Abstract
Lacazia loboi is the last of the classical fungal pathogens to remain a taxonomic enigma, primarily because it has resisted cultivation and only causes cutaneous and subcutaneous infections in humans and dolphins in the New World tropics. To place it in the evolutionary tree of life, as has been done for the other enigmatic human pathogens Pneumocystis carinii and Rhinosporidium seeberi, we amplified its 18S small-subunit ribosomal DNA (SSU rDNA) and 600 bp of its chitin synthase-2 gene. Our phylogenetic analysis indicated that L. loboi is the sister taxon of the human dimorphic fungal pathogen Paracoccidioides brasiliensis and that both species belong with the other dimorphic fungal pathogens in the order Onygenales. The low nucleotide variation among three P. brasiliensis 18S SSU rDNA sequences contrasts with the surprising amount of nucleotide differences between the two sequences of L. loboi used in this study, suggesting that the nucleic acid epidemiology of this hydrophilic pathogen will be rewarding.
Lobomycosis (Jorge Lobo's disease) is a chronic cutaneous and subcutaneous disease that manifests itself by the development of numerous nodular (para-keloidal) lesions on the bodies of its victims (1, 23). The occurrence of most para-keloidal lesions in cooler areas of the body had led to the speculation that its etiologic agent does not grow well at 37°C (11, 12). Human infections are known to occur only in Latin American countries. A human case, apparently acquired in Venezuela, was recently reported in a United States male (7). Besides humans, the disease has also been recorded in two species of dolphins in the endemic areas and around the coasts of Florida and the Gulf of Mexico (1, 23). In addition to the many Latin American cases, two anomalous European cases involving a bottle-nosed dolphin and an aquarium attendant also have been described (26). This apparently obligate pathogen has yet to be cultured in vitro, and it is identified in host tissue by its morphology. Thus, little or nothing is known about its biological and epidemiological characteristics. Its in vivo phenotype consists of unicellular, thick-walled yeast-like cells that occur singly as well as in branched and unbranched chains of 3 or more cells connected by short tubules. The cells measure 5 to 12 μm in diameter and are readily detected with most fungal stains.
The taxonomic identity of the agent of lobomycosis has been contentious primarily because it has never been successfully cultivated. This frustrating fact has led to the use of several names to designate this obligate pathogen. These include the binomials Glenosporella loboi, Glenosporopsis amazonica, Loboa loboi, Lobomyces loboi, and Paracoccidioides loboi (2, 10–12, 18). Although its legitimacy has long been questioned, Loboa loboi is the most widely used name (1, 8, 23, 25). Most recently, Taborda et al. (27) proposed the binomial Lacazia loboi, arguing that previous designations were taxonomically invalid. Based on its yeast-like morphology in infected tissues (8, 25) and on studies utilizing DNA based probes (15), the etiologic agent of lobomycosis has been assumed to be a member of the kingdom Fungi. Conversely, due to its inability to grow in culture and the unresponsiveness of the host to most antifungal drugs, it has been speculated that this pathogen could be “an obligate parasite of some lower animal forms” (25). In this article, we report that phylogenetic analysis, using L. loboi's 18S small-subunit ribosomal DNA (SSU rDNA) and 600 bp of the chitin synthase-2 (CHS-2) gene (CHS2) from the genomic DNA of its yeast-like cells, places this unique pathogen within the systemic dimorphic fungal pathogens.
MATERIALS AND METHODS
Collection of tissue containing yeast-like cells of L. loboi.
Biopsied tissues with L. loboi yeast cells were collected from two Brazilian human patients with lobomycosis. For diagnostic purposes, half of the collected tissues were fixed in formalin and then sectioned and stained by the hematoxylin and eosin and Gomori's silver stains. The other half were used for culture and DNA extraction. Due to the fact that (i) this human pathogen is intractable to culture, (ii) the tissue forms of this disease agent resemble those of paracoccidioidomycosis (caused by a dimorphic fungus readily isolated in the laboratory), and (iii) cases of concomitant lobomycosis and paracoccidioidomycosis have been diagnosed (17), samples from both patients were cultured on Sabouraud and blood agars.
