Abstract
Wastewater-based surveillance (WBS) for SARS-CoV-2 RNA is a promising complementary approach to monitor community viral circulation. A myriad of factors, however, can influence RNA concentrations in wastewater, impeding its epidemiological value. This article aims to provide an overview and discussion of factors up to the sampling stage that impact SARS-CoV-2 RNA concentration estimates in wastewater. To this end, a systematic review was performed in three databases (MEDLINE, Web of Science and Embase) and two preprint servers (MedRxiv and BioRxiv). Two authors independently screened and selected articles published between January 1, 2019 and May 4, 2021. A total of 22 eligible articles were included in this systematic review. The following factors up to sampling were identified to have an influence on SARS-CoV-2 RNA concentrations in wastewater and its interpretation: (i) shedding-related factors, including faecal shedding parameters (i.e. shedding pattern, recovery, rate, and load distribution), (ii) population size, (iii) in-sewer factors, including solid particles, organic load, travel time, flow rate, wastewater pH and temperature, and (iv) sampling strategy. In conclusion, factors influencing SARS-CoV-2 RNA concentration estimates in wastewater were identified and research gaps were discussed. The identification of these factors supports the need for further research on WBS for COVID-19.
Abbreviations: WBS, wastewater-based surveillance
Keywords: COVID-19, Wastewater-based surveillance, Sewage, Epidemiology, Public health
Graphical abstract
1. Introduction
From the beginning of the COVID-19 pandemic up until December 21, 2021, there have been approximately 275 million reported cases and more than 5.3 million associated deaths worldwide (Worldometer, 2021). Optimal monitoring of viral circulation is essential to mitigate the spread or resurgence of SARS-CoV-2. Currently, monitoring is predominantly done through wide clinical testing of symptomatic patients and by indicated testing of risk contacts and travelers. However, such strategy is limited for several reasons. Firstly, clinical test reports are systematically biased by under-detection of asymptomatic cases and depend on contact tracing efficiency, patient ability to recognize COVID-19 symptoms, and the willingness and resources to test (Girum et al., 2020). Secondly, testing strategies have been highly variable between regions and within regions over time leading to uncertainties in comparability. Thirdly, delays in reporting of cases further contribute to the need of additional surveillance tools. Moreover, daily reported cases have been encompassing positive tests from a range of specimen dates and have been containing anomalies. Hence, its results should not be seen as a golden standard for true community-wide infection levels. Lastly, clinical testing is associated with a high financial cost and is inefficient for the detection of sporadic cases of COVID-19.
As a result, wastewater-based surveillance (WBS) was proposed as a complementary system to clinical epidemiology. Given that both asymptomatic and symptomatic cases can shed SARS-CoV-2 RNA via feces (Cevik et al., 2021; Park et al., 2021), it was assumed that spatiotemporal variation in RNA concentrations in wastewater would better approximate the true community infection dynamics of COVID-19 compared to clinical testing. Around 40% of infected individuals shed SARS-CoV-2 RNA in their feces and can contribute to viral loads in the sewer network (Parasa et al., 2020). In addition, WBS can potentially be used as an early warning system and prevent disease outbreaks in defined communities if high temporal resolution is followed by timely analysis and reporting. It is a non-invasive, community-wide surveillance system which reduces selection bias since sub-clinical infections are also being detected (Thompson et al., 2020). Several countries have successfully implemented WBS as a complementary system to clinical testing and acquired relevant qualitative and (semi-)quantitative data on community infection levels (Medema et al., 2020b; Agrawal et al., 2021; Fernandez-Cassi et al., 2021; Hasan et al., 2021; Hillary et al., 2021; Sciensano, 2021).
Although sample processing and analytical methodology of wastewater analysis markedly improved, there remain important uncertainties on WBS for COVID-19. In this review, we focus on factors up to the sampling stage of WBS that may affect downstream SARS-CoV-2 RNA concentrations and its interpretation. These factors may encompass uncertainties on a population level (e.g., number of people in the catchment area), viral shedding characteristics of the population, in-sewer factors (e.g., dilution effects, wastewater properties), and factors related to the sampling strategy. The variability introduced by these factors minimizes the representativeness and predictivity of wastewater estimates. This review does not cover post-sampling factors, such as sample processing and analytical methodology, which are known to be major factors influencing quantitative results. Readers interested in post-sampling factors are referred to relevant literature described elsewhere (Ahmed et al., 2020b; Kitajima et al., 2020; Medema et al., 2020a; Alygizakis et al., 2021).
We conducted a systematic review to identify influencing factors up to the sampling stage by retrieving original research articles from three databases (MEDLINE, Embase and Web Of Science Core Collection) and, additionally, from two preprint servers MedRxiv and BioRxiv. This review presents a qualitative synthesis of the factors reported and its evidence. Additionally, studies examining the relationship between SARS-CoV-2 RNA concentrations in wastewater and clinical- or diagnostic-based epidemiological markers (e.g., cases, positivity rate, hospitalizations or deaths), from now on referred to as clinical indicators, were reported here to demonstrate large variability in correlation estimates. Importantly, intrinsic variability in case reports will continue to distort any relationship with WBS data. Adjustment for determining factors of WBS may decrease unexplained variability and improve its usefulness. Lastly, recommendations are provided to facilitate the application of WBS for COVID-19 detection and surveillance.
2. Methods
A systematic review was conducted to review factors affecting SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage. Given the large heterogeneity between WBS studies and various factors studied by different methodologies, we could not adhere strictly to PRISMA guidelines (Page et al., 2021). The main objective of this study was to identify and discuss all possible influencing factors up to sampling. A search was performed on May 4, 2021 in three databases (MEDLINE, Embase and Web of Science Core Collection). Additionally, two preprint servers MedRxiv and BioRxiv were screened to accommodate with the high publishing rate of COVID-19 articles. Articles published between January 1, 2019 and May 4, 2021 were eligible for inclusion. The included preprints were revisited on December 21, 2021 to verify revisions upon Journal acceptance and warn on the validity of unrefereed preprints. Supplementary Table 1 shows the search strategy used in the MEDLINE database.
