Abstract
Background:
An appropriate personalized molecular testing ensures the most efficacious treatment in lung cancer. It is still controversial whether younger lung adenocarcinoma (LUAD) patients have different molecular features compared with their older counterparts. MicroRNAs have been involved in lung cancer and their altered expression has been suggested as a potential biomarker in the pathogenesis, diagnosis, prognosis, and therapy of LUAD.
Materials and Methods:
To analyze putative differences in miR-25 expression between young (with age ≤50 years) and old adenocarcinoma patients, we quantified miR-25 levels with NanoString technology in 88 LUAD specimens. We further investigated a cohort of 309 LUAD patients from the cancer genome atlas (TCGA) database to test our hypothesis.
Results:
miR-25 expression was upregulated in young LUAD patients in comparison to the older ones (P = 0.03) in our series. The analysis of public database TCGA confirmed our results, which miR-25 differentially expressed in the two aged groups (P = 0.0009). Moreover, a consequential pairing of miR-25 with a target region in phosphatase and tensin homolog (PTEN) 3’ untranslated region (UTR) and actually low PTEN expression seemed to be associated with high miR-25 (P = 0.001) in young patients.
Conclusions:
The interaction of miR-25 and PTEN in young LUAD may define a subgroup of patients, highlighting the concept of molecular testing in different age subtypes.
Keywords: Lung adenocarcinoma, MicroRNAs, miR-25, phosphatase and tensin homolog, young
INTRODUCTION
Despite lung cancer is one of the most common cancers and the leading cause of cancer-related death worldwide[1] the mechanisms controlling lung tumorigenesis remain unclear. An appropriate personalized molecular testing ensures the most efficacious treatment in lung cancer. It is still controversial whether younger lung adenocarcinoma (LUAD) patients have different molecular features compared with their older counterparts.[2]
MicroRNAs (miRNAs) are a class of small noncoding RNA acting in regulation of a multiplicity of target genes through mRNA degradation or translation repression.[3] MiRNAs have been involved in lung cancer, and their altered expression has been suggested as a potential biomarker in LUAD. However, few data are known regarding the different expression level of these small RNAs in the young compared to old LUAD patients.
MiR-25 is a member of the miR-106b-25 family, with miR-106b and miR-93. The role of miR-25 on cancer can be oncogenic or tumor suppressive in different kinds of cancer.[4] In particular, the exact miR-25 involvement in LUAD is still understood as well as if miR-25 may have diverse effects depending on age.
In the current study, we assayed miR-25 expression level by NanoString platform in young and old LUAD patients surgically resected at the Unit of Thoracic Surgery in Pisa (Italy); furthermore, to better investigate the potential role of this miRNA in the two age subgroups on a larger population, we analyzed a cohort of 309 LUAD patients from the cancer genome atlas (TCGA) database.
Phosphatase and tensin homolog (PTEN) is a negative regulator of phosphatidylinositol 3-kinase (PI3K) signaling pathway, one of the most important in cell growth regulation, with a crucial role in cancer.[5] Impaired PTEN function due to genetic and epigenetic alterations leads to uncontrolled activation of downstream signaling pathway, necessary in the cancer phenotype development.[6] In addition, recently, some miRNAs have been reported to regulate cell proliferation by targeting PTEN.[7,8] We used bioinformatics tools to verify if PTEN was a potential miR-25 predicted target. The interaction of miR-25 and PTEN in young LUAD may define a subgroup of patients, highlighting the concept of molecular testing in different age subtypes.
MATERIALS AND METHODS
Patients and tumor characteristics
We retrospectively selected 88-LUAD patients who underwent surgical resection at the Unit of Thoracic Surgery in the Department of Surgical, Medical, Molecular Pathology, and Critical Area at Pisa University. We used a cutoff value of 50 years to divide young (50 years or less), from old patients (range: 51–88 years). The diagnoses were formulated in formalin-fixed and paraffin-embedded (FFPE) histological samples, according to the World Health Organization classifications, describing identifiable predominant pattern present (lepidic, acinar, papillary, solid, and micropapillary), and invasive component (minimally invasive adenocarcinoma, adenocarcinoma in situ, or an invasive adenocarcinoma).[9,10] Molecular analysis (miRNA isolation and expression) was performed after the selection of the most representative paraffin blocks of the tumor tissues. The clinicopathological characteristics were available for all patients. The study was conducted in accordance with the Helsinki Declaration of 1975, as revised in 2000.
