Abstract
Protein abundance data of drug-metabolizing enzymes and transporters (DMETs) are broadly applicable to the characterization of in vitro and in vivo models, in vitro to in vivo extrapolation (IVIVE), and interindividual variability prediction. However, the emerging need of DMET quantification in small sample volumes such as organ-on a chip effluent, organoids, and biopsies requires ultrasensitive protein quantification methods. We present an ultrasensitive method that relies on an optimized sample preparation approach involving acetone precipitation coupled with a microflow-based liquid chromatography-tandem mass spectrometry (μLC-MS/MS) for the DMET quantification using limited sample volume or protein concentration, i.e., liver tissues (1–100 mg), hepatocyte counts (~4000 to 1 million cells), and microsomal protein concentration (0.01–1 mg/ml). The method was applied to quantify DMETs in differential tissue S9 fractions (liver, intestine, kidney, lung, and heart) and cryopreserved human intestinal mucosa (i.e., CHIM). The method successfully quantified >75% of the target DMETs in the trypsin digests of 1 mg tissue homogenate, 15,000 hepatocytes, and 0.06 mg/ml microsomal protein concentration. The precision of DMET quantification measured as the coefficient of variation across different tissue weights, cell counts, or microsomal protein concentration was within 30%. The method confirmed significant extrahepatic abundance of non-cytochrome P450 enzymes such as dihydropyridine dehydrogenase (DPYD), epoxide hydrolases (EPXs), arylacetamide deacetylase (AADAC), paraoxonases (PONs), and glutathione S-transferases (GSTs). The ultrasensitive method developed here is applicable to characterize emerging miniaturized in vitro models and small volume biopsies. In addition, the differential tissue abundance data of the understudied DMETs will be important for physiologically-based pharmacokinetic (PBPK) modeling of drugs.
Keywords: proteomics, liquid chromatography-mass spectroscopy, physiology-based pharmacokinetic modeling, drug-metabolizing enzymes, transporters, In vitro to in vivo extrapolation
1. INTRODUCTION
In vitro to in vivo extrapolation (IVIVE) and prediction of interindividual variability in drug metabolism and transport are important for prospective assessment of in vivo pharmacokinetics and potential toxicity of drugs. Human cell-based models and subcellular fractions (microsomes, cytosol, and S9 fractions) are considered superior to the preclinical animal models for the prediction of human drug metabolism, transport, and drug-drug interactions (DDIs) due to the large inter-species differences in these processes.1–4 During the early drug development stage, the in vitro data are utilized for IVIVE and physiologically-based pharmacokinetic (PBPK) modeling is used to assist in clinical trial design, e.g., the first-in-human dose prediction. In the later stages of drug development, PBPK modeling is used for the prediction of inter-individual variability in pharmacokinetics, e.g., in special populations such as patients with liver and kidney dysfunctions.5
For precise scaling of in vitro data using PBPK modeling, differences in the protein abundance between in vitro models and human tissues should be taken into consideration.6 Similarly, inter-individual variability in drug metabolism and transport can be predicted using the protein abundance data of drug-metabolizing enzymes and transporters (DMETs) in human tissues.7 Protein abundance data can be generated using immuno-quantification methods such as Western blotting, but these techniques are associated with several limitations such as non-availability of specific antibodies, low throughput, semi-quantitative nature, poor sensitivity, reproducibility, and higher overall cost.8 Quantitative proteomics by liquid chromatography-triple quadrupole mass spectrometry (LC-MS/MS) addresses these limitations.9 Quantitative proteomics has emerged as for high throughput and selective quantification of DMETs 9,10 that allows distinguishing highly homologues proteins, e.g., CYP3A4, CYP3A5 versus CYP3A7.11 In targeted quantitative proteomics, a protein sample is digested using proteases such as trypsin, and unique surrogate peptides are quantified in multiple reaction monitoring (MRM) mode using a triple quadrupole mass spectrometer. We and others have used this technique for the quantification of various DMETs (CYPs, non-CYPs, and transporters) and applied it in translational pharmacology.12–17 Notable applications of quantitative proteomics in translational pharmacology include i) characterization of interindividual variability in DMET abundance, i.e., the effect of age, sex, disease state, and genotype, ii) characterization of in vitro models including subcellular localization of DMETs, iii) IVIVE of drug metabolism and transport, and iv) differential tissue abundance of drug-metabolizing enzymes (DMEs) across human tissues.16,18–21
Although quantitative proteomics offers multiplex quantification of proteins (i.e., simultaneous analysis of 30–100 proteins in a single LC-MS/MS injection), sensitivity is the major limitation of the conventional LC-MS/MS approach. In particular, the quantification of DMETs is challenging in small sample volumes. For example, protein extraction from 96-well plate culture or 3D-culture (e.g., organ-on-a-chip) often yields samples with a limited total protein concentration. Similarly, biopsy samples that are precious and often non-renewable, are available only in low volume.22 Further, a variable protein with a dynamic expression profile across the population cannot be quantified in the low expressers.7 Similarly, quantitative proteomics analysis of subcellular fractions with low yield (e.g., brain endothelial cells, purified plasma membranes, and exosomes isolated from biofluids) requires greater analytical sensitivity.
Enrichment of proteins using techniques such as immunoprecipitation or stable isotope labeling and capture by antibodies (SISCAPA)23 greatly improve protein quantification sensitivity, but these methods are often cumbersome, expensive, and do not support multiplexing. Moreover, quantification of low abundant proteins with poor signal to noise ratio by conventional LC-MS/MS is highly variable leading to large inter-laboratory variation.24,25 To address these limitations, we developed an ultrasensitive method for DMET quantification that can be applied to low quantities of tissue and cell samples using an optimized sample preparation and microflow LC (flow rate of 3 μl/min) coupled to a triple quadrupole MS by an IonKey interphase, i.e., μLC-MS/MS (Waters, Milford, MA). Varying quantities of samples were used: i.e., 1–100 mg of liver tissue, 4000– 1 million hepatocytes, and 10–1000 μg/ml microsomal protein of the human liver and intestine. We then applied this method for the quantification of DMETs to determine differential tissue abundance and regional variability in the intestine. The μLC-MS/MS method has crucial advantages of the low volume requirements of protein quantification as compared to the conventional LC-MS/MS method and can be applied for the high-throughput quantification of samples such as extracts from 96 or 384 well plates, organ-on-a-chip model, solid or liquid biopsies, and dry-blood spot samples.
2. MATERIALS AND METHODS
2.1. Materials
Ammonium bicarbonate (ABC, 98% purity), bovine serum albumin (BSA), dithiothreitol (DTT), iodoacetamide (IAA), and trypsin (MS grade) were procured from Thermo Fisher Scientific (Rockford, IL). Stable isotope-labeled (SIL) peptides and synthetic unlabeled peptides were purchased from Thermo Fisher Scientific (Rockford, IL) and New England Peptides (Boston, MA), respectively. Mem-PER Plus membrane protein extraction kit, Pierce bicinchoninic acid (BCA) protein assay kit, Optima MS-grade acetonitrile, chloroform, methanol, and formic acid were procured from Fisher Scientific (Fair Lawn, NJ).