Initial screening was performed independently by different operators, based on the title and abstract. Only original articles mentioning the assessment of SARS-CoV-2 (RNA) in wastewater were selected for further analysis. Also, they independently performed full-text screening for in- and exclusion criteria, and conflicts were resolved by mutual agreement.
The inclusion criteria were as follows: (i) original research articles, (ii) examining a factor/phenomenon up to the sampling stage with a possible effect on SARS-CoV-2 RNA concentrations in wastewater, independent of infection levels. Additionally, original research articles investigating an association between clinical data and WBS data were retrieved from the search to perform a supplementary analysis. Articles published in a non-English language or in the form of a short communication, letter, abstract, proceeding, opinion, review or book chapter were excluded.
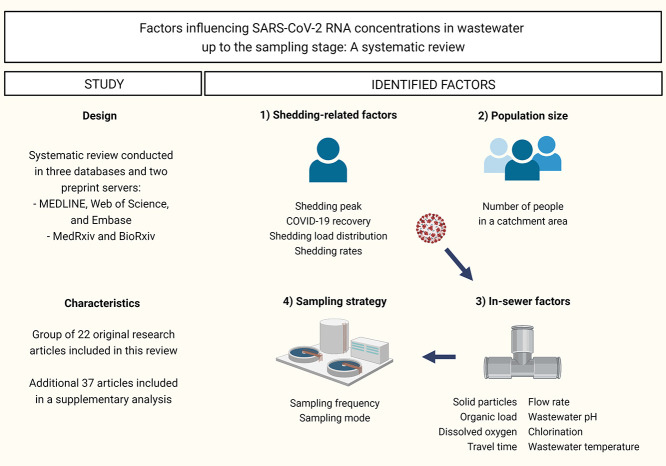
Discrepancies regarding article selection were resolved during a consensus meeting with a senior reviewer (LL). Qualitative synthesis of factors was carried out, leading to four categories: i) shedding-related factors, ii) population size, iii) in-sewer factors and iv) sampling strategy. The findings from included studies are reported accordingly in the results section and compiled in Table 1 . As a supplementary analysis, a synthesis of articles examining the association between WBS data and clinical indicators was provided (Supplementary File).
Table 1.
Summary of identified factors that may impact SARS-CoV-2 RNA concentrations in wastewater or its interpretation.
| Factor | Expected impact on SARS-CoV-2 RNA concentrations or relevance | References |
|---|---|---|
| i) Shedding-related factors | ||
| Shedding peak | Early infections have a predominant influence on RNA loads. | Wu et al., 2022 |
| COVID-19 recovery | Reduction in RNA loads, but contribution to residual noise in RNA concentrations. |
McMahan et al., 2021; Wu et al., 2022 |
| Shedding load distribution | Shedding load distribution may possibly be used to estimate total infections. | Huisman et al., 2021 (preprint) |
| Shedding rate | Shedding rates can be calculated for a specific community, but may vary depending on underlying population. | Schmitz et al., 2021 |
| Incontinence aids | High-risk populations may not contribute to RNA load when incontinence aids are used. | Acosta et al., 2021 |
| ii) Population size | ||
| Population size | Positive relationship between population size and RNA concentrations. | Wilder et al., 2021 |
| Population size | Positive relationship between population size and RNA detection rates. | Wu et al., 2021 |
| iii) In-sewer factors | ||
| Solid particles | Distribution of viral RNA between solid phase and aqueous phase. Possible reduction in aqueous phase due to solid particles. | Jørgensen et al., 2020 (preprint); Fores et al., 2021; Graham et al., 2021; Weidhaas et al., 2021; Westhaus et al., 2021 |
| Organic matter | Reduced RNA concentrations in aqueous phase for increasing organic load, dependent on physicochemical properties (e.g., humic-like substances). |
Hong et al., 2021; Petala et al., 2021 |
| Dissolved oxygen | Increased RNA concentrations in aqueous phase with increasing dissolved oxygen. | Petala et al., 2021 |
| Flow rate | Increased or reduced concentrations. |
Wilder et al., 2021; Vallejo et al., 2022 |
| Travel time | Reduced RNA concentrations. | Weidhaas et al., 2021 |
| Wastewater pH | Possible reduced concentrations at low pH with high concentrations of volatile fatty acids. | Hong et al., 2021 |
| Chlorination | Reduced RNA concentrations, depending on fulfilling of chlorine demand. | Zhang et al., 2020; Hemalatha et al., 2021 |
| Wastewater temperature | Possible reduced RNA concentrations. |
Ahmed et al., 2020a; Bardi and Oliaee, 2021 |
| Ambient temperature | Possible reduced concentrations, situation specific. | Arora et al., 2020; Hart and Halden, 2020; Hong et al., 2021; Vallejo et al., 2022 |
| iv) Sampling strategy | ||
| Sampling frequency | At least two or three non-consecutive samples a week to minimize noise in trends. | Feng et al., 2021; Graham et al., 2021; Huisman et al., 2021 (preprint) |
| Sampling mode | 24-h composite flow-proportional sampling of influent wastewater to capture RNA loads adequately. |
Ort et al., 2010 (not included in review) |
The expected impact on SARS-CoV-2 RNA concentrations is given for a relative increase in quantitative factors. For categorical factors, the expected effect or relevance is briefly mentioned.