MicroRNA isolation
MiR-25 was isolated from FFPE tissues after deparaffinization and manual macrodissection of the neoplastic area, using the miRNeasy FFPE kit (QIAGEN Inc., Hilden, Germany), accordingly to the manufacturer's instructions. After quality and quantity evaluation with a NanoDrop ND-1000 (ThermoScientific, USA) spectrophotometer, the samples were stored at −80°C until used.
miR-25 expression by nanostring
MiR-25 expression was evaluated by the nCounter assay, with a reporter and capture probe specific for miR-25, in accordance with the manufacturer's instructions (NanoString Technologies, Seattle, WA, USA). Nanostring nSolver (version 2.5) software was used for the analysis of raw NanoString counts.
The cancer genome atlas database
We extracted IlluminaHiSeq miR-25 expression plan together with the corresponding clinicopathological characteristics for 309 adenocarcinoma patients (LUAD) from the TCGA data portal (http://tcga. cancer.gov/; accessed October 2017).
Statistical analysis
Chi-square and Student's unpaired t-test was applied using JMP10 software (SAS) to determine the association between miR-25 expression and clinicopathological parameters. A P < 0.05 was considered statistically significant.
MiR-25 target prediction and pathway analysis
We used different databases such as miRanda (http://www.microrna.org/) and TargetScan 6.2 (http://www.microRNA.org/), to predict putative miR-25 targets.
RESULTS
Patients and tumor characteristics
Eighty-eight patients with LUAD were enrolled in this study, 56 males and 32 females. The cutoff value of 50 years was used to identify younger LUAD patients and the older counterparts. The different subtypes of adenocarcinoma were characterized as follows: Lepidic (29/88, 33%), solid (26/88, 29.5%), acinar (22/88, 25%), and papillary (11/88, 12.5%) variants. Their stages were classified as I in 40 cases (17 IA, 23 IB), as II in 22 cases (13 IIA, 9 IIB), as III-IV in 26 cases (23 IIIA, 1 IIIB, and 2 IV).
miR-25 expression and clinicopathological characteristics
miR-25 expression was evaluated using the nSolver Software version 2.5 (nanoString Technologies, Seattle, Washington) and compared with our patients’ clinicopathological characteristics. The samples were divided into high and low expression groups based on the median fold-change value (6583 for miR-25). High miR-25 expression was significantly associated with younger (P = 0.01) [Table 1]; t-test confirmed a higher mir-25 level (7531.54 ± 563.39) in younger than in older patients (5844.13 ± 563.39) (P = 0.03) [Figure 1].
Table 1.
MicroRNAs-25 expression level and clinicopathological characteristics in 88 lung adenocarcinoma patients
| Variables | Low | High | P |
|---|---|---|---|
| Age (years) | |||
| ≤50 | 16 | 28 | 0.01 |
| >50 | 28 | 16 | |
| Gender | |||
| Male | 31 | 25 | 0.18 |
| Female | 13 | 19 | |
| ADC previous pattern | |||
| Lepidic | 15 | 14 | 0.05 |
| Solid | 13 | 13 | |
| Acinar | 7 | 15 | |
| Papillar | 9 | 2 | |
| T | |||
| T1 | 10 | 12 | 0.79 |
| T2 | 21 | 18 | |
| T3-T4 | 13 | 16 | |
| N | |||
| N0 | 22 | 25 | 0.41 |
| N1 | 6 | 9 | |
| N2 | 12 | 6 | |
| Nx | 4 | 4 | |
| Stage | |||
| I | 20 | 20 | 1 |
| II | 11 | 11 | |
| III-IV | 13 | 13 |
ADC=Apparent diffusion coefficient
Figure 1.
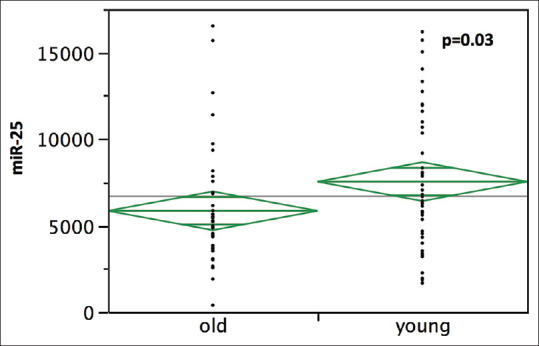
Comparison of miR-25 expression level in 44 young (with age =50 years) and 44 old lung adenocarcinoma patients
The cancer genome atlas data analysis
A cohort of 309 LUAD patients from TCGA database was analyzed, to identify putative differences in miR-25 expression levels between young and old adenocarcinoma patients on a larger population. Twenty-three patients out of 309 TCGA cases showed an age of 50 years or less; in this younger group, miR-25 levels were higher (10901.9 ± 967.38) than in the older counterparts (7528.3 ± 274.33) (P = 0.0009) [Figure 2].