DNA isolation, PCR protocol, and sequencing of L. loboi 18S SSU rDNA and chitin synthase partial gene.
The tissues from both patients were aseptically collected and transported without fixatives to the laboratory. The infected tissues, containing L. loboi yeast cells, were ground under liquid nitrogen within a few minutes of collection. The DNA from the ground samples was treated with sodium dodecyl sulfate and proteinase K digestion and then extracted with phenol and chloroform. Amplification of the 18S SSU rDNA gene was accomplished by PCR using the oligonucleotide forward primer NS1 (13, 29), 5′GTAGTCATATGCTTGTCTC3′. The NS8 reverse primer, 5′TCCGCAGGTTCACC(TA)ACGGA3′, was degenerated per the method of Issakainen et al. (16). The L. loboi CHS2 gene from patient 1 was amplified by PCR per the methods of Chen-Wu et al. (9) and Bowen et al. (3). The amplicons from both molecules were ligated into pCR 2.1-TOPO vectors (Invitrogen, Carlsbad, Calif.), purified, and then sequenced using BigDye Terminator chemistry in an ABI Prism 310 genetic analyzer (Perkin-Elmer, Foster City, Calif.). In addition to the above cultures, the sterility of the ground samples was also checked prior to phenol DNA extraction on Sabouraud and blood agars.
Phylogenetic analysis.
Phylogenetic analyses were conducted with the computer software program Phylogenetic Analysis Using Parsimony (version 4.0b.4a; D. L. Swofford, Illinois Natural History Survey, Champlain). Neighbor-joining analysis of the 18S SSU rDNA sequences used a maximum-likelihood multiple-hit correction with an empirical transition/transversion ratio, empirical base frequencies, a gamma distribution of 0.5, and four categories of variation. In the chitin synthase gene phylogenetic tree, conditions were identical, except that the transition/transversion ratio was 2/1. With both molecules, 1,000 bootstrap-resampled data sets analyzed by both neighbor-joining and parsimony methods (heuristic) were used to assess branch support.
Nucleotide sequence accession numbers.
The sequences for L. loboi 18S SSU rDNA and the chitin synthase partial gene were submitted to GenBank under the accession numbers AF238301, AF255331, and AF238303.
RESULTS
Clinical, histological, and laboratory findings.
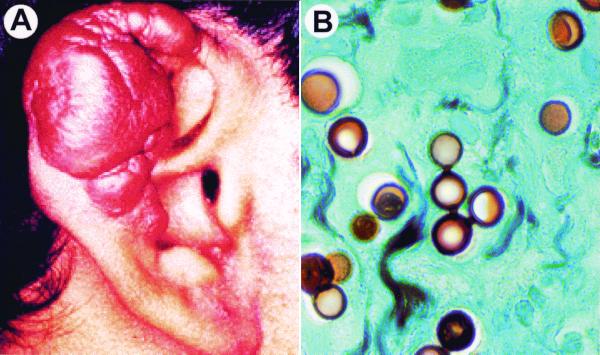
The patients in this study were from two different locations in Acre, Brazil, a northwestern area of the country. Clinical symptoms related to paracoccidioidomycosis were not detected in the two patients with lobomycosis (no mucous membranes were involved). Since the primary focus of Paracoccidioides brasiliensis infections is the lungs, chest X rays were carried out on the patients. Both patients showed no lung involvement. Para-keloidal lesions typical of lobomycosis were observed in these patients (Fig. 1A). Their lesions were restricted to different areas of the ears. Silver-stained sections of the para-keloidal lesions showed single yeast-like cells as well as branched and unbranched chains of three or more cells connected by short tubules (Fig. 1B). The size of the yeast-like cells was found to be uniform, a typical feature of L. loboi's parasitic form. Cultures of the infected tissues on Sabouraud and blood agar incubated at 25°C and 37°C for more than 1 month were consistently negative.