3. Results
3.1. Study characteristics
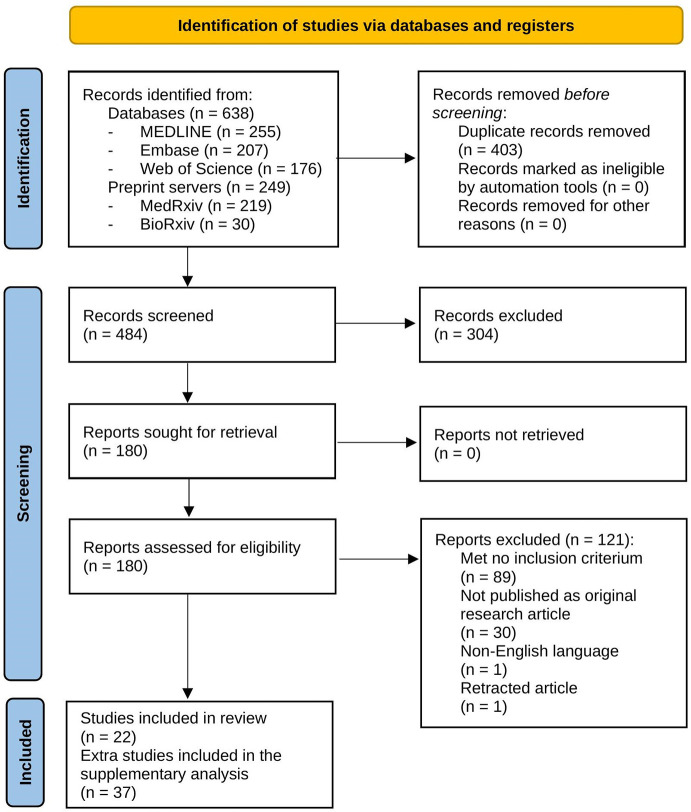
The literature search resulted in 887 hits: 255 from MEDLINE, 207 from Embase, 176 from Web of Science, 219 from MedRxiv and 30 from BioRxiv. Authors reached consensus about the title/abstract screening and full-text screening in 82% and 79% of the articles, respectively, counting for a group of 59 articles (Fig. 1 ). Of these 59 articles, 22 were included in this systematic review. The remaining 37 articles were reported in the supplementary analysis (Supplementary File).
Fig. 1.
PRISMA flowchart of the systematic review (Page et al., 2021).
Geographic distribution of all 59 studies showed that 24 were conducted in North America, 15 in Asia, 13 in Europe, three in Australia, and three in South America. One study could not report a location because the authors developed a theoretical model and did not sample wastewater. Out of the 59 articles, 50 were related to sampling of wastewater or sludge on a macro-level in wastewater treatment plants (WWTPs). Eight articles were reports of the concentration of SARS-CoV-2 RNA in wastewater on a building-level, in either hospitals or university dorms, or related to pumping stations; and 14 articles included samples of wastewater from both hospital sewers/manholes and WWTPs.
A total of 50 studies were related to sampling of influent/raw wastewater, 13 studies of effluent wastewater, eight studies of solids/sludge and eight studies of hospital wastewater. It should be explicitly emphasized that WWTP-effluents should not be used for WBS purposes due to significant changes to the viral RNA concentrations caused by primary or secondary treatment steps.
A total of 25 studies used only composite samples (42%), 15 articles used only grab samples (25%), and 11 articles used both grab and composite samples (19%). Out of 36 studies using composite samplers, 32 used 24-h samplers (89%), one only applied 6-h samplers (3%), one only 8-h samplers (3%), and two did not report composite sampling duration (6%). Furthermore, eight studies used flow-proportional composite samplers (22%), nine used time-proportional composite samplers (25%), and 20 did not explicitly report the type of composite sampling (56%). Seven articles (12%) did not specify sampling methods at all.
3.2. Identified factors
3.2.1. Shedding-related factors
The influence of faecal shedding parameters on WBS data was studied by several research groups. Wu et al. (2022) optimized a SARS-CoV-2 shedding function to relate back-dated daily incident cases with wastewater titers. As a result, a shedding peak of SARS-CoV-2 on the second day of viral shedding, which lasts for 3–4 days and is followed by prolonged shedding of lower levels of viral RNA, was suggested. This shedding burst may occur even before respiratory symptoms develop and could coincide with gastro-intestinal symptomatology. According to McMahan et al. (2021), patient's recovery evoked a reduction in viral RNA mass rate release in wastewater (copies/day). However, even after recovery, faecal shedding may persist for several weeks contributing to noise in WBS data (McMahan et al., 2021; Wu et al., 2022). A preprint by Huisman et al. (2021) suggests that the shedding load distribution could be used to maximize the fit between reproductive number estimates derived from wastewater and clinical data. Furthermore, Schmitz et al. (2021) calculated mean faecal shedding rates, from clinical and wastewater data obtained from a university dorm community, which may be used to estimate the total number of infected individuals. Yet, no external validation has been performed to substantiate its significance. Also, shedding rates may differ between populations as age seems to be an important determinant of viral shedding kinetics. Omori et al. (2021) showed that individuals older than 80 years had a higher relative contribution compared to individuals younger than 19 years. Increased peak viral loads in the elderly may be partly due to immunosenescence (Vellas et al., 2020), a condition that is frequently found in frail elderly (Lang et al., 2010). Of note, use of incontinence aids in high-risk populations (e.g., diapers and sanitary pads) prevents SARS-CoV-2 RNA from introduction in sewage as was discussed by Acosta et al., 2021. This should be considered as an important consideration for WBS, especially for building-level surveillance, such as in hospitals and residential care facilities.
In conclusion, accurate estimation of shedding rates within specific populations will be necessary for prediction of COVID-19 infections. Within a catchment area of interest, the population distributions of shedding pattern, recovery, rate and load distribution are determining factors for the interpretation of SARS-CoV-2 RNA concentrations and may depend on clinical and demographic characteristics of the population.
3.2.2. Population size
Population size in the catchment area can be associated with the SARS-CoV-2 RNA concentration in wastewater. Wilder et al. (2021) demonstrated a linear association (p < 0.010) between the RNA concentration and population size in the catchment area in New-York (USA), based on census data. Also, Wu et al. (2021) found a positive correlation between RNA detection rates in wastewater and population sizes of catchment areas. Although the association between population size and viral RNA concentrations may be driven by absolute number of COVID-19 cases, it is important to state that RNA concentrations may also be affected independently. As population size increases, sewage flow increases due to increased water usage causing dilution of viral RNA (Wu et al., 2021).
As the total covered population size is a critical parameter for WBS, population size estimates should be interpreted with great care. The geographical coverage of a sewer network does not necessarily align with the number of residents in a given jurisdiction. This requires detailed mapping of sewer networks onto different postal codes.
Even more so, the total population size within a catchment area cannot be assumed as being static. Significant day-to-day variability due to commuting and tourism has been shown. Especially during the COVID-19 pandemic, population fluxes are being severely impacted. Monitoring the fluxes of individuals in the catchment area is vital to interpret viral concentrations.
