Abstract
Both intrinsic and acquired resistance to amphotericin B have been documented for Candida lusitaniae. Amphotericin B remains the drug of choice for many critical fungal infections, and the detection of resistance is essential to monitor treatment effectively. The limitations of the National Committee for Clinical Laboratory Standards (NCCLS) reference methodology for detection of amphotericin B resistance are well documented, and several alternative methods have been proposed. Etest assays with RPMI and antibiotic medium 3 (AM3) agar were compared to the NCCLS M27-A broth macrodilution method using AM3 for amphotericin B resistance testing with 49 clinical isolates of C. lusitaniae. The panel included nine isolates with known or presumed resistance to amphotericin B on the basis of in vivo and/or in vitro data. The distribution of amphotericin B MICs by Etest with RPMI ranged from 0.032 to 16 μg/ml and was bimodal. All of the putatively resistant isolates were inhibited by amphotericin B at ≥0.38 μg/ml and could be categorized as resistant using this breakpoint. Etest with AM3 yielded a broader amphotericin B MIC range (0.047 to 32 μg/ml), and there were six putatively resistant isolates for which MICs were >1 μg/ml. The separation of putatively susceptible and resistant isolates was less obvious. Broth macrodilution with AM3 generated a unimodal distribution of MICs (ranging from 0.032 to 2 μg/ml) and failed to discriminate most of the putatively resistant isolates at both 24 and 48 h. Etest using RPMI and, to a lesser extent, using AM3 provided better discrimination between amphotericin B-resistant and -susceptible isolates of C. lusitaniae.
Amphotericin B is the drug of choice for many systemic fungal infections (7). Although rare, treatment failures associated with resistance to amphotericin B or resistance to amphotericin B and azoles in both immunocompromised and immunocompetent patients have been documented (3, 13, 18). Among the non-Candida albicans species, Candida lusitaniae is of special interest, owing to its uncommon susceptibility pattern (1, 5, 9). Rapidly acquired resistance to amphotericin B has been described or suspected, and some strains of C. lusitaniae may be intrinsically resistant (6, 15). The detection of resistance to amphotericin B is essential for treatment of C. lusitaniae-associated infections. In the past 10 years, there have been major advances in antifungal susceptibility testing, as illustrated by several methodology documents published by the National Committee for Clinical Laboratory Standards (NCCLS) (11). Although standardized NCCLS methods and MIC interpretive breakpoints are now available for azole and flucytosine susceptibility testing of yeasts, amphotericin B testing is still under investigation (4, 11). The methodology initially proposed by the NCCLS, i.e., a broth macrodilution procedure using RPMI 1640 medium, did not consistently detect amphotericin B-resistant isolates, nor does the microdilution format (17). Proposed alternative media and methods include the use of antibiotic medium 3 (AM3) with the NCCLS method and/or the Etest predefined gradient method to better discriminate between amphotericin B-susceptible and -resistant Candida isolates (16, 17, 19).
This study assessed the performance of Etest using both RPMI and AM3 agars for amphotericin B susceptibility testing of C. lusitaniae. Etest results were compared to results of the NCCLS broth macrodilution method using AM3 broth. The challenge panel of strains tested included nine strains known or presumed to be amphotericin B resistant on the basis of in vivo and/or in vitro data.