Figure 2.
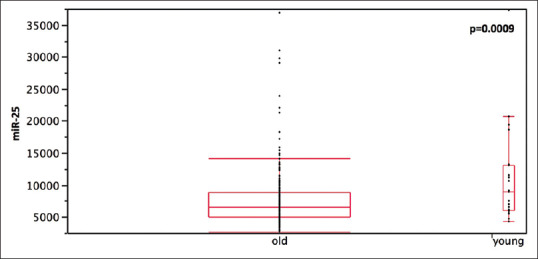
Comparison of miR-25 expression level in young (with age ≤ 50 years) and old lung adenocarcinoma patients extracted from the cancer genome atlas database
MicroRNA regulation prediction analysis
Bioinformatic tools showed that most of the predicted miR-25 targets are involved in critical pathways such as cell cycle regulation. Interestingly, we found a consequential pairing of miR-25 with a target region in PTEN 3’ UTR [Figure 3].
Figure 3.
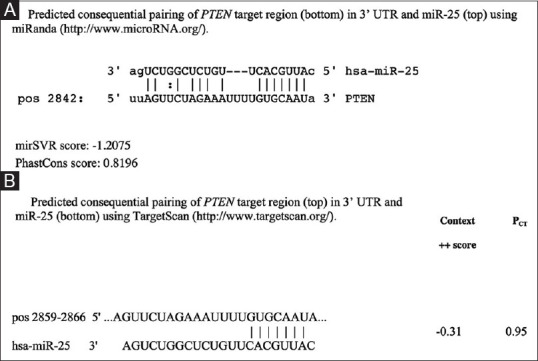
Predicted consequential pairing of phosphatase and tensin homolog target region (top) and miR-25-3p (bottom) using miRanda (http://www.microRNA.org/) in upper panel A, and TargetScan (http://www.targetscan.org/) in lower panel B
Phosphatase and tensin homolog expression
The samples were divided into high and low PTEN expression groups based on the median fold-change value (1.53 e + 7 for PTEN). Low PTEN levels were significantly associated with high miR-25 expression; samples with high PTEN showed lower miR-25 levels (6917 ± 378) than samples with low PTEN (8603 ± 369) (t-test, P = 0.001) [Figure 4]. Moreover, most of the young patients (17/23) showed lower PTEN levels when compared with their older counterparts (P = 0.02).
Figure 4.
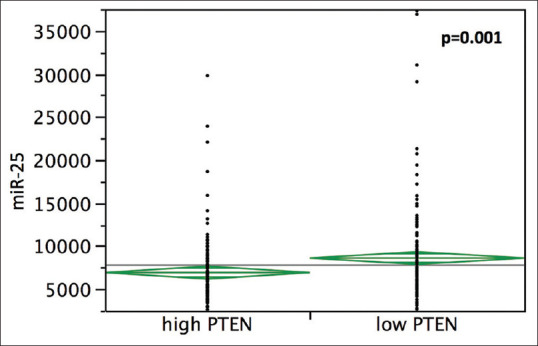
Relationship between miR-25-3p and phosphatase and tensin homolog expression level in 309 lung adenocarcinoma samples from the cancer genome atlas database
DISCUSSION
In the last decade, lung cancer incidence has been rising, and the age at diagnosis continues to decrease.[11] However, it remains controversial whether younger patients could represent a separate clinicopathological entity when compared to their old counterparts. We used the cutoff age of 50 years to define young lung cancer patients, which is commonly used to separate the two aged groups.[12,13,14,15]
Aberrant microRNA expression is involved in lung cancer regulation, affecting tumor suppression as well as oncogenesis, indicating their importance in the regulation of lung cancer,[16] but few studies have been conducted on miRNAs expression profile in the two age groups.[17] MiR-25 may have different roles on the pathogenesis of several types of cancer, including small and nonsmall-cell lung cancer (NSCLC).[18,19] The present study aimed to better investigate the expression of miR-25 in young and old lung cancer patients. In detail, the miR-25 levels were compared between ADC patients younger and older than 50 years, and we found that miR-25-3p was significantly upregulated in the young patients in our series of 88 patients. A cohort of 309 LUAD patients from the TCGA database confirmed our findings on a larger population and also using a different transcriptome-based technologies such as Illumina HiSeq quantification. Our study demonstrated higher miR-25 levels in young in comparison to old LUAD patients; however, Noren Hooten et al.[20] demonstrated changes in miRNA expression with human aging and Peña-Chilet et al.[21] suggested that miRNAs signature have the potential to be age-related in breast cancer.