To this end, multiple studies used pepper mild mottle virus (PMMoV) as human faecal load marker for internal standardization (Acosta et al., 2021; D'Aoust et al., 2021; Feng et al., 2021; Whitney et al., 2021). In addition, crAssphage (Wilder et al., 2021), creatinine (Westhaus et al., 2021), and total nitrogen (Yaniv et al., 2021) were identified in this review to normalize for sewage strength (i.e. faecal content per volume of sewage).
3.2.3. In-sewer factors
In-sewer factors are major sources of variability in concentrations of SARS-CoV-2 RNA. The load and physiochemical properties of solid particles and organic matter impact the distribution of SARS-CoV-2 RNA in wastewater. Weidhaas et al. (2021) showed that 9 ± 12 m/m% SARS-CoV-2 RNA (n = 8) was found in the influent solid fraction of wastewater. Fores et al. (2021) observed that on average 23% of SARS-CoV-2 N1 genes (n = 9) remains in the solid phase of wastewater after ultrafiltration. Furthermore, a preprint by Jørgensen et al. (2020) indicates the presence of RNA in the solid phase in about half of the wastewater analyses which were positive for SARS-CoV-2 RNA (n = 77). In some of these analyses RNA could be detected in the solid phase while the aqueous phase was negative. According to Westhaus et al. (2021), the concentration of SARS-CoV-2 RNA is one log higher in the solid phase than in the aqueous phase of influent wastewater. Solid phase analyses may thus complement aqueous phase analyses and should be performed to exclude false negatives.
Petala et al. (2021) reported that SARS-CoV-2 RNA copy numbers in aqueous samples are negatively correlated with specific UV absorbance divided by dissolved oxygen (DO). The specific absorbance was defined as the ratio of UV254 over total dissolved organic carbon (DOC), as a measure for the concentration of humic-like substances (UV254) in the DOC. This association may be explained by humic-like substances providing additional binding sites to SARS-CoV-2 RNA onto solid matter, which would have a negative down-stream effect on aqueous-phase quantification. Conversely, increased DO may reduce viral adsorption to solids through viral inactivation and bacterial-mediated viral destruction, potentially reversing this distribution of viral RNA between solid and aqueous phases. In line with this study, a negative association between N1 gene copy number (n = 31) and DOC was observed in liquid phase samples from a hospital septic tank and sludge tank, but not with N2 (n = 26) and N3 (n = 23) gene copy numbers by Hong et al. (2021).
Altogether, organic matter seems to have a downstream negative impact on SARS-CoV-2 RNA quantification in the aqueous phase. Next to adsorption-related processes, another explanation could be the phenomenon of PCR-inhibition caused by organic matter. Further studies should evaluate whether in-sewer organic matter impacts quantification and whether correction is indicated for a given methodology.
The influx of stormwater, rainwater, increased household water consumption and water from other sources causes dilution of wastewater, possibly leading to a lower or absent SARS-CoV-2 RNA concentrations as these are typically measured at trace levels, close to the lower limit of quantification (LLOQ). Wilder et al. (2021) observed increased concentrations of SARS-CoV-2 RNA at a sampling date with a significantly decreased flow (p = 0.045) compared to a previous sampling date. However, no COVID-19 incidences were provided for comparison between both dates. Furthermore, McMahan et al. (2021) and Fernandez-Cassi et al. (2021) used mass rate (copies/day) instead of concentration (copies/L) to compensate for sewage flow. Vallejo et al. (2022) observed a weak positive correlation (r = 0.32) between mean flow of wastewater and daily mean viral load. Hence, the effect of flow rate on SARS-CoV-2 RNA concentrations in wastewater can be manifold. RNA concentrations can increase when flow significantly decreases, possibly through reduced SARS-CoV-2 RNA dilution (less rainfall, water consumption). However, in combined sewer networks a higher flow rate due to more rainwater may be associated with shorter travel times, resulting in mitigated RNA degradation. Also, increased flow rates can be the result of more individuals contributing to the catchment area of interest. Lastly, dilution could reduce matrix effects in wastewater and possibly explain positive correlations with RNA concentrations. Correction for source-specific dilution may be indicated for combined sewers, while correction for overall dilution is expected to be appropriate for strictly sanitary sewers.
As expected, longer in-sewer travel times could lead to enhanced decline of SARS-CoV-2 RNA in wastewater because of decay during transit, which was observed in the study conducted by Weidhaas et al. (2021).
The effect of pH on sewage samples was examined by two articles: Hong et al. (2021) observed in a hospital septic tank and sludge tank a negative association between pH and the N1 gene copy number in loglinear regression model with total organic carbon, conductivity and turbidity, suggesting that (extreme) alkaline environments cause degradation of SARS-CoV-2 RNA in wastewater. Wastewater pH was a non-significant predictor for N2 and N3 gene copy number. Secondly, total absence of SARS-CoV-2 genes was observed in wastewater reactors with a pH lower than 5.4 and a volatile fatty acids concentration above 2000 mg/L (Bardi and Oliaee, 2021). Even though no thorough analysis of a pH effect was available in this review, RNA estimates from samples with extreme pH should be interpreted with caution.
For safety reasons chlorination of wastewater is being performed by facilities such as hospitals as a pre-treatment step before sewage discharge into the sewer network. The effect of chlorination was considered by several studies, typically in the context of biosafety of WWTP effluents. According to Hemalatha et al. (2021), treating 1 L wastewater samples with 20 mL of a 0.1% sodium hypochlorite solution to decrease pathogenicity, has no effect on the SARS-CoV-2 RNA concentration. The study performed by Zhang et al. (2020) illustrated that a dosage of 6700 g/m3 sodium hypochlorite during one hour and a half resulted in complete removal of SARS-CoV-2 RNA in effluents of hospital septic tanks, in contrast to 800 g/m3 sodium hypochlorite. It is postulated that embedding of viral particles in stool may inhibit the effect of chlorination and extend the release of SARS-CoV-2 in the aqueous phase. In line with these results, Arora et al. (2021) argue that chlorination only influences the concentration of SARS-CoV-2 RNA if the chlorine demand is fulfilled. Beyond the chlorine demand free chlorine residuals appear in the matrix upon additional chlorination. The chlorine demand is relatively high in wastewater due to the high level of organic impurities which may react with chlorine. Nonetheless, chlorination as pre-treatment step may have a detrimental effect on SARS-CoV-2 RNA concentrations, especially relevant when sampling in the context of facility-level WBS such as in hospitals.
