Abstract
Background
The Alphapapillomavirus 9 (α-9 HPV) is a member of the Alphapapillomavirus genus and Papillomaviridae family. These viruses are almost all carcinogenic HPV, which is closely related to 75% of invasive cervical cancer worldwide, and has a high prevalence in Sichuan. The carcinogenic function is mainly realized by its E6 oncoprotein.
Methods
Cell samples were collected by cervical scraped for HPV detecting and typing. HPV-16, HPV-31, HPV-33, HPV-52, HPV-58 5 α-9 genus HPV subtype positive samples were selected, their E6 gene was sequenced and analyzed. The positive selection sites of HPV E6 genes were estimated by PAML 4.8 server. The secondary and tertiary structure of E6 protein were predicted by PSIPred and Swiss-model. The T-cell antigen epitopes of E6 protein were predicted by IEDB.
Results
α-9 HPV has a high prevalence in Sichuan, China. From 2012 to 2017, 18,067 cell cervical samples were collected, and 3135 were detected with α-9 HPV infection. Among which, 250 cases HPV-16 E6, 96 cases HPV-31 E6, 216 cases HPV-33 E6, 288 cases HPV-52 E6 and 405 cases HPV-58 E6 were successfully amplified, 17, 6, 6, 13, and 4 non-synonymous nucleotide mutations were respectively detected in HPV-16, 31, 33, 52, and 58 E6, 7 positive selection sites of α-9 HPV E6 were selected out (D32E of HPV-16 E6, K35N, K93N and R145I of HPV-33 E6, K93R of HPV-52 E6, K93N and R145K of HPV-58 E6). The structure and antigen epitopes of E6 protein with amino acid substitution differ from those of wild-type E6 protein, especially for the mutation located in the E6 positive selection site.
Conclusions
HPV E6 nucleotide non-synonymous mutation in the positive selection site influence the protein structure and decrease the antigen epitopes affinity of the E6 protein overall, making it more difficult for the HPV-infected cells to be detected by the immune system, and enhancing the HPV adaptability to the environment. Mutations influence the validity of HPV clinical diagnostic probes, the polymorphism analysis of α-9 HPV E6 enrich the data of HR-risk HPV in Sichuan China, and the detection probes designed with the polymorphism data in mind can improve the efficiency of clinical detection; Mutations influence epitopes affinity, the association of E6 polymorphism and epitope affinity can improve the design of therapeutic vaccine with good immunity and high generality antigen epitope; The above study all provide a good theoretical basis for the prevention and treatment of HPV-related diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-021-01728-4.
Keywords: Human Papillomavirus, Alphapapillomavirus 9 genus HPV, E6 gene polymorphisms, Protein structure, Positive selection site, Antigen epitope
Introduction
Cervical cancer is the second most common cancer among women aged 15–44, 99.7% of the cervical cancers were found to be associated with high-risk (HR) Human Papilloma Virus (HPV) persistent infection [1]. The three main genera are α, β, γ, of which α genus is associated with anal and oral mucosal infection. α-9 genus HPV is almost all carcinogenic HPV, causing 75% invasive cervical cancer worldwide, and its carcinogenicity is mainly realized by E6 and E7 early proteins encoded by HPV E6 and E7. The carcinogenicity of HPV E6 protein is more evident than that of E7 protein, in terms of the cell cycle changes and the efficiency of HPV infected cells to permanent biochemical transformations [2, 3]. Without E7, E6 can connect and ubiquitin degradation the p53 protein via E6AP, interfere the cell cycle, activate telomerase and reverse transcriptase to accumulate the mutations, for infected cells immortalization and maintain immortalization, which is closely related to the function of HPV immortalization, cell transformation, and carcinogenesis [4–7].
At present, there is no specific drug for HPV treatment and mainly relies on the body's immune system to detect and eliminate the virus. Human leukocyte antigen (HLA) has the function of recognizing itself target by recognizing and stimulating CD8+ cytotoxic T lymphocytes (CTL), CD4+ helper T lymphocytes (Th) as well as binding antigen polypeptide to regulate the immune response, control and eliminate HPV infection [8–11]. The antigen epitopes are composed of specific amino acid sequences, are the targets of immune rejection [12–14]. HPV E6 protein has been considered as a potential target for the activation of T cells in immune response strategies and maybe an ideal target for HPV therapeutic vaccines [15, 16].
HPV is a high infectious and mutable virus, with different epidemic trends and mutation types in different regions and populations [17]. The polymorphism of HPV E6 oncogene is strong, its non-synonymous mutation changed E6 protein amino acid composition, which may relate to the differences in immune response and pathogenicity [18]. Some mutant strains can even fix their genes by mutations, enhancing their adaptability to the environment and changing its infection rate [19]. For example, L83V (L90V) of HPV-16 E6 in Swedish and Italian populations as well as D25E (D32E) of HPV-16 E6 in Japanese populations have been proven to be associated with the progression of cervical cancer [20–23]. In the current HPV vaccine design targeting E6 protein, the E6 mutants have almost never been considered. In the epitope-specific vaccine designed by Kelly L, the body's long-active T-cell response to HPV-18 was induced by targeting the reference sequence of HPV E6 and E7. Due to the relatively rapid mutation rate of HPV, the host response capacity, malignant tumor prevention and therapeutic efficacy of vaccine were changed great [15].
HPV has strong regional and population differences, the prevalence of α-9 HPV and the harmfulness of E6 oncoprotein are extremely high, and E6 polymorphism is closely related to the difference of immunogenicity, adaptability, and pathogenicity. Therefore, it’s urgent to study the genetic diversity, positive selection sites, antigen epitope, the protein structure of α-9 HPV E6 for providing data to realize the effective prevention and control of the disease in this region.
Materials and methods
Samples resource
The study was ethically approved by the Education and Research Committee and Ethics Committee of Sichuan University, Sichuan, China. Eighteen thousand sixty-seven specimens were randomly collected from January 2012 to December 2017 in Chengdu Women and Children's Center Hospital, Chengdu Jinjiang District Women and Children's Hospital, Angel Women's and Children's Hospital, Affiliated Hospital of Sichuan Reproductive Health Research Center, Sichuan Reproductive Health Research Center Affiliated Hospital, Shuangnan Hospital, Chengdu Song zi niao Sterility Hospital, Infertility Hospital Affiliated to Chengdu Medical College and Chengdu Jinsha hospital. Before sample collection, written informed consent was obtained from all patients or their guardians, and patient privacy is strictly protected. The cell specimens were collected randomly by cervical scraped and placed in − 20 °C antiseptic buffer (9 g NaCl, 10 g C6H5CO2Na, 1 L H2O).
Genomic DNA extraction and HPV typing
HPV DNA was extracted and evaluated using the Human Papillomavirus Genotyping Kit For 23 Types (Yaneng Bio, Shenzhen, China) according to the manufacturer’s guidelines.
PCR amplification and variant identification
The primers of α-9 HPV E6 were designed by PRIMER version 5.0 and NCBI (National Center for Biotechnology Information) Primer Blast based on the reference sequences, the primers and reference sequences used for the molecular characterization analysis of α-9 HPV E6 were shown in Additional file 1: Table S1 and synthesized by TSINGKE (Chengdu, China). The PCR reaction system consists of 5 µl HPV DNA, 13.1 µl ddH2O, 1 µl primers, 0.4 µl TransTaq DNA polymerase, 2.5 µl dNTPs, and 3 µl buffer. The reaction conditions were shown in Additional file 1: Table S1. The PCR products were visualized by gel electrophoresis in 2% agarose gel (Sangon Biotech Co., Ltd.). The target products of E6 were purified and sequenced by TSINGKE at least twice (Chengdu, China).
Sequence analysis
Genetic polymorphisms analysis of α-9 HPV E6 gene
The successfully amplified sequences was sequenced, and the sequences were analyzed by NCBI BLAST, Premier5, and DNAMAN5.2.2. Nucleotide mutations of α-9 HPV E6 sequence were determined according to the reference sequence in GenBank (Additional file 1: Table S1). Chi-square test was used to confirm the significance of data differences, and P < 0.05 was considered as significant differences between the data.
Selective pressure analysis of α-9 HPV E6
Phylogenetic Analysis by Maximum Likelihood 4.8 (PAML 4.8, http://abacus.gene.ucl.ac.uk/software/paml.html) was used to calculate the ratio (ω = dN/dS) between non-synonymous mutation rate (dN) and synonymous mutation rate (dS) to determine the α-9 HPV E6 gene-positive selection sites.
Amino acid composition and protein structure analysis of α-9 HPV E6
Mega6.0 software was used to translate the E6 nucleotide sequence into the E6 protein sequence. PSIPred (http://bioinf.cs.ucl.ac.uk/psipred/) and Swiss-model were used to analyze the secondary and tertiary structure of E6 protein.
T-cell antigen epitopes predicted analysis of α-9 HPV E6 protein
According to the Chinese major histocompatibility complex database (dbMHC) average frequency of HLA alleles, 13 HLA-I and 6 HLA-II alleles were selected (Additional file 1: Table S2). Based on the selected HLA alleles, the T-lymphocyte epitopes of α-9 HPV E6 protein were predicted by IEDB resource (http://www.iedb.org/). According to the method recommended by IEDB, lower the percentile rank (PR) of antigen epitopes is better the affinity, peptides with PR < 1.0 for HLA-I and peptides with PR < 5.0 for HLA-II were deemed to meaningful as well as selected for further analysis.
Results
The prevalence of α-9 HPV in Sichuan
Out of 18,067 samples, 6092 positive results were detected and 4466 were HR HPV, all of which belonged to α genus. The HPV positive samples of α-1, α-3, α-5, α-6, α-7, α-8, α-9, α-10, α-11 are 167 (2.74%), 25 (0.41%), 137 (2.25%), 438 (7.19%), 571 (9.37%,), 413 (6.78%), 3270 (53.68%), 1021 (16.76%), 50 (0.82%) respectively (Fig. 1). α-9 HPV accounting for 73.22% of HR HPV positive samples. Due to the small positive sample sizes of HPV-35 and HPV-67, five other HPV type (HPV-16, HPV-31, HPV-33, HPV-52, HPV-58) were selected for subsequent studies.
Fig. 1.
