Abstract
Purpose:
Clinical trials with immune checkpoint inhibition in sarcomas have demonstrated minimal response. Here, we interrogated the tumor microenvironment (TME) of two contrasting soft-tissue sarcomas (STS), rhabdomyosarcomas and undifferentiated pleomorphic sarcomas (UPS), with differing genetic underpinnings and responses to immune checkpoint inhibition to understand the mechanisms that lead to response.
Experimental Design:
Utilizing fresh and formalin-fixed, paraffin-embedded tissue from patients diagnosed with UPS and rhabdomyosarcomas, we dissected the TME by using IHC, flow cytometry, and comparative transcriptomic studies.
Results:
Our results demonstrated both STS subtypes to be dominated by tumor-associated macrophages and infiltrated with immune cells that localized near the tumor vasculature. Both subtypes had similar T-cell densities, however, their in situ distribution diverged. UPS specimens demonstrated diffuse intratumoral infiltration of T cells, while rhabdomyosarcomas samples revealed intratumoral T cells that clustered with B cells near perivascular beds, forming tertiary lymphoid structures (TLS). T cells in UPS specimens were comprised of abundant CD8+ T cells exhibiting high PD-1 expression, which might represent the tumor reactive repertoire. In rhabdomyosarcomas, T cells were limited to TLS, but expressed immune checkpoints and immunomodulatory molecules which, if appropriately targeted, could help unleash T cells into the rest of the tumor tissue.
Conclusions:
Our work in STS revealed an immunosuppressive TME dominated by myeloid cells, which may be overcome with activation of T cells that traffic into the tumor. In rhabdomyosarcomas, targeting T cells found within TLS may be key to achieve antitumor response.
Introduction
Sarcomas are a heterogeneous group of bone and soft-tissue tumors affecting both children and adults. In the realm of malignant soft-tissue sarcomas (STS), the most prevalent types are undifferentiated pleomorphic sarcomas (UPS) in adults and rhabdomyosarcomas in children (1). Both diseases are deforming and difficult to treat with poor 5-year survival outcomes of 52%–62% in patients with high-grade UPS, and 60% in patients with rhabdomyosarcoma (1-4). There are several subtypes of rhabdomyosarcoma. The two most common subtypes affecting patients are alveolar (ARMS) and embryonal (ERMS) rhabdomyosarcoma. ARMS occurs more commonly in adolescents with tumor cells containing the characteristic chromosomal translocations t(2;13) or t(1;13), and are commonly referred to as “fusion positive” rhabdomyosarcoma. ERMS occurs mainly in younger children, carries a better prognosis, is characterized by LOH at 11p15 locus, and does not carry any characteristic chromosomal translocations, thus being termed “fusion negative” rhabdomyosarcoma (1, 3, 5). For the past four decades, the backbone of sarcoma therapy has been unchanged, consisting of surgery, radiation, and cytotoxic drugs. Improvement in survival outcomes have come to a standstill (5).
The breakthrough of chimeric antigen receptor T cells in pre B-cell acute lymphoblastic leukemia and checkpoint inhibitors in melanoma, non-small cell lung cancer, and numerous other solid tumor types, are the exemplar of successful immunotherapy and its potential to be utilized for all tumors (6). Immunotherapy in sarcomas has thus far had limited success. Trials in patients with osteosarcoma with high dose IL2, IFNα, and immunomodulatory agent muramyl tripeptide have not yielded statistically significant responses (7-12). Similarly, vaccines with peptides spanning fusion genes unique to specific sarcoma subtypes, including SYT-SSX in synovial sarcoma, EWS/FLI-1 in Ewing Sarcoma, and PAX3/FKHR (now known as PAX3/FOXO1) in ARMS, have mainly been ineffective (13-15). In patients with synovial sarcoma, adoptive T-cell therapy targeting cancer testis antigen, NY-ESO-1, has been promising, but applicability toward other sarcomas has been unsuccessful (16, 17). The most recent Alliance trial investigating nivolumab with or without ipilimumab in metastatic sarcomas demonstrated low response rates of 5% (of 43 patients) with monotherapy nivolumab and 16% (of 42 patients) with nivolumab and ipilimumab combined. This small cohort of responders included patients with UPS and leiomyosarcoma. Checkpoint inhibition therapy has not shown to have any effect in rhabdomyosarcoma (18-20).
The basis of successful immunotherapy lays in the ability of the immune system to recognize cancer as foreign and attack, and is dependent on genetics, host and environmental factors, and makeup of the tumor microenvironment (TME; refs. 6, 21, 22). To learn and improve on past immunotherapy trials in sarcomas, we need to understand the immune niche that sustains the tumor and how it supports tolerance, which consequently leads to cancer immune escape. To date, there has not been a comprehensive study into the sarcoma immune TME with the exception of the recently published study by Petitprez and colleagues, associating B cells with survival and immunotherapy response in adult sarcomas (23). Much of the published literature solely focuses on presence of PD-1/PD-L1 expression with one recent article correlating expression of PD-1 and CD56 with worse overall survival (24-28). Despite these works, there is still little insight into other mechanisms of immune escape. Hence, it is imperative to learn about factors influencing the immune set point in sarcomas to determine what druggable targets can be used to mount antitumor immunity (21).
The objective of our work was to perform an in depth interrogation of the immunobiology of prototypic STS with the goal of identifying immunotherapy agents that would match the immune characteristics of specific TMEs, thus providing more precise treatments for individual patients and cancer types. We specifically concentrated on UPS and rhabdomyosarcoma given their prevalence and more importantly their contrasting biology and response to immune checkpoint blockade (ICB), UPS is one of the few sarcomas that have shown complete responses to checkpoint inhibition, while rhabdomyosarcoma shows no response whatsoever (18-20). Our in-depth comparison of these two TMEs revealed a similar immune niche that was dependent on angiogenesis and predominated by myeloid cells, but differed in their spatial distribution of immune cells with UPS characterized by diffuse infiltration of T cells and rhabdomyosarcoma marked by T cells clustered in tertiary lymphoid structures (TLS). These differing characteristics may be key to understanding tumor immunity in patients with STS.
Materials and Methods
Patient selection and tumor samples
Tumor tissue was collected at the Johns Hopkins Hospital (Baltimore, MD) from patients with UPS, ERMS, ARMS, and leiomyosarcoma. This study was conducted in accordance with the ethical principles stated in the Belmont Report and the U.S. Common Rule. It was approved by the Johns Hopkins Institutional Review Board (Baltimore, MD) and all samples were obtained in accordance with the Health Insurance and Accountability Act. This study was conducted under an institutional review board–approved protocol with a waiver of consent.