2.2. Sample procurement
The demographic information of the donors is provided in Supplementary Table 1. Human liver tissue was provided for research by Corning (New York, NY). The human hepatocytes were procured from BioIVT (Baltimore, MD) and the pooled microsomes and S9 fractions were purchased from Sekisui XenoTech (Kansas City, KS). Cryopreserved human intestinal mucosa (CHIM) samples were procured from IVAL (Columbia, MD) and the pooled tissue fractions were from our previous study.26 The use of these samples has been classified as non-human subject research. All the samples were de-identified and the tissue source was anonymous to the researcher. This study did not involve any human research, thus waiving the need for ethical review and informed consent.
2.3. Sample preparation for protein digestion
Human liver tissue (n=2 donors) was individually homogenized by a hand-held homogenizer with plastic probes according to a previously published protocol.7 Briefly, the tissue was homogenized in different volumes (100–500 μl) of the permeabilization buffer (Mem-PER Plus membrane protein extraction kit) to allow complete homogenization, mixed, and incubated for 30 min (4º C) at 300 rpm using Compact Digital Rocker (Thermo Scientific, Waltham, MA). The homogenate was centrifuged at 16,000 xg for 15 min (4 ºC) and the non-membrane fraction was separated. The resultant pellet was re-suspended with gentle mixing in sodium dodecyl sulfate (SDS) and the membrane solubilization buffer of the kit (1:1 ratio, v/v) and protease inhibitor cocktail (PI). The sample was incubated for 60 min at 300 rpm (15 ºC) using Thermo Scientific Compact Digital Rocker before centrifugation at 16,000 xg for 15 min (4 ºC). The supernatant containing the membrane proteins was separated. The samples were subjected to total protein quantification using BCA assay before the digestion. All samples were diluted to a fixed 1 mg/ml concentration before protein digestion.
Individual hepatocytes (n = 10) were pooled and diluted to 1 million cells/ml stock concentration. Nine different concentrations ranging from 4000 to 1 million cells/ml were prepared by serial dilution (Figure 1). The hepatocyte suspension was centrifuged at 4000 xg for 15 min (4 ºC), and the pellet was re-suspended in SDS: membrane solubilization buffer (1:1 ratio, v/v) and protease inhibitor was added with gentle mixing. The resultant hepatocyte homogenate samples were incubated for 60 min at 300 rpm (15 ºC) and centrifuged at 16,000 xg for 15 min (4 ºC). Eighty μL of the supernatant was used for trypsin digestion. Separately, serial dilutions of HLM and HIM was performed using 1 mg/ml of human serum albumin (HSA) as a sample dilution matrix to obtain a range of protein concentrations from 0.01 to 1 mg/ml (Figure 1). The method was also applied to quantify DMETs in the S9 fractions from different human tissues (liver, intestine, kidney, lung, and heart) and the CHIM samples from our previous studies.26,27 Standard stable labeled peptides (average concentration range, ~2–5000 fmol on-column) were spiked into the human hepatocyte homogenate matrix to determine the linearity, range, accuracy, precision, and lower limit of quantification (LLOQ) of the μLC-MS/MS method.
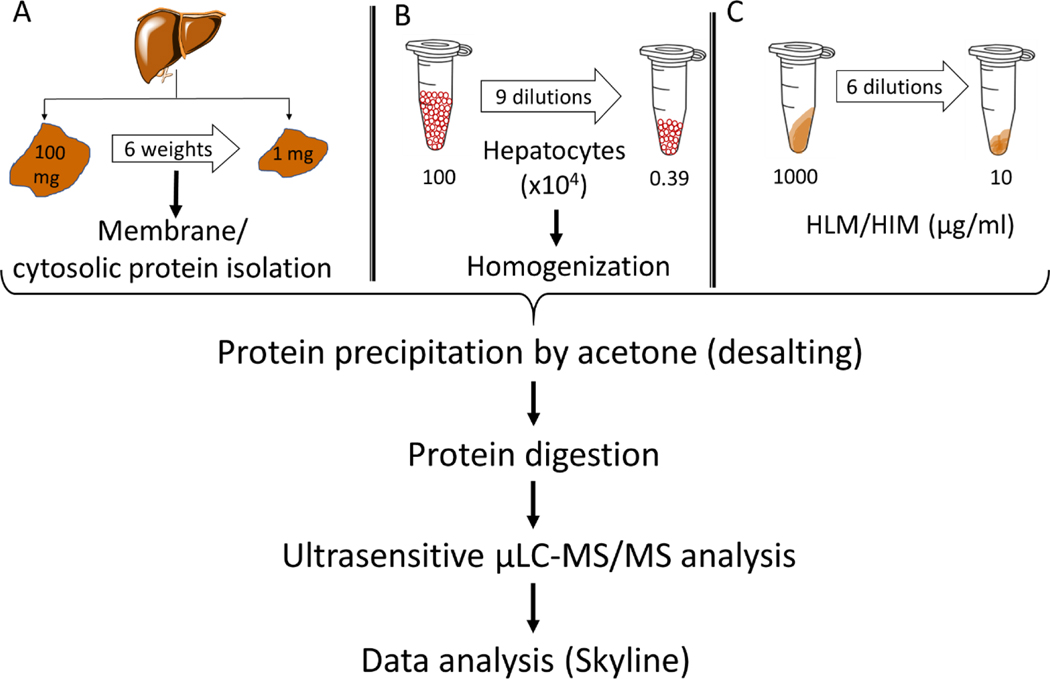
Figure 1:
Sample preparation workflow for the quantification of DMETs in samples isolated from six different human liver tissue weights ranging 1–100 mg (A), 3900 to 1 million cells/ml human hepatocytes (B) HLM and HIM concentrations ranging from 10 to 1000 μg/ml (C). For tissue samples, membrane and non-membrane fractions were isolated and a uniform final protein concentration of 1 mg/ml was used for further analysis. For hepatocytes, the cell lysate was directly used for the further processing, whereas HLM and HIM samples were diluted using human serum albumin (HSA) to keep the total protein concentration at 1 mg/ml. All samples were denatured, reduced, alkylated, and desalted using acetone prior to trypsin digestion. The analysis was performed using μLC-MS/MS at a mobile phase flow rate of 3 μl/min. Data analysis was carried out by Skyline.