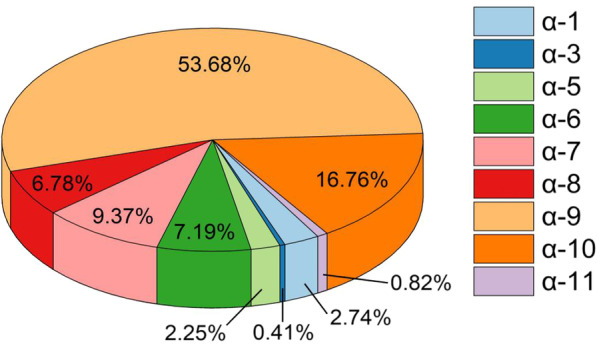
Distribution of different HPVs positive samples from 2012 to 2017
Nucleotide polymorphisms and selective pressure analysis of α-9 HPV E6
250 HPV-16 E6 were successfully amplified, 162 (64.80%) HPV-16 E6 samples were variants, and 17 non-synonymous mutations were detected. 96 HPV-31 E6 were successfully amplified, 68 (70.80%) variants and 6 non-synonymous mutations were detected. 216 HPV-33 E6 were successfully amplified, 76 (35.19%) variants and 6 non-synonymous mutations were detected. 288 HPV-52 E6 were successfully amplified, 250 (86.80%) variants and 13 non-synonymous mutations were detected. 405 HPV-58 E6 were successfully amplified, 356 (87.90%) variants and 4 non-synonymous mutations were detected. Details of α-9 HPV E6 nucleotide polymorphisms were shown in Tables 1, 2, 3, 4 and 5. All the sequences were submitted to the GenBank, and accession numbers were obtained. (HPV16E6: MZ803036-MZ803058, HPV31E6: MZ803026-MZ803035, HPV33E6: MZ576479-MZ576485, HPV52E6: MZ803059-MZ803078, HPV58E6: MZ803079-MZ803087).
Table 1.
Nucleotide mutation and amino acid substitution in HPV-16 E6
| HPV-16 E6 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| Location | 2 | 12 | 30 | 49 | 94 | 96 | 96 | 96 | 101 | 103 | 103 | 111 | 185 | 192 | 194 | 196 | 253 | 268 | 313 | 360 | 452 |
| Mutation | T–A | G–A | G–A | A–G | G–A | T–A | T–G | T–C | T–G | T–G | T–C | T–C | G–T | G–T | A–G | C–G | C–T | T–G | G–C | A–C | G–C |
| Frequency (%) | 0.80 | 3.20 | 0.80 | 0.80 | 0.80 | 0.80 | 49.6 | 0.80 | 0.80 | 0.80 | 0.80 | 1.60 | 0.80 | 0.80 | 1.60 | 0.80 | 0.80 | 5.60 | 0.80 | 5.60 | 0.80 |
| Substitution | M1K | – | – | R17G | D32N | D32E | D32E | D32E | I34R | L35V | L35V | – | R62I | – | N65S | P66A | H85Y | L90V | D105H | E120D | R151T |
Compared with the HPV-16 E6 reference sequence (NC001526), the mutations are marked with the corresponding bases and amino acid, and those without changes are replaced with a dash (–). No. means the number of nucleotide mutations, Location means the sites of nucleotide mutations, Mutation means the style of nucleotide mutations, Frequency (%) means the percentage of nucleotide mutations, Substitution means the amino acid substitution that occurred by nucleotide mutations
Table 2.
Nucleotide mutation and amino acid substitution in HPV-31 E6
| HPV-31 E6 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| Location | 27 | 69 | 141 | 178 | 190 | 194 | 205 | 219 | 228 | 297 | 321 | 368 | 413 |
| Mutation | T–A | C–T | T–C | C–T | A–G | A–G | T–C | A–G | T–C | G–A | A–G | A–G | C–T |
| Frequency (%) | 8.33 | 27.08 | 4.17 | 31.25 | 4.17 | 2.08 | 4.17 | 16.67 | 2.08 | 31.25 | 27.08 | 4.17 | 35.42 |
| Substitution | – | – | – | H60Y | T64A | K65R | F69L | – | – | – | – | K123R | A138V |
Compared with the HPV-31 E6 reference sequence (J04353), the mutations are marked with the corresponding bases and amino acid, and those without changes are replaced with a dash (–). No. means the number of nucleotide mutations, Location means the sites of nucleotide mutations, Mutation means the style of nucleotide mutations, Frequency (%) means the percentage of nucleotide mutations, Substitution means the amino acid substitution that occurred by nucleotide mutations
Table 3.
Nucleotide mutation and amino acid substitution in HPV-33 E6
| HPV-33 E6 | ||||||||
|---|---|---|---|---|---|---|---|---|
| No | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Location | 105 | 165 | 221 | 256 | 279 | 338 | 434 | 441 |
| Mutation | A–C | A–G | G–C | A–C | A–C | A–G | G–T | T–C |
| Frequency (%) | 19.44 | 7.87 | 3.24 | 7.41 | 19.44 | 7.41 | 15.28 | 11.57 |
| Substitution | K35N | – | S74T | N86H | K93N | Q113R | R145I | – |
Compared with the HPV-33 E6 reference sequence (M12732.1), the mutations are marked with the corresponding bases and amino acid, and those without changes are replaced with a dash (–). No. means the number of nucleotide mutations, Location means the sites of nucleotide mutations, Mutation means the style of nucleotide mutations, Frequency (%) means the percentage of nucleotide mutations, Substitution means the amino acid substitution that occurred by nucleotide mutations
Table 4.
Nucleotide mutation and amino acid substitution in HPV-52 E6
| HPV-52 E6 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| Location | 61 | 82 | 93 | 118 | 136 | 136 | 230 | 237 | 249 | 255 | 265 | 270 | 277 | 277 | 278 | 315 | 315 | 366 | 381 | 412 | 429 |
| Mutation | G–A | C–T | G–C | A–C | C–G | C–T | G–A | T–C | G–T | G–A | G–A | G–A | A–C | A–G | A–G | T–G | T–A | C–A | T–A | G–A | A–G |
| Frequency (%) | 0.70 | 0.70 | 0.70 | 0.70 | 0.70 | 2.10 | 0.70 | 0.70 | 85.40 | 9.70 | 0.70 | 0.70 | 1.40 | 2.10 | 84.00 | 0.70 | 0.70 | 0.70 | 0.70 | 0.70 | 2.80 |
| Substitution | E21K | – | – | – | L46V | L46V | R77K | – | – | – | E89K | – | K93R | K93R | K93R | I105M | I105M | N122K | N127I | E138K | – |
Compared with the HPV-52 E6 reference sequence (NC001592), the mutations are marked with the corresponding bases and amino acid, and those without changes are replaced with a dash (–). No. means the number of nucleotide mutations, Location means the sites of nucleotide mutations, Mutation means the style of nucleotide mutations, Frequency (%) means the percentage of nucleotide mutations, Substitution means the amino acid substitution that occurred by nucleotide mutations
Table 5.
Nucleotide mutation and amino acid substitution in HPV-58 E6
| HPV-58 E6 | ||||||||
|---|---|---|---|---|---|---|---|---|
| No | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Location | 78 | 94 | 150 | 198 | 258 | 279 | 286 | 434 |
| Mutation | C–T | G–C | A–G | C–T | C–A | A–C | T–C | G–A |
| Frequency (%) | 1.73 | 0.25 | 0.49 | 58.52 | 0.25 | 27.41 | 0.25 | 3.95 |
| Substitution | – | E32Q | – | – | D86E | K93N | – | R145K |
Compared with the HPV-58 E6 reference sequence (D90400), the mutations are marked with the corresponding bases and amino acid, and those without changes are replaced with a dash (–). No. means the number of nucleotide mutations, Location means the sites of nucleotide mutations, Mutation means the style of nucleotide mutations, Frequency (%) means the percentage of nucleotide mutations, Substitution means the amino acid substitution that occurred by nucleotide mutations
Calculated by Codeml software using Naive NEB and Bayes Empirical Bayes models, seven positive selection sites of α-9 HPV E6 were detected, there were D32E of HPV-16 E6, K35N, K93N, R145I of HPV-33 E6, K93R of HPV-52 E6, K93N, R145K of HPV-58 E6. In contrast, no reliable HPV-31 E6 positive selection site was selected out (Table 6).
Table 6.
Positive section site of α-9 HPV E6
| Model | HPV-16 | HPV-31 | HPV-33 | HPV-52 | HPV-58 |
|---|---|---|---|---|---|
| M7 | NA | NA | NA | NA | NA |
| M8 | 32D** | NA | 35 K**, 93 K**, 145R** | 93 K** | 93 K**, 145R** |
M7 means NEB (Naive Empirical Bayes) model, M8 means BEB (Bayes Empirical Bayes) model. When the posterior probability was ≥ 0.9, the BEB method was used to identify the positive selection sites. P < 0.05 indicates that the results of M8 model are reliable, two asterisks means a posteriori probability ≥ 0.99, and NA means not apply
The protein structure analysis of α-9 HPV E6
Nucleotides non-synonymous mutation changed the amino acid composition of protein, which affects the structure of the protein, while the protein function is mainly realized by its structures. With the help of Mega6.0, PSIPred and Swiss-model, the primary, secondary, and tertiary structure difference of α-9 HPV E6 protein reference and mutation sequence were revealed.