FIG. 1.
The clinical features encountered in lobomycosis are of importance to differentiate between paracoccidioidomycosis and infections caused by L. loboi. (A) Characteristic para-keloidal nodules of lobomycosis (patient 1) used in this study to extract genomic DNA from its in vivo yeast forms. (B) Tissue section obtained from the patient in panel A. Note the uniform cell sizes and the tubules connecting the chains of L. loboi yeast cells, two important features of its in vivo morphology (Gomori-methenamine silver stain; magnification, ×800).
Phylogenetic analysis with L. loboi's 18S SSU rDNA and chitin synthase-2 partial gene.
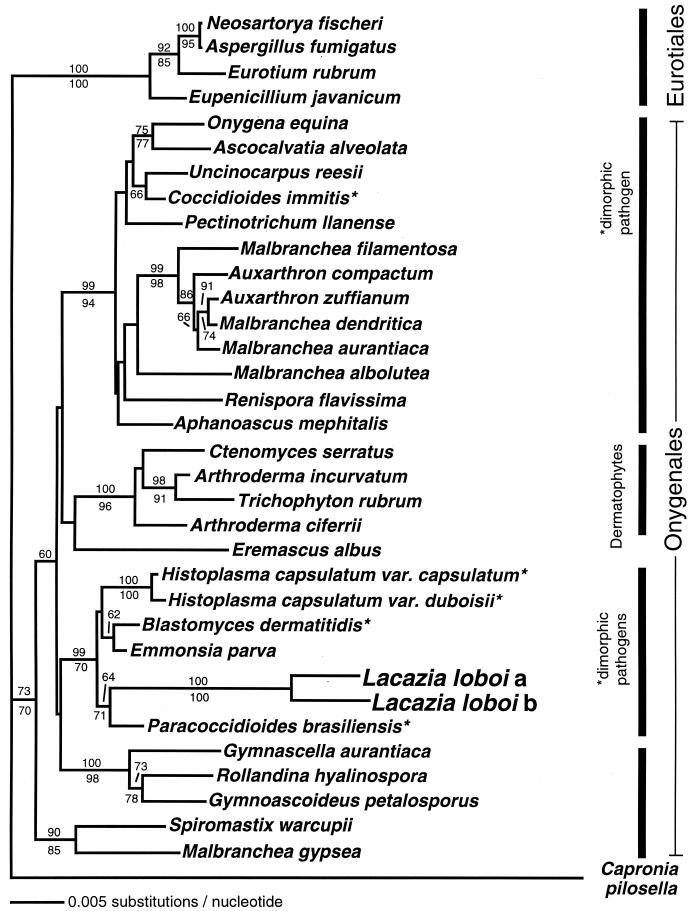
Using 18S rDNA primers NS1 and NS8, 1,768 bp from patient 1 (AF238301) and 1,769 bp from patient 2 (AF255331) were amplified. A 600-bp fragment of L. loboi's chitin synthase gene was amplified from patient 1 (AF238303). To ensure that the L. loboi 18S rDNA sequences were compared broadly, they were first used in a BLAST (Basic Local Alignment Search Tool) search of GenBank holdings that showed their closest matches to be ascomycetous fungi. The L. loboi sequences were then aligned with 235 ascomycete and basidiomycete fungal sequences (provided by J. L. Platt, Plant and Microbial Biology, University of California at Berkeley) and subjected to neighbor-joining analysis that showed them to lie among the Onygenales. A second neighbor-joining analysis using 36 sequences representing ascomycete diversity gave the same result. Figure 2 shows a phylogenetic analysis of the two L. loboi 18S SSU rDNA sequences, the 28 Onygenales judged closest to L. loboi sequences by a BLAST search of GenBank, and outgroup sequences from four Eurotiales plus one Chaetothryiales. The analyses showed that the L. loboi sequences form a sister clade to the dimorphic human fungal pathogen P. brasiliensis. The two L. loboi and P. brasiliensis sequences were in a strongly supported clade with the other Onygenales. This clade included the pathogenic ascomycetes capable of causing systemic diseases in mammals: Ajellomyces dermatitidis (=Blastomyces dermatitidis) and two varieties of Ajellomyces capsulatus (=Histoplasma capsulatum var. capsulatum and duboisii). This clade also includes Emmonsia parva, another dimorphic pathogen of mammals, but does not include Coccidioides immitis, the remaining species of the dimorphic systemic pathogens.
