. pylori in gastric biopsies.
It is well known that the stability of H. pylori in biopsy material during transport is crucial for culturing the pathogen (24, 26). In all 53 specimens from which Helicobacter was cultured (Table 1, groups I, II, and V), the determination of CFU was carried out. As shown in Table 1, group I comprises 36 gastric biopsy specimens for which H. pylori was detected by all three methods. Table 1 also reveals that the bacterial load of viable H. pylori in these biopsy specimens (311.2 ± 110.2 CFU) was higher than those for groups II and V. In group II (41.2 ± 8.8 CFU), the urease test gave false-negative results, whereas FISH and culturing were able to identify H. pylori. This indicates that the reliability of the urease test might be dependent on the quantity of viable Helicobacter in gastric biopsy specimens. This assumption is supported by the determination of the number of H. pylori CFU in group V, because in all four biopsy specimens fewer than 10 CFU were cultured, leading to false-negative urease tests. In seven biopsy specimens of group III (Table 1), H. pylori was detected by FISH and the urease test but not by culturing. The reason for these culture-negative results in combination with positive urease tests was the fact that all seven biopsy specimens were heavily contaminated with bacteria of the upper respiratory tract, leading to an overgrowth on agar plates of microorganisms other than H. pylori.
In seven biopsy specimens of group IV (Table 1), H. pylori was detected by FISH but not by culturing or the urease test. An explanation for this result is that during transportation and preparation of these particular gastric biopsy specimens, the number of viable H. pylori bacteria had dropped below the detection limit for culturing and the urease test.
In four biopsy specimens of group V, FISH failed to detect H. pylori, probably due to a very low primary bacterial load in these specimens (4.0 ± 1.4 CFU). Thus, culturing appears to be a useful method for the diagnosis of an H. pylori infection in these cases.
Determination of the clarithromycin susceptibility of H. pylori.
All 49 H. pylori isolates which were detected by FISH and culturing underwent antimicrobial testing of susceptibility to clarithromycin (Fig. 1b). As shown in Table 2, a complete correlation between phenotypic determination by E-test and genotypic determination by FISH of the clarithromycin susceptibility was obtained: 37 strains were clarithromycin sensitive. For 26 of these 37 isolates (70.3%), a MIC of 0.016 μg/ml was determined, whereas the MIC of clarithromycin for the remaining 11 H. pylori strains ranged from 0.023 to 0.19 μg/ml.
TABLE 2.
Clarithromycin susceptibility and resistance of 49 H. pylori isolates as determined by the E-test and FISH using a mixture of the oligonucleotide probes ClaR1, ClaR2, and ClaR3 for hybridization
| No. of strains | MIC(s) (μg/ml) | Presence of resistance genotype | Result of testinga |
|---|---|---|---|
| 26 | 0.016 | Negative | S |
| 1 | 0.023 | Negative | S |
| 4 | 0.032 | Negative | S |
| 2 | 0.047 | Negative | S |
| 1 | 0.064 | Negative | S |
| 1 | 0.094 | Negative | S |
| 1 | 0.125 | Negative | S |
| 1 | 0.19 | Negative | S |
| 1 | 32 | Positive | R |
| 7 | >256 | Positive | R |
| 4 | 0.016, >256 | Negative, positive | S, R |
Results of both the E-test and genotyping. Clarithromycin susceptibility (S) and resistance (R) are shown.
The majority of the eight macrolide-resistant H. pylori isolates (seven strains; 87.5%) had point mutations in their 23S rRNA peptidyltransferase region that correlated with a high MIC of clarithromycin (>256 μg/ml). However, for one isolate resistant to this drug, the MIC was 32 μg/ml.
In another four biopsy specimens, a mixture of clarithromycin-sensitive and -resistant H. pylori strains was identified (Fig. 1c). For all macrolide-sensitive isolates from these four specimens, a MIC of 0.016 μg/ml was determined, whereas for all four clarithromycin-resistant strains, the MIC of clarithromycin was >256 μg/ml (Table 2). Interestingly, the ratios of sensitive to resistant H. pylori strains in these gastric biopsy specimens differed, ranging from ∼200:1 to ∼1:1 (Fig. 1c), ∼1:50, and ∼1:200, as determined by FISH (data not shown).