The impact of temperature on RNA degradation was studied by Ahmed et al. (2020a) in different aqueous matrices. The first-order degradation constant increases with temperature in untreated and autoclaved wastewater as well as in dechlorinated tap water. Increasing temperature also showed to have a negative impact on SARS-CoV-2 RNA concentrations in batch reactors during anaerobic co-digestion of sewage sludge and its effect may synergize with higher organic load concentrations (Bardi and Oliaee, 2021) and longer in-sewer travel times (Hart and Halden, 2020). As to ambient temperature, SARS-CoV-2 RNA was detected in wastewater from an aerobic biological open-air hospital tank in Jeddah (Saudi Arabia), at extreme ambient temperatures up to 46.3 °C (Hong et al., 2021). In line with this study, SARS-CoV-2 RNA could be found in wastewater at periods with up to 45 °C ambient temperature in Rajasthan, India (Arora et al., 2020). However, on a quantitative level, Hart and Halden (2020) suggested, through computational analysis, that an ambient temperature of 20 °C leads to reduced detectability of SARS-CoV-2 RNA compared to lower ambient temperatures (5–10 °C) through increased decay, and recommended to adjust for seasonal variations. Also, mean ambient temperature showed a weak negative correlation with daily mean viral load (r = −0.39) in a study by Vallejo et al. (2022), suggestive for increased loss of signal with increasing temperature.
It is important to realize that the overall effects of seasonal weather conditions may be confounded by seasonal variations in populations sizes and behavior. For example, high ambient temperature could lead to increased environmental degradation of SARS-CoV-2 RNA, while coinciding with increasing loads of viral RNA in wastewater due to higher influxes of individuals during summer at tourist locations.
3.2.4. Sampling strategy
A sampling frequency of three times a week showed comparable results in estimating the reproductive number based on wastewater surveillance (Rww) as compared to daily sampling. Yet, increased variability of Rww estimates was observed when sampling was less frequent (Huisman et al., 2021, preprint). Feng et al. (2021) showed that a resolution of at least two non-consecutive samples a week is necessary to ensure adequate accuracy for trend analysis of clinical cases. Similarly, Graham et al. (2021) observed loss of significance of linear regression models, adjusted for autoregression and technical errors, with once weekly or fortnightly samples of wastewater solids to predict case counts. All studies on sampling frequency applied flow-proportional 24-h composite sampling.
Regarding sampling mode, it should be emphasized that from a traditional WBS perspective flow-proportional sampling is more appropriate compared to time- and volume proportional sampling in order to capture WBS biomarker loads more accurately (Ort et al., 2010). In case of volume- or time-proportional sampling modes, it is imperative to operate in high sampling frequencies. As described earlier, 44% of the included studies used grab samples and 12% did not report sampling strategy at all. Grab samples can introduce important variability with regards to diurnal variation and time of defecation and should be replaced by 24-h composite samplers for WBS purposes (Ahmed et al., 2021; Bivins et al., 2021).
4. Discussion
Clinical- and diagnostic-based epidemiology for COVID-19 has been deployed successfully during the ongoing pandemic. However, this strategy suffers from several important limitations including selection bias. Wastewater-based surveillance (WBS) has been explored as a complementary system to monitor viral community circulation. In this review, we present factors up to the sampling stage that may influence SARS-CoV-2 RNA concentrations in wastewater or its interpretation, and possibly impede the potential of WBS if not corrected for. Although the effect of sampling processing and analytical methodology is not covered in this review, we would like to stipulate its extreme importance. All steps involved in WBS beyond the sampling stage including sampling handling, pre-treatment and analysis can be major sources of variability. These steps should be performed as consistent as possible and in accordance to state-of-the-art findings on this matter. Still, WBS data is prone to significant uncertainties due to its inherent complexity. This study has classified the origins of this uncertainty into four major categories, of relevance for WBS of SARS-CoV-2 genes.
Firstly, the importance of SARS-CoV-2 faecal shedding parameters on WBS is demonstrated. The reviewed studies provide evidence that overall, the incident cases of COVID-19 are main drivers of viral load. Nonetheless, persistent shedding at lower-levels after the early shedding peak distorts cumulative WBS measures, especially during declining incidence rates. Furthermore, it is expected that WBS is most sensitive during the start of a surge and when overall RNA loads are at lower baseline levels. We encourage future studies to unravel the relationship between viral load and faecal shedding and investigate characteristics associated with faecal shedding parameters. For example, the severity of COVID-19 infections may influence the amount of RNA shedding, with overall higher viral loads in adult patients with severe COVID-19 (Kwon et al., 2020; Liu et al., 2020).
In this review, we have not identified studies investigating the impact of SARS-CoV-2 variants on wastewater RNA concentrations, which may alter viral shedding loads. Infections with Delta variant (B.1.617.2) showed markedly increased viral loads on first positive oropharyngeal samples compared to lineage A/B infections (Li et al., 2021). We encourage further study of faecal/urine shedding of different SARS-CoV-2 variants with an urgent emphasis on Omicron (B.1.1.529), to allow comparability of historical WBS data with current estimates, which is necessary for statistical modelling of WBS data.
Furthermore, the impact of the ongoing vaccination strategy on WBS was not identified in this review. Vaccinees have significantly reduced viral loads after infection with SARS-CoV-2 (Levine-Tiefenbrun et al., 2021; McEllistrem et al., 2021). Ongoing vaccination may lead to reduced shedding and RNA concentrations in wastewater. Yet, we expect that the importance of WBS to monitor viral circulation will most likely increase among the vaccinated population as they could show reduced symptomatology, possibly complicating clinical testing strategies.