IHC analysis
Formalin-fixed, paraffin-embedded (FFPE) tissue blocks from 39 UPS, 27 ERMS, 24 ARMS, and 11 leiomyosarcoma specimens were cut into 5-μm sections and mounted onto plus-charge glass slides. Each case was stained with hematoxylin and eosin (H&E), CD3, CD8, CD163, Foxp3, CD20, CSF-1R, PD-L1, and CD31 markers. H&E slides were reviewed by a pathologist to demarcate areas of tumor and areas of normal tissue. Whole slides (20×) were then digitally scanned (Scanscope XT) and analyzed with the Indica Labs HALO pathology software. The whole slide was used for digital image analysis with areas of tumor manually delineated into the software program. Immune cell density for each slide was calculated in HALO by digital analysis of intratumoral immune cell count and tumor tissue area. For each slide, we used the same algorithm to calculate immune cell density and manually reviewed each case to ensure consistency and accuracy.
Laser capture microdissection and RNA extraction
FFPE blocks were cut into 10- μm sections and mounted onto polyethylene naphthalate membrane glass slides. Slides were then stained with H&E. Utilizing the Leica LMD 7000 System, areas of interest on the slide were captured with laser capture microdissection. These areas were then collected in tissue lysis buffer and extracted for RNA. We used Roche High Pure Paraffin Kit for RNA extraction and followed the manufacturer's instructions.
RNA-sequencing and gene expression analysis
Transcriptome sequencing was performed using the Ion Torrent RNA Next-Generation Sequencing platform. This genomic analysis plugin aligns the raw sequence reads to a human reference genome that contains 20,802 RefSeq transcripts (hg19 Ampliseq Transcriptome_ERCC_V1.fasta) using the Torrent Mapping Alignment Program. Then, the number of reads mapped per gene are counted to generate raw counts files and normalized reads per gene per million mapped reads (RPM) files. Gene set enrichment analysis (GSEA) from the Broad Institute was used to analyze enriched gene sets from published works in our data (29-33). CIBERSORT deconvolution was utilized to analyze immune cell composition in our data but also published gene sets from the Gene Expression Omnibus (34, 35). We utilized GSE75885 from Delespaul and colleagues and GSE108022 from Shern and colleagues (36, 37).
Isolation of tumor-infiltrating lymphocytes
Sarcoma specimens were collected and digested with Enzymatic Cocktail (Roche, 0.1% DNaseI and Liberase 400 μg/mL). Mononuclear cells were enriched with Percoll Density Gradient (GE Healthcare) and leukocytes were retrieved and cryopreserved, banked in liquid nitrogen until further analysis.
Multiparameter flow cytometry
Tumor-infiltrating lymphocytes (TIL) from UPS and leiomyosarcoma cases were analyzed by multiparameter flow cytometry (MFC) using lymphoid markers (CD45 APC-Cy7, CD3 BV786, CD4 PE-CF594, Foxp3 V450, and CD8 PerCp-Cy5.5), intracellular cytokines (IFNγ AF700 and IL10 BV711), and checkpoint expression (PD- 1 PeCy7, LAG-3 FITC, and TIM-3 PE). For intracellular cytokine staining, cells were first stimulated with eBiosciences phorbol-12-myristate-13-acetate + ionomycin and GolgiStop for 3 hours followed by staining with IFNγ and IL10. Data were analyzed using BDdiva and FCS Express Software.
Results
Gene expression data from publicly available datasets support a stronger M2 tumor-associated macrophage signature in UPS and higher B-cell signature in rhabdomyosarcoma
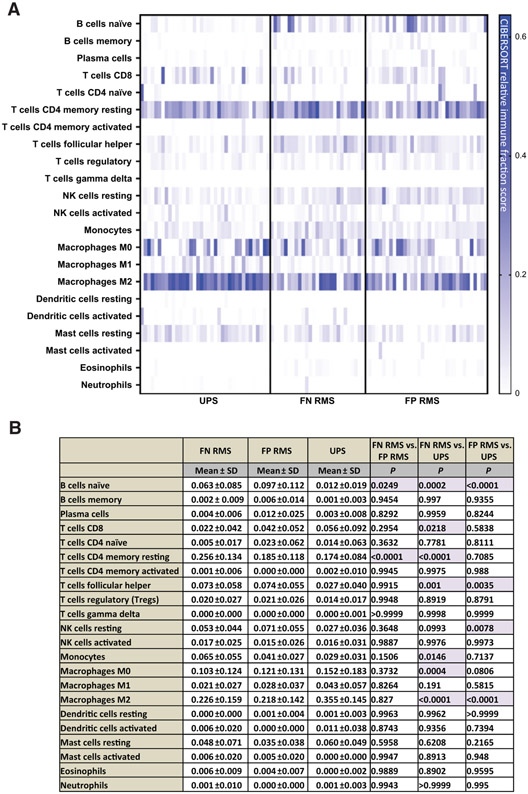
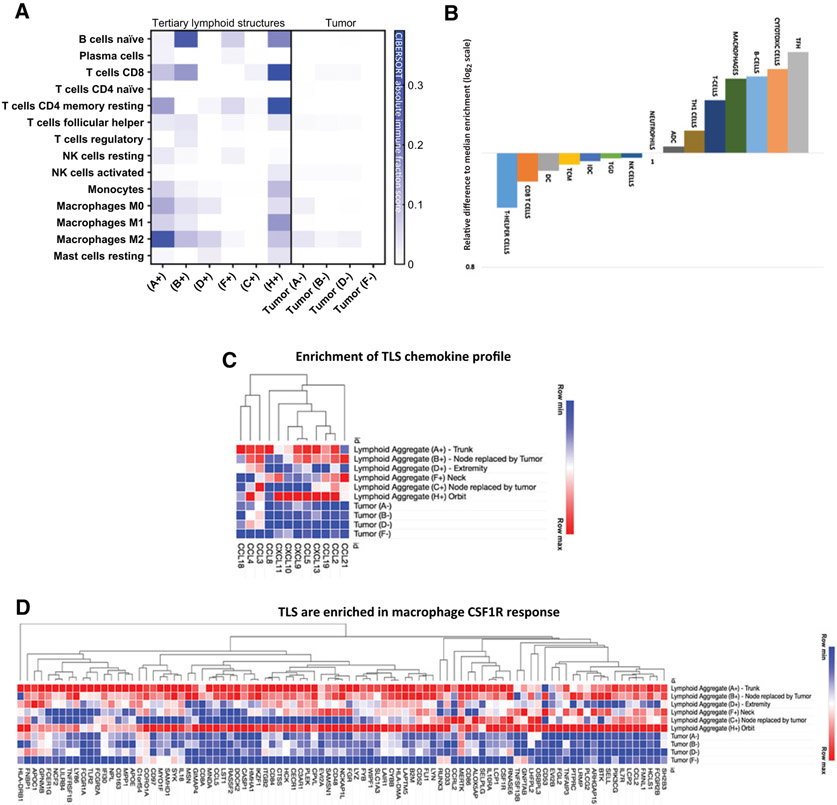
We started our initial survey of the STS TME by looking at published RNA gene expression datasets for UPS (Delespaul and colleagues) and rhabdomyosarcoma (Shern and colleagues) specimens (36, 37). We utilized the CIBERSORT deconvolution algorithm to compare the immune cell composition in these two different sarcoma subtypes and give insight into TME factors that may influence immunotherapy response. We utilized CIBERSORT relative immune fraction score to compare immune cell composition between two different cohorts of publicly available data. Interestingly, we found UPS and rhabdomyosarcoma specimens to both have a strong myeloid signature characterized by high immune fraction scores of M0 and M2 macrophages, but UPS specimens had higher M2 macrophage immune fraction scores compared with rhabdomyosarcoma samples (Fig. 1A and B).