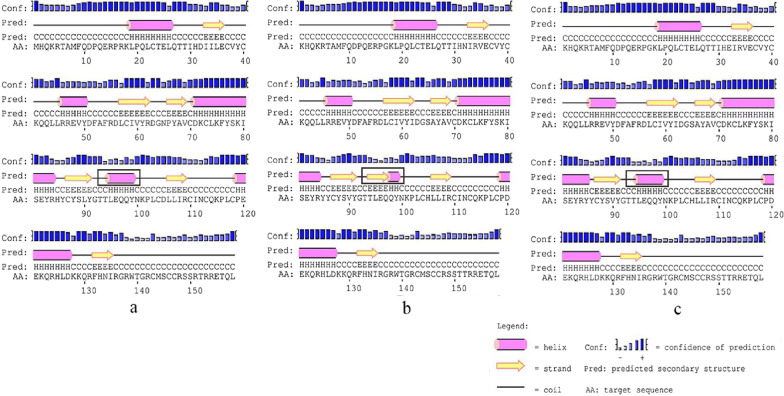
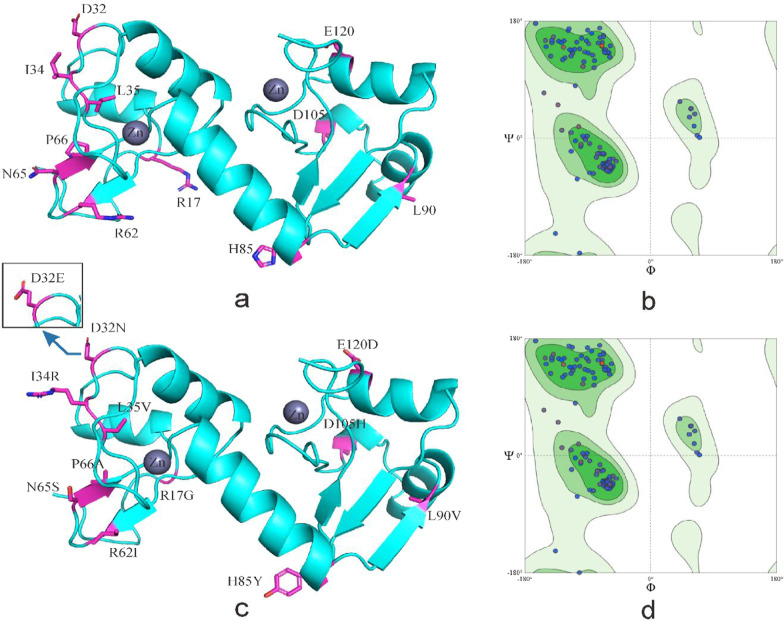
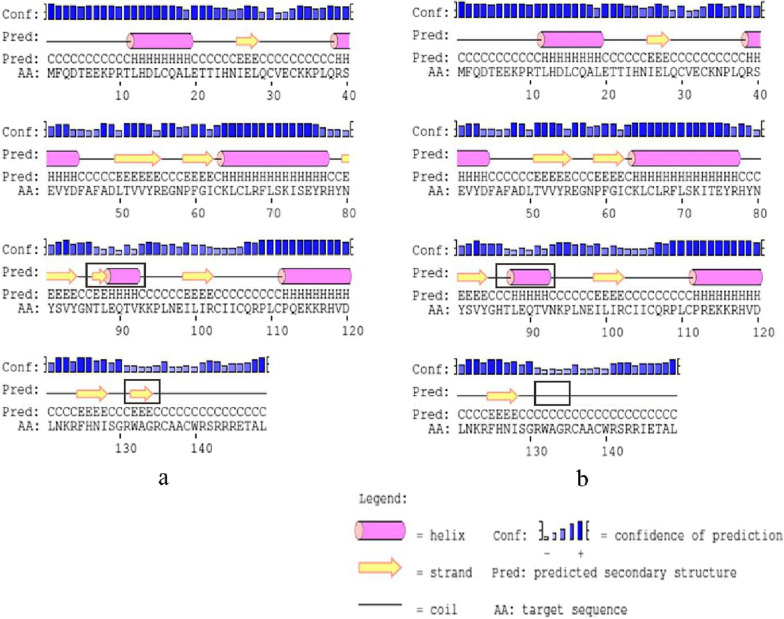
In HPV-16 E6, I34R, L35V, R62I, P66A and L90V all located in β-fold, E120D, D32N, D32E located in the periphery of the spatial protein structure and close to the active region of znic granules. The amino acid number in the α-helix and β-sheet regions are different in protein reference and mutation sequence. Details are shown in Figs. 2 and 3.
Fig. 2.
Secondary structure of HPV-16 E6 comparing reference to the variant sequence. Note a is the secondary structure pattern diagram constructed based on HPV-16 E6 reference sequence; b is the secondary structure pattern diagram constructed based on HPV-16 E6-1 mutation sequence; c is the secondary structure pattern diagram constructed based on HPV-16 E6-2 mutation sequence, b and c are different in 32th amino acid. The black boxes are the difference areas between the reference and mutation sequence secondary structure.
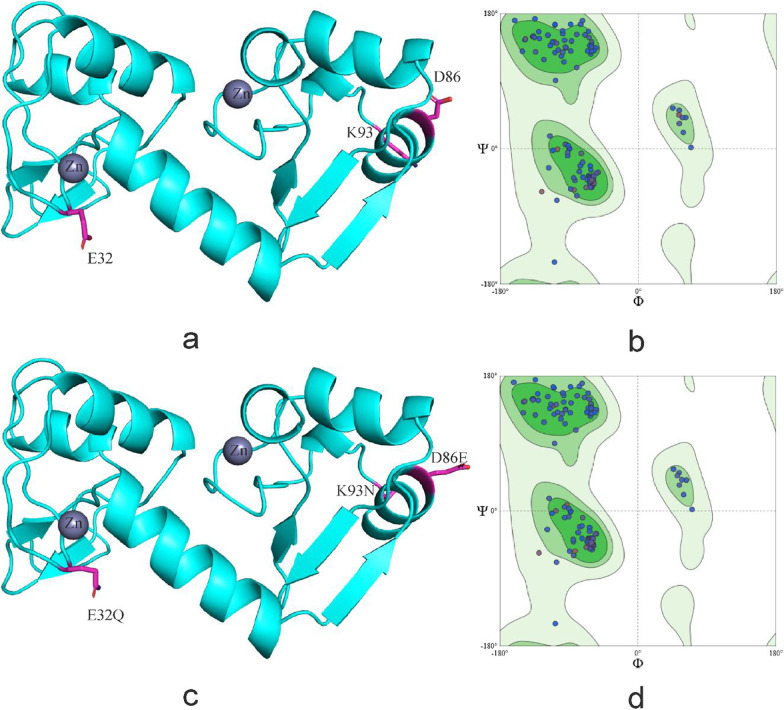
Fig. 3.
Tertiary structure of HPV-16 E6 comparing reference to the variant sequence. Note a is the homology modeling structure of HPV-16 E6 reference sequence; b is the larlarian diagram of HPV-16 E6 reference sequence homology modeling; c is homology modeling structure of HPV-16 E6 mutation sequence; d is the larchian diagram of HPV-16 E6 mutation sequence homology modeling
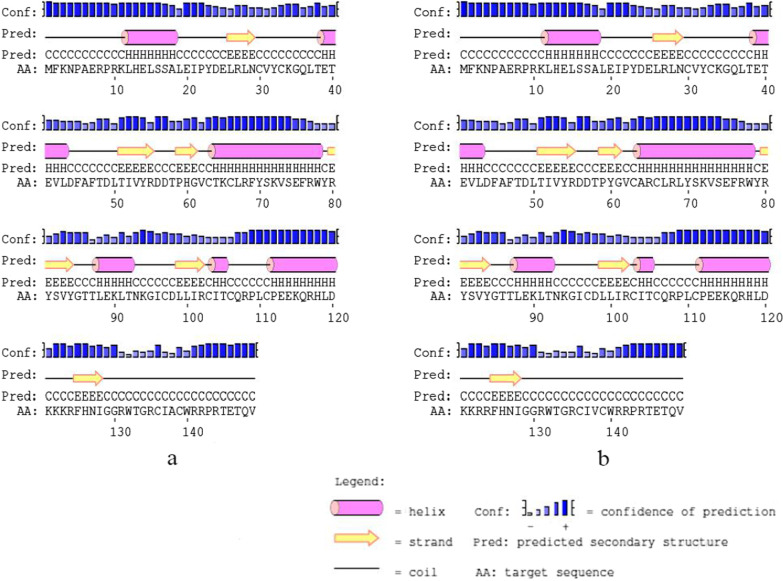
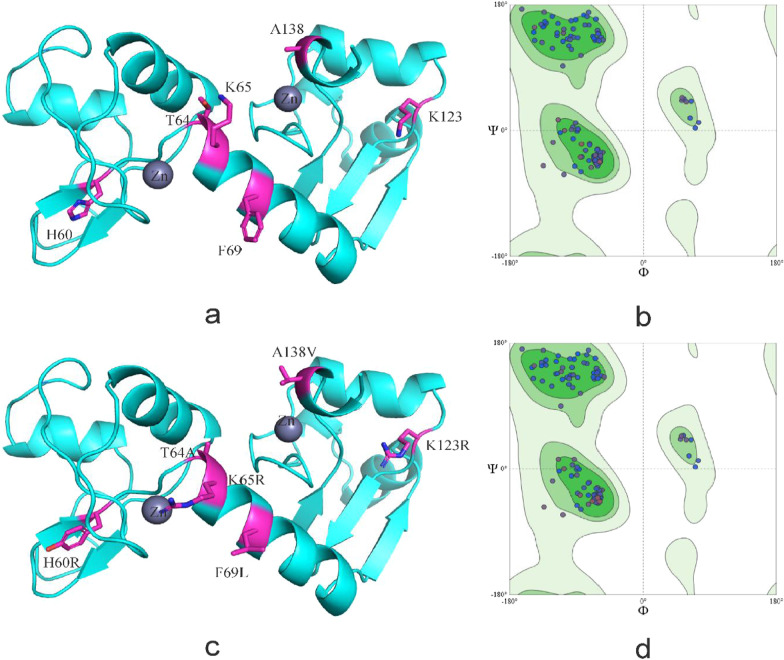
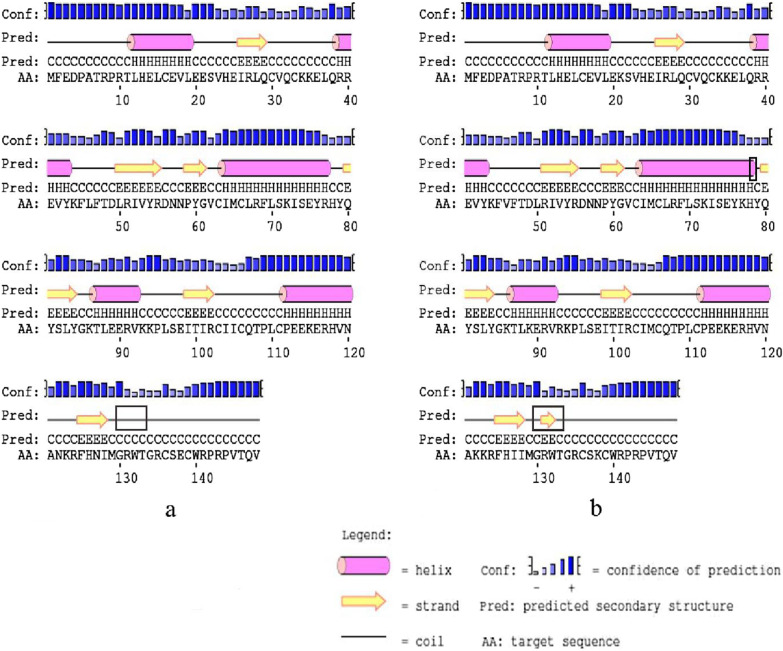
In HPV-31 E6, T64A, K65R, and F69L located in α-helix, T60Y located in β-sheet region, and K123R, A138V located in the coil. Amino acid substitution has no influence on the secondary and tertiary structure (Figs. 4, 5).
Fig. 4.
Secondary structure of HPV-31 E6 comparing reference to the variant sequence. Note a is the secondary structure pattern diagram constructed based on HPV-31 E6 reference sequence; b is the secondary structure pattern diagram constructed based on HPV-31 E6 mutation sequence
Fig. 5.
Tertiary structure of HPV-31 E6 comparing reference to the variant sequence. Note a is the homology modeling structure of HPV-31 E6 reference sequence; b is the larlarian diagram of HPV-31 E6 reference sequence homology modeling; c is homology modeling structure of HPV-31 E6 mutation sequence; d is the larchian diagram of HPV-31 E6 mutation sequence homology modeling
S74T and Q113R located in α-helix of HPV-33 E6 protein, K93N located on the outer edge of E6 protein and near the zinc granule, the above amino acid substitutions all located in the active region of the protein. Amino acid substitution changed the number of amino acids in the α-helix and β-sheet region, as well as made the E6 protein show more contact with the environment (Figs. 6, 7).