Secondly, we highlight population size as key factor for the interpretation of SARS-CoV-2 RNA concentrations in wastewater and stipulated the need for internal standardization, for example via PMMoV-correction. Adequate correction for sewage strength is essential to further improve WBS through a possible more accurate estimation of viral loads. Dynamic anthropogenic markers currently being used in WBS for COVID-19 are presented. Other indicators such as ammonium (Been et al., 2014), pharmaceutical markers (Lai et al., 2011) and a Bayesian model based on mass loads of pharmaceuticals and personal care products (O'Brien et al., 2014) could also act as markers for sewage strength and population fluxes as they showed promising results in traditional WBS studies.
Thirdly, we present in-sewer factors that could influence RNA concentrations and its distribution between wastewater phases. These include solid particles, organic matter, sewage flow, travel time, pH and temperature. Other variables that could be of influence but were not identified in this review may include the concentration and activity of RNases, influence of micro-organisms, and effect of biofilms. Furthermore, the state in which the SARS-CoV-2 genome is present in the wastewater system is essential, as the genome might be present as cell debris. We suggest that solid phase analyses should complement negative aqueous phase samples, particularly in building-level surveillance and at times of low viral circulation. Lastly, we illustrate the importance of high temporal resolution to infer trends from wastewater and the need for flow-proportional composite sampling of untreated wastewater.
As this review cannot claim to be an exhaustive listing, additional factors can introduce unexplained variability as well. For example, disconnected sewer systems (e.g. septic tanks) reduce the number of persons connected to the sewer network. This is especially important for WWTPs in rural areas. Also, the sewer structure (i.e., rising main vs gravitational sewers) determines if primarily aerobic or anaerobic processes take place, relevant for SARS-CoV-2 in-sewer decay.
To gain insights into the need to adjust wastewater RNA estimates, we reviewed studies that associated wastewater with clinical indicators. The supplementary analysis demonstrates that correlation and regression coefficients show high variability between studies, suggestive for unexplained variability, and that few studies have already adjusted for factors to optimize correlations between both systems. Regarding correlation studies, a considerable risk for reporting bias should be considered as studies with negative or no correlation may not report their (quantitative) results.
The advantages of this review were its systematic approach, inclusion and revisiting of preprints, and the validation of the research gap by reviewing association studies between WBS data and clinical indicators. The main limitations of this study were the difficulty to compare included studies due to large intrinsic heterogeneity of WBS systems and differences in methodology. Some preprints were not accepted or published by a Journal at the revisiting date, thus prone to errors or inaccurate interpretation due to the lack of peer-review. These unrefereed preprints were explicitly labelled as preprint. Next, it is also likely that several factors remain unidentified or have not been studied to date, given the complexity of WBS as a system. Finally, we want to re-emphasize that post-sampling factors are key contributors to variation in RNA concentrations (Ahmed et al., 2020b; Kitajima et al., 2020; Medema et al., 2020a; Alygizakis et al., 2021; Boogaerts et al., 2021) but were not discussed in this review as this was beyond the scope of this article.
5. Future recommendations
We encourage future studies to monitor and adjust for the identified factors contributing to uncertainties in wastewater measurements of SARS-CoV-2 RNA when possible and to further investigate these and other factors that may distort estimates of WBS. Firstly, the lessons learnt from traditional WBS should be implemented. Good practices would include the consequent use of 24-h composite flow-proportional influent wastewater samples. On a WWTP-level, effluents should not be considered for WBS purposes at any given time. Additional research is needed to find the optimal way to perform normalization of WBS data for a given population, and how to improve comparability of WBS data between influents of WWTPs and standard operating procedures. Adjusting for dynamic anthropogenic markers is especially important in areas with high commuting and tourism activities, thus its impact on correlation estimates may vary. Regarding building-level analysis, the use of incontinence aids and effect of pre-treatment procedures should be considered and accounted for. Factors with a negative impact are expected to be more influential on building-level WBS with trace levels of SARS-CoV-2 RNA. Hence, complementary solid phase analyses should be carried out to reduce the chance of false negative results. As the effect size of some factors and interactions between them is still uncertain, further modelling studies should investigate these factors in a multiple regression framework. Correlation and regression analyses should also be conducted in line with time series analysis principles. For example, the phenomenon of autocorrelation and non-stationarity of data should be accounted for when modelling WBS data. Lastly, it is emphasized that even after adjusting for all determinant factors of WBS, an association with diagnostic and clinical indicators can be absent due to intrinsic variation of clinical epidemiology.
Funding
This work was funded by The Flemish Agency of Care and Health, which has granted funding to Lies Lahousse, Tim Boogaerts, Siel Van den Bogaert, Alexander van Nuijs and Peter Delputte to facilitate wastewater-based epidemiology for COVID-19 in Flanders (project number: GE0-1GPFZJA-WT). This work was also funded by the Research Council (BOF) of the University of Antwerp (project number: FFB200184), which granted funding to Tim Boogaerts and Siel Van den Bogaert.
Ethics approval
Not applicable.
Data availability
Not applicable.
CRediT authorship contribution statement
Xander Bertels: Conceptualization, Methodology, Investigation, Writing – Original Draft; Phaedra Demeyer: Investigation; Siel Van den Bogaert: Writing – Review & Editing; Tim Boogaerts: Writing – Review & Editing; Alexander L.N. van Nuijs: Writing – Review & Editing; Peter Delputte: Writing – Review & Editing; Lies Lahousse: Conceptualization, Methodology, Funding acquisition, Resources, Supervision, Writing – Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by The Flemish Agency of Care and Health, which has granted funding to Lies Lahousse, Tim Boogaerts, Siel Van den Bogaert, Alexander van Nuijs and Peter Delputte to facilitate wastewater-based surveillance for COVID-19 in Flanders (project number: GE0-1GPFZJA-WT). This work was also funded by the Research Council (BOF) of the University of Antwerp (project number: FFB200184), which granted funding to Tim Boogaerts and Siel Van den Bogaert. The graphical abstract was created with BioRender.com.
Editor: Kevin V. Thomas
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.153290.