2.4. Trypsin digestion and μLC-MS/MS analysis
Samples were digested by trypsin using a previously described protocol28 with modified protein precipitation and desalting protocol using acetone. Briefly, protein samples were mixed with ABC buffer (100 mM, pH 7.8), DTT (250 mM), and BSA (0.02 mg/mL), followed by denaturation for 10 min at 95 °C. After cooling to the room temperature for 10 min, the protein mixture was alkylated by IAA (500 mM) in dark for 30 min. The sample was then subjected to protein precipitation by adding ice-cold acetone and incubation at −80 ˚C for 1 hr. The proteins were recovered by centrifugation at 16,000 xg for 10 min. The resultant protein pellet was dried, washed with 500 μL ice-cold methanol, and dried under vacuum for 30 min. The dried pellet was resuspended in ABC buffer (50 mM, pH 7.8) and the digestion was performed by trypsin (20 μL; protein/trypsin ratio ~80:1) for 16 h at 37 °C with gentle shaking (300 rpm). The digestion was quenched by adding 5 μL of 0.5% formic acid and centrifuged at 16,000 xg for 10 min (4 ºC). The samples were stored in – 80 ˚C freezer prior to μLC-MS/MS analysis. A cocktail of SIL peptides (internal standard) was added to the samples (Table S3). A previously characterized sample (a pool of human intestine and liver tissue extracts) with the known abundance of all the studied proteins was used as a calibrator cocktail.
The samples were analyzed using an M-class Waters UPLC system coupled with Waters Xevo TQ-XS μLC-MS/MS instrument supported by ionKey interphase. The peptides were separated on iKey BEH C18 column (130 A°, 1.7 μm, 150μm * 50 mm) and nano Ease Symmetry C18 trap column (300μm * 50 mm) (Waters, Milford, MA). The optimized μLC-MS/MS acquisition parameters are provided in Table S2.
2.5. Data analysis
μLC-MS/MS data analysis was performed on Skyline 20.2 (University of Washington, Seattle, WA). The targeted peptide peaks were identified by matching the retention time and fragmentation patterns with the externally added SIL peptide cocktail. The analyte peptide peak area was normalized by SIL peptide area to address any instrument-related technical variability, whereas BSA was used as an internal protein standard to address artifacts in the trypsin digestion. The absolute peptide abundance was quantification by using the previously characterized pooled sample with known DMET abundance as the calibrator. If case of non-availability of the light peptide standards, the DMET quantification data are expressed as relative abundance, i.e., peak area ratios normalized to per mg of total protein digest.
RESULTS
Effect of tissue weight on DMET abundance quantification
We first evaluated the ability of the μLC-MS/MS method to detect DMETs in crude non-membrane and membrane fractions of small tissue amounts ranging in weight from 1, 5, 10, 25, 50, and 100 mg. The total protein amount recovered was proportional (R2 = 0.92) to the tissue weights (Figure S1). A total of 55 DMETs were detected and quantified in each sample (Figure 2) when a fixed protein amount (i.e., 80 μg) extracted from 1–100 mg tissue weights was digested. The DMETs detected include 10 CYPs, 8 UGTs, 24 non CYPs, and 12 transporters (Table S4, S5, and S6). A few membrane-bound proteins were detected in the non-membrane fraction, whereas some cytosolic proteins were detected in the membrane fraction, likely due to the contamination during the crude fractionation. When a protein was detected in both membrane and non-membrane fractions (e.g., CES1), the data were reported from the subcellular fraction with higher enrichment.
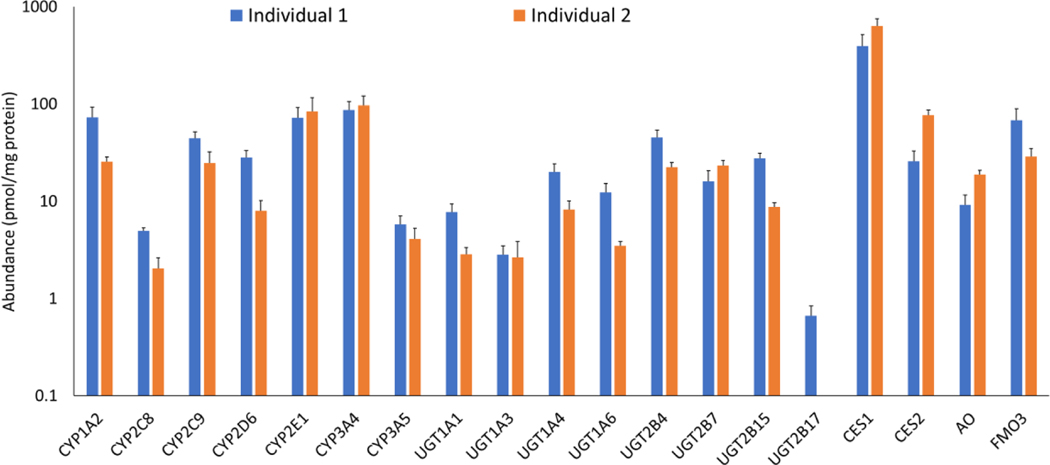
Figure 2:
Protein abundance of CYPs, UGTs, and other non-CYPs in samples (membrane or non-membrane fractions) isolated from differential liver tissue weights ranging from 1–100 mg. CYPs, UGTs, and FMO3 were quantified in the membrane fractions, whereas CESs and AO were quantified in the non-membrane fraction. Data are presented as mean and standard deviation of the protein abundance values (pmol/mg protein) calculated across six tissue weights.
Overall these data suggest that all the target DMETs can be quantified in a 1 mg liver tissue sample using the optimized sample preparation and μLC-MS/MS method. Similarly, the quantification of DMETs revealed that in general all major proteins were reproducibly quantifiable in the sample extracted from 1–100 mg tissue with a few exceptions. For example, the cumulative % coefficient of variation of individual DMETs quantified in 1 mg/ml protein extracted from different weights of liver tissue was within 30%. The variability observed across samples is perhaps due to the differential zonal abundance of DMETs in liver tissue, particularly when small tissue weights were used. Significant between-subject variability was observed for several proteins, and UGT2B17 was only detected in one sample likely due to the presence of non-expresser genotype (gene deletion).7 Among the enzymes quantified at the absolute peptide levels, the order of mean hepatic abundance (n=2 donors) of CYPs and UGTs in the membrane fraction was: CYP3A4 > 2E1 > 1A2 > 2C9 > 2D6 > 3A5 > 2C8 and UGT2B4 > 2B7 > 2B15 > 1A4 > 1A6 > 1A1 > 1A3 > 2B17, respectively. The order of protein abundance of enzymes quantified in the non-membrane fraction was CES1 > CES2 > FMO3 > AO. Our data on the order of different UGTs and CYPs quantification was generally in-line with reported data with a few exceptions29,30. For example, the abundance values of highly polymorphic DMEs (UGT1A1 and UGT2B17) were different in our samples as compared to the literature values. Such variability in DMET abundance data across studies can be due to other factors such as the number of samples, use of different peptides, and different methods (Western blot vs LC-MS/MS), as discussed by us previously6. In addition to the DMEs, we detected 12 transporter proteins in the membrane fraction (Table S6) which includes P-gp (ABCB1), MRP3 (ABCC3), OCT1 (SLC22A1), OCT3 (SLC22A3), OAT2 (SLC22A7), OAT4 (SLC22A11), CNT1 (SLC28A1), CNT-3 (SLC28A3), OATP1B1 (SLCO1B3), OATP1B3 (SLCO1B3), and OATP2B1 (SLCO2B1).