Fig. 6.
Secondary structure of HPV-33 E6 comparing reference to the variant sequence. Note a is the secondary structure pattern diagram constructed based on HPV-33 E6 reference sequence; a is the secondary structure pattern diagram constructed based on HPV-33 E6 mutation sequence. The black boxes are the difference areas between the reference and mutation sequence secondary structure
Fig. 7.
Tertiary structure of HPV-33 E6 comparing reference to the variant sequence. Note a is the homology modeling structure of HPV-33 E6 reference sequence; b is the larlarian diagram of HPV-33 E6 reference sequence homology modeling; c is homology modeling structure of HPV-33 E6 mutation sequence; d is the larlarian diagram of HPV-33 E6 mutation sequence homology modeling
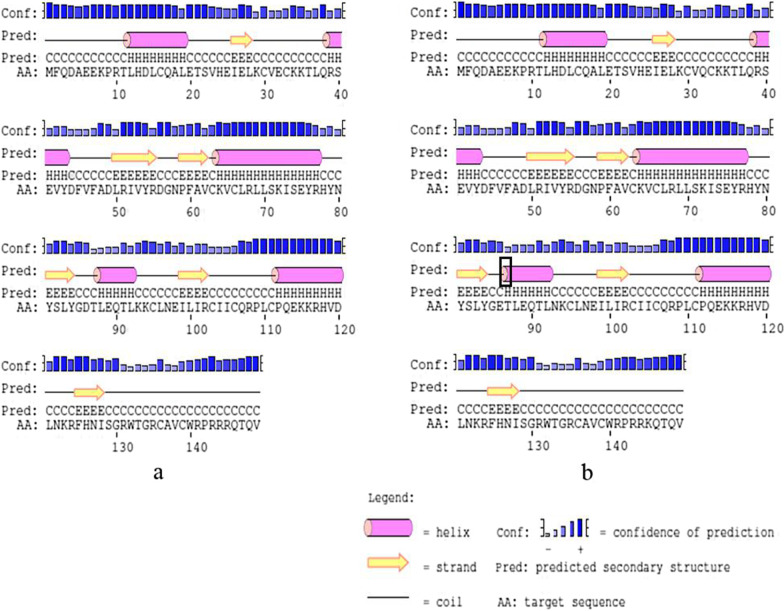
R77K, E89K of HPV-52 E6 located in α-helix, N127I located in the β-sheet region, K93R situated on the outer edge of E6 protein and close to the zinc granules, all the amino acid substitutions found in the active region of the protein. Amino acid substitution increased the number of amino acids in the α-helix and β-sheet region, and the number of buried amino acids decreased (Figs. 8, 9).
Fig. 8.
Secondary structure of HPV-52 E6 comparing reference to the variant sequence. Note a is the secondary structure pattern diagram constructed based on HPV-52 E6 reference sequence; b is the secondary structure pattern diagram constructed based on HPV-52 E6 mutation sequence. The black boxes are the difference areas between the reference and mutation sequence secondary structure
Fig. 9.
Tertiary structure of HPV-52 E6 comparing reference to the variant sequence. Note a is the homology modeling structure of HPV-52 E6 reference sequence; b is the larlarian diagram of HPV-52 E6 reference sequence homology modeling; c is homology modeling structure of HPV-52 E6 mutation sequence; d is the larlarian diagram of HPV-52 E6 mutation sequence homology modeling
E32Q, D86E, K93N and R145K are located in the coil of HPV-58 E6 protein, E32Q, K93N situated on the outer edge of E6 protein, and close to the zinc granule, belonging to the active region of E6 protein. Amino acid substitution increased the number of amino acids in the α-helix region and decreased the number of amino acids in the coil region (Figs. 10, 11).
Fig. 10.
Secondary structure of HPV-58 E6 comparing reference to the variant sequence. Note a is the secondary structure pattern diagram constructed based on HPV-58 E6 reference sequence; b is the secondary structure pattern diagram constructed based on HPV-58 E6 mutation sequence. The black boxes are the difference areas between the reference and mutation sequence secondary structure
Fig. 11.
Tertiary structure of HPV-58 E6 comparing reference to the variant sequence. Note a is the homology modeling structure of HPV-58 E6 reference sequence; b is the larlarian diagram of HPV-58 E6 reference sequence homology modeling; c is homology modeling structure of HPV-58 E6 mutation sequence; d is the larlarian diagram of HPV-58 E6 mutation sequence homology modeling
The antigen epitopes analysis of α-9 HPV E6 protein
In HPV-16 E6 reference sequence, 97 HLA-I and 25 HLA-II epitopes were selected out, and epitope prediction results of variants were different, details were shown in Additional file 1: Tables S3 and S4. M1K made epitope affinity increase; R17G, D32N, D32E, I34R, L35V, P66A, H85Y and L90V changed epitope number and affinity; E120D and R151T made new epitopes appear. The effection of amino-acid substitution on HPV-16 E6 epitopes were summarized in Table 7.
Table 7.
Effect of Amino-acid substution on T-cell epitopes of HPV-16 E6
| Substution | HPV-16 E6 | ||
|---|---|---|---|
| Epitopes | Alles | Effection | |
| M1K | 1-9KHQKRTAMF | HLA-A*24:02 | Better affinity |
| R17G | 13-22QERPGKLPQL | HLA-B*40:01 | Better affinity |
| 9-19FQDPQERPGKL | HLA-B*13:01 | ||
| 9-19FQDPQERPGKL | HLA-C*08:01 | Affinity decreased | |
| 8-17MFQDPQERPR | HLA-A*33:03 | Disappear | |
| 17-26RKLPQLCTEL | HLA-C*01:02 | ||
| 9-19FQDPQERPRKL | HLA-C*03:04 | ||
| 7-17AMFQDPQERPR | HLA-A*33:03 | ||
| D32E | 26-34LQTTIHEII | HLA-B*13:01 | Better affinity |
| 29-39TIHEIILECVY | HLA-B*15:02 | Affinity decreased | |
| 29-37TIHEIILEC | HLA-A*02:01 | ||
| 29-38TIHEIILECV | HLA-A*02:01 | ||
| 24-33TELQTTIHEI | HLA-B*40:01 | New epitopes | |
| 25-33ELQTTIHEI | HLA-B*13:01 | ||
| 32-39EIILECVY | HLA-B*15:02 | ||
| 25-33ELQTTIHEI | HLA-A*02:01 | ||
| 31-39HEIILECVY | HLA-B*15:02 | ||
| 26-33LQTTIHEI | HLA-B*13:01 | ||
| 29-39TIHDIILECVY | HLA-B*46:01 | Disappear | |
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*02:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*02:01 | ||
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*05:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*02:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*05:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*05:01 | ||
| D32N | 26-34LQTTIHNII | HLA-B*13:01 | Better affinity |
| 24-35TELQTTIHNIIL | HLA-B*40:01 | ||
| 24-33TELQTTIHNI | HLA-B*40:01 | New epitopes | |
| 27-35QTTIHNIIL | HLA-C*03:04 | ||
| 29-39TIHDIILECVY | HLA-B*15:02 | Disappear | |
| 29-37TIHDIILEC | HLA-A*02:01 | ||
| D32N | 29-38TIHDIILECV | HLA-A*02:01 | Disappear |
| 29-39TIHDIILECVY | HLA-B*46:01 | ||
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*02:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*02:01 | ||
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*05:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*02:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*05:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*05:01 | ||
| I34R | 29-39TIHDIRLECVY | HLA-B*15:02 | Better affinity |
| 29-39TIHDIRLECVY | HLA-B*46:01 | Affinity decreased | |
| 24-35TELQTTIHDIRL | HLA-B*40:01 | ||
| 25-34ELQTTIHDIR | HLA-A*33:03 | New epitopes | |
| 29-37TIHDIILEC | HLA-A*02:01 | Disappear | |
| 26-34LQTTIHDII | HLA-B*13:01 | ||
| 29-38TIHDIILECV | HLA-A*02:01 | ||
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*02:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*02:01 | ||
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*05:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*02:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*05:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*05:01 | ||
| L35V | 29-39TIHDIIVECVY | HLA-B*15:02 | Better affinity |
| 29-38TIHDIIVECV | HLA-A*02:01 | ||
| 29-39TIHDIIVECVY | HLA-B*46:01 | ||
| 29-37TIHDIIVEC | HLA-A*02:01 | ||
| 35-45VECVYCKQQLL | HLA-B*40:01 | New epitopes | |
| 24-35TELQTTIHDIIL | HLA-B*40:01 | Disappear | |
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*02:01 | ||
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*02:01 | ||
| 25-39ELQTTIHDIILECVY | HLA-DQA1*01:01/DQB1*05:01 | ||
| 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*02:01 | ||
| L35V | 26-40LQTTIHDIILECVYC | HLA-DQA1*01:01/DQB1*05:01 | Disappear |
| 24-38TELQTTIHDIILECV | HLA-DQA1*01:01/DQB1*05:01 | ||
| P66A | 59-67IVYRDGNAY | HLA-B*15:02 | Better affinity |
| 59-67IVYRDGNAY | HLA-B*46:01 | ||
| 59-67IVYRDGNAY | HLA-C*03:02 | ||
| 66-76AYAVCDKCLKF | HLA-A*24:02 | ||
| 61-69YRDGNAYAV | HLA-A*11:01 | Affinity decreased | |
| 59-67IVYRDGNAY | HLA-C*08:01 | ||
| 56-67DLCIVYRDGNAY | HLA-B*15:02 | New epitopes | |
| 57-67LCIVYRDGNAY | HLA-B*46:01 | ||
| 60-67VYRDGNAY | HLA-B*15:02 | ||
| 58-67CIVYRDGNAY | HLA-B*15:02 | ||