Figure 1.
A, CIBERSORT immune deconvolution analysis in published UPS gene sets compared with rhabdomyosarcoma gene sets. Fusion status of rhabdomyosarcoma is also included. Heatmaps are representative of CIBERSORT relative immune fraction scores. B, CIBERSORT relative immune fraction score means with SD of sarcoma subtypes. Comparison of means analyzed via two-way ANOVA, Tukey multiple comparisons test.
Highlighted cells are statistically significant with P < 0.005.
Abbreviations: SD = Standard Deviation; FN RMS: Fusion Negative Rhabdomyosarcoma, FP RMS: Fusion Positive Rhabdomyosarcoma; UPS: Undifferentiated Pleomorphic Sarcoma.
In the T-cell compartment, both UPS and rhabdomyosarcoma specimens had high CD4+ memory resting T-cell immune fraction scores, however, fusion negative (FN) rhabdomyosarcoma (FN RMS) samples had significantly higher immune cell fraction scores compared with both fusion positive (FP) rhabdomyosarcoma (FP RMS) and UPS (Fig. 1A and B). CD8+ T-cell immune fraction scores were significantly higher in UPS compared with FN RMS, but there was no difference between the other subtypes (Fig. 1A and B). Rhabdomyosarcoma samples had higher naïve B cell and T-follicular helper cells (TFH) immune fraction scores compared with UPS samples, and FP RMS samples had higher naïve B-cell immune fraction scores compared with FN RMS samples (Fig. 1A and B). To explore where in the tumor M2 macrophage and B-cell signatures arose, we looked at the tissue to better characterize the immune milieu.
Tumor-infiltrating immune cells remain in proximity to tumor vasculature
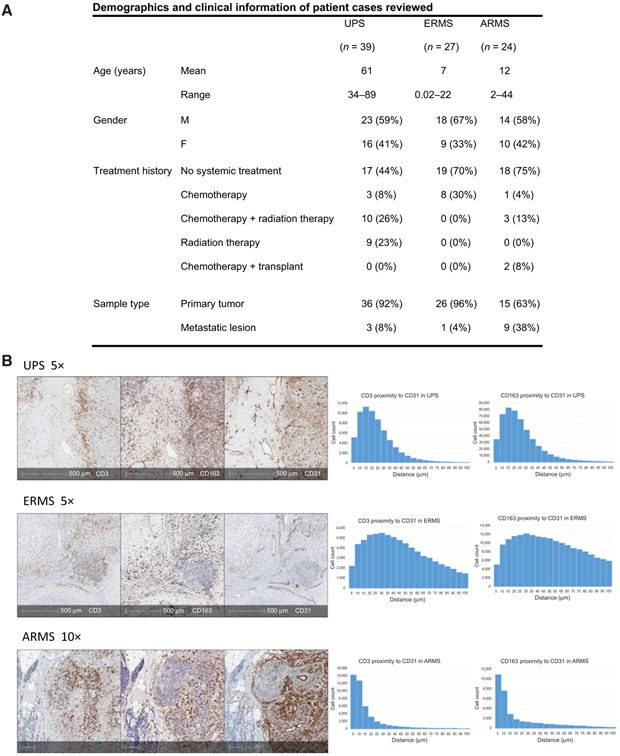
We reviewed 90 STS cases with our pathologists to define tumor and immune cell geography. Patient demographics and clinical information of cases reviewed are in Fig. 2A. Through IHC analysis, we were able to capture in situ relationships between immune cells. We found the majority of T cells (CD3+) and tumor associated macrophages (CD163+ TAM) to be in close proximity to endothelial cells (CD31+; Fig. 2B). This pattern was seen in UPS, and the two subsets of rhabdomyosarcoma—ARMS and ERMS. To quantify and confirm our gross findings, we utilized digital image proximity analysis to calculate the distance of CD3+ and CD163+ cells in relation to CD31+ cells. Figure 2B exhibits three cases that are representative of our patient cohort and demonstrates that majority of T cells and TAMs are found within 20–40 μm of endothelial cells in sarcoma subtypes analyzed.
Figure 2.
A, Patient demographics and clinical history of all cases reviewed. B, Three representative cases illustrating tumor vasculature and immune cell infiltration. IHC slides stained with CD3 (T cells), CD163 (TAMs), and CD31 (endothelial cells) provide a geographic overview of UPS, ERMS, and ARMS specimens (left). In all three subtypes, the majority of T cells and TAMs cluster near endothelial cells. Corresponding histograms represent proximity analysis with HALO pathology software measuring distance (mm) between CD3+ cells and CD31+ cells, as well as between CD163+ cells and CD31+ cells (right). Results demonstrate the majority of T cells and TAMs to be found within 40 μm to tumor endothelial cells in UPS, ERMS, and ARMS.