Quantification of DMETs in hepatocytes
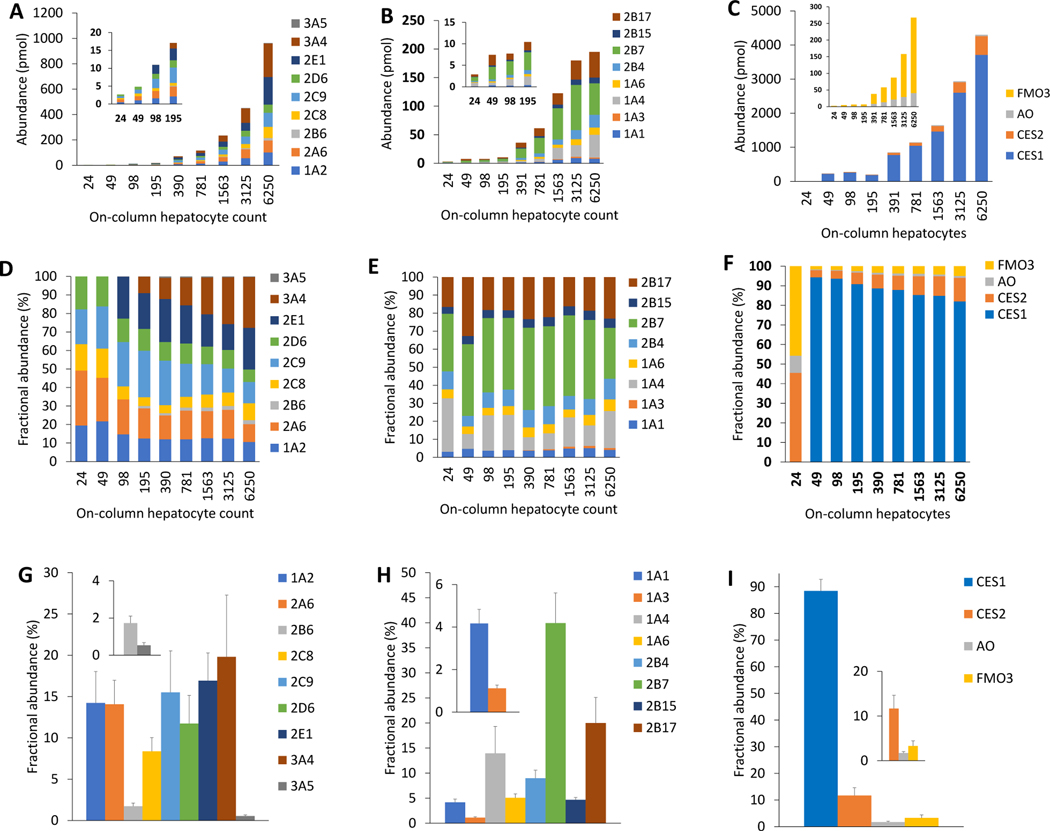
The sensitivity, linearity, and reproducibility of the μLC-MS/MS method to quantify DMETs in human hepatocytes were assessed by using different starting cell counts, i.e., 4000 to 1 million cells (24–6250 cells on-column; Figure 1). Unlike tissue protein extracts described above, the total protein concentration using BCA assay was not sensitive to the low volumes of hepatocyte samples. Therefore, the DMET abundance data for these samples were directly compared with the hepatocyte count. Out of 43 target proteins, 25 DMEs were detected and reproducibly quantified in the protein homogenate of 4000 hepatocytes (24 cell on-column; Table S7–S9 and Figure 3). The abundance of DMEs for which absolute peptide levels were quantified, was proportional to the cell count (R2 > 0.99; Figure 3) except for UGTs (R2 > 0.82). The sum of protein abundance data of UGTs was linear up to 1500 cell count on-column (Figure S4), but hepatocyte counts more than 1500 showed saturation of signal intensity likely due to inefficient protein digestion or plateauing of MS response (Figure S4). The following CYPs and UGTs were quantified in 24 hepatocytes on-column: CYP1A2, 2A6, 2C8, 2C9, and 2D6, and UGT1A1, 1A4, 1A6, 2B4, 2B7, 2B15, and 2B17 (Figure 3). The minimum on-column hepatocyte count needed for quantification of DPYD, ADH1C, CES1, PON1, PON2, PON3, GSTA1, GSTM4, SULT1A2, and SULT2B1 was: 98, 195, 390, 98, 98, 98, 49, 390, 98, and 49 cells, respectively (Table S8). MRP2 (ABCC2) and OATP2B1 (SLCO2B1) were detectable in the hepatocyte homogenate of 31,000 cells (195 cells on-column), whereas OATP1B3 (SLCO1B3) and OCT1 (SLC22A1) were detectable in the extract isolated from 62,000 cells (390 cells on-column). P-gp (ABCB1) was detectable in the extract from 125,000 hepatocytes (781 cells on-column) (Table S9). These data suggest that the studied DMETs can be quantified in 96- or 384-plate cell culture models or organ-on-a-chip model.
Figure 3:
Protein abundance (pmol) of CYPs (A) UGTs (B), and other non-CYPs (C) in 24–6250 cells human hepatocytes on-column. Cell-count normalized fractional abundance (%) abundance of CYPs (D), UGTs (E), and other non-CYPs (F) calculated by dividing the abundance value by the on-column hepatocyte cell count. Fractional abundance (%) of CYPs (G), UGTs (H), and other non-CYPs (I) reported as means and standard deviations of the cell-count normalized values. CYP3A4, CYP2E1, CYP2B6, CES1, and UGT1A3 were not detected in low cell count. The figure insets are zoomed data for low abundance proteins.