DISCUSSION
H. pylori is one of the most widespread infectious agents in humans, affecting nearly one-third of the world population (5). Since its cure prevents peptic ulcer disease, chronic gastritis, and possibly mucosa-associated lymphoid tissue lymphoma (22), the importance of accurate methods in the microbiology laboratory for detection of this pathogen can hardly be overestimated. Noninvasive clinical tests like the urea breath test and tests based on serology are well-established screening procedures which help to reduce the cost and workload of invasive endoscopy (12, 21). However, antimicrobial susceptibility testing of H. pylori obtained from gastric biopsies is essential prior to the first therapy and after the first treatment failure, since macrolide resistance in this pathogen is considered a main reason for failure of antibiotic eradication therapy (6).
The macrolide clarithromycin is a key component of the most recent treatment recommendations. The triple therapies include a proton pump inhibitor and either amoxicillin and clarithromycin or metronidazole and clarithromycin (5). In developed countries, the rates of prevalence of H. pylori resistance against these antibiotics are 11 to 70% for metronidazole resistance (3), up to 15% for clarithromycin resistance (13), and less than 1% for resistance to amoxicillin (23). In contrast to results with metronidazole, resistance to clarithromycin always correlates with a significant decrease in therapeutic efficacy, from ∼90% to ∼0 to 5% (5, 10). Resistance to clarithromycin in clinical H. pylori isolates is caused predominantly by distinct point mutations within the peptidyltransferase region of the 23S rRNA (A2143G, A2144G, and A2143C) (15, 17, 23). Recently we have reported the successful application of FISH for simultaneous detection of H. pylori and the respective 23S rRNA point mutations responsible for macrolide resistance (20). The aim of this study was to directly compare the highly specific FISH technique to conventional culturing under daily routine working conditions. As a method for indirect detection of H. pylori, we included the urease test in this comparative study. This last test was the fastest and easiest method but revealed the lowest sensitivity (73.7%) for detection of H. pylori in gastric biopsy specimens. False-negative results using the urease test were probably due to a relatively small amount of viable H. pylori bacteria in gastric biopsy specimens, since all 17 urease-negative specimens from which the pathogen was cultured (Table 1, groups II and V) revealed fewer than 60 CFU on agar plates (Table 1, groups II and V). In contrast, in all 36 urease-positive biopsy specimens (Table 1, group I), we could detect more than 100 CFU of H. pylori. False-positive results were not obtained with the relatively nonspecific urease test.
Significant higher sensitivities could be achieved by using either culturing (82.7%) or FISH (94.4%). Interestingly, out of 67 specimens positive for H. pylori, 14 biopsy samples tested positive for the pathogen as determined by FISH but negative as determined by culturing. This discrepancy might be due to the fact that the stability of Helicobacter in biopsy material during transport is a limiting factor for culturing (7, 8, 17) but does not necessarily result in small amounts of rRNA which would lead to the failure of FISH to detect H. pylori. In fact, recent investigations from our institute revealed that bacteria which have been killed by antibiotic drugs can nevertheless be detected by FISH (9). In our study, the period of time during the endosocopic procedure and the preparation of the biopsies in the microbiology laboratory was very short (1 to 3 h). The above-mentioned advantage of FISH over culturing might be even more evident if longer transport distances or a delayed transport occurred.
Group IV (Table 1) comprises seven gastric biopsy specimens in which H. pylori was detected by FISH but not by culturing or the urease test. We can exclude false-positive results, since we had carefully evaluated the specificity of the oligonucleotide probe Hpy-1 in a recent publication (20). Probe Hpy-1 does not bind to various species from the ɛ subclass of Proteobacteria, such as Helicobacter spp. other than Helicobacter pylori, Campylobacter spp., or Wolinella succinogenes. The specificity of the probe was further analyzed by hybridizing Lactobacillus lactis, Streptococcus mutans, and Proteus vulgaris, which are sometimes recovered from the human stomach. No binding of the probe was observed. Thus, we conclude that probe Hpy-1 is highly specific and discriminates between closely related bacterial species that may be present in human gastric samples. In addition to the specificity of the oligonucleotide probe, the microscopic shape (curved rods, long spirals, S shapes, or seagull wing shapes) of bacteria from group IV was characteristic. This shape was visualized not only by fluorescent probes but also by staining with DAPI.