TAMs predominate the sarcoma microenvironment, but spatial distribution of immune cells differs between UPS and rhabdomyosarcoma
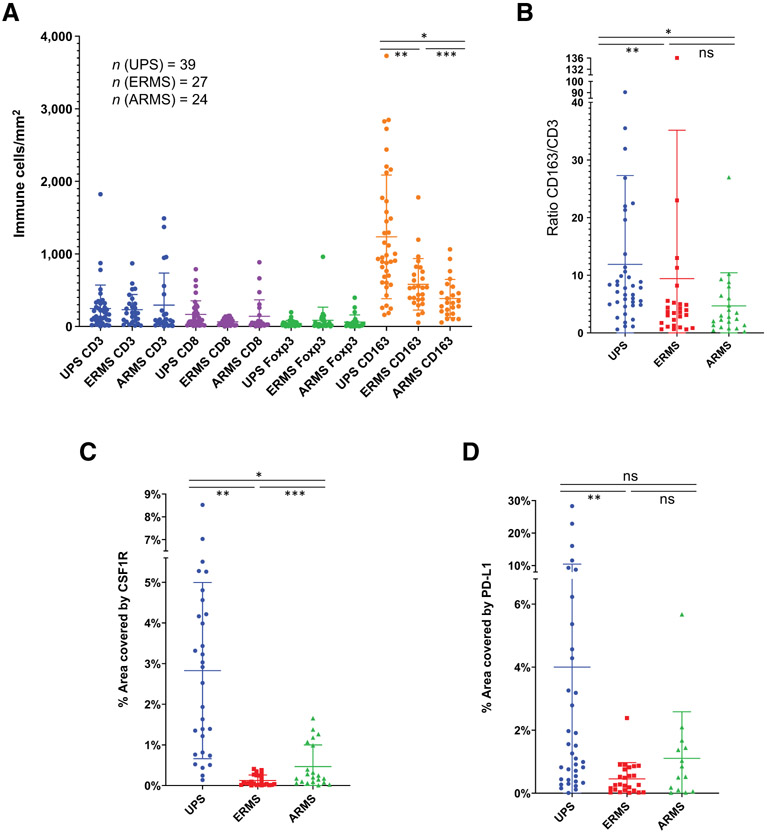
Using digital image analysis, we analyzed the densities of T cells (CD3+), CD8+ T cells (CD8+), T-regulatory cells (Foxp3+), TAMs (CD163+), and PD-L1 expression in complete tumor sections of UPS, ERMS, and ARMS specimens. Similar to the strong macrophage signature detected through our immune deconvolution analysis of publicly available data, our digital image analysis revealed that in all three sarcomas TAMs (CD163+) predominated the sarcoma immune microenvironment with the highest intratumor density, but differed depending on subtype: UPS had more TAMs compared with ERMS, followed by ARMS (Fig. 3A; Supplementary S1A). To account for total immune cell density, we compared the ratio of CD163+/CD3+ densities among the different sarcoma subtypes and found UPS specimens to have the highest ratio compared with both rhabdomyosarcoma subtypes, supporting its higher density of TAMs. There was no significant difference between CD163+/CD3+ ratios of rhabdomyosarcoma subtypes, ERMS and ARMS (Fig. 3B). With the strong TAM signature, we also tested colony-stimulating factor-1 receptor (CSF1R) expression in the same specimens and noted its highest expression in UPS followed by ARMS, and then ERMS specimens (Fig. 3C). CSF1R plays an important role in differentiation and survival of tissue macrophages and is a marker of immunosuppressive macrophages, which are thought to sustain an anti-inflammatory niche for tissue healing and tumor growth (38, 39). The higher CSF1R expression in UPS specimens is not surprising as CSF1R expression positively correlates with density of TAMs (CD163+; Supplementary Fig. S1B). Interestingly, however, ARMS specimens have higher CSF1R expression compared with ERMS specimens despite similar CD163+/CD3+ ratios (Fig. 3B and C). Our immune deconvolution data from Fig. 1 supports the strong immunosuppressive macrophage signature observed in UPS specimens with higher immune fraction of M2 macrophages in UPS compared with rhabdomyosarcoma.
Figure 3.
Immune cell densities. A, Densities (immune cells/mm2) for CD3 (T cells), CD8 (cytotoxic T cells), Foxp3 (T-regulatory cells), and CD163 (TAMs) were plotted in UPS, ERMS, and ARMS specimens. TAMS have the strongest presence in all sarcomas, with UPS having the greatest density, followed by ERMS, and then ARMS (*, P < 0.0001; **, P = 0.0003; ***, P = 0.0072 by paired t test). B, Normalization of CD163 densities to CD3 densities with CD163/CD3 ratio among the three different STS subtypes. CD163/CD3 ratio in UPS specimens are higher than rhabdomyosarcoma subtypes, both ARMS and ERMS, but there is no difference between ARMS and ERMS (*, P = 0.0009; **, P = 0.0010). C, Percent surface area covered by CSF1R was measured in the three subtypes. UPS specimens have the highest percent of CSF1R expression, followed by ARMS, and then ERMS (*, P < 0.0001; **, P < 0.0001; ***, P = 0.01 by paired t test). D, Mean surface area covered by PD-L1 was highest in UPS specimens, but only statistically significant when compared with mean of ERMS specimens (**, P < 0.0001). ns, nonsignificant (P > 0.05).
Mean T-cell (CD3+), T-regulatory cell (Foxp3+), and CD8+ T-cell densities were not significantly different among the three sarcoma subtypes (P > 0.05 in paired t test analysis; Fig. 3A; Supplementary S1A). Mean T-regulatory (Foxp3+) cell densities were lower than mean CD8+ T-cell densities, but no statistical significance could be concluded (Fig. 3A; Supplementary S1A). We also looked at PD-L1 expression and found it to be highest in UPS specimens and much lower in ARMS and ERMS specimens. However, mean PD-L1 expression was only significantly higher in UPS specimens when compared with ERMS specimens (Fig. 3D).
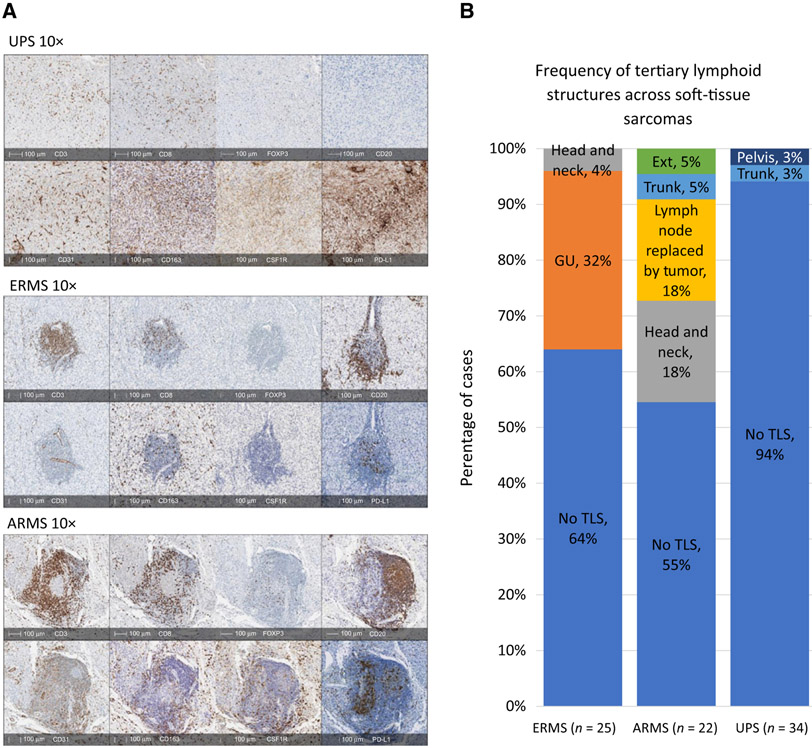
Despite the observation that all three sarcomas had similar T-cell (CD3+) densities, their spatial distribution was markedly different. In UPS specimens, CD3+ and CD8+ T cells were diffusely present in the tumor with some colocalization near CD31+ endothelial cells. In ERMS and ARMS specimens, however, the majority of CD3+ and CD8+ T cells appeared in aggregates with B cells (CD20+), forming TLS located near perivascular beds. Immune cells comprising TLS were the main source of PD-L1 expression for rhabdomyosarcoma specimens (Fig. 4A). To confirm this finding, we reexamined and visually quantified all PD-L1 slides to investigate cell origin of PD-L1 expression. We found in UPS specimens, PD-L1 expression mainly originated from tumor cells and immune cells (specifically TAMs) throughout the specimen, while in rhabdomyosarcoma specimens, PD-L1 expression stemmed mainly from TLS immune cells and to a lesser extent tumor cells (Fig. 4A).