The collection of 47 clinical isolates used included 38 colonizing isolates recovered from a variety of specimens from 37 patients, immunocompromised and immunocompetent, who had not received antifungal prophylaxis or treatment. These strains were considered putatively susceptible to amphotericin B. Nine isolates were used to represent amphotericin B-resistant strains. Three of them were kindly provided by J. Rex (University of Texas Medical School, Houston). One isolate (2887) was proven resistant in an animal model, and the other two isolates (Y533 and Y534) were presumed resistant on the basis of in vivo and in vitro data (17). The other six were selected because the associated clinical data and results obtained by multiple in vitro testing suggested resistance. They were obtained from stool (6856-1 and 6856-2), blood (6-103), and urine (2-367, 679, and 787). All but one (2-367) were recovered from patients during amphotericin B prophylaxis or treatment. All isolates were identified to the species level by the ID 32C biochemical system (bioMérieux, Marcy l'Etoile, France). C. lusitaniae CBS 6936 was the amphotericin B-susceptible reference strain, while Candida tropicalis IP 1275-81 and C. albicans ATCC 38248 were used as amphotericin B-resistant reference strains. Candida parapsilosis ATCC 22019 was included in each test run to ensure quality control compliance. Strains were maintained on Sabouraud dextrose agar slants and stored at 4°C. Strains were subcultured twice on Sabouraud dextrose agar before being tested. The broth macrodilution procedure was done according to NCCLS M27-A guidelines using AM3 broth (Difco Laboratories, Detroit, Mich.) supplemented with 2% glucose (Difco) (11). MICs were read at 24 and 48 h. The MIC was defined as the lowest drug concentration preventing any visible growth. Etest (AB BIODISK, Solna, Sweden) was used according to the manufacturer's recommendations (Etest technical guide number 4. Antifungal susceptibility of yeasts. AB BIODISK, Solna, Sweden.) Two medium formulations supplemented with 2% glucose (Difco) and 1.5% agar (Difco) were used: RPMI 1640 (American Bioorganics, Buffalo, N.Y.) and AM3 (Difco). The inoculum suspensions were adjusted spectrophotometrically at 530 nm to match the turbidity of a 0.5 McFarland standard. Agar plates were inoculated with a cotton swab, and they were allowed to dry for at least 15 min before the Etest strips were applied. Etest agar plates were incubated at 35°C and read at 48 h. The lowest drug concentration on the Etest strip inhibiting 100% growth was defined as the MIC. A MIC breakpoint of resistance of ≥0.38 μg/ml was chosen for Etest with RPMI (2). The reproducibilities of the Etest using RPMI and AM3 were determined by testing C. lusitaniae 2887, 6856-1, and 6856-2 in 10 experiments.
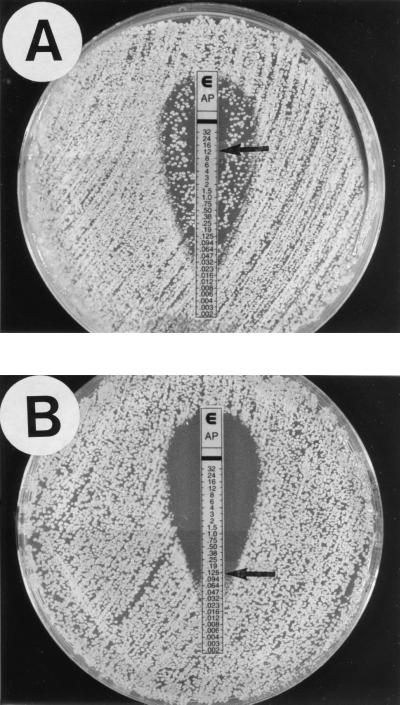
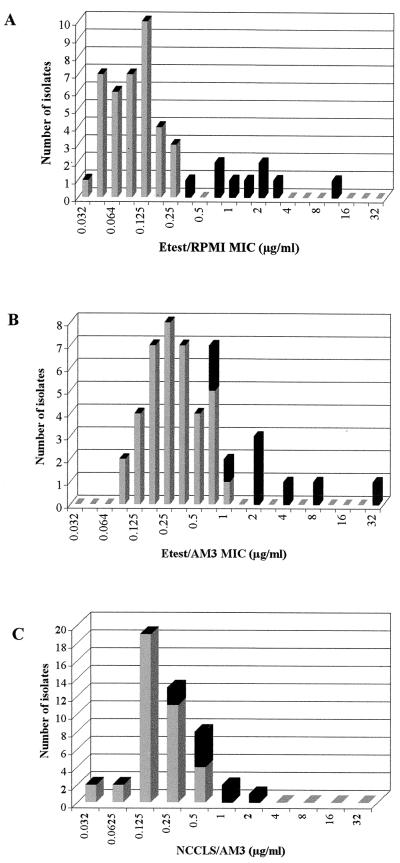
With Etest, a 48-h reading made it easy to select MICs and to visualize macro- and microcolonies sometimes present within the inhibition ellipse and around the endpoint, respectively (Fig. 1). MIC ranges obtained with Etest using RPMI or AM3 agar were broader than those of the reference method (Fig. 2). With Etest using RPMI, amphotericin B MIC ranges for the putatively susceptible and resistant isolates did not overlap (Fig. 2A). All putatively resistant isolates were inhibited by drug concentrations of ≥0.38 μg/ml and were easily categorized as resistant to