FIG. 2.
Neighbor-joining tree of aligned 18S rDNA sequences of two L. loboi individuals (a, patient 1; b, patient 2), all 28 available Onygenales sequences, and four Eurotiales species and one Chaetothyriales (Capronia pilosella) as outgroups. Together, the two L. loboi sequences form the sister clade to another Latin American endemic human pathogen, P. brasiliensis. The distance between the two L. loboi isolates and the distance to its neighbor are remarkably large. Numbers above and below the branches are percentages of bootstrap-resampled data sets supporting the branch as obtained by neighbor-joining and parsimony analyses, respectively. The only dimorphic human pathogen not in the clade with L. loboi is C. immitis. There was no topological conflict between this neighbor-joining tree and the consensus of 312 most parsimonious trees found in a heuristic search with 1,000 random additions of taxa, in part because the parsimony consensus tree was more poorly resolved. The scale bar represents 0.005 nucleotide substitutions per nucleotide. The organisms used in this tree and the GenBank accession numbers of their 18S rDNA sequences are as follows: Aphanoascus mephitalis, AB015779; Arthroderma ciferrii, AB015769; Arthroderma incurvatum, AB015770; Ascocalvatia alveolata, AB015782; Aspergillus fumigatus, AB008401; Auxarthron compactum, AB015767; Auxarthron zuffianum, AZU29395; B. dermatiditis, X59420; B. dermatitidis, M55624 and M63096 (two strains); C. pilosella, U42473; E. parva (=Chrysosporium parvum), U29390; C. immitus, X58571; Ctenomyces serratus, U29391; Eremascus albus, M83258; Eupenicillium javanicum, U21298; Eurotium rubrum, U00970; Gymnascella aurantiaca, AB015772; Gymnoascoideus petalosporus, U29392; H. capsulatum var. capsulatum, X58572; H. capsulatum var. duboisii, Z75306; L. loboi a, AF238301; L. loboi b, AF255331; Malbranchea albolutea, L28063; Malbranchea aurantiaca, AB015786; Malbranchea dendritica, U29389; Malbranchea filamentosa, L28065; Malbranchea gypsea, L28066; Neosartorya fischeri, U21299; Onygena equina, U45442; P. brasiliensis, AF227151, AF238302, and AF241655; Pectinotrichum llanense, AB015783; Renispora flavissima, U29393, Rollandina hyalinospora, AB015775; Spiromastix warcupii, AB015768; Trichophyton rubrum, X58570; and Uncinocarpus reesii, U29394. The three B. dermatitidis sequences are identical except for one or two ambiguous nucleotide positions. The three P. brasiliensis sequences are identical except for two gaps and one nucleotide difference in sequence AF227151.