Figure 4.
Organization of immune cells and frequency of TLS. A, IHC stains shows diffuse distribution of T cells (CD3+ and CD8+) in UPS with no B cells (CD20+) present. In ERMS and ARMS, T cells (CD3+ and CD8+) cluster together with B cells (CD20+) forming TLS. TAMS (CD163+) are diffusely distributed in all sarcomas. CSF1R appears stronger in UPS and ARMS. PD-L1 is present through all sarcomas but stronger in UPS. B, The frequency of TLS in each sarcoma subtype varies and originates from different anatomical sites.
For the purpose of this article, we define TLS as a cluster of B cells (CD20+) and T cells (CD3+) located near endothelial cells (CD31+; ref. 40). We used this criteria to determine TLS frequency in our sarcoma samples and found at least one or more TLS to be present in 6% of UPS, 36% of ERMS, and 45% of ARMS specimens. In ERMS, of the tumors with TLS present (n = 9), 88% (n = 8) came from genital urinary organs (bladder and testicles) with the other being from a head and neck tumor (n = 1). In ARMS, of the 10 samples with TLS present, the most common sites of tumor were head and neck (n = 4) and lymph nodes replaced by tumor (n = 4; Fig. 4B).
We consider B cells to be a surrogate marker of intratumoral TLS presence based on our IHC investigations demonstrating that B cells were mainly found in TLS of rhabdomyosarcoma and not elsewhere in the tumor tissue (Fig. 4A). The difference in naïve B-cell immune fraction scores from our immune deconvolution analysis shown in Fig. 1 also corroborates our in situ findings that TLS are more frequently found in ARMS (FP RMS; 45% cases have TLS), followed by ERMS (FN RMS; 36% cases have TLS), and then UPS (6% cases have TLS; Fig. 4B).
The immune cell composition of rhabdomyosarcoma is predominantly determined by intratumoral TLS
We implemented LCM methodology to isolate the rhabdomyosarcoma TLS with subsequent RNA extraction, which was utilized for gene expression profiling via RNA-sequencing (Supplementary Fig. S2). We were able to successfully sequence six different tumor specimens with TLS and four paired specimens of tumor without TLS for comparison purposes. To analyze the immune signatures generated from different regions of specimens, we implemented the CIBERSORT platform and determined the cell type abundance and expression from bulk tissues looking specifically at tumor immune cell composition. We employed CIBERSORT absolute immune fraction score to capture overall immune content and allow for better comparison of immune cell infiltration among our samples (34, 35). We observed that tumor areas without TLS had low absolute immune cell fraction scores in all immune cell types with the exception of M0 and M2 macrophages (Fig. 5A). This observation concurs with our IHC analysis showing myeloid cells to be the predominant immune cell lineage throughout the tumor tissue of rhabdomyosarcoma, as well as our larger immune deconvolution analysis of publicly available data confirming the strong M0 and M2 macrophage signature in UPS and rhabdomyosarcoma. On the other hand, intratumoral TLS regions were rich in naive B cells, CD4 memory resting T cells, TFH, and macrophages. Among our samples, specimen H+ (an orbital tumor) had the highest absolute CIBERSORT score for T cells (CD8 and CD4 memory resting cells) compared with the other samples (Fig. 5A). Our results suggest that in rhabdomyosarcoma, tumor-infiltrating immune cells, specifically lymphocytes, are found within TLS regions and not elsewhere in the tumor.
Figure 5.
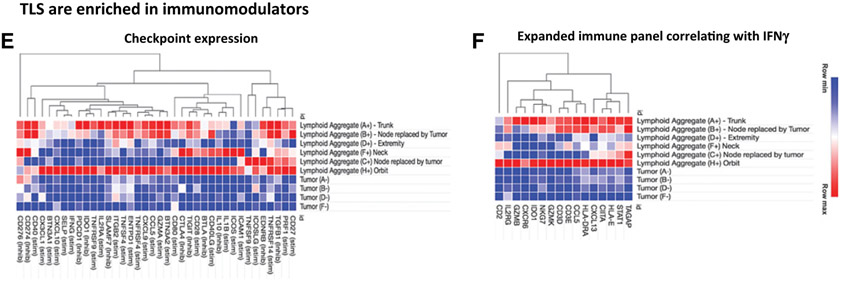
A, CIBERSORT absolute immune fraction scores generated from immune deconvolution analysis comparing our six rhabdomyosarcoma intratumoral TLS samples and four paired rhabdomyosarcoma tumor samples without TLS. Immune cell content in rhabdomyosarcoma comes from TLS regions and not areas of tumor devoid of TLS. Location of six intratumoral TLS samples: trunk (A); node replaced by tumor (B); extremity (D); neck (F); node replaced by tumor (C); and orbit (H). (+) denotes areas of tumor with TLS and (−) are areas of tumor devoid of TLS. B, Gene expression profiling of TLS. rhabdomyosarcomas intratumoral TLS are more enriched with gene sets reflective of TLS. All genes shown have FDR < 25%: b) NES of gene sets representative of different immune phenotypes normalized to the median NES show TLS regions to be more enriched in TFH and B cells, compared with tumor areas devoid of TLS. ADC, activated dendritic cells, NK cells, natural killer cells; TGD, T gamma delta cells; IDC, immature dendritic cells; TCM, T central memory cells; DC, dendritic cells. C-F, Heatmaps are representative of core enrichment genes from different gene sets that are more enriched in TLS regions compared with areas of tumor devoid of TLS. TLS regions are more enriched in gene sets representative of TLS chemokine profile, CSF1R response, checkpoint expression, and expanded immune panel corresponding with IFNγ response, compared with areas of tumor devoid of TLS.