| 57-67LCIVYRDGNAY | HLA-B*15:02 | ||
| 60-69VYRDGNPYAV | HLA-A*24:02 | Disappear | |
| H85Y | 85-95YYCYSLYGTTL | HLA-A*24:02 | Better affinity |
| 77-86YSKISEYRYY | HLA-B*46:01 | ||
| 82-96EYRYYCYSLYGTTLE | HLA-DRB1*15:02 | ||
| 80-94ISEYRYYCYSLYGTT | HLA-DRB1*15:02 | ||
| 79-93KISEYRYYCYSLYGT | HLA-DRB1*15:02 | ||
| 81-95SEYRYYCYSLYGTTL | HLA-DRB1*15:02 | ||
| 78-92SKISEYRYYCYSLYG | HLA-DRB1*15:02 | ||
| 83-97YRYYCYSLYGTTLEQ | HLA-DRB1*15:02 | ||
| 77-91YSKISEYRYYCYSLY | HLA-DRB1*15:02 | ||
| 82-96EYRYYCYSLYGTTLE | HLA-DPA1*01:03/DPB1*04:01 | ||
| 83-97YRYYCYSLYGTTLEQ | HLA-DPA1*01:03/DPB1*04:01 | ||
| 77-85YSKISEYRY | HLA-C*03:02 | New epitopes | |
| 77-85YSKISEYRY | HLA-B*46:01 | ||
| 77-85YSKISEYRY | HLA-B*15:02 | ||
| 77-85YSKISEYRY | HLA-B*58:01 | ||
| 77-86YSKISEYRYY | HLA-C*03:02 | ||
| 76-85FYSKISEYRY | HLA-A*24:02 | ||
| H85Y | 80-94ISEYRYYCYSLYGTT | HLA-DPA1*01:03/DPB1*04:01 | New epitopes |
| 84-98RYYCYSLYGTTLEQQ | HLA-DPA1*01:03/DPB1*04:01 | ||
| 79-93KISEYRYYCYSLYGT | HLA-DPA1*01:03/DPB1*04:01 | ||
| 85-99YYCYSLYGTTLEQQY | HLA-DPA1*01:03/DPB1*04:01 | ||
| 84-98RYYCYSLYGTTLEQQ | HLA-DRB1*15:02 | ||
| 85-99YYCYSLYGTTLEQQY | HLA-DRB1*15:02 | ||
| 72-86KCLKFYSKISEYRYY | HLA-DRB1*12:02 | ||
| 81-95SEYRYYCYSLYGTTL | HLA-DPA1*01:03/DPB1*04:01 | ||
| 81-90SEYRYYCYSL | HLA-B*40:01 | Affinity decreased | |
| 82-90EYRYYCYSL | HLA-A*24:02 | ||
| 80-88ISEYRHYCY | HLA-C*03:02 | Disappear | |
| 77-86YSKISEYRHY | HLA-C*03:02 | ||
| 76-86FYSKISEYRHY | HLA-A*24:02 | ||
| L90V | 86-95YCYSVYGTTL | HLA-C*03:04 | Better affinity |
| 88-95YSVYGTTL | HLA-C*01:02 | ||
| 88-95YSVYGTTL | HLA-C*03:02 | ||
| 88-95YSVYGTTL | HLA-C*03:04 | ||
| 85-95HYCYSVYGTTL | HLA-A*24:02 | ||
| 89-99SVYGTTLEQQY | HLA-B*46:01 | ||
| 89-101SVYGTTLEQQYNK | HLA-A*11:01 | ||
| 86-95YCYSVYGTTL | HLA-A*24:02 | ||
| 87-95CYSVYGTTL | HLA-A*24:02 | ||
| 89-99SVYGTTLEQQY | HLA-A*11:01 | ||
| 89-99SVYGTTLEQQY | HLA-C*03:02 | New epitopes | |
| 89-97SVYGTTLEQ | HLA-A*11:01 | ||
| 89-99SVYGTTLEQQY | HLA-B*15:02 | Affinity decreased | |
| 82-90EYRHYCYSL | HLA-A*24:02 | Disappear | |
| 81-90SEYRHYCYSL | HLA-B*40:01 | ||
| 82-96EYRHYCYSLYGTTLE | HLA-DPA1*01:03/DPB1*04:01 | ||
| 83-97YRHYCYSLYGTTLEQ | HLA-DPA1*01:03/DPB1*04:01 | ||
| E120D | 118-126CPDEKQRHL | HLA-C*08:01 | New epitopes |
| E120D | 118-126CPDEKQRHL | HLA-C*03:04 | New epitopes |
| R151T | 150-158STTRRETQL | HLA-C*01:02 | New epitopes |
| 144-154MSCCRSSTTRR | HLA-A*33:03 | ||
| 150-158STTRRETQL | HLA-C*03:04 | ||
125 HLA-I and 43 HLA-II epitopes of HPV-31 E6 reference sequence was selected out, and epitope of variants were different (Additional file 1: Tables S5, S6). H60Y, K65R changed epitope number and affinity, T64A decreased epitope number, and K123R, A138V made new epitope appear. The effection of amino-acid substitution on HPV-31 E6 epitopes were summarized in Table 8.
Table 8.
Effect of Amino-acid substution on T-cell epitopes of HPV-31 E6
| Substution | HPV-31 E6 | ||
|---|---|---|---|
| Epitopes | Alles | Effection | |
| H60Y | 52-62IVYRDDTPYGV | HLA-A*02:01 | Affinity decreased |
| 54-62YRDDTPYGV | HLA-C*08:01 | ||
| 52-60IVYRDDTPY | HLA-B*15:02 | New epitopes | |
| 51-60TIVYRDDTPY | HLA-B*15:02 | ||
| 52-60IVYRDDTPY | HLA-B*46:01 | ||
| 52-60IVYRDDTPY | HLA-C*03:02 | ||
| 57-65DTPYGVCTR | HLA-A*33:03 | ||
| 53-62VYRDDTPHGV | HLA-A*24:02 | Disappear | |
| 55-65RDDTPHGVCTK | HLA-A*11:01 | ||
| T64A | 55-65RDDTPHGVCTK | HLA-A*11:01 | Disappear |
| K65R | 65-76RCLRFYSKVSEF | HLA-A*24:02 | Better affinity |
| 57-65DTPHGVCTR | HLA-A*33:03 | New epitopes | |
| 65-79RCLRFYSKVSEFRWY | HLA-DPA1*01:03/DPB1*04:01 | ||
| 55-65RDDTPHGVCTK | HLA-A*11:01 | Disappear | |
| K123R | 117-125RHLDKKRRF | HLA-A*24:02 | New epitopes |
| A138V | 132-140WTGRCIVCW | HLA-B*58:01 | New epitopes |
109 HLA-I and 41 HLA-II epitopes of HPV-33 E6 reference sequence was selected out, epitope of variants were different (Additional file 1: Tables S7, S8). K35N decreased epitope number and affinity; S74T, N86H, K93N and R145I changed epitope number and affinity; Q113R increased epitope affinity. The effection of amino-acid substitution on HPV-33 E6 epitopes were summarized in Table 9.
Table 9.
Effect of Amino-acid substution on T-cell epitopes of HPV-33 E6
| Substution | HPV-33 E6 | ||
|---|---|---|---|
| Epitopes | Alles | Effection | |
| K35N | 35-43KPLQRSEVY | HLA-C*03:02 | Disappear |
| 35-49NPLQRSEVYDFAFAD | HLA-DQA1*01:01/DQB1*02:01 | Affinity decreased | |
| S74T | 68-76RFLSKITEY | HLA-B*46:01 | Better affinity |
| 69-76FLSKITEY | HLA-B*46:01 | ||
| 69-77FLSKITEYR | HLA-A*33:03 | ||
| 68-77RFLSKITEYR | HLA-A*33:03 | ||
| 68-76RFLSKITEY | HLA-B*15:02 | ||
| 68-76RFLSKITEY | HLA-A*24:02 | ||
| 68-76RFLSKITEY | HLA-C*03:02 | ||
| 69-76FLSKITEY | HLA-C*03:02 | ||
| 62-76ICKLCLRFLSKISEY | HLA-DRB1*14:01 | Disappear | |
| 69-79FLSKISEYRHY | HLA-B*15:02 | ||
| 74-83SEYRHYNYSV | HLA-B*40:01 | ||
| 73-81ITEYRHYNY | HLA-C*03:02 | Affinity decreased | |
| 63-77CKLCLRFLSKITEYR | HLA-DRB1*12:02 | ||
| 64-78KLCLRFLSKITEYRH | HLA-DRB1*12:02 | ||
| 62-76ICKLCLRFLSKITEY | HLA-DRB1*12:02 | ||
| 61-75GICKLCLRFLSKITE | HLA-DRB1*12:02 | ||
| 65-79LCLRFLSKITEYRHY | HLA-DRB1*12:02 | ||
| 60-74FGICKLCLRFLSKIT | HLA-DRB1*12:02 | ||
| 66-80CLRFLSKITEYRHYN | HLA-DRB1*12:02 | ||
| 64-78KLCLRFLSKITEYRH | HLA-DRB1*14:01 | ||
| 63-77CKLCLRFLSKITEYR | HLA-DRB1*14:01 | ||
| 65-79LCLRFLSKITEYRHY | HLA-DRB1*14:01 | ||
| N86H | 81-88YSVYGHTL | HLA-C*03:02 | Better affinity |
| 81-88YSVYGHTL | HLA-C*01:02 | ||
| 81-88YSVYGHTL | HLA-C*03:04 | ||
| 81-88YSVYGHTL | HLA-C*08:01 | ||
| 80-88NYSVYGHTL | HLA-A*24:02 | ||
| 78-88HYNYSVYGHTL | HLA-A*24:02 | ||
| 79-88YNYSVYGHTL | HLA-A*24:02 | ||
| N86H | 82-92SVYGHTLEQTV | HLA-A*02:01 | Better affinity |
| 83-92VYGHTLEQTV | HLA-A*24:02 | Affinity decreased | |
| 84-92YGHTLEQTV | HLA-C*08:01 | ||
| 77-88RHYNYSVYGHTL | HLA-A*24:02 | New epitopes | |
| 82-90SVYGHTLEQ | HLA-A*11:01 | ||
| 81-88YSVYGHTL | HLA-B*46:01 | ||
| 80-88NYSVYGHTL | HLA-C*01:02 | ||
| N86H/K93N | 86-94HTLEQTVNK | HLA-A*11:01 | Better affinity |
| 85-94GHTLEQTVNK | HLA-A*11:01 | ||
| 86-94HTLEQTVNK | HLA-A*33:03 | ||
| 83-94VYGHTLEQTVNK | HLA-A*11:01 | Affinity decreased | |
| 86-96HTLEQTVNKPL | HLA-B*40:01 | New epitopes | |
| 82-93SVYGNTLEQTVK | HLA-A*11:01 | Disappear | |
| K93N | 88-96LEQTVNKPL | HLA-B*40:01 | Better affinity |
| 85-94GNTLEQTVNK | HLA-A*11:01 | ||
| 86-94NTLEQTVNK | HLA-A*11:01 | ||
| 86-94NTLEQTVNK | HLA-A*33:03 | Affinity decreased | |
| 91-102TVNKPLNEILIR | HLA-A*33:03 | ||
| 83-94VYGNTLEQTVNK | HLA-A*11:01 | ||
| 82-94SVYGNTLEQTVNK | HLA-A*11:01 | ||
| 91-99TVNKPLNEI | HLA-C*08:01 | New epitopes | |
| 91-99TVNKPLNEI | HLA-B*13:01 | ||
| 91-99TVNKPLNEI | HLA-C*03:04 | ||
| 91-99TVNKPLNEI | HLA-C*01:02 | ||
| 87-96TLEQTVNKPL | HLA-B*40:01 | ||
| 82-93SVYGNTLEQTVK | HLA-A*11:01 | Disappear | |
| Q113R | 113-121REKKRHVDL | HLA-B*40:01 | Better affinity |
| R145I | 141-149RSRRIETAL | HLA-C*03:04 | Better affinity |
| 141-149RSRRRETAL | HLA-C*01:02 | Disappear | |
95 HLA-I and 50 HLA-II epitopes of HPV-52 E6 reference sequence was selected out, epitope of variants were different (Additional file 1: Tables S9, S10). E21K, L46V, E89K, K93R and N127I changed epitopes number and affinity; 105 M increased epitope affinity; N122K decreased epitopes number and E138K decreased epitope affinity. The effection of amino-acid substitution on HPV-52 E6 epitopes were summarized in Table 10.