amphotericin B. AM3 agar used in the Etest generated an upward shift of MICs by 1 to 2 dilutions, and the amphotericin B MICs for six susceptible isolates were similar to those of three resistant isolates (0.75 to 1 μg/ml) (Fig. 2B). The NCCLS method generated a unimodal distribution of MICs, with a 2-dilution overlap for putatively susceptible and resistant isolates (Fig. 2C). A similar unimodal distribution, but with a 1-dilution overlap, was observed after 24 h of incubation (data not shown). Table 1 outlines results from two experiments for the set of amphotericin B-resistant isolates and reference strains. Whatever the test method, MICs for the two experiments were within 1 twofold dilution. For the four reference strains, resistance or susceptibility to amphotericin B was detected by all three methods. For the two resistant reference strains, the same high amphotericin B MICs (≥16 μg/ml) were found with Etest on both media. Among clinical isolates, only for C. lusitaniae 6856-2 were high amphotericin B MICs found with Etest on both test media. Regardless of the test method, results for C. lusitaniae Y534 were at the lower end of the MIC range for the set of amphotericin B-resistant isolates. Percentage agreements between Etest and the NCCLS method for susceptible and resistant isolates are shown in Table 2. For the susceptible isolates, the Etest results correlated well with the reference MICs, whatever the test medium. For the resistant isolates, the low agreements were due to low MICs by the NCCLS method and high MICs by Etest. The reproducibilities of Etest using RPMI and AM3 were excellent, with 100% of MICs within ±1 dilution for the three isolates tested (data not shown).
FIG. 1.
Etest amphotericin B reading pattern on RPMI for C. lusitaniae. (A) Macrocolonies inside the inhibition ellipse (MIC = 12 μg/ml); (B) microcolonies at the endpoint (MIC = 0.125 μg/ml).
FIG. 2.
Distribution of amphotericin B MICs obtained by Etest with RPMI (A), by Etest with AM3 (B), and by the broth macrodilution method with AM3 (C) for 47 isolates of C. lusitaniae. ■, resistant isolates;  , susceptible isolates.
, susceptible isolates.
TABLE 1.
Ranges of MICs of amphotericin B by Etest with RPMI and AM3 and by the NCCLS broth macrodilution method from two separate experiments
| Isolate | Putative statusa | Amphotericin B MIC (μg/ml)
|
||
|---|---|---|---|---|
| Etest using:
|
NCCLS broth macrodilution using AM3 | |||
| RPMI | AM3 | |||
| C. lusitaniae 2887 | Resistant | 1.5–2 | 4–12 | 0.5–1 |
| C. lusitaniae 2-367 | Resistant | 0.5–0.75 | 2–4 | 0.5–0.5 |
| C. lusitaniae 6-103 | Resistant | 0.75–1 | 8–12 | 0.5–1 |
| C. lusitaniae 6856-1 | Resistant | 0.38–0.38 | 0.5–0.75 | 0.25–0.5 |
| C. lusitaniae 6856-2 | Resistant | 8–12 | >32–>32 | 1–2 |
| C. lusitaniae Y533 | Resistant | 0.38–1.5 | 1.5–2 | 0.5–0.5 |
| C. lusitaniae Y534 | Resistant | 0.25–0.75 | 0.75–0.75 | 0.25–0.25 |
| C. lusitaniae 679 | Resistant | 1–2 | 1–2 | 0.25–0.5 |
| C. lusitaniae 787 | Resistant | 1–3 | 2–4 | 0.5–0.5 |
| C. tropicalis IP 1275-81 | Resistant | 16–16 | 16–16 | 1–2 |
| C. albicans ATCC 38248 | Resistant | 12–16 | >32–>32 | 1–1 |
| C. lusitaniae CBS 6936 | Susceptible | 0.125–0.125 | 0.094–0.125 | 0.125–0.125 |
| C. parapsilosis ATCC 22019 | Susceptible | 0.38–0.25 | 0.125–0.125 | 0.25–0.25 |
Putative susceptibility status was assigned on the basis of isolate source, clinical data, and any known in vivo or in vitro studies of that isolate.