DMET quantification in small initial protein amount
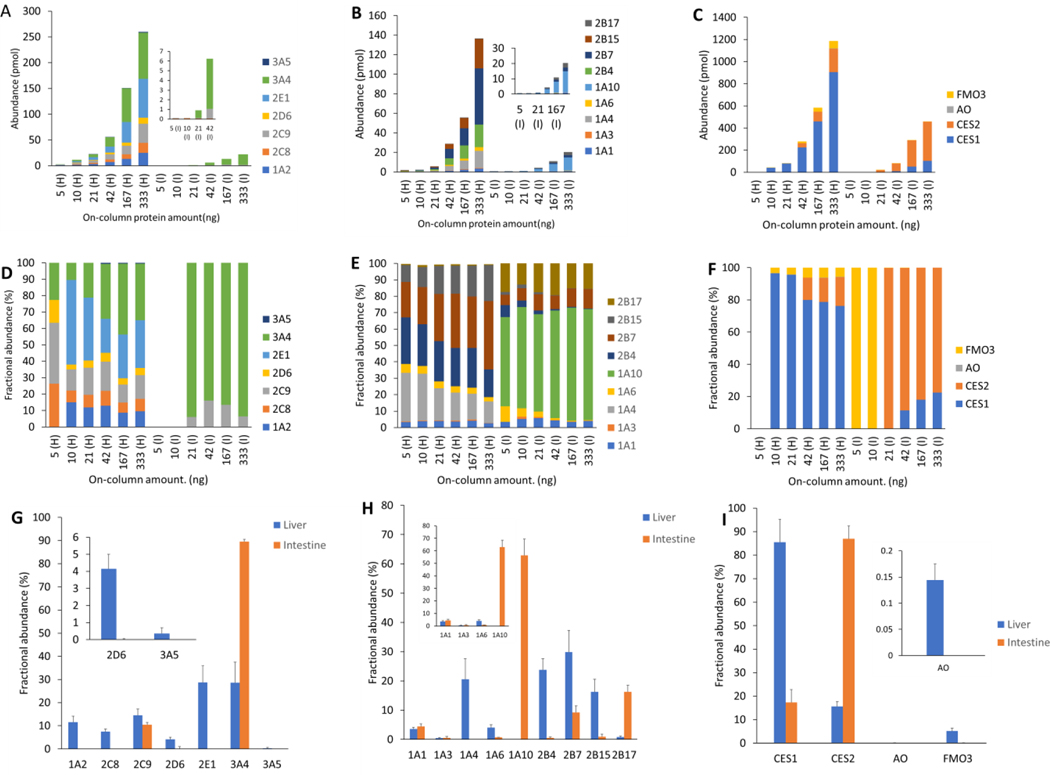
We further tested the sensitivity and reproducibility of the method to detect DMETs using a large range of initial protein concentrations of HLMs and HIMs (5 to 333 ng on-column). All major targeted DMEs were quantifiable in more than 10 ng on-column protein amount with a few of them reproducibly quantified in 5 ng on-column (Figure 4). Although drug transporters are expressed in the plasma membrane, we detected some of the transporters in the pooled microsomal samples with > 20 ng on-column protein amount. A total of 9 CYPs, 9 UGTs, 16 other non-CYPs, and 9 transporters, were detected in HLM (Table S10- S12). Similarly, although a limited number of proteins were detected in HIM as compared to HLM, DMETs were quantifiable in 83 ng on-column total HIM protein. A total of 5 CYPs, 7 UGTs, 10 other non-CYPs, and 4 transporters, were detected in HIM. UGT2B15 was only detected in HLM whereas UGT1A10 was exclusively detected in HIM. BCRP (ABCG2) was only detected in HIM in 5 ng on-column protein amount, whereas OATP1B1, 1B3, and 2B1 (SLCO1B1, 1B3, and 2B1) were only detected in HLM. Total CYPs or UGT abundance in HLM or HIM was proportional to the on-column microsomal protein amount, R2 >0.92 (Figure S5). The order of absolute abundance of CYPs and UGTs in HLM was consistent with the crude membrane data from liver tissues (Figure 2) discussed above. The relative quantification data for the DMETs for which the calibrator values were not available are shown in Supplementary Table S10–S12.
Figure 4:
Protein abundance in human liver (H) and intestine (I) microsomes. Protein abundance (pmol/mg) of CYPs (A) UGTs (B), and other non-CYPs (C) in different microsomal on-column protein amount ranging from 5–333 ng. Protein amount normalized fractional abundance (%) of CYPs (D), UGTs (E), and other non-CYPs (F). Fractional abundance (%) of CYPs (G), UGTs (H), and other non-CYPs (I) reported as means and standard deviations of the protein normalized abundance values. The protein amount normalized abundance was calculated by dividing the abundance value by the on-column microsomal protein amount. The figure insets are zoomed data for low abundance proteins.
DMET protein quantification in CHIM cells-based model
We have previously quantified UGTs and SULTs in CHIM samples using a conventional LC-MS/MS method.27 In addition to these proteins, our newer method also detected all major intestinal CYPs (05), ADHs (02), AADAC, and transporters (08) (Figure S2). The order of CYP and transporter abundance in the CHIM model was CYP3A4 > 2C9 > 1A2 > 2C18 > 2D6 and OATP2B1 (SLCO2B1) > MRP2 (ABCC2) > MRP3 (ABCC3) > P-gp (ABCB1) > BCRP (ABCG2), respectively. Similar to our previous study,27 we investigated the regional expression of DMETs along the small intestine using sucrase-isomaltase (SI) and villin-1 (VIL1) as membrane markers and fatty acid-binding protein (FABP2) as cytosolic markers of human intestinal villi (Figure S2). While the DMET protein abundance did not show any trend, the marker-normalized expression data suggest that the abundance of most of the DMETs decreases from the duodenum to the ileum (Figure S2).
Differential tissue abundance of DMEs
We have recently performed a comprehensive analysis of major non-CYP enzymes in human tissue S9 fractions using a conventional LC-MS/MS method.26 The pooled samples from this study were used to quantify DMETs using the current μLC-MS/MS method. A total of 53 DMETs were detected in the liver, intestine, kidney, lung, or heart S9 fraction (Figure S3). The liver showed the highest expression of 40 DMETs, whereas 11 and 2 DMETs were most abundant in the intestine and kidney, respectively. In particular, UGT2B17, CES2, GSTP1, SULT1A3, SULT1B1, and P-gp (ABCB1) were expressed with > 6-fold higher abundance in the intestine than the liver. UGT1A9 and MATE1 (SLC47A1) were >1.5 and 4-fold higher in the kidney than the liver.
DISCUSSION
We present here an optimized sample preparation approach involving acetone precipitation coupled with a sensitive μLC-MS/MS method for simultaneous quantification of DMETs in small starting sample volumes of human liver tissue (1–100 mg), hepatocytes (~4000 to 1 million cells), and microsomal concentrations (0.01–1 mg/ml). The method was validated for linearity, range, accuracy, precision, and LLOQ (Table S13). Our method allowed the quantification of 43 CYPs, UGTs, other non-CYP enzymes, and 13 transporters. The %CV across different tissue weights, cell counts, or microsomal protein concentrations was within 30%. The μLC-MS/MS method presented here is significantly sensitive as it allowed quantification DMETs in 4000 cells, 1 mg liver tissue, and 0.01 mg/ml microsomes as compared to the conventional LC-MS/MS method 26 that generally requires a minimum of 50,000 cells, 30 mg of liver tissue, and 1 mg/ml microsomes. The method was applied to characterize differential tissue abundance and regional abundance in human gut mucosa (CHIM). Quantitative data for some proteins, e.g., GSTs, AADAC, EPHXs, PONs, and DYDP, are reported for the first time.