Table 10.
Effect of Amino-acid substution on T-cell epitopes of HPV-52 E6
| Substution | HPV-52 E6 | ||
|---|---|---|---|
| Epitopes | Alles | Effection | |
| E21K | 17-27EVLEKSVHEIR | HLA-A*33:03 | Better affinity |
| 18-26VLEKSVHEI | HLA-B*13:01 | Affinity decreased | |
| 18-26VLEKSVHEI | HLA-A*02:01 | ||
| 19-28LEKSVHEIRL | HLA-B*40:01 | ||
| 18-26VLEKSVHEI | HLA-C*08:01 | ||
| 18-26VLEKSVHEI | HLA-C*01:02 | ||
| 19-26LEKSVHEI | HLA-B*40:01 | ||
| 10-21RTLHELCEVLEK | HLA-A*11:01 | New epitopes | |
| 20-28EESVHEIRL | HLA-B*40:01 | Disappear | |
| 82-93SLYGKTLEERVK | HLA-A*11:01 | ||
| 18-28VLEESVHEIRL | HLA-A*02:01 | ||
| 9-23PRTLHELCEVLEESV | HLA-DQA1*01:01/DQB1*02:01 | ||
| 10-24RTLHELCEVLEESVH | HLA-DQA1*01:01/DQB1*02:01 | ||
| L46V | 46-55VFTDLRIVYR | HLA-A*33:03 | Better affinity |
| 42-51VYKFVFTDLR | HLA-A*33:03 | ||
| 45-55FVFTDLRIVYR | HLA-A*33:03 | ||
| 41-51EVYKFVFTDLR | HLA-A*33:03 | ||
| 45-54FVFTDLRIVY | HLA-B*46:01 | ||
| 40-47REVYKFVF | HLA-B*40:01 | ||
| 46-54VFTDLRIVY | HLA-B*46:01 | ||
| 42-50VYKFVFTDL | HLA-A*24:02 | ||
| 45-54FVFTDLRIVY | HLA-C*03:02 | ||
| 46-54VFTDLRIVY | HLA-C*03:02 | ||
| 45-54FVFTDLRIVY | HLA-B*15:02 | Affinity decreased | |
| 44-52KFVFTDLRI | HLA-A*24:02 | ||
| 45-53FVFTDLRIV | HLA-A*02:01 | ||
| 41-55EVYKFVFTDLRIVYR | HLA-DPA1*01:03/DPB1*04:01 | ||
| 39-53RREVYKFVFTDLRIV | HLA-DPA1*01:03/DPB1*04:01 | ||
| 43-57YKFVFTDLRIVYRDN | HLA-DPA1*01:03/DPB1*04:01 | ||
| 38-52QRREVYKFVFTDLRI | HLA-DPA1*01:03/DPB1*04:01 | ||
| 37-51LQRREVYKFVFTDLR | HLA-DPA1*01:03/DPB1*04:01 | ||
| L46V | 44-58KFVFTDLRIVYRDNN | HLA-DPA1*01:03/DPB1*04:01 | Affinity decreased |
| 36-50ELQRREVYKFVFTDL | HLA-DPA1*01:03/DPB1*04:01 | ||
| 41-55EVYKFVFTDLRIVYR | HLA-DRB1*14:01 | ||
| 42-56VYKFVFTDLRIVYRD | HLA-DRB1*14:01 | ||
| 43-57YKFVFTDLRIVYRDN | HLA-DRB1*14:01 | ||
| 40-54REVYKFVFTDLRIVY | HLA-DRB1*14:01 | ||
| 41-55EVYKFVFTDLRIVYR | HLA-DRB1*12:02 | ||
| 46-54VFTDLRIVY | HLA-A*24:02 | New epitopes | |
| 40-50REVYKFVFTDL | HLA-B*40:01 | ||
| 45-53FVFTDLRIV | HLA-B*46:01 | ||
| 45-53FVFTDLRIV | HLA-C*03:02 | ||
| 45-53FVFTDLRIV | HLA-C*03:04 | ||
| 40-54REVYKFLFTDLRIVY | HLA-DQA1*01:01/DQB1*05:01 | Disappear | |
| 39-53RREVYKFLFTDLRIV | HLA-DQA1*01:01/DQB1*05:01 | ||
| 41-55EVYKFLFTDLRIVYR | HLA-DQA1*01:01/DQB1*05:01 | ||
| 42-56VYKFLFTDLRIVYRD | HLA-DQA1*01:01/DQB1*05:01 | ||
| 42-56VYKFLFTDLRIVYRD | HLA-DRB1*12:02 | ||
| 40-54REVYKFLFTDLRIVY | HLA-DQA1*01:01/DQB1*02:01 | ||
| 41-55EVYKFLFTDLRIVYR | HLA-DQA1*01:01/DQB1*02:01 | ||
| 40-54REVYKFLFTDLRIVY | HLA-DRB1*12:02 | ||
| 44-58KFLFTDLRIVYRDNN | HLA-DRB1*14:01 | ||
| 43-57YKFLFTDLRIVYRDN | HLA-DRB1*12:02 | ||
| E89K | 82-92SLYGKTLKERV | HLA-A*02:01 | Affinity decreased |
| 86-94KTLKERVKK | HLA-A*11:01 | ||
| 85-94GKTLKERVKK | HLA-A*11:01 | ||
| 82-93SLYGKTLKERVK | HLA-A*11:01 | ||
| 41-55EVYKFVFTDLRIVYR | HLA-DPA1*01:03/DPB1*04:01 | ||
| 39-53RREVYKFVFTDLRIV | HLA-DPA1*01:03/DPB1*04:01 | ||
| 43-57YKFVFTDLRIVYRDN | HLA-DPA1*01:03/DPB1*04:01 | ||
| 38-52QRREVYKFVFTDLRI | HLA-DPA1*01:03/DPB1*04:01 | ||
| 37-51LQRREVYKFVFTDLR | HLA-DPA1*01:03/DPB1*04:01 | ||
| E89K | 44-58KFVFTDLRIVYRDNN | HLA-DPA1*01:03/DPB1*04:01 | Affinity decreased |
| 36-50ELQRREVYKFVFTDL | HLA-DPA1*01:03/DPB1*04:01 | ||
| 42-56VYKFVFTDLRIVYRD | HLA-DRB1*14:01 | ||
| 41-55EVYKFVFTDLRIVYR | HLA-DRB1*14:01 | ||
| 43-57YKFVFTDLRIVYRDN | HLA-DRB1*14:01 | ||
| 40-54REVYKFVFTDLRIVY | HLA-DRB1*14:01 | ||
| 41-55EVYKFVFTDLRIVYR | HLA-DRB1*12:02 | ||
| 82-91SLYGKTLKER | HLA-A*11:01 | Better affinity | |
| 82-91SLYGKTLKER | HLA-A*33:03 | ||
| 75-89EYRHYQYSLYGKTLK | HLA-DPA1*01:03/DPB1*04:01 | ||
| 81-89YSLYGKTLK | HLA-A*11:01 | New epitopes | |
| 79-89YQYSLYGKTLK | HLA-A*11:01 | ||
| 80-89QYSLYGKTLK | HLA-A*11:01 | ||
| 82-94SLYGKTLEERVKK | HLA-A*11:01 | Disappear | |
| 84-94YGKTLEERVKK | HLA-A*11:01 | ||
| 40-54REVYKFLFTDLRIVY | HLA-DQA1*01:01/DQB1*05:01 | ||
| 39-53RREVYKFLFTDLRIV | HLA-DQA1*01:01/DQB1*05:01 | ||
| 41-55EVYKFLFTDLRIVYR | HLA-DQA1*01:01/DQB1*05:01 | ||
| 42-56VYKFLFTDLRIVYRD | HLA-DQA1*01:01/DQB1*05:01 | ||
| 42-56VYKFLFTDLRIVYRD | HLA-DRB1*12:02 | ||
| 40-54REVYKFLFTDLRIVY | HLA-DQA1*01:01/DQB1*02:01 | ||
| 41-55EVYKFLFTDLRIVYR | HLA-DQA1*01:01/DQB1*02:01 | ||
| 40-54REVYKFLFTDLRIVY | HLA-DRB1*12:02 | ||
| 44-58KFLFTDLRIVYRDNN | HLA-DRB1*14:01 | ||
| 43-57YKFLFTDLRIVYRDN | HLA-DRB1*12:02 | ||
| K93R | 85-94GKTLEERVRK | HLA-A*11:01 | Affinity decreased |
| 82-94SLYGKTLEERVRK | HLA-A*11:01 | ||
| 84-94YGKTLEERVRK | HLA-A*11:01 | ||
| 88-96LEERVRKPL | HLA-B*40:01 | ||
| 82-93SLYGKTLEERVR | HLA-A*33:03 | New epitopes | |
| 91-99RVKKPLSEI | HLA-B*13:01 | Disappear | |
| K93R | 82-93SLYGKTLEERVK | HLA-A*11:01 | Disappear |
| I105M | 97-105SEITIRCIM | HLA-B*40:01 | Better affinity |
| N122K | 120-128NANKRFHNI | HLA-C*08:01 | Disappear |
| 120-128NANKRFHNI | HLA-C*03:04 | ||
| N127I | 127-135IIMGRWTGR | HLA-A*33:03 | Affinity decreased |
| 124-132RFHIIMGRW | HLA-A*24:02 | ||
| 126-135HIIMGRWTGR | HLA-A*33:03 | Better affinity | |
| 125-135FHIIMGRWTGR | HLA-A*33:03 | ||
| 120-128NANKRFHII | HLA-C*03:04 | ||
| 120-128NANKRFHII | HLA-C*08:01 | ||
| 120-131NANKRFHIIMGR | HLA-A*33:03 | New epitopes | |
| 121-135ANKRFHIIMGRWTGR | HLA-DRB1*12:02 | ||
| 123-137KRFHIIMGRWTGRCS | HLA-DRB1*12:02 | ||
| 122-136NKRFHIIMGRWTGRC | HLA-DRB1*12:02 | ||
| 120-134NANKRFHIIMGRWTG | HLA-DRB1*12:02 | ||
| 119-133VNANKRFHIIMGRWT | HLA-DRB1*12:02 | ||
| 124-138RFHIIMGRWTGRCSE | HLA-DRB1*12:02 | ||
| 124-132RFHNIMGRW | HLA-B*58:01 | Disappear | |
| E138K | 132-140WTGRCSKCW | HLA-B*58:01 | Affinity decreased |
113 HLA-I and 44 HLA-II epitopes of HPV-58 E6 reference sequence was selected out, epitope of variants were different (Additional file 1: Tables S11, S12). D86E, K93N changed epitopes number and affinity, and R145K changed HLA-I epitope. The effection of amino-acid substitution on HPV-58 E6 epitopes were summarized in Table 11.