The online prediction software’s, such as miRanda, TargetScan, and others, identified different putative targets of miR-25, but few reports have investigated the downstream signaling pathway of miR-25. Wang et al.[22] demonstrated that miR-25 acts as oncogene in osteosarcoma cells by directly targeting the cell cycle inhibitor p27. Tissue miR-25 expression was also involved in female LUAD patients[23] and, recently, Ding et al. showed that miR-25 enhances cell migration and invasion in nonsmall cell lung cancer cells through extracellular signal-regulated kinase (ERK) signaling pathway by inhibiting Kruppel-like factor 4 (KLF4).[24]
PTEN is a main controller of PI3K signaling pathway with a central role in several tumors, including lung cancer.[25] The results concerning the prognostic value of PTEN downregulation seem to be contradictory, with association to unfavorable survival in NSCLC patients in some papers,[26,27,28] and the opposite conclusions in others.[29] However, Xiao et al.[30] suggested that PTEN expression levels might also add information on the effect of treatment for patients with NSCLC. In our work, we observed that lung cancer in young and old patients may be influenced by different regulatory mechanisms since we found PTEN downregulated in the younger group, probably due to distinct age-related mi-R25 expression. Young LUAD patients with decreased expression of PTEN due to high miR-25 levels may represent a subtype of lung cancer and might benefit from individualized treatment plans.
CONCLUSIONS
Our study provides new insights into the role of miR-25 in LUAD, in particular in young patients; the interaction of miR-25, and PTEN in young LUAD patients may define a subgroup of patients, highlighting the concept of molecular testing in different age subtypes. Further validations are needed to better define if an age-based genomic signature could be used as a tool in the personalized therapy of LUAD.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Subramanian J, Morgensztern D, Goodgame B, Baggstrom MQ, Gao F, Piccirillo J, et al. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: A surveillance, epidemiology, and end results (SEER) analysis. J Thorac Oncol. 2010;5:23–8. doi: 10.1097/JTO.0b013e3181c41e8d. [DOI] [PubMed] [Google Scholar]
- 3.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sárközy M, Kahán Z, Csont T. A myriad of roles of miR-25 in health and disease. Oncotarget. 2018;9:21580–612. doi: 10.18632/oncotarget.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noorolyai S, Shajari N, Baghbani E, Sadreddini S, Baradaran B. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;698:120–8. doi: 10.1016/j.gene.2019.02.076. [DOI] [PubMed] [Google Scholar]
- 6.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–50. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong ZH, Zhou F, Shi C, Xiang T, Zhou CK, Wang QQ, et al. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24:9. doi: 10.1186/s11658-018-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, Su Z, Gu W, Shen X, Zhao Q, Shi L, et al. MiR-19b and miR-20a suppress apoptosis, promote proliferation and induce tumorigenicity of multiple myeloma cells by targeting PTEN. Cancer Biomark. 2019;24:279–89. doi: 10.3233/CBM-182182. [DOI] [PubMed] [Google Scholar]
- 9.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, et al. The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 11.Liu NS, Spitz MR, Kemp BL, Cooksley C, Fossella FV, Lee JS, et al. Adenocarcinoma of the lung in young patients: The M.D. Anderson experience. Cancer. 2000;88:1837–41. doi: 10.1002/(sici)1097-0142(20000415)88:8<1837::aid-cncr12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 12.Ramalingam S, Pawlish K, Gadgeel S, Demers R, Kalemkerian GP. Lung cancer in young patients: Analysis of a surveillance, epidemiology, and end results database. J Clin Oncol. 1998;16:651–7. doi: 10.1200/JCO.1998.16.2.651. [DOI] [PubMed] [Google Scholar]