The unparallel advantages of quantitative proteomics over immunoquantification (e.g., Western blotting) is now well recognized.11 Quantitative DMET proteomics has emerged as a key tool in translational pharmacology, which has shown wide applications in IVIVE, prediction of inter-individual variability (e.g., the effect of age, sex, and genotype), in vitro model characterization, inter-tissue and inter-species differences in DMET abundance, and the assessment of induction or suppression of DMETs.31 The paradigm shift in drug discovery strategies towards biologics is also supported by robust, selective, and high-throughput quantitative LC-MS technologies. Further, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) effort of the National Cancer Institute (NCI) has accelerated the understanding of the molecular basis of cancer through the application of quantitative proteomics. However, the sensitivity of quantitative proteomics to determine protein levels in small sample volumes or cell counts is often a critical challenge and continuous improvements in the methodology are warranted for reproducible protein quantification when the quantity of sample is limited.
The ability of quantitative proteomics to detect proteins in small sample quantities depends on the sensitivity of the LC-MS method, which is a result of a combination of factors such as surrogate peptide response, protein enrichment during sample preparation, inherent MS instrument sensitivity, and the mobile phase flow rate (nano, micro, or conventional flow). The intrinsic response of a surrogate peptide cannot be further improved if the ionization and collision energy are already optimized. Moreover, for DMETs, only 1–3 unique surrogate peptides are available for the quantification because of the membrane-bound nature of most of these proteins. In addition, these proteins are closely homologous with overlapping amino acid sequences. Therefore, improving sample preparation and decreasing LC-MS flow are the only available options for increasing MS sensitivity. Here, we optimized our sample preparation protocol with ice-cold acetone wash, which helped us to remove salts before the trypsin digestion. Further, we used small sample volume (1 μL) and lower mobile phase flow rate (3 μL/min) to reduce the matrix effect or noise. We present an optimized sample preparation coupled with a sensitive microflow-based LC-MS/MS method to enable DMET protein quantification in small sample volumes. Our method allowed reliable quantification of proteins in 1 mg liver tissue sample, 24 on-column hepatocytes, and 1 ng on-column microsomal protein, which can be applied for DMET protein quantification in biopsy samples, low cell counts (96–384 well plate cell extracts and organ on a chip), and liquid biopsy (exosomes) samples.
Interindividual variability in drug disposition is one of the major challenges that lead to variable pharmacokinetics and pharmacodynamics of drugs. Tissue abundance of DMETs when coupled with PBPK and PBPK-PD can estimate inter-individual variability in drug disposition.7 However, due to the need for large tissue quantity (~100 mg) in the conventional quantitative proteomics experiment, these experiments are only possible in the case of the post-mortem tissue samples except in the case when the tissue is utilized for the transplant purpose. The post-mortem tissue samples are associated with several limitations, including contamination of pathological or scarred tissue, variable protein degradation due to inconsistent sample collection protocol, and the potential effect of medications. Moreover, the availability of tissue samples from special populations such as neonates, pregnant women, and disease conditions is a critical challenge. The conventional tissue sample archiving also requires greater freezer space. These problems can be resolved by the use of tissue biopsy from non-pathological tissues that can be collected on a larger scale from a patient undergoing surgeries. The quantitative proteomics method developed in this study can be applied to tissue biopsy samples to support PBPK modeling for stratifying patients undergoing surgeries based on their potential to clear drugs. In particular, such an approach can be used in precision dosing of the narrow therapeutic index drugs, e.g., antineoplastic agents. The sensitive μLC-MS/MS based quantitative proteomics analysis of tumor biopsies can determine disease progression, heterogenicity in drug resistance, and interindividual variability in efficacy or toxicity.32 Recently, research on liquid biopsy (e.g., exosomes) has shown promises for precision medicine due to its non-invasive applications in real-time disease progression, drug efficacy and toxicity monitoring, and drug-drug interactions.33–36 The limitation of the poor signal in the conventional quantitative proteomics due to the low circulating concentration of these extracellular vesicles in biofluids can be addressed by our μLC-MS/MS method.
Rapid advances in in vitro absorption, distribution, metabolism, and transport (ADMET) technologies including advanced 2D and 3D models to screen drug molecules for their metabolism and toxicity are allowing speedy drug development. In vitro systems offer a myriad of advantages including high throughputness and cheaper alternate to animal models to select the best candidates for the clinic. The μLC-MS/MS method can quantify DMETs in 24 cells on-column and thus can characterize these advanced models. For example, using our method, one vial of human hepatocytes (equivalent to 5 million cells) can generate 1000–1300 wells of reactions for CYP induction assay, that can screen hundreds of compounds. The advanced 3D models or organ-on-a-chip models recapitulate in vivo human biology and microenvironment for reliable IVIVE.37,38 Suh models bring a revolution in the in vitro models due to recent advancements in the micro physiological field and microfluidics science39 and these models exhibit gene expression patterns, nutrient accesses, and microenvironment closer to in vivo system.40 The protein characterization in these models is often not possible due to the low initial cell count (5,000–20,000 cells), which can be resolved through the sample preparation and μLC-MS/MS method presented here.
In the last decade, DMET protein quantification has been reported by multiple laboratories in different human tissue fractions such as HLM, HIM, and hepatocytes. The quantitative DMET data presented here using a small sample volume is in agreement with the published data (Table S14).41–43 However, while the relative abundance of DMETs in our study was consistent across different tissue weights or sample amounts, some differences in the protein abundance were observed perhaps due to the non-homogenous expression in the liver. In that case, smaller sample sizes are prone to more variability and it is important that sample is taken from multiple tissue sites to address zonal variability.
Inter-laboratory variability in DMET protein abundance data has been a cause of concern for the reliable use of these data in PBPK modeling.44 This variability is generally related to sample preparation of variable enrichment during subcellular fractionation (HLM or membrane preparation) requires scaling factors needed for reliable IVIVE. For example, microsomal protein per gram of liver (MPPGL) is used for scaling microsomal data, however, MPPGL is highly variable 10–150 with a mean of around 40 mg.44 Therefore, DMET protein quantification in tissue homogenate has been recommended as a plausible solution to this problem, but tissue lysate generally shows a poor signal in comparison to the enriched fractions. Since a high mobile phase flow rate is often associated with greater noise and ion suppression, the μLC-MS/MS method provides a potential solution to quantify DMETs directly in tissue homogenate, cell homogenate, or crude membranes. Further, improvements in the analytical micro-sampling (<100 μL) techniques, e.g., dry blood spot (DBS), and micro-dialysis for cerebrospinal fluid (CSF) sampling, require sensitive μLC-MS/MS method for quantification.
There are some limitations of this study. We only quantified clinically relevant DMETs and the major focus of our study was on DMEs. We were unable to quantify a few targeted proteins (e.g., UGT1A9) due to technical problems in SIL peptide quantification. Absolute peptide abundance data is not reported here for target proteins due to the non-availability of standard peptides. Finally, the scope of this manuscript does not include functional activity analysis. Nevertheless, we tested here for the first time the ability of μLC-MS/MS to quantify DMETs in a small sample volume.