Table 11.
Effect of Amino-acid substution on T-cell epitopes of HPV-58 E6
| Substution | HPV-58 E6 | ||
|---|---|---|---|
| Epitopes | Alles | Effection | |
| D86E | 81-88YSLYGETL | HLA-C*03:04 | better affinity |
| 84-92YGETLEQTL | HLA-C*08:01 | ||
| 81-88YSLYGETL | HLA-C*08:01 | ||
| 80-88NYSLYGETL | HLA-A*24:02 | ||
| 79-88YNYSLYGETL | HLA-A*24:02 | ||
| 78-88HYNYSLYGETL | HLA-A*24:02 | ||
| 83-92LYGETLEQTL | HLA-A*24:02 | Affinity decreased | |
| 82-91SLYGETLEQT | HLA-A*02:01 | ||
| 84-92YGETLEQTL | HLA-C*03:04 | ||
| 81-88YSLYGETL | HLA-C*01:02 | ||
| 82-92SLYGETLEQTL | HLA-B*13:01 | ||
| 85-92GETLEQTL | HLA-B*40:01 | New epitopes | |
| 81-88YSLYGETL | HLA-C*03:02 | ||
| 84-92YGETLEQTL | HLA-C*01:02 | ||
| 84-92YGDTLEQTL | HLA-C*03:02 | Disappear | |
| D86E/K93N | 85-94GETLEQTLNK | HLA-A*11:01 | Better affinity |
| 86-94ETLEQTLNK | HLA-A*11:01 | ||
| 82-94SLYGETLEQTLNK | HLA-A*11:01 | Affinity decreased | |
| 84-94YGETLEQTLNK | HLA-A*11:01 | ||
| 85-96GETLEQTLNKCL | HLA-B*40:01 | New epitopes | |
| 86-94ETLEQTLNK | HLA-A*33:03 | ||
| 82-93SLYGDTLEQTLK | HLA-A*11:01 | Disappear | |
| K93N | 88-96LEQTLNKCL | HLA-B*40:01 | Better affinity |
| 85-94GDTLEQTLNK | HLA-A*11:01 | ||
| 84-94YGDTLEQTLNK | HLA-A*11:01 | Affinity decreased | |
| 82-94SLYGDTLEQTLNK | HLA-A*11:01 | ||
| 91-99TLNKCLNEI | HLA-A*02:01 | New epitopes | |
| 82-93SLYGDTLEQTLK | HLA-A*11:01 | Disappear | |
| R145K | 137-145AVCWRPRRK | HLA-A*11:01 | New epitopes |
| 137-145AVCWRPRRR | HLA-A*33:03 | Disappear | |
Discussion
Cervical cancer is the second major malignant tumor in women in childbearing age and seriously threatens women's health. HR-HPV persistent infection is closely related to the occurrence and development of cervical cancer and other malignant diseases. α-9 HPV is almost all carcinogenic and associated with 75% cervical cancers. Sichuan is a multi-ethnic mixed residence area with a high prevalence rate of α-9 HPV. From 2012 to 2017, α-9 HPV positive samples accounted for 53.68% of all HPV positive samples and 73.22% of high-risk HPV positive samples, showing an increasing trend.
α-9 genus HPV is highly prevalent and pathogenic; its carcinogenicity is mainly realized through E6 oncoproteins guided by the E6 gene through triggering the immortalization of infected cells. As an early gene of the virus, HPV E6 has a high mutation risk. In Sichuan, 21, 13, 8, 21, 8 nucleotide mutations were detected in HPV-16, HPV-31, HPV-33, HPV-52, HPV-58 E6 respectively, and among them, 17, 6, 6, 13, 4 were non-synonymous mutation.
Gene non-synonymous mutations change the amino acid composition, and structure of the protein, as well as the functions of protein, are mainly realized by its structures. HPV E6 consists of one N-terminal (residues 1–36), one C-terminal (residues 147–158) and two Zinc fingers (residues 37–73 and 110–146, CxxC-(29x)-CxxC) three domains. The two Zinc finger binding domains form a deep pocket, which can mediate the most important tumor suppressor protein p53 ubiquitination degradation by binding to the "LXXLL" sequence of E6AP protein [24, 25]. 145–149 were PDZ domain-containing combined region that was the target of E6 protein for cellular transformation and the carboxy-terminal half being principally involved in p53 binding [26]. K93N of HPV-33 E6, K93R of HPV-52 E6, and K93N of HPV-58 E6 are located at the outer edge of E6 protein and near the zinc granule [27]. The N86H, R145I of HPV-33 E6 and D86E, R145K of HPV-58 E6 occurred in the same positions; K93N of HPV-33 E6, K93R of HPV-52 E6 and K93N of HPV-58 E6 all located in the 93rd of the E6 protein; those amino acid substitutions located in protein active region, can cause the E6 terminal and the trend of the carboxyl end structure disorder. Those protein conformational changes may lead to the differences in their ability to bind to the host p53 protein and other potential proteins, thus affecting the pathogenicity of α-9 HPV [29].
Positive selection sites make the gene frequency of the corresponding amino acid increasingly stable and enhance the species' adaptability to the environment [28]. According to the calculation, the positive selection site of HPV-16 E6 was D32E (128/250); HPV-33 E6 were K35N (42/216), K93N (42/216), R145I (33/216); HPV-52 E6 was K93R (252/288); HPV-58 E6 were K93N (111/405), R145K (16/405); These positive selection sites all belong to its high frequency non-synonymous mutation, suggesting that these positive selection sites, which contribute to the adaptation of α-9 HPV E6, have been widely spread.
HPV E6 protein plays a key role in cervical cancer development. During HPV infection, the immune system will treat E6 protein as an antigen presentation to eliminate HPV infection and reduce the occurrence risk of HPV-related diseases with the help of body immunity [30]. Some specific mutations in HPV E6 may lead to the differences in the infection ability and pathogenicity of the virus. Positive selection sites of HPV-16 E6 D32E, D32N located in protein outer edge and next to the zinc granules; 6 HLA-II epitopes disappeared due to D32E/D32N; In Japan, D32E has been confirmed to be associated with the development of cervical cancer [31]; T-cell antigen epitopes affinity reduced due to D32E, D32N, that may lead to the persistent infection of virus and promote the development of cervical cancer. Positive selection sites K35N and K93N of HPV-33 E6 are close to the zinc granules, while R145I located in the E6 PDZ binding domain; K35N and R145I made 35-43KPLQRSEVY for HLA-C*03:02 and 141-149RSRRRETAL for HLA-C*01:02 disappear respectively, K93N changed the epitope number and affinity; Above three positive selection sites of HPV-33 E6 located in E6 protein active region, affect the protein conformation, function and reduced the immunogenicity of the peptide containing the above sites to a certain amount. The positive selection site K93R of HPV-52 E6 changed the epitope number and decreased the affinity of excellent epitopes. K93N and R145I of HPV-58 E6 reduced the affinity of excellent HLA-I antigen epitopes. Those positive selection sites reduced the immunogenicity of E6 overall, which may make HPV-infected cells more difficult to be detected by the immune system, and enhance HPV adaptability to the environment. No positive selection site was selected out in HPV-31 E6, and the high-frequency non-synonymous mutation sites enhanced the affinity and number of E6 epitopes, which may relate to its extremely low prevalence.
Studies have found that mutations affect the efficiency of HPV vaccine [32], the protein structure and antigen epitope bioinformatics prediction method were introduced to analyze the influence of HPV E6 mutation on protein conformational and immunogenicity. We discussed the relationship between protein structure, positive selection site, antigen epitope and pathogenicity of α-9 HPV E6 protein in Sichuan was discussed for the first time. Amino acid substitution in positive selection sites may affect the virus infection efficiency, immunogenicity, and pathogenicity by altering their T-cell epitopes affinity to improve the survival ability of α-9 HPV as well as an adaptation to evolution. These results help explore the relationship between HPV E6 polymorphism and HPV affection capacity and its action mechanism to improve the therapeutic vaccine of α-9 HPV in Sichuan regions of China.
Conclusion
α-9 HPV is extremely prevalent in Sichuan, China. The positive selection site K93N of HPV-33 E6, K93R of HPV-52 E6 and K93N of HPV-58 E6 all occurred in the 93rd amino acid of E6 protein. N86H, R145I (positive selection site) of HPV-33 E6 and D86E, R145K (positive selection site) of HPV-58 E6 occurred in the same location of E6. α-9 HPV E6 positive selection sites that adaptive to the environment D32E, K35N, K93N, R145I, K93R, R145K have been widely spread, they all located in the E6 protein active region and altered their protein structure, as well as overall reduce the immunogenicity of the E6 protein, so that HPV infected cells are more difficult to be detected by the immune system and enhance the adaptability of α-9 HPV to the environment.
E6 mutations in positive selection sites may affect the virus infection efficiency, immunogenicity by altering their protein structure, epitopes affinity to improve the survival ability of HPV.
Supplementary Information
Additional file 1: Table S1. Primers and PCR condition used for the molecular characterization of α-9 HPV E6. Table S2. Average frequency of HLA-I and HLA-II alleles (> 5%) across the Chinese population. Table S3. The HLA-I predicted epitopes of HPV-16 E6. Table S4. The HLA-II predicted epitopes of HPV-16 E6. Table S5. The HLA-I predicted epitopes of HPV-31 E6. Table S6. The HLA-II predicted epitopes of HPV-31 E6. Table S7. The HLA-I predicted epitopes of HPV-33 E6. Table S8. The HLA-II predicted epitopes of HPV-33 E6. Table S9. The HLA-I predicted epitopes of HPV-52 E6. Table S10. The HLA-II predicted epitopes of HPV-52 E6. Table S11. The HLA-I predicted epitopes of HPV-58 E6. Table S12. The HLA-II predicted epitopes of HPV-58 E6.
Acknowledgements
The authors would like to thank the following hospitals for the sample collection: Sichuan Reproductive Health Research Center Affiliated Hospital, Chengdu Song ziniao Sterility Hospital, Infertility Hospital Affiliated to Chengdu Medical College, Angel Women's and Children's Hospital, and Chengdu Jinsha hospital.