TABLE 2.
Agreement between amphotericin B MICs for 47 C. lusitaniae isolates by Etest and NCCLS AM3 broth macrodilution
| Test method | % Agreement with NCCLS AM3 broth macrodilution
|
|||
|---|---|---|---|---|
| Susceptible isolates (n = 38)
|
Resistant isolates (n = 9)
|
|||
| ±1 dilution | ±2 dilutions | ±1 dilution | ±2 dilutions | |
| Etest with AM3 | 65.8 | 94.7 | 11.1 | 66.7 |
| Etest with RPMI | 81.6 | 100 | 66.7 | 88.9 |
More than ever, the need for a reliable and clinically relevant method for amphotericin B susceptibility testing of yeasts is important. Previous studies have demonstrated that replacement of RPMI with AM3 broth for the NCCLS method and the Etest provided more clinically significant amphotericin B results than did the standard reference methodology (8, 16, 17, 19). This study provides additional evidence of the superiority of Etest over the NCCLS broth dilution method, even with AM3, for detection of resistance to amphotericin B among isolates of C. lusitaniae (16, 19). Our results, however, are slightly different from those of others who have shown that Etest performed similarly with either RPMI medium or AM3 (16, 19). In our study, RPMI agar provided the best discrimination between putatively susceptible and resistant isolates of C. lusitaniae. All but one of the putatively resistant isolates were consistently categorized as amphotericin B resistant, with the interpretive breakpoint MIC of ≥0.38 μg/ml recently proposed by Clancy and Nguyen (2) on the basis of MIC distribution stratified by response to therapy. With Etest using AM3, MICs of amphotericin B for resistant isolates were high but the separation of susceptible and resistant strains was incomplete. One possibility is that some susceptible C. lusitaniae isolates have intrinsically reduced susceptibility to amphotericin B. The NCCLS reference method with AM3 broth failed to discriminate susceptible and resistant isolates at either 24 or 48 h. The few studies that have evaluated the use of AM3 with the NCCLS method have provided contradictory results, especially about whether the MIC should be read after 24 or 48 h to better predict microbiological failure (8, 10, 12, 17). Since AM3 is not a standardized medium, the lot-to-lot and brand-to-brand variability alone can give discrepant results (10, 11, 12).
In conclusion, this study confirms the findings of Pfaller et al. (16), who recommended the use of Etest with standardized RPMI medium supplemented with 2% glucose as the most sensitive and reliable means for detecting amphotericin B resistance. In spite of the drawbacks of AM3, the elevated Etest amphotericin B MICs for some susceptible C. lusitaniae isolates with AM3 should be further investigated by additional techniques to determine if the isolates have low-level intrinsic resistance or heteroresistance (14).
Acknowledgments
We thank AB BIODISK for providing the Etest strips and the RPMI 1640 agar plates.