When comparing immune cell fraction scores from published rhabdomyosarcoma datasets in Fig. 1A with our laser dissected specimens, we detected the same strong content of naïve B cells, CD4 memory cells, and TFH cells in our six intratumoral TLS samples, but no resemblance in our tumor areas devoid of TLS (Figs. 1A and 5A). We think this reflects the different methods of RNA isolation. The published rhabdomyosarcoma gene expression sets were generated utilizing RNA isolated from whole-tumor sections and does not take into account the regional distribution of immune infiltrates; it is unclear where in the tumor the immune signatures arose from. In our laser dissected specimens, we purposely aimed to extract RNA from specific TLS regions. Our results reveal the lymphocytic signature of rhabdomyosarcoma arises from intratumoral TLS regions.
Rhabdomyosarcoma intratumoral TLS express secondary lymphoid organ signatures as well as numerous immunomodulatory molecules
Implementing GSEA, we examined immune gene sets representing immune cell types (Bindea and colleagues), TLS canonical chemokine/chemokine receptor axes (Coppola and colleagues), macrophage CSF1R response (Beck and colleagues), immune checkpoint expression (Thorsson and colleagues), and IFNγ response (Ayers and colleagues), to determine differentially expressed genes in intratumoral TLS compared with tumor tissue devoid of TLS (29-33). Figure 5B reflects immune cell type composition of TLS and shows 15 different immune gene sets that were preferentially enriched in the intratumoral TLS with a FDR < 25%. Applying GSEA normalized enrichment scores (NES), we then compared each gene group with the median NES to visualize which immune cell types dominated in the TLS. Our results exhibited TLS enriched in gene sets representative of TFH, cytotoxic T cells, B cells, and macrophages which are the specific cell populations present in secondary lymphoid organs. We also examined chemokine profiles that are indicative of ectopic lymphoid neogenesis and found our intratumoral TLS areas to be augmented in specific chemokines and adhesion molecule genes associated with B and T cell presence compared with tumor tissue devoid of TLS (Fig. 5C).
We also looked at immunomodulatory genes in the intratumoral TLS that could serve as markers for selecting targeted immunotherapies in these patients. Given the predominant myeloid cell signature found on IHC analysis and our enumeration of cell subsets from transcriptomic data, we compared the rhabdomyosarcomas gene expression profiles with a set of genes that reflect macrophage CSF1R response. We found the intratumoral TLS to be more enriched in macrophage CSF1R response compared with tumor tissue devoid of TLS (Fig. 5D; FDR < 25%). We also looked at a panel of immune checkpoint and immunoregulatory genes, as well as an expanded immune panel that correlates with IFNγ signature and found the intratumoral TLS to be more enriched in these two gene sets compared with tumor tissue devoid of TLS (Fig. 5E and F; FDR < 25%).
TIL from UPS specimens are characterized by CD4+ non-T-regulatory cells (CD4+/FOX3−) and CD8+/PD-1− cells that produce IFNγ
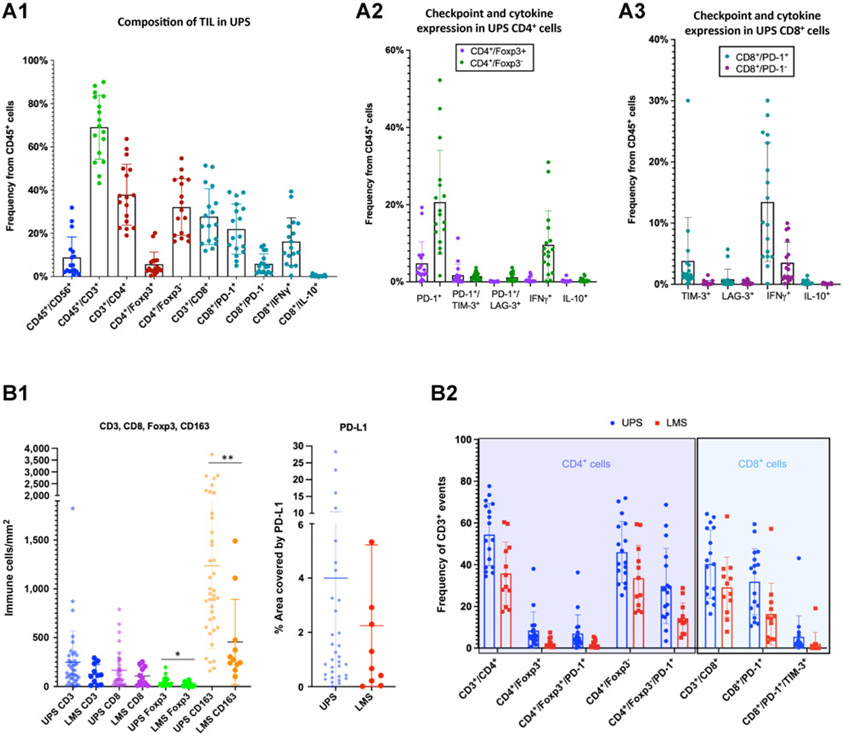
To better characterize the diffuse T cells observed in situ in UPS, we used MFC to further interrogate viable immune cell phenotypes existent in UPS specimens by immunophenotyping TIL. We discovered UPS TIL were composed mainly of CD4+ and CD8+ T cells, and less so natural killer cells (CD45+/CD56+; Fig. 6A.1). Of the CD4+ T-cell population, the majority were non-T-regulatory cells (CD4+/FOX3−) that also expressed PD-1 and were capable of producing IFNγ (Fig. 6A.2). The CD8+ T-cell population was comprised mostly of CD8+/PD-1+ T cells that were capable of generating IFNγ (Fig. 6A.3). The CD8+/PD-1+ population split into a dichotomous group of PD-1+ high and PD-1+ low CD8+ T cells. In CD8+ cells, the amount of IFNγ production positively corresponds with the degree of PD-1+ expression (Supplementary Fig. S3C). A portion of these CD8+/PD-1+ high T cells also expressed other exhaustion checkpoint molecules such as LAG-3 (3.7% of CD8+/PD-1+ cells) or TIM-3 (15.8% of CD8+/PD-1+ cells; Fig. 6A.3). This CD8+/PD-1+ high group with coexpression of other immune checkpoints might represent the tumor reactive repertoire against UPS tumor cells and conceivably represent the group of patients with UPS who respond to checkpoint inhibition.
Figure 6.