Appendix A. Supplementary data
Supplementary material
References
- Acosta N., Bautista M.A., Hollman J., McCalder J., Beaudet A.B., Man L., Waddell B.J., Chen J., Li C., Kuzma D., Bhatnagar S., Leal J., Meddings J., Hu J., Cabaj J.L., Ruecker N.J., Naugler C., Pillai D.R., Achari G., Ryan M.C., Conly J.M., Frankowski K., Hubert C.R., Parkins M.D. A multicenter study investigating SARS-CoV-2 in tertiary-care hospital wastewater. Viral burden correlates with increasing hospitalized cases as well as hospital-associated transmissions and outbreaks. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11(1):5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., Simpson S.L., Thomas K.V., Verhagen R., Kitajima M., Mueller J.F., Korajkic A. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alygizakis N., Markou A.N., Rousis N.I., Galani A., Avgeris M., Adamopoulos P.G., Scorilas A., Lianidou E.S., Paraskevis D., Tsiodras S., Tsakris A., Dimopoulos M.A., Thomaidis N.S. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: protocols and future perspectives. Trends Anal. Chem. 2021;134 doi: 10.1016/j.trac.2020.116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Nag A., Sethi J., Rajvanshi J., Saxena S., Shrivastava S.K., Gupta A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 2020;82(12):2823–2836. doi: 10.2166/wst.2020.540. [DOI] [PubMed] [Google Scholar]
- Arora S., Nag A., Rajpal A., Tyagi V.K., Tiwari S.B., Sethi J., Sutaria D., Rajvanshi J., Saxena S., Shrivastava S.K., Srivastava V., Gupta A.B., Kazmi A.A., Kumar M. Imprints of lockdown and treatment processes on the wastewater surveillance of SARS-CoV-2: a curious case of fourteen plants in Northern India. Water-Sui. 2021;13(16) doi: 10.3390/w13162265. [DOI] [Google Scholar]
- Bardi M.J., Oliaee M.A. Impacts of different operational temperatures and organic loads in anaerobic co-digestion of food waste and sewage sludge on the fate of SARS-CoV-2. Process. Saf. Environ. Prot. 2021;146:464–472. doi: 10.1016/j.psep.2020.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been F., Rossi L., Ort C., Rudaz S., Delemont O., Esseiva P. Population normalization with ammonium in wastewater-based epidemiology: application to illicit drug monitoring. Environ. Sci. Technol. 2014;48(14):8162–8169. doi: 10.1021/es5008388. [DOI] [PubMed] [Google Scholar]
- Bivins A., North D., Wu Z., Shaffer M., Ahmed W., Bibby K. Within- and between-day variability of SARS-CoV-2 RNA in municipal wastewater during periods of varying COVID-19 prevalence and positivity. ACS ES&T Water. 2021;1(9):2097–2108. doi: 10.1021/acsestwater.1c00178. [DOI] [Google Scholar]
- Boogaerts T., Jacobs L., De Roeck N., Van den Bogaert S., Aertgeerts B., Lahousse L., van Nuijs A.L.N., Delputte P. An alternative approach for bioanalytical assay optimization for wastewater-based epidemiology of SARS-CoV-2. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.148043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2(1):e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Juni P., MacKenzie A.E., Delatolla R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS ES&T Water. 2021;1(8):1955–1965. doi: 10.1021/acsestwater.1c00160. [DOI] [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Banziger C., Cariti F., Tunas Corzon A., Ganesanandamoorthy P., Lemaitre J.C., Ort C., Julian T.R., Kohn T. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fores E., Bofill-Mas S., Itarte M., Martinez-Puchol S., Hundesa A., Calvo M., Borrego C.M., Corominas L.L., Girones R., Rusinol M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girum T., Lentiro K., Geremew M., Migora B., Shewamare S. Global strategies and effectiveness for COVID-19 prevention through contact tracing, screening, quarantine, and isolation: a systematic review. Trop. Med. Health. 2020;48(1):91. doi: 10.1186/s41182-020-00285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., Alsafar H., Yousef A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemalatha M., Kiran U., Kuncha S.K., Kopperi H., Gokulan C.G., Mohan S.V., Mishra R.K. Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: comprehensive study. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., Gaze W.H., Paterson S., Burke T., Connor T.R., McDonald J.E., Malham S.K., Jones D.L. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P.Y., Rachmadi A.T., Mantilla-Calderon D., Alkahtani M., Bashawri Y.M., Al Qarni H., O'Reilly K.M., Zhou J. Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: to what extent of the outbreak can surveillance of wastewater tell us? Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman J.S., Scire J., Caduff L., Fernandez-Cassi X., Ganesanandamoorthy P., Kull A., Scheidegger A., Stachler E., Boehm A.B., Hughes B., Knudson A., Topol A., Wigginton K.R., Wolfe M.K., Kohn T., Ort C., Stadler T., Julian T.R. medRxiv; 2021. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen A.C.U., Gamst J., Hansen L.V., Knudsen I.I.H., Jensen S.K.S. medRxiv; 2020. Eurofins Covid-19 Sentinel™ Wastewater Test Provide Early Warning of a Potential COVID-19 Outbreak. [DOI] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J.S., Kim J.Y., Kim M.C., Park S.Y., Kim B.N., Bae S., Cha H.H., Jung J., Kim M.J., Lee M.J., Choi S.H., Chung J.W., Shin E.C., Kim S.H. Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am. J. Trop. Med.. Hyg. 2020;103(6):2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.Y., Ort C., Gartner C., Carter S., Prichard J., Kirkbride P., Bruno R., Hall W., Eaglesham G., Mueller J.F. Refining the estimation of illicit drug consumptions from wastewater analysis: co-analysis of prescription pharmaceuticals and uncertainty assessment. Water Res. 2011;45(15):4437–4448. doi: 10.1016/j.watres.2011.05.042. [DOI] [PubMed] [Google Scholar]