REFERENCES
- 1.Alexander B D, Perfect J R. Antifungal resistance trends towards the year 2000. Drugs. 1997;54:667–678. doi: 10.2165/00003495-199754050-00002. [DOI] [PubMed] [Google Scholar]
- 2.Clancy C J, Nguyen M H. Correlation between in vitro susceptibility determined by E test and response to therapy with amphotericin B: results from a multicenter prospective study of candidemia. Antimicrob Agents Chemother. 1999;43:1289–1290. doi: 10.1128/aac.43.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conly J, Rennie R, Johnson J, Farah S, Hellman L. Disseminated candidiasis due to amphotericin B-resistant Candida albicans. J Infect Dis. 1992;165:761–764. doi: 10.1093/infdis/165.4.761. [DOI] [PubMed] [Google Scholar]
- 4.Espinel-Ingroff A, Barchiesi F, Hazen K C, Martinez-Suarez J V, Scalise G. Standardization of antifungal susceptibility testing and clinical relevance. Med Mycol. 1998;36:68–78. [PubMed] [Google Scholar]
- 5.Favel A, Michel-Nguyen A, Chastin C, Trousson F, Penaud A, Regli P. In-vitro susceptibility patterns of Candida lusitaniae isolates and evaluation of the Etest method. J Antimicrob Chemother. 1997;39:591–596. doi: 10.1093/jac/39.5.591. [DOI] [PubMed] [Google Scholar]
- 6.Guinet R, Chanas J, Goullier A, Bonnefoy G, Ambroise-Thomas P. Fatal septicemia due to amphotericin B-resistant Candida lusitaniae. J Clin Microbiol. 1983;18:443–444. doi: 10.1128/jcm.18.2.443-444.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauffman C A, Carver P L. Antifungal agents in the 1990s. Current status and future developments. Drugs. 1997;53:539–549. doi: 10.2165/00003495-199753040-00001. [DOI] [PubMed] [Google Scholar]
- 8.Law D, Moore C B, Denning D W. Amphotericin B resistance testing of Candida spp.: a comparison of methods. J Antimicrob Chemother. 1997;40:109–112. doi: 10.1093/jac/40.1.109. [DOI] [PubMed] [Google Scholar]
- 9.Lewis R E, Klepser M E. The changing face of nosocomial candidemia: epidemiology, resistance, and drug therapy. Am J Health-Syst Pharm. 1999;56:525–533. [PubMed] [Google Scholar]
- 10.Lozano-Chiu M, Nelson P W, Lancaster M, Pfaller M A, Rex J H. Lot-to-lot variability of antibiotic medium 3 used for testing susceptibility of Candida isolates to amphotericin B. J Clin Microbiol. 1997;35:270–272. doi: 10.1128/jcm.35.1.270-272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, NCCLS document M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 12.Nguyen M H, Clancy C J, Yu V L, Yu Y C, Morris A J, Snydman D R, Sutton D A, Rinaldi M G. Do in vitro susceptibility data predict the microbiologic response to amphotericin B? Results of a prospective study of patients with Candida fungemia. J Infect Dis. 1998;177:425–430. doi: 10.1086/514193. [DOI] [PubMed] [Google Scholar]
- 13.Nolte F S, Parkinson T, Falconer D J, Dix S, Williams J C, Gilmore, Geller R, Wingard J R. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob Agents Chemother. 1997;41:196–199. doi: 10.1128/aac.41.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peyron F, Favel A, Guiraud-Dauriac H, El Mzibri M, Chastin C, Duménil G, Regli P. Evaluation of a flow cytofluorometric method for rapid determination of amphotericin B susceptibility of yeast isolates. Antimicrob Agents Chemother. 1997;41:1537–1540. doi: 10.1128/aac.41.7.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller M A, Messer M A, Hollis R J. Strain delineation and antifungal susceptibilities of epidemiologically related and unrelated isolates of Candida lusitaniae. Diagn Microbiol Infect Dis. 1994;20:127–133. doi: 10.1016/0732-8893(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller M A, Messer M A, Bolmström A. Evaluation of Etest for determining in vitro susceptibility of yeast isolates to amphotericin B. Diagn Microbiol Infect Dis. 1998;32:223–227. doi: 10.1016/s0732-8893(98)00120-5. [DOI] [PubMed] [Google Scholar]
- 17.Rex J H, Cooper C R, Jr, Merz W G, Galgiani J N, Anaissie E J. Detection of amphotericin B-resistant Candida isolates in a broth-based system. Antimicrob Agents Chemother. 1995;39:906–909. doi: 10.1128/aac.39.4.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterling T R, Gasser R A, Ziegler A. Emergence of resistance to amphotericin B during therapy for Candida glabrata infection in an immunocompetent host. Clin Infect Dis. 1996;23:187–188. doi: 10.1093/clinids/23.1.187. [DOI] [PubMed] [Google Scholar]
- 19.Wanger A, Mills K, Nelson P W, Rex J H. Comparison of Etest and National Committee for Clinical Laboratory Standards broth macrodilution method for antifungal susceptibility testing: enhanced ability to detect amphotericin B-resistant Candida isolates. Antimicrob Agents Chemother. 1995;39:2520–2522. doi: 10.1128/aac.39.11.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]