- 13.Sekine I, Nishiwaki Y, Yokose T, Nagai K, Suzuki K, Kodama T. Young lung cancer patients in Japan: Different characteristics between the sexes. Ann Thorac Surg. 1999;67:1451–5. doi: 10.1016/s0003-4975(99)00171-x. [DOI] [PubMed] [Google Scholar]
- 14.Radzikowska E, Roszkowski K, Głaz P. Lung cancer in patients under 50 years old. Lung Cancer. 2001;33:203–11. doi: 10.1016/s0169-5002(01)00199-4. [DOI] [PubMed] [Google Scholar]
- 15.Boldrini L, Giordano M, Lucchi M, Melfi F, Fontanini G. Expression profiling and microRNA regulation of the LKB1 pathway in young and aged lung adenocarcinoma patients. Biomed Rep. 2018;9:198–205. doi: 10.3892/br.2018.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3–20. doi: 10.1016/j.mam.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Giordano M, Boldrini L, Servadio A, Niccoli C, Melfi F, Lucchi M, et al. Differential microRNA expression profiles between young and old lung adenocarcinoma patients. Am J Transl Res. 2018;10:892–900. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Z, Liu J, Wang C, Wang Y, Jiang Y, Guo M. MicroRNA-25 regulates small cell lung cancer cell development and cell cycle through cyclin E2. Int J Clin Exp Pathol. 2014;7:7726–34. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Wu Y, Meng Q, Xia Z. Elevated microRNA-25 inhibits cell apoptosis in lung cancer by targeting RGS3. In vitro Cell Dev Biol Anim. 2016;52:62–7. doi: 10.1007/s11626-015-9947-2. [DOI] [PubMed] [Google Scholar]
- 20.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peña-Chilet M, Martínez MT, Pérez-Fidalgo JA, Peiró-Chova L, Oltra SS, Tormo E, et al. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14:529. doi: 10.1186/1471-2407-14-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang XH, Cai P, Wang MH, Wang Z. microRNA-25 promotes osteosarcoma cell proliferation by targeting the cell-cycle inhibitor p27. Mol Med Rep. 2014;10:855–9. doi: 10.3892/mmr.2014.2260. [DOI] [PubMed] [Google Scholar]
- 23.Xu FX, Su YL, Zhang H, Kong JY, Yu H, Qian BY. Prognostic implications for high expression of MiR-25 in lung adenocarcinomas of female non-smokers. Asian Pac J Cancer Prev. 2014;15:1197–203. doi: 10.7314/apjcp.2014.15.3.1197. [DOI] [PubMed] [Google Scholar]
- 24.Ding X, Zhong T, Jiang L, Huang J, Xia Y, Hu R. miR-25 enhances cell migration and invasion in non-small-cell lung cancer cells via ERK signaling pathway by inhibiting KLF4. Mol Med Rep. 2018;17:7005–16. doi: 10.3892/mmr.2018.8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgescu MM. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1:1170–7. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Chen H, Liao Y, Chen N, Liu T, Zhang H, et al. Expression and clinical evidence of miR-494 and PTEN in non-small cell lung cancer. Tumour Biol. 2015;36:6965–72. doi: 10.1007/s13277-015-3416-0. [DOI] [PubMed] [Google Scholar]
- 27.Li XB, Yang Y, Zhang HQ, Yue WT, Zhang TM, Lu BH, et al. High levels of phosphatase and tensin homolog expression predict favorable prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2015;19:2231–9. [PubMed] [Google Scholar]
- 28.Sun Y, Li D, Lv XH, Hua SC, Han JC, Xu F, et al. Roles of osteopontin and matrix metalloproteinase-7 in occurrence, progression, and prognosis of nonsmall cell lung cancer. J Res Med Sci. 2015;20:1138–46. doi: 10.4103/1735-1995.172980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inamura K, Togashi Y, Nomura K, Ninomiya H, Hiramatsu M, Okui M, et al. Up-regulation of PTEN at the transcriptional level is an adverse prognostic factor in female lung adenocarcinomas. Lung Cancer. 2007;57:201–6. doi: 10.1016/j.lungcan.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Xiao J, Hu CP, He BX, Chen X, Lu XX, Xie MX, et al. PTEN expression is a prognostic marker for patients with non-small cell lung cancer: A systematic review and meta-analysis of the literature. Oncotarget. 2016;7:57832–40. doi: 10.18632/oncotarget.11068. [DOI] [PMC free article] [PubMed] [Google Scholar]