A, MFC analysis of UPS TILs. A1, Live CD45+ cells are composed mainly of T-cells (CD45+/CD3+) at a frequency of 69% ± 15% and less so NK cells (CD45+/CD56+) at a frequency of 9% ± 9%. T cells break down to CD4+ T cells (38% ± 14% of CD45+ cells) and CD8+ T cells (28% ± 13% of CD45+ cells). CD4+ T cells are comprised of more non-T-regulatory cells (CD4+/Foxp3−;32% ± 13% of CD45+ cells) and less T-regulatory cells (CD4+/Foxp3+; 6% ± 6% of CD45+ cells). More CD8+ T cells express PD-1: CD8+/PD-1+ cells make up 22% ± 12% of CD45+ cells while CD8+/PD-1− cells make up 6% ± 5% of CD45+ cells. There is a group of CD8+/PD-1 high cells that may represent the tumor reactive repertoire. Many CD8+ T cells produce IFNγ (CD8+/IFNγ+ cells comprise 16% ± 11% of CD45+ cells) and almost no IL10 (CD8+/IL10+ cells comprise 0.4% ± 0.4% of CD45 cells). A2, Frequency of CD4+ T-cell subpopulations show non-T-regulatory cells (CD4+/Foxp3−) to express more PD-1 (21% ± 13% of CD45+ cells) and IFNγ (10% ± 9% of CD45+ cells) compared with T-regulatory cells (CD4+/Foxp3+; CD4+/Foxp3+/PD-1+ comprise 5% ± 6% of CD45 cells and CD4+/Foxp3+/IFNγ+ comprise 0.5% ± 0.7% of CD45+ cells). CD4+ subpopulation frequencies are found in Supplementary Fig. S3A. A3, Frequency of CD8+ T-cell subpopulations show the majority of CD8+/PD-1+ cells are capable of producing IFNγ (13% ± 10% of CD45+ cells). Fewer CD8+/PD-1− cells produce IFNγ (4% ± 3% of CD45+ cells). PD-1 expression positively correlates with IFNγ production (Supplementary Fig. S3C). A small population of CD8+/PD-1+ cells also express TIM-3 (4% ± 7% of CD45+ cells). CD8+ subpopulation frequencies are found in Supplementary Fig.S3B. B, Comparison of immune cell composition and T-cell phenotypes between UPS and leiomyosarcoma via IHC and MFC analyses. B1, IHC analysis demonstrates a higher mean density of CD3+ and CD8+ T cells in UPS, however P values were not significant. Mean density of T-regulatory cells (Foxp3+) is significantly higher in UPS compared with leiomyosarcoma specimens and once again, similar to comparisons with rhabdomyosarcoma specimens, UPS specimens have significantly higher mean density of TAMs compared with leiomyosarcoma specimens (*, P = 0.0459; **, P = 0.0008). Mean PD-L1 expression is higher in UPS compared with leiomyosarcoma specimens, but P value was not significant. B2, Comparison of T-cell phenotypes characterized by MFC in UPS and leiomyosarcoma TIL show higher frequency of T-regulatory cells (CD4+/Foxp3+) and non-T-regulatory cells (CD4+/Foxp3−) in UPS specimens. Both T-regulatory and non-T-regulatory cells have higher expression of PD-1+ in UPS compared with leiomyosarcoma TIL. UPS TIL also have higher frequency of CD8+ T-cells (CD3+/CD8+), as well as CD8+ T cells that express PD-1+ and both PD-1+/TIM-3+.
Higher density of TAMs, and higher levels of TIL infiltration and PD-1 checkpoint expression found in UPS compared with leiomyosarcoma specimens
Finally, we compared the immune TME of UPS and leiomyosarcoma. Patient demographics of leiomyosarcoma specimens are in Supplementary Fig. S4. UPS and leiomyosarcoma are two very common STS with complex and unbalanced karyotypes that lead to severe genome instability resulting in multiple genomic aberrations (41). Despite their similar genetic underpinnings, they differ dramatically in their responses to ICB with the majority of clinical responses occurring in UPS (20). To compare the immune cell composition of these two differing sarcomas, we explored publicly available leiomyosarcoma and UPS gene datasets from Delespaul and colleagues and performed CIBERSORT immune deconvolution analysis (37). We found UPS samples to have significantly higher relative immune fraction scores of M0 and M2 macrophage signatures compared with leiomyosarcoma samples (Supplementary Fig. S5).
We also performed IHC and MFC analyses of freshly isolated immune cells from leiomyosarcoma specimens, which we then compared with immune phenotypes ofUPS specimens (Fig. 6B.1 and 6B.2). By IHC analysis, we found a significantly higher density of T-regulatory cells (Foxp3+) and TAMs (CD163+) in UPS compared with leiomyosarcoma specimens (Fig. 6B.1). Interestingly, the dominating myeloid signature of TAMs, by IHC, and M2 macrophages, by immune deconvolution analysis, in both leiomyosarcoma and UPS TMEs is similar to what we see in rhabdomyosarcoma specimens (Figs. 1 and 3A). PD-L1 expression by IHC analysis is also higher in UPS compared with leiomyosarcoma specimens, although this is not statistically significant (Fig. 6B.1). It is notable that these IHC findings are similarly appreciated in our MFC analysis. We identified a higher frequency of T regulatory (CD4+/Foxp3+), non-T-regulatory (CD4+/Foxp3−), and CD8+ T cells by MFC analysis in UPS compared with leiomyosarcoma TIL. Remarkably, CD4+ and CD8+ TIL in UPS cases exhibited higher expression of immunoregulatory molecules PD-1 and TIM-3 (Fig. 6B.2). These findings, along with the higher PD-L1 expression seen in IHC analysis, indicate a higher degree of T-cell activation and exhaustion in TIL of UPS specimens. This is interesting because many investigations in different types of cancers are now reporting an association between densities and level of expression of PD-1+ CD4+ and CD8+ TIL with better responses to anti-PD-1 therapy (42-44).
Discussion
In our in-depth analysis of two contrasting STS with different biology and response to immune checkpoint inhibition: UPS, genetically complex sarcomas and some responsive to ICB, and rhabdomyosarcoma, some of which are translocation driven sarcomas and unresponsive to ICB; we identified common immunosuppressive factors that dampen antitumor immunity but also differing attributes that could potentially overcome immunosuppression and lead to antitumor activity. The common predominate immune signature in both UPS and rhabdomyosarcoma were myeloid cells. On a protein expression level, TAMs (CD163+) were expressed at the highest density in UPS and rhabdomyosarcoma tumors. Similarly, on a gene expression level, M2 macrophages had the strongest signature in UPS and rhabdomyosarcoma gene expression datasets. We postulate the intratumoral myeloid compartment to consist of M2 TAMs that are sustaining an immunosuppressive TME and promoting tumor growth and proliferation. These observations for sarcomas correlate with breast and bladder cancer discoveries, in which TAMs are thought to exert an immunosuppressive effect and have been associated with poor prognoses (45-48). In rhabdomyosarcoma specifically, the combination of M2 macrophage genes detected, along with the increased CSF1R expression in ARMS compared with ERMS, could indicate a stronger M2 signature leading to differences in their survival outcomes (patients with ERMS have better 5-year survival outcomes than patients with ARMS; ref. 5). However, further studies investigating functional properties of rhabdomyosarcoma TAMs and their correlation with survival data are needed to confirm this relationship.