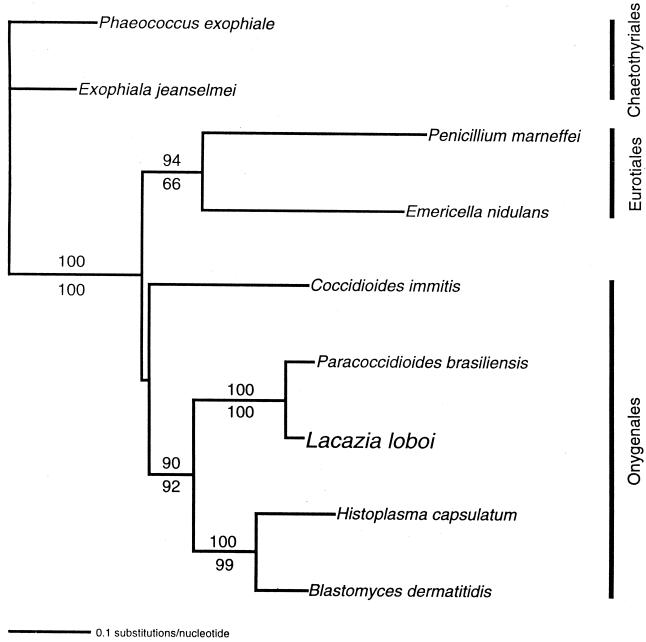
To challenge the SSU rDNA result with a different DNA region and to see if the remarkably long branches leading to and between the L. loboi sequences were typical of other DNA sequences, we turned to the gene coding for the CHS protein. Fortunately, many fungal CHS gene sequences are available in GenBank. Figure 3 shows the comparison of L. loboi's CHS2 nucleotide sequences with those of other Onygenales in parsimony and neighbor-joining analyses of L. loboi and eight other fungal 600-bp CHS2 nucleotide sequences. In these analyses, L. loboi was also the sister taxon of P. brasiliensis, and both fungi were again close relatives of the dimorphic systemic fungal pathogens B. dermatitidis and H. capsulatum. The relatively long branches leading to L. loboi in the analysis of 18S rDNA were not found in the analysis of CHS2.
FIG. 3.
Neighbor-joining tree of aligned CHS2 nucleotide sequences of one L. loboi strain, other dimorphic human pathogens, and two Eurotiales and two Chaetothyriales species as outgroup taxa. The L. loboi sequence is the sister taxon to P. brasiliensis and a member of the strongly supported clade of all dimorphic human pathogens except C. immitis. Changes in the parameters of maximum-likelihood multiple-hit correction affected the placement of C. immitis but not of the other taxa. Parsimony analysis (branch and bound) gave one tree with the same topology as the neighbor-joining tree. Numbers above and below the branches are percentages of bootstrap-resampled data sets supporting the branch as obtained by neighbor-joining and parsimony analyses, respectively. The scale bar represents 0.1 nucleotide substitution per nucleotide. The organisms used in this tree and the GenBank accession numbers of the CHS nucleotide sequences are as follows: B. dermatitidis M82943, C. immitis U60213, Emericella nidulans M82941, Exophiala jeanselmei M82945, H. capsulatum M82949, L. loboi AF238303, P. brasiliensis Y09231, Penicillium marneffei U60516, Phaeococcus exophiale M82953.
DISCUSSION
Phylogenetic analyses using the two 18S SSU rDNA molecules indicate that L. loboi is a distinct and novel species phylogenetically close to but fundamentally different from P. brasiliensis. Of interest to this study were findings that placed the dimorphic fungal pathogens within the Onygenales (4–6, 14, 21) and also a recent study that connected P. brasiliensis with the same order (22). These studies and the sequences that they provided made it possible for us to place L. loboi within the classical dimorphic fungal pathogens. Clustal alignment (Sequence Navigator, version 1.0.1; Perkin-Elmer) of the three P. brasiliensis 18S SSU rDNA sequences available in GenBank (one of them sequenced by us during this analysis [AF238302]) revealed that with the exception of nucleotides at the extreme 5′ and 3′ ends, our sequence is identical to AF241655 and differs from AF227151 at three nucleotide positions in the middle of the molecule (two gaps and one transition). In all other sequences used to make the tree in Fig. 2, these three nucleotide positions were invariant, suggesting that they could be sequencing artifacts. The close similarity or identity of the three P. brasiliensis sequences makes it very unlikely that the L. loboi sequences are variants of P. brasiliensis and stands in contrast to the large variation between our two L. loboi sequences (4–6). This finding further supports the concept that L. loboi is closely related to P. brasiliensis but with enough differences to stand as an independent species. Our DNA analyses suggest that L. loboi evolved among the dimorphic systemic fungal pathogens of the order Onygenales (Fig. 2 and 3), making it a reasonable hypothesis that L. loboi is a dimorphic fungus with a mycelial stage in nature as well as a yeast-like stage in the host.