In summary, the proteomics method presented here can be applied for high-throughput and sensitive analysis of DMETs in small volumes of tissue or homogenate and enriched fractions. The approach has the ability to validate gene expression and activity data in these systems. The differential tissue abundance data for multiple proteins presented here will be important for PBPK modeling.
Supplementary Material
Acknowledgement
Authors would like to thank BioIVT Inc (Baltimore, MD) and IVAL (Columbia, MD) for providing human hepatocytes and intestinal mucosa (CHIM) used in this study.
Funding information
This work was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH grant (R01.HD081299) and the Department of Pharmaceutical Sciences, Washington State University, Spokane, WA.
Abbreviations:
- DMETs
drug-metabolizing enzymes and transporters
- DMEs
drug-metabolizing enzymes
- IVIVE
In vitro to in vivo extrapolation
- PBPK
physiologically-based pharmacokinetic
- μLC-MS/MS
microflow-based liquid chromatography-tandem mass spectrometry
Footnotes
Conflict of interest
Authors declare no conflict of interest
REFERENCES
- 1.Li AP 2020. In Vitro Human Cell–Based Experimental Models for the Evaluation of Enteric Metabolism and Drug Interaction Potential of Drugs and Natural Products. Drug Metabolism and Disposition 48(10):980–992. [DOI] [PubMed] [Google Scholar]
- 2.Venkatakrishnan K, von Moltke LL, Greenblatt DJ 2001. Human drug metabolism and the cytochromes P450: application and relevance of in vitro models. The Journal of Clinical Pharmacology 41(11):1149–1179. [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Lechon MJ, Castell JV, Donato MT 2007. Hepatocytes—the choice to investigate drug metabolism and toxicity in man: in vitro variability as a reflection of in vivo. Chemico-biological interactions 168(1):30–50. [DOI] [PubMed] [Google Scholar]
- 4.Ahire D, Sinha S, Brock B, Iyer R, Mandlekar S, Subramanian M 2017. Metabolite identification, reaction phenotyping, and retrospective drug-drug interaction predictions of 17-deacetylnorgestimate, the active component of the oral contraceptive norgestimate. Drug Metabolism and Disposition 45(6):676–685. [DOI] [PubMed] [Google Scholar]
- 5.Sharma S, Ahire D, Prasad B 2020. Utility of Quantitative Proteomics for Enhancing the Predictive Ability of Physiologically Based Pharmacokinetic Models Across Disease States. The Journal of Clinical Pharmacology 60:S17–S35. [DOI] [PubMed] [Google Scholar]
- 6.Bhatt DK, Prasad B 2018. Critical issues and optimized practices in quantification of protein abundance level to determine interindividual variability in DMET proteins by LC-MS/MS proteomics. Clinical Pharmacology & Therapeutics 103(4):619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatt DK, Basit A, Zhang H, Gaedigk A, Lee S-b, Claw KG, Mehrotra A, Chaudhry AS, Pearce RE, Gaedigk R 2018. Hepatic abundance and activity of androgen-and drug-metabolizing enzyme UGT2B17 are associated with genotype, age, and sex. Drug Metabolism and Disposition 46(6):888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aebersold R, Burlingame AL, Bradshaw RA. 2013. Western blots versus selected reaction monitoring assays: time to turn the tables?, ed.: ASBMB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sechi S, Sechi S. 2007. Quantitative proteomics by mass spectrometry. ed.: Springer. [Google Scholar]
- 10.Prasad B, Vrana M, Mehrotra A, Johnson K, Bhatt DK 2017. The promises of quantitative proteomics in precision medicine. Journal of pharmaceutical sciences 106(3):738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad B, Achour B, Artursson P, Hop CE, Lai Y, Smith PC, Barber J, Wisniewski JR, Spellman D, Uchida Y 2019. Toward a consensus on applying quantitative liquid chromatography-tandem mass spectrometry proteomics in translational pharmacology research: A white paper. Clinical Pharmacology & Therapeutics 106(3):525–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couto N, Al-Majdoub ZM, Gibson S, Davies PJ, Achour B, Harwood MD, Carlson G, Barber J, Rostami-Hodjegan A, Warhurst G 2020. Quantitative proteomics of clinically relevant drug-metabolizing enzymes and drug transporters and their intercorrelations in the human small intestine. Drug Metabolism and Disposition 48(4):245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad B, Bhatt DK, Johnson K, Chapa R, Chu X, Salphati L, Xiao G, Lee C, Hop CE, Mathias A 2018. Abundance of phase 1 and 2 drug-metabolizing enzymes in alcoholic and hepatitis C cirrhotic livers: a quantitative targeted proteomics study. Drug Metabolism and Disposition 46(7):943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Feteisi H, Achour B, Rostami-Hodjegan A, Barber J 2015. Translational value of liquid chromatography coupled with tandem mass spectrometry-based quantitative proteomics for in vitro–in vivo extrapolation of drug metabolism and transport and considerations in selecting appropriate techniques. Expert Opinion on Drug Metabolism & Toxicology 11(9):1357–1369. [DOI] [PubMed] [Google Scholar]
- 15.Basit A, Radi Z, Vaidya VS, Karasu M, Prasad B 2019. Kidney cortical transporter expression across species using quantitative proteomics. Drug Metabolism and Disposition 47(8):802–808. [DOI] [PubMed] [Google Scholar]
- 16.Billington S, Salphati L, Hop CE, Chu X, Evers R, Burdette D, Rowbottom C, Lai Y, Xiao G, Humphreys WG 2019. Interindividual and regional variability in drug transporter abundance at the human blood–brain barrier measured by quantitative targeted proteomics. Clinical Pharmacology & Therapeutics 106(1):228–237. [DOI] [PubMed] [Google Scholar]
- 17.Al Feteisi H, Achour B, Barber J, Rostami-Hodjegan A 2015. Choice of LC-MS methods for the absolute quantification of drug-metabolizing enzymes and transporters in human tissue: a comparative cost analysis. The AAPS journal 17(2):438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li CY, Hosey-Cojocari C, Basit A, Unadkat JD, Leeder JS, Prasad B 2019. Optimized renal transporter quantification by using aquaporin 1 and aquaporin 2 as anatomical markers: application in characterizing the ontogeny of renal transporters and its correlation with hepatic transporters in paired human samples. The AAPS journal 21(5):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladumor MK, Bhatt DK, Gaedigk A, Sharma S, Thakur A, Pearce RE, Leeder JS, Bolger MB, Singh S, Prasad B 2019. Ontogeny of Hepatic Sulfotransferases and Prediction of Age-Dependent Fractional Contribution of Sulfation in Acetaminophen Metabolism. Drug Metabolism and Disposition 47(8):818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu M, Saxena N, Vrana M, Zhang H, Kumar V, Billington S, Khojasteh C, Heyward S, Unadkat JD, Prasad B 2018. Targeted LC-MS/MS proteomics-based strategy to characterize in vitro models used in drug metabolism and transport studies. Analytical chemistry 90(20):11873–11882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar V, Yin J, Billington S, Prasad B, Brown CD, Wang J, Unadkat JD 2018. The importance of incorporating OCT2 plasma membrane expression and membrane potential in IVIVE of metformin renal secretory clearance. Drug Metabolism and Disposition 46(10):1441–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo T, Kouvonen P, Koh CC, Gillet LC, Wolski WE, Röst HL, Rosenberger G, Collins BC, Blum LC, Gillessen S 2015. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nature medicine 21(4):407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW 2004. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). Journal of proteome research 3(2):235–244. [DOI] [PubMed] [Google Scholar]