- Lang P.O., Mitchell W.A., Lapenna A., Pitts D., Aspinall R. Immunological pathogenesis of main age-related diseases and frailty: role of immunosenescence. Eur. Geriatr. Med. 2010;1(2):112–121. doi: 10.1016/j.eurger.2010.01.010. [DOI] [Google Scholar]
- Levine-Tiefenbrun M., Yelin I., Katz R., Herzel E., Golan Z., Schreiber L., Wolf T., Nadler V., Ben-Tov A., Kuint J., Gazit S., Patalon T., Chodick G., Kishony R. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat. Med. 2021;27(5):790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
- Li B., Deng A., Li K., Hu Y., Li Z., Xiong Q., Liu Z., Guo Q., Zou L., Zhang H., Zhang M., Ouyang F., Su J., Su W., Xu J., Lin H., Sun J., Peng J., Jiang H., Zhou P., Hu T., Luo M., Zhang Y., Zheng H., Xiao J., Liu T., Che R., Zeng H., Zheng Z., Huang Y., Yu J., Yi L., Wu J., Chen J., Zhong H., Deng X., Kang M., Pybus O.G., Hall M., Lythgoe K.A., Li Y., Yuan J., He J., Lu J. medRxiv; 2021. Viral Infection and Transmission in a Large, Well-traced Outbreak Caused by the SARS-CoV-2 Delta Variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20(6):656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEllistrem M.C., Clancy C.J., Buehrle D.J., Lucas A., Decker B.K. Single dose of a mRNA SARS-CoV-2 vaccine is associated with lower nasopharyngeal viral load among nursing home residents with asymptomatic COVID-19. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan C.S., Self S., Rennert L., Kalbaugh C., Kriebel D., Graves D., Colby C., Deaver J.A., Popat S.C., Karanfil T., Freedman D.L. COVID-19 wastewater epidemiology: a model to estimate infected populations. Lancet Planet Health. 2021;5(12):e874–e881. doi: 10.1016/S2542-5196(21)00230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- O'Brien J.W., Thai P.K., Eaglesham G., Ort C., Scheidegger A., Carter S., Lai F.Y., Mueller J.F. A model to estimate the population contributing to the wastewater using samples collected on census day. Environ. Sci. Technol. 2014;48(1):517–525. doi: 10.1021/es403251g. [DOI] [PubMed] [Google Scholar]
- Omori R., Miura F., Kitajima M. medRxiv; 2021. Age-dependent Association Between SARS-CoV-2 Cases Reported by Passive Surveillance and Viral Load in Wastewater. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort C., Lawrence M.G., Reungoat J., Mueller J.F. Sampling for PPCPs in wastewater systems: comparison of different sampling modes and optimization strategies. Environ. Sci. Technol. 2010;44(16):6289–6296. doi: 10.1021/es100778d. [DOI] [PubMed] [Google Scholar]
- Page M.J., Moher D., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., Chou R., Glanville J., Grimshaw J.M., Hrobjartsson A., Lalu M.M., Li T., Loder E.W., Mayo-Wilson E., McDonald S., McGuinness L.A., Stewart L.A., Thomas J., Tricco A.C., Welch V.A., Whiting P., McKenzie J.E. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. 2021;372 doi: 10.1136/bmj.n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., Repici A., Sharma P. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3(6) doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Lee C.W., Park D.I., Woo H.Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.J., Joo E.J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2021;19(7):1387–1394 e1382. doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petala M., Dafou D., Kostoglou M., Karapantsios T., Kanata E., Chatziefstathiou A., Sakaveli F., Kotoulas K., Arsenakis M., Roilides E., Sklaviadis T., Metallidis S., Papa A., Stylianidis E., Papadopoulos A., Papaioannou N. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. Case study: the city of Thessaloniki in Greece. Sci. Total Environ. 2021;755(Pt 1) doi: 10.1016/j.scitotenv.2020.142855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciensano SARS-CoV-2 wastewater surveillance in Belgium. 2021. https://www.sciensano.be/nl/pershoek/sars-cov-2-afvalwateronderzoek-belgie-ons-afvalwater-bevestigt-de-dalende-trend (accessed 16/7/2021)
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo J.A., Trigo-Tasende N., Rumbo-Feal S., Conde-Pérez K., López-Oriona Á., Barbeito I., Vaamonde M., Tarrío-Saavedra J., Reif R., Ladra S., Rodiño-Janeiro B.K., Nasser-Ali M., Cid Á., Veiga M., Acevedo A., Lamora C., Bou G., Cao R., Poza M. Modeling the number of people infected with SARS-COV-2 from wastewater viral load in Northwest Spain. Sci. Total Environ. 2022;811 doi: 10.1016/j.scitotenv.2021.152334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellas C., Delobel P., de Souto Barreto P., Izopet J. COVID-19, virology and geroscience: a perspective. J. Nutr. Health Aging. 2020;24(7):685–691. doi: 10.1007/s12603-020-1416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney O.N., Kennedy L.C., Fan V.B., Hinkle A., Kantor R., Greenwald H., Crits-Christoph A., Al-Shayeb B., Chaplin M., Maurer A.C., Tjian R., Nelson K.L. Sewage, salt, silica, and SARS-CoV-2 (4S): an economical kit-free method for direct capture of SARS-CoV-2 RNA from wastewater. Environ. Sci. Technol. 2021;55(8):4880–4888. doi: 10.1021/acs.est.0c08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worldometer COVID-19 live statistics. 2021. https://www.worldometers.info/coronavirus/ accessed 21/12/2021.
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., McElroy K.A., Rhode S.F., Matus M., Wuertz S., Thompson J., Alm E.J. Wastewater surveillance of SARS-CoV-2 across 40 U.S. states from February to June 2020. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Shagan M., Lewis Y.E., Kramarsky-Winter E., Weil M., Indenbaum V., Elul M., Erster O., Brown A.S., Mendelson E., Mannasse B., Shirazi R., Lakkakula S., Miron O., Rinott E., Baibich R.G., Bigler I., Malul M., Rishti R., Brenner A., Friedler E., Gilboa Y., Sabach S., Alfiya Y., Cheruti U., Davidovich N., Moran-Gilad J., Berchenko Y., Bar-Or I., Kushmaro A. City-level SARS-CoV-2 sewage surveillance. Chemosphere. 2021;283 doi: 10.1016/j.chemosphere.2021.131194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Not applicable.