The phylogenetic affinities between L. loboi and P. brasiliensis revealed during these analyses serve to explain the conflicting morphological taxonomic interpretations that have plagued studies on L. loboi. For instance, phylogenetic similarities between L. loboi and P. brasiliensis could well explain why the etiologic agent of paracoccidioidomycosis occasionally occurs as chains of small yeast-like cells connected by tubules in tissue sections (8). It also could explain why in 1930 Jorge Lobo (19, 20) stated that the etiologic agent of lobomycosis was probably a fungus similar to P. brasiliensis and why Fonseca and Lacaz (12) placed the etiologic agent of lobomycosis in the genus Paracoccidioides as P. loboi. Those conclusions are in sharp contrast with the one proposed by Haubold et al. (15). Those investigators stated that the L. loboi found in dolphins was related to the darkly pigmented (phaeoid, dematiaceous) fungus Cladosporium sphaerospermum. Our phylogenetic analyses, however, placed L. loboi distant from the black fungi, suggesting that, although L. loboi possesses the pigment melanin in its cell walls (28), it is not a phaeoid fungus. This is not unusual, since many fungi, including the human and animal pathogen Cryptococcus neoformans, have melanin in their cell walls and yet are not classified as black fungi (24).
Although L. loboi and P. brasiliensis are close relatives, there are significant differences between the two fungi, not only at the DNA level but also in the diseases that they cause. While P. brasiliensis clearly is a geophilic dimorphic fungus acquired through inhalation and eventually disseminating from lungs to other organs, L. loboi is always localized in the cutaneous and subcutaneous tissues (because it is apparently intolerant to 37°C temperature) and it is believed to be acquired through trauma. Moreover, because the disease is also found in dolphins and most humans are infected in or around aquatic habitats, it has been postulated that L. loboi is a hydrophilic microorganism (1). These facts are in contrast with the clinical and epidemiological features of the infections caused by P. brasiliensis and the other pathogenic dimorphic Onygenales. Examples of pathogenic fungi with similar phylogenetic backgrounds and different clinical signs are well known. A good example of this dichotomy is H. capsulatum var. capsulatum, which is acquired from the environment via the lungs, and H. capsulatum var. farciminosum, which is passed from animal to animal via yeast-like cells produced in skin lesions (8, 8a, 25, 25a). The ability to grow at 37°C appears to be ancestral in the clade containing members of the genera Blastomyces, Emmonsia, Histoplasma, Lacazia, and Paracoccidioides. If L. loboi lost the ability to grow at 37°C, as is implied by the locations of its lesions on ears and other cooler areas of the body (2, 11, 12), this early divergent feature could well explain why L. loboi causes only cutaneous and subcutaneous infections.
The biggest surprise in this study was finding only 98% similarity between the two L. loboi 18S SSU rDNA sequences. A study of more strains will show if this variation foreshadows different populations of L. loboi or simply reflects rapid substitution in this molecule, as might be inferred from the minor variation between the isolates found in the CHS2 gene. Our analyses indicated that this obligate pathogen has not been previously classified in any of the taxa examined in this study. Therefore, the binomial Lacazia loboi, recently introduced for the etiologic agent of lobomycosis, is valid. The use of molecular procedures to study L. loboi initiates a new chapter in the study of this pathogen. It is anticipated that studies using molecular approaches will be essential in understanding L. loboi's biological and epidemiological features and its intractability to culture. This could lead to the development of new drugs and/or vaccines to treat and prevent lobomycosis in the areas of endemicity and beyond.