Abbreviations
- CC
Cervical cancer
- HPV
Human Papillomavirus
- HLA
Human leukocyte antigen
- CTL
Cytotoxic T lymphocytes
- Th
Helper T lymphocytes
- NCBI
National Center for Biotechnology Information
- dN
Non-synonymous mutation rate
- dS
Synonymous mutation rate
- dbMHC
Major histocompatibility complex database
- PR
Percentile rank
- HR-HPV
High-risk human papilloma virus
- MEGA 6
Molecular Evolutionary Genetics Analysis version 6.0
Authors' contributions
JH, XD and QL conceived and designed the study; JH, XD, TL, XW, YC and SM collected the samples; JP, YC, YS and YL performed the experiments. JH and XD wrote the manuscript. All authors read, edited, and approved the final manuscript for submission.
Funding
This work was funded by Key Scientific Research Foundation Projects of Sichuan Province (No. 2018JY0601). The fund is used for research design, data collection, analysis, interpretation, and manuscript writing.
Availability of data and materials
All data generated or analyzed during this study are included in this article and GeneBank.
Declarations
Ethics approval and consent to participate
The present study was approved by the Education and Research Committee and the Ethics Committee of Sichuan University (Chengdu, China; approval number SCU20100196494). Before sample collection, a written informed consent was obtained from the patients or their guardians, and the privacy of patient/study was protected carefully.
Consent to publication
All patients participating in this study have consent for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiaoyu He, Email: 1061355567@qq.com.
Qiufu Li, Email: lqf1192069072@126.com.
Shiyu Ma, Email: 895686227@qq.com.
Tianjun Li, Email: 403873142@qq.com.
Yuning Chen, Email: 646118358@qq.com.
Yiran Liu, Email: 532154290@qq.com.
Yanru Cui, Email: 512927123@qq.com.
Jianying Peng, Email: 2271644005@qq.com.
Yunfan Shi, Email: 2811829836@qq.com.
Xia Wei, Email: 531197860@qq.com.
Xianping Ding, Email: brainding@scu.edu.cn.
References
- 1.Parikesit AA. HPV Bioinformatics: in silico detection, drug design and prevention agent development. 2012.
- 2.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70:3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, Schiffman M, Herrero R, DeSalle R, Anastos K, Segondy M, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS ONE. 2011;6:e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malecka KA, Fera D, Schultz DC, Hodwadekar S, Reichman M, Donover PS, et al. identification and characterizatio of small molecule human papillomavirus E6 inhibitors. Am Chem Soc Chem Biol. 2014;7:1603–1612. doi: 10.1021/cb500229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanier K, M’Hamed Ould Sidi AO, Boulade-Ladame C, Rybin V, Chappelle A, Atkinson A, et al. Solution structure analysis of the HPV16 E6 oncoprotein reveals a self-association mechanism required for E6-mediated degradation of p53. Structure. 2012;20:604–617. doi: 10.1016/j.str.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Fehrmann F, Laimins LA. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene. 2003;22:5201–5207. doi: 10.1038/sj.onc.1206554. [DOI] [PubMed] [Google Scholar]
- 8.Kohaar I, Hussain S, Thakur N, Tiwari P, Nasare V, Batra S, et al. Association between human leukocyte antigen class II alleles and human papillomavirus-mediated cervical cancer in Indian women. Hum Immunol. 2009;70:222–229. doi: 10.1016/j.humimm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Brady CS, Bartholomew JS, Burt DJ, Duggan-Keen MF, Glenville S, Telford N, et al. Multiple mechanisms underlie HLA dysregulation in cervical cancer. Tissue Antigens. 2000;55:401–411. doi: 10.1034/j.1399-0039.2000.550502.x. [DOI] [PubMed] [Google Scholar]
- 10.King A, Hiby SE, Gardner L, Joseph S, Bowen JM, Verma S, et al. Recognition of trophoblast HLA class I molecules by decidual NK cell receptors—a review. Placenta. 2000;21(Suppl A):S81–S85. doi: 10.1053/plac.1999.0520. [DOI] [PubMed] [Google Scholar]
- 11.Schiff MA, Apple RJ, Lin P, Nelson JL, Wheeler CM, Becker TM. HLA alleles and risk of cervical intraepithelial neoplasia among Southwestern American Indian Women. Hum Immunol. 2005;66:1050–1056. doi: 10.1016/j.humimm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Thompson DA, Zacny V, Belinsky GS, Classon M, Jones DL, Schlegel R, et al. The HPV E7 oncoprotein inhibits tumor necrosis factor alpha-mediated apoptosis in normal human fibroblasts. Oncogene. 2001;20:3629–3640. doi: 10.1038/sj.onc.1204483. [DOI] [PubMed] [Google Scholar]
- 13.Piersma SJ, Welters MJP, Van Der Hulst JM, Kloth JN, Kwappenberg KMC, Trimbos BJ, et al. Human papilloma virus specific T cells infiltrating cervical cancer and draining lymph nodes show remarkably frequent use of HLA-DQ and -DP as a restriction element. Int J Cancer. 2008;122:486–494. doi: 10.1002/ijc.23162. [DOI] [PubMed] [Google Scholar]
- 14.Chan PKS, Cheung JLK, Cheung T-H, Lin C, Tam AOY, Chan DPC, et al. HLA-B alleles, high-risk HPV infection and risk for cervical neoplasia in southern Chinese women. Int J Cancer. 2006;118:1430–1435. doi: 10.1002/ijc.21528. [DOI] [PubMed] [Google Scholar]
- 15.Smith KL, Tristram A, Gallagher KM, Fiander AN, Man S, Park H, et al. Epitope specificity and longevity of a vaccine-induced human T cell response against HPV18. Int Immunol. 2004;17:167–176. doi: 10.1093/intimm/dxh197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang A, Farmer E, Wu TC, Hung CF. Perspectives for therapeutic HPV vaccine development. J Biomed Sci. 2016;23:1–19. doi: 10.1186/s12929-015-0217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calleja-macias IE, Villa LL, Prado JC, Allan B, Williamson A, Collins RJ, et al. Worldwide genomic diversity of the high-risk human papillomavirus types 31, human papillomavirus type 16 Worldwide genomic diversity of the high-risk human Papillomavirus types 31, 35, 52, and 58, four close relatives of human papillomavirus type 16. J Virol. 2005;79:13630–13640. doi: 10.1128/JVI.79.21.13630-13640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stöppler MC, Ching K, Stöppler H, Clancy K, Schlegel R, Icenogle J. Natural variants of the human papillomavirus type 16 E6 protein differ in their abilities to alter keratinocyte differentiation and to induce p53 degradation. J Virol. 1996;70:6987–6993. doi: 10.1128/jvi.70.10.6987-6993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu EQ, Zha X, Yu XH, Zhang GN, Wu YG, Fan Y, et al. Profile of physical status and gene variation of human papillomavirus 58 genome in cervical cancer. J Gen Virol. 2009;90:1229–1237. doi: 10.1099/vir.0.008227-0. [DOI] [PubMed] [Google Scholar]
- 20.Kukimoto I, Muramatsu M. Genetic variations of human papillomavirus type 16: implications for cervical carcinogenesis. Jpn J Infect Dis. 2015;68:169–175. doi: 10.7883/yoken.JJID.2014.584. [DOI] [PubMed] [Google Scholar]
- 21.Cai HB, Chen CC, Ding XH. Human papillomavirus type 16 E6 gene variations in Chinese population. Eur J Surg Oncol. 2010;36:160–163. doi: 10.1016/j.ejso.2009.07.186. [DOI] [PubMed] [Google Scholar]
- 22.Shang Q, Wang Y, Fang Y, Wei L, Chen S, Sun Y, et al. Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in identification of cervical carcinomas in patients in northeast China. J Clin Microbiol. 2011;49:2656–2663. doi: 10.1128/JCM.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zehbe I, Mytilineos J, Wikström I, Henriksen R, Edler L, Tommasino M. Association between human papillomavirus 16 E6 variants and human leukocyte antigen class I polymorphism in cervical cancer of Swedish women. Hum Immunol. 2003;64:538–542. doi: 10.1016/S0198-8859(03)00033-8. [DOI] [PubMed] [Google Scholar]
- 24.Nominé Y, Charbonnier S, Ristriani T, Stier G, Masson M, Cavusoglu N, et al. Domain substructure of HPV E6 oncoprotein: biophysical characterization of the E6 C-terminal DNA-binding domain. Biochemistry. 2003;42:4909–4917. doi: 10.1021/bi026980c. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas M, Glaunsinger B, Pim D, Javier R, Banks L. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene. 2001;20:5431–5439. doi: 10.1038/sj.onc.1204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbosa MS, Lowy DR, Schiller JT. Papillomavirus polypeptides E6 and E7 are zinc-binding proteins. J Virol. 1989;63:1404–1407. doi: 10.1128/jvi.63.3.1404-1407.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyengar VK, Reeve HK, Eisner T. Paternal inheritance of a female moth’s mating preference. Nature. 2002;419:830–832. doi: 10.1038/nature01027. [DOI] [PubMed] [Google Scholar]
- 29.Chen AA, Heideman DAM, Boon D, Chen Z, Burk RD, De Vuyst H, et al. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology. 2014;448:356–362. doi: 10.1016/j.virol.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh AK, Rath SK, Misra K. Identification of epitopes in Indian human papilloma virus 16 E6: a bioinformatics approach. J Virol Methods. 2011;177:26–30. doi: 10.1016/j.jviromet.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Simas B, Vinicius M, Batista DA, Crovella S, Pavla A, Diniz A, et al. Infection, genetics and evolution novel E6 and E7 oncogenes variants of human papillomavirus type 31 in Brazilian women with abnormal cervical cytology. Infect Genet Evol. 2013;16:13–18. doi: 10.1016/j.meegid.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Fleury MJJ, Touzé A, Coursaget P. Human papillomavirus type 16 pseudovirions with few point mutations in L1 major capsid protein FG loop could escape actual or future vaccination for potential use in gene therapy. Mol Biotechnol. 2014;56:479–486. doi: 10.1007/s12033-014-9745-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers and PCR condition used for the molecular characterization of α-9 HPV E6. Table S2. Average frequency of HLA-I and HLA-II alleles (> 5%) across the Chinese population. Table S3. The HLA-I predicted epitopes of HPV-16 E6. Table S4. The HLA-II predicted epitopes of HPV-16 E6. Table S5. The HLA-I predicted epitopes of HPV-31 E6. Table S6. The HLA-II predicted epitopes of HPV-31 E6. Table S7. The HLA-I predicted epitopes of HPV-33 E6. Table S8. The HLA-II predicted epitopes of HPV-33 E6. Table S9. The HLA-I predicted epitopes of HPV-52 E6. Table S10. The HLA-II predicted epitopes of HPV-52 E6. Table S11. The HLA-I predicted epitopes of HPV-58 E6. Table S12. The HLA-II predicted epitopes of HPV-58 E6.
Data Availability Statement
All data generated or analyzed during this study are included in this article and GeneBank.