With conventional culturing, we were able to detect H. pylori in 4 out of 67 biopsy specimens (Table 1, group V) for which FISH failed to identify the pathogen. In fact, even the expanded examination of more than three tissue sections from these four biopsy specimens (see Material and Methods) by fluorescence microscopy did not lead to the identification of H. pylori by FISH. Thus, very low quantities of bacteria in whole biopsy specimens (4.0 ± 1.4 CFU) as determined by culturing (Table 1, group V) were a limiting factor for FISH detection of H. pylori in 4-μm-thick tissue sections derived from these specimens.
In comparison with the phenotypic clarithromycin resistance measurement by the E-test, the analysis of genotypic clarithromycin resistance revealed 100% correlation. In our study, three different oligonucleotide probes were used to detect the most predominant point mutations within the peptidyl transferase region of the 23S rRNA leading to macrolide resistance. However, Occhialini et al. have reported a small number of clarithromycin-resistant H. pylori strains with different point mutations (14). These strains should be addressed in future investigations using corresponding oligonucleotides.
Gastric biopsy specimens of four patients harbored two different H. pylori strains: one clarithromycin-sensitive strain (hybridization with probe Hpy-1) and one clarithromycin-resistant strain hybridizing with the probe mixture of Hpy-1–ClaR1–ClaR2–ClaR3 (Fig. 1c). In one biopsy specimen, the ratio of sensitive to resistant H. pylori strains was approximately 200:1. The identification of the minority population of clarithromycin-resistant strains was much easier by FISH than by E-test. The former technique allowed distinguishing of susceptible (green) and resistant (yellow) strains at a glance by fluorescence microscopy (Fig. 1c), whereas the latter method revealed only very few and small isolated colonies, for which the MIC was >256 μg/ml, which were barely detectable on the agar plate. Thus, FISH is a more reliable technique for the identification of a few clarithromycin-resistant H. pylori strains hidden behind a majority of clarithromycin-susceptible isolates. It is tempting to speculate that the identification of clarithromycin-resistant strains in a mixture of susceptible and resistant organisms has clinical relevance. Clarithromycin treatment of patients colonized with both strains could lead to the selection of the resistant strain and subsequent failure of the eradication therapy.
In conclusion, the combined application of the conventional culturing method and FISH significantly increases the sensitivity in detection of H. pylori in gastric biopsy specimens. Since gastroduodenoscopy is an expensive and, for the patient, stressful procedure, both clinicians and microbiologists should be interested in choosing the best techniques for the identification of Helicobacter in this biopsy tissue. In our opinion, culturing is mandatory, since isolated strains of H. pylori can be used for susceptibility testing with all relevant antibiotic drugs and for epidemiological studies. FISH appears to be a method highly complementary to culturing, since the detection of H. pylori and its clarithromycin susceptibility is reliable for biopsy specimens containing nonviable bacteria. In addition, FISH is rapid and cost-effective (about $20 per biopsy) and is easy to implement without the need of special equipment or facilities. Thus, FISH in combination with conventional culturing is appropriate for daily routine work in the microbiology laboratory.
ACKNOWLEDGMENTS
H.R. was supported by the AIDS-Stipendienprogramm from the Bundesministerium für Bildung, Wissenschaft, Forschung and Technologie of Germany.
REFRENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser M J. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology. 1992;102:720–727. doi: 10.1016/0016-5085(92)90126-j. [DOI] [PubMed] [Google Scholar]
- 3.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Infect Dis. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EUROGAST Study Group. An international association between Helicobacter infection and gastric cancer. Lancet. 1993;341:1359–1362. [PubMed] [Google Scholar]
- 5.European Helicobacter Pylori Study Group. Current European concepts in the management of Helicobacter pylori infection. The Maastricht Consensus Report. Gut. 1997;41:8–13. doi: 10.1136/gut.41.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham D Y. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272–1277. doi: 10.1016/s0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 7.Han S W, Flamm R, Hachem C Y, Kim H Y, Clarridge J E, Evans D G, Beyer J, Drnec J, Graham D Y. Transport and storage of Helicobacter pylori from gastric mucosal biopsies and clinical isolates. Eur J Clin Microbiol Infect Dis. 1995;14:349–352. doi: 10.1007/BF02116531. [DOI] [PubMed] [Google Scholar]
- 8.Heep M, Scheibl K, Degrell A, Lehn N. Transport and storage of fresh and frozen gastric biopsy specimens for optimal recovery of Helicobacter pylori. J Clin Microbiol. 1999;37:3764–3766. doi: 10.1128/jcm.37.11.3764-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogardt M, Trebesius K, Geiger A M, Hornef M, Rosenecker J, Heesemann J. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J Clin Microbiol. 2000;38:818–825. doi: 10.1128/jcm.38.2.818-825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazell S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 11.Kempf V A J, Trebesius K, Autenrieth I B. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J Clin Microbiol. 2000;38:830–837. doi: 10.1128/jcm.38.2.830-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laheij R J F, Straatman H, Hansen J B M J, Verbeek A L M. Evaluation of commercially available Helicobacter pylori kits: a review. J Clin Microbiol. 1998;36:2803–2809. doi: 10.1128/jcm.36.10.2803-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Megraud F. Antibiotic resistance in Helicobacter pylori infection. Br Med Bull. 1998;54:207–216. doi: 10.1093/oxfordjournals.bmb.a011671. [DOI] [PubMed] [Google Scholar]
- 14.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsonnet J, Hansen S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 16.Pina M, Occhialini A, Monteiro L, Doermann H P, Megraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siu L K, Leung W K, Cheng A F, Sung J Y, Ling T K, Ling J M, Ng E K, Lau J Y, Chung S C. Evaluation of a selective transport medium for gastric biopsy specimens to be cultured for Helicobacter pylori. J Clin Microbiol. 1998;36:3048–3050. doi: 10.1128/jcm.36.10.3048-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Graham D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trebesius K, Harmsen D, Rakin A, Schmelz J, Heesemann J. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J Clin Microbiol. 1998;36:2557–2564. doi: 10.1128/jcm.36.9.2557-2564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trebesius K, Panthel K, Strobel S, Vogt K, Faller G, Kirchner T, Kist M, Heesemann J, Haas R. Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridisation. Gut. 2000;46:608–614. doi: 10.1136/gut.46.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaira D, Malfertheiner P, Megraud F, Axon A T, Deltenre M, Hirschl A M, Gasbarrini G, O'Morain C, Garcia J M, Quina M, Tytgat G N. Diagnosis of Helicobacter pylori infection with a new non-invasive antigen-based assay. HpSA European study group. Lancet. 1999;354:30–33. doi: 10.1016/s0140-6736(98)08103-3. [DOI] [PubMed] [Google Scholar]
- 22.van der Hulst R W M, Rauws E A J, Köycü B, Keller J J, Tyssen J G P, Bruno M, Tytgat G N J. Prevention of ulcer recurrence after successful eradication of Helicobacter pylori infection: a prospective long term follow-up study. Gastroenterology. 1997;113:1082–1086. doi: 10.1053/gast.1997.v113.pm9322501. [DOI] [PubMed] [Google Scholar]
- 23.van Zwet A A, Vandenbroucke-Grauls C M, Thijs J C, van der Wouden E J, Gerrits M M, Kusters J G. Stable amoxicillin resistance in Helicobacter pylori. Lancet. 1998;352:1595. doi: 10.1016/s0140-6736(98)00064-6. [DOI] [PubMed] [Google Scholar]
- 24.Veenendaal R A, Lichtendahl-Bernards A T, Pena A S, Endtz H P, van Boven C P A, Lamers C B H W. Effect of transport medium and transportation time on culture of Helicobacter pylori from gastric biopsy specimens. J Clin Pathol. 1993;46:561–563. doi: 10.1136/jcp.46.6.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia H X, Keane C T, Omorain C A. Determination of the optimal transport system for Helicobacter pylori cultures. J Med Microbiol. 1993;39:334–337. doi: 10.1099/00222615-39-5-334. [DOI] [PubMed] [Google Scholar]