ACKNOWLEDGMENTS
We thank Araujo Opromolla for the use of his facilities and resources to isolate L. loboi's genomic DNA.
The study was supported by the Medical Technology Program, College of Natural Science, Michigan State University, and the Miller Institute for Basic Research in Science at the University of California, Berkeley.
REFERENCES
- 1.Ajello L. Ecology and epidemiology of hydrophilic infectious fungi and parafungi of medical mycological importance: a new category of pathogens. In: Ajello L, Hay R J, editors. Topley & Wilson's microbiology and microbial infections, medical mycology. 9th ed. Vol. 4. London, England: Arnold; 1998. pp. 67–73. [Google Scholar]
- 2.Borelli D. Lobomicose: nomen de su agente (revision crìtica) Med Cut. 1968;3:151–156. [Google Scholar]
- 3.Bowen A R, Chen-Wu J L, Momany M, Young R, Szaniszlo P J, Robbins P W. Classification of fungal chitin synthases. Proc Natl Acad Sci USA. 1992;89:519–523. doi: 10.1073/pnas.89.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman B, Taylor J W, White T J. Molecular evolution of the fungi: human pathogens. Mol Biol Evol. 1992;9:893–904. doi: 10.1093/oxfordjournals.molbev.a040766. [DOI] [PubMed] [Google Scholar]
- 5.Bowman B H, Taylor J W. Molecular phylogeny of pathogenic and non-pathogenic Onygenales. In: Reynolds D R, Taylor J W, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, United Kingdom: CAB International; 1993. pp. 169–178. [Google Scholar]
- 6.Bowman B H, White T J, Taylor J W. Evolutionary relationships of human pathogenic fungi: multiple origins of pathogenicity in the fungal order Onygenales. Mol Phylog Evol. 1996;6:89–96. [Google Scholar]
- 7.Burns R A, Roy J S, Woods C, Padhye A A, Warnock D W. Report of the first human case of lobomycosis in the United States. J Clin Microbiol. 2000;38:1283–1285. doi: 10.1128/jcm.38.3.1283-1285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler F W, Kaplan W, Ajello L. Color atlas and text of the histopathology of mycotic diseases. Chicago, Ill: YearBook; 1980. pp. 73–75. [Google Scholar]
- 8a.Chandler F W, Kaplan W, Ajello L. Color atlas and text of the histopathology of mycotic diseases. Chicago, Ill: YearBook; 1980. pp. 88–91. [Google Scholar]
- 9.Chen-Wu J L, Swicker J, Bowen A R, Robbins P W. Expression of chitin synthase genes during yeast and hyphal growth phases of Candida albicans. Mol Microbiol. 1992;6:497–502. doi: 10.1111/j.1365-2958.1992.tb01494.x. [DOI] [PubMed] [Google Scholar]
- 10.Ciferri R, Acevedo P C, Campos S, Carneiro L S. Taxonomy of Jorge Lobo's disease fungus. Inst Micol Univ Recife. 1956;53:1–21. [Google Scholar]
- 11.Fonseca O F, Leao A E A. Contribuçao para o conhecimento des granulomatoses blastomycoides. O agente etiologico da doença de Jorge Lôbo. Rev Med Cirug Brasil. 1940;48:147–158. [Google Scholar]
- 12.Fonseca O J M, Lacaz C S. Estudo de culturas isoladas de blastomicose queloidiforme (doença de Jorge Lobo). Denominação ao seu agente etiològico. Rev Inst Med Trop São Paulo. 1971;13:225–251. [PubMed] [Google Scholar]
- 13.Gargas A, DePriest P T. A nomenclature for fungal PCR primers with examples from intron-containing SSU rDNA. Mycologia. 1996;88:745–748. [Google Scholar]