- 24.Razavi M, Anderson NL, Pope ME, Yip R, Pearson TW 2016. High precision quantification of human plasma proteins using the automated SISCAPA Immuno-MS workflow. New biotechnology 33(5):494–502. [DOI] [PubMed] [Google Scholar]
- 25.Harwood MD, Achour B, Neuhoff S, Russell MR, Carlson G, Warhurst G, Rostami-Hodjegan A 2016. In vitro–in vivo extrapolation scaling factors for intestinal P-glycoprotein and breast cancer resistance protein: part I: a cross-laboratory comparison of transporter-protein abundances and relative expression factors in human intestine and Caco-2 cells. Drug Metabolism and Disposition 44(3):297–307. [DOI] [PubMed] [Google Scholar]
- 26.Basit A, Neradugomma NK, Wolford C, Fan PW, Murray B, Takahashi RH, Khojasteh SC, Smith BJ, Heyward S, Totah RA 2020. Characterization of Differential Tissue Abundance of Major Non-CYP Enzymes in Human. Molecular Pharmaceutics 17(11):4114–4124. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Wolford C, Basit A, Li AP, Fan PW, Murray BP, Takahashi RH, Khojasteh SC, Smith BJ, Thummel KE 2020. Regional proteomic quantification of clinically relevant non-cytochrome P450 enzymes along the human small intestine. Drug Metabolism and Disposition 48(7):528–536. [DOI] [PubMed] [Google Scholar]
- 28.Prasad B 2019. Quality-by-design (QBD) approach to address rigor and reproducibility in dmet quantitative proteomics. Drug Metabolism and Pharmacokinetics 34(1):S70. [Google Scholar]
- 29.Gröer C, Busch D, Patrzyk M, Beyer K, Busemann A, Heidecke C, Drozdzik M, Siegmund W, Oswald S 2014. Absolute protein quantification of clinically relevant cytochrome P450 enzymes and UDP-glucuronosyltransferases by mass spectrometry-based targeted proteomics. Journal of pharmaceutical and biomedical analysis 100:393–401. [DOI] [PubMed] [Google Scholar]
- 30.Sato Y, Nagata M, Tetsuka K, Tamura K, Miyashita A, Kawamura A, Usui T 2014. Optimized methods for targeted peptide-based quantification of human uridine 5′-diphosphate-glucuronosyltransferases in biological specimens using liquid chromatography–tandem mass spectrometry. Drug metabolism and disposition 42(5):885–889. [DOI] [PubMed] [Google Scholar]
- 31.Bhatt DK, Mehrotra A, Gaedigk A, Chapa R, Basit A, Zhang H, Choudhari P, Boberg M, Pearce RE, Gaedigk R 2019. Age-and genotype-dependent variability in the protein abundance and activity of six major uridine diphosphate-glucuronosyltransferases in human liver. Clinical Pharmacology & Therapeutics 105(1):131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres R, Vesuna S, Levene MJ 2014. High-resolution, 2-and 3-dimensional imaging of uncut, unembedded tissue biopsy samples. Archives of Pathology and Laboratory Medicine 138(3):395–402. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, Shao Y, Zheng S 2020. Application of exosomes as liquid biopsy in clinical diagnosis. Signal transduction and targeted therapy 5(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi G, Ignatiadis M 2019. Promises and pitfalls of using liquid biopsy for precision medicine. Cancer research 79(11):2798–2804. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigues D, Rowland A 2019. From Endogenous Compounds as Biomarkers to Plasma-Derived Nanovesicles as Liquid Biopsy; Has the Golden Age of Translational Pharmacokinetics-Absorption, Distribution, Metabolism, Excretion-Drug–Drug Interaction Science Finally Arrived? Clinical Pharmacology & Therapeutics 105(6):1407–1420. [DOI] [PubMed] [Google Scholar]
- 36.Achour B, Al-Majdoub ZM, Grybos-Gajniak A, Lea K, Kilford P, Zhang M, Knight D, Barber J, Schageman J, Rostami-Hodjegan A 2020. Liquid Biopsy Enables Quantification of the Abundance and Interindividual Variability of Hepatic Enzymes and Transporters. Clinical Pharmacology & Therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langhans SA 2018. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Frontiers in pharmacology 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaroch K, Jaroch A, Bojko B 2018. Cell cultures in drug discovery and development: The need of reliable in vitro-in vivo extrapolation for pharmacodynamics and pharmacokinetics assessment. Journal of Pharmaceutical and Biomedical Analysis 147:297–312. [DOI] [PubMed] [Google Scholar]
- 39.Amin R, Knowlton S, Hart A, Yenilmez B, Ghaderinezhad F, Katebifar S, Messina M, Khademhosseini A, Tasoglu S 2016. 3D-printed microfluidic devices. Biofabrication 8(2):022001. [DOI] [PubMed] [Google Scholar]
- 40.Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z 2017. Modeling physiological events in 2D vs. 3D cell culture. Physiology 32(4):266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 41.Schaefer O, Ohtsuki S, Kawakami H, Inoue T, Liehner S, Saito A, Sakamoto A, Ishiguro N, Matsumaru T, Terasaki T 2012. Absolute quantification and differential expression of drug transporters, cytochrome P450 enzymes, and UDP-glucuronosyltransferases in cultured primary human hepatocytes. Drug Metabolism and Disposition 40(1):93–103. [DOI] [PubMed] [Google Scholar]
- 42.Kawakami H, Ohtsuki S, Kamiie J, Suzuki T, Abe T, Terasaki T 2011. Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. Journal of pharmaceutical sciences 100(1):341–352. [DOI] [PubMed] [Google Scholar]
- 43.Drozdzik M, Busch D, Lapczuk J, Müller J, Ostrowski M, Kurzawski M, Oswald S 2018. Protein abundance of clinically relevant drug-metabolizing enzymes in the human liver and intestine: a comparative analysis in paired tissue specimens. Clinical Pharmacology & Therapeutics 104(3):515–524. [DOI] [PubMed] [Google Scholar]
- 44.Wilson Z, Rostami-Hodjegan A, Burn J, Tooley A, Boyle J, Ellis S, Tucker G 2003. Inter-individual variability in levels of human microsomal protein and hepatocellularity per gram of liver. British journal of clinical pharmacology 56(4):433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.