- 14.Gueho E, Leclerc M C, de Hoog G S, Dupont B. Molecular taxonomy and epidemiology of Blastomyces and Histoplasma species. Mycoses. 1997;40:69–81. doi: 10.1111/j.1439-0507.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 15.Haubold E M, Aronson J F, Cowan D F, McGinnis M R, Cooper C R., Jr Isolation of fungal rDNA from bottlenose dolphin skin infected with Loboa loboi. Med Mycol. 1998;36:263–267. [PubMed] [Google Scholar]
- 16.Issakainen J, Jalava J, Eerola E, Campbell C K. Relatedness of Pseudoallescheria, Scedosporium, and Graphium proparte based on SSU rDNA sequences. J Med Vet Mycol. 1997;35:389–398. [PubMed] [Google Scholar]
- 17.Lacaz C S, Ferri R G, Raphael A, Fava-Neto C, Minami P S, Castro R M, Dillon N L. Blastomicose queloideana asociada a blastomicose sulamericana. Registro de um caso. Hospital (Rio de Janeiro) 1967;71:7–11. [PubMed] [Google Scholar]
- 18.Langeron M, Vanbreuseghem R. Mycologie humaine et animale. Techniques. Paris, France: Mason et Cie; 1952. pp. 490–491. [Google Scholar]
- 19.Lobo J O. Nova especie de blastomicose. Brasil Med. 1930;44:1227. [Google Scholar]
- 20.Lobo J O. Um caso de blastomicose produzido por uma espécie nova, encontrada en Recife. Rev Med Pernambuco. 1931;1:763–765. [Google Scholar]
- 21.Pan S, Sigler L, Cole G T. Evidence for a phylogenetic connection between Coccidioides immitis and Uncinocarpus reesii (Onygenaceae) Microbiology. 1994;140:1481–1494. doi: 10.1099/00221287-140-6-1481. [DOI] [PubMed] [Google Scholar]
- 22.Peterson S W, Sigler L. Molecular genetic variation in Emmonsia crescens and Emmonsia parva, etiologic agents of adiaspiromycosis, and their phylogenetic relationship to Blastomyces dermatitidis (Ajellomyces dermatitidis) and other systemic fungal pathogens. J Clin Microbiol. 1998;36:2918–2925. doi: 10.1128/jcm.36.10.2918-2925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pradinaud R. Loboa loboi. In: Ajello L, Hay R J, editors. Topley & Wilson's microbiology and microbial infections, medical mycology. 9th ed. Vol. 4. London, England: Arnold; 1998. pp. 67–73. [Google Scholar]
- 24.Pulverer C, Korth H. Cryptococcus neoformans: Pigment Bildung aus verschiedenen Polyphenolen. Med Microbiol Immunol. 1971;175:46–51. doi: 10.1007/BF02121290. [DOI] [PubMed] [Google Scholar]
- 25.Rippon J W. Medical mycology. 3rd ed. Philadelphia, Pa: W. B. Saunders Company; 1988. pp. 353–361. [Google Scholar]
- 25a.Rippon J W. Medical mycology. 3rd ed. Philadelphia, Pa: W. B. Saunders Company; 1988. pp. 381–432. [Google Scholar]
- 26.Symmers W S. A possible case of Lobo's disease acquired in Europe from a bottle nosed dolphin. Bull Soc Path Exot. 1983;46:777–784. [PubMed] [Google Scholar]
- 27.Taborda P R, Taborda V A, McGinnis M R. Lacazia loboi gen. nov., comb. nov., the etiologic agent of lobomycosis. J Clin Microbiol. 1999;37:2031–2033. doi: 10.1128/jcm.37.6.2031-2033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taborda P R, Taborda V A, McGinnis M R. Constitutive melanin in the cell wall of the etiologic agent of Lobo's disease. Rev Inst Med Trop São Paulo. 1999;41:9–12. doi: 10.1590/s0036-46651999000100003. [DOI] [PubMed] [Google Scholar]
- 29.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. Orlando, Fla: Academic Press; 1990. pp. 315–322. [Google Scholar]