In both rhabdomyosarcoma and UPS, the majority of myeloid cells were found in close proximity (within 40 μm) to tumor endothelial cells. This is noteworthy as TAMs are thought to modulate tumor angiogenesis and promote metastasis through a distinct group of CD14+ monocytes expressing endothelial tyrosine kinase receptor, TIE2, that are found near endothelial cells and known as perivascular TAMs or TEMs (TIE2-expressing monocytes; refs. 49-52). We plan to explore this in the future by profiling freshly isolated sarcoma myeloid cells by MFC and with gene expression analysis. In mice studies, CSF1 is thought to stimulate TIE2 expression in monocytes and increase monocytic production of VEGF, causing increased tumor vascular formation and tumor growth. Interestingly, on GSEA of the rhabdomyosarcoma intratumoral TLS, we discovered a higher CSF1R response signature in the perivascular TLS compared with the tumor stroma devoid of TLS. This observation led us to believe that myeloid cells surrounding the rhabdomyosarcoma TLS are perivascular TAMs that are circumventing T-cell effector response from the TLS. Targeting these perivascular TAMs with agents such as CSF1R inhibitors/antibodies may inhibit monocyte–macrophage survival, negate their proangiogenic effects, and disrupt the immunosuppressive niche (38, 53, 54). In STS, dissolution of the immunosuppressive mechanisms described may lead to enhanced responses to immune checkpoint inhibition.
In addition to the pervasive immunosuppressive TAM signature in the STS TME, we reason the in situ T-cell distribution in the tumor is influential of immunotherapy responsiveness. In UPS tumors, the diffuse intratumoral T-cell infiltration may represent a population of activated effector T cells that are ready to attack when unleashed with immune checkpoint inhibition. This is reflected in the higher frequency of CD4+ and CD8+ T cells that express PD-1 in UPS compared with the molecularly similar, yet less ICB responsive, leiomyosarcoma. We believe this population of activated effector T cells is represented by the CD8+/PD-1high/IFNγ+ cells seen on MFC analysis of UPS TILs, and may in fact represent the cohort of patients with UPS who will respond to checkpoint blockade.
When we contrast this with rhabdomyosarcoma tumors, their T cells seem to be trapped in TLS, with surrounding perivascular TAMs. Immune deconvolution analysis of published datasets supports our IHC findings that TLS dominate the rhabdomyosarcoma T-cell compartment. Rhabdomyosarcoma samples have higher immune fraction scores of naïve B cells and TFH compared with UPS samples (Fig. 1A and B). TLS have been reported in a multitude of solid tumors including sarcomas and mainly associated with positive prognoses and responses to ICB (23, 55-57). TLS are ectopic lymphoid organs that develop in nonlymphoid tissues at sites of chronic inflammation including tumors. They culminate in germinal center formation and function as protected sites of antigen presentation to T and B cells, promoting an adaptive immune response (40, 58). Interestingly, the genomic immune signatures generated from our rhabdomyosarcoma TLS revealed existence of cytotoxic T cells capable of mounting an IFNγ response and concomitant expression of multiple immunomodulatory molecules, suggesting presence of effector T cells that, if freed from the oppressive TAM niche, may also respond to the ICB approach. In rhabdomyosarcoma, we postulate exploiting the perivascular TLS is key to generating antitumor immunity.
Data are not shown, but we did not find any correlations between T-cell density, TAM density, or frequency of TLS, with clinical outcomes. In UPS specimens, we did notice a decrease in TAM PD-L1 expression in samples obtained after radiation or radiation and chemotherapy treatment, however, this was not associated with clinical outcome. These relationships may be better elucidated with a larger number of patients and longer follow-up period.
Our findings reveal the heterogeneity and complexity of the immune microenvironment across quintessential sarcomas and affirm the necessity to define the most important mechanisms of immune exclusion to target in order to increase the probability of success. We lack predictive biomarkers of response to ICB in sarcomas, such as detection of PD-L1 on patient tumors with IHC. High tumor mutational burden (TMB) is a predictor of response to immune checkpoint inhibition, but STS as a group fall low on the TMB scale with a median of 2.5 mutations/Mb (59). Despite this and the pervasive immunosuppressive niche sustained by TAMs, certain patients with UPS still respond to ICB (19, 20). We speculate that the strong cytotoxic T-cell immune signature as well as the diffuse distribution of CD8+ T cells intratumorally can overcome the immunosuppressive niche and tip the balance toward antitumor immunity. Potential strategies for strengthening immune responses could implicate the elimination of TAMs and myeloid-derived suppressor cells in the TME of sarcomas via antimyeloid agents or via metabolic reprogramming therapies, and increase lymphocyte trafficking by normalizing the tumor vasculature via antiangiogenic drugs, which if used in combination with immune checkpoint inhibition may translate to clinical responses in patients with STS.
Supplementary Material
Translational Relevance.
Treatment paradigm for sarcomas has been unchanged for the past four decades with survival outcomes plateauing and patients with HR disease facing an abysmal prognosis. SARC028, trial testing pembrolizumab in soft-tissue sarcoma, demonstrated responses in undifferentiated pleomorphic sarcomas (UPS) and dedifferentiated liposarcomas. Alliance A091401, trial combining ipilimumab and nivolumab, showed responses in more histologies, but overall survival rates similar to standard chemotherapy. Here, we compare the immune tumor microenvironment of two molecularly distinct sarcomas: the genetically complex, immune checkpoint responsive (ICR), UPS, and the genetically simple, fusion-driven, poorly ICR, rhabdomyosarcoma, to identify factors that may contribute to their immunotherapy responsiveness. These two subtypes represent the main genomic aberrancies observed in sarcomas. Results show both tumors are dominated by tumor-associated macrophages (TAM). T cells in UPS are diffusely distributed, while T cells in rhabdomyosarcomas cluster with B cells near perivascular beds forming tertiary lymphoid structure. Our findings suggest targeting the myeloid compartment and tumor angiogenesis could overcome the immunosuppressive niche sustained by TAMs and lead to potential therapeutic targets.
Acknowledgments
This work was supported by Johns Hopkins Hospital, Bloomberg-Kimmel Institute for Immunotherapy; Bloomberg Philanthropies, BMS II-ON, Pediatric Cancer Research Foundation, Rally Foundation, Infinite Love for Kids Fighting Cancer in memory of Mia Rose McCaffrey Forever 6, Open Hands Overflowing Hearts, and Giant Food.
Footnotes
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
G. Cojocaru is an employee/paid consultant for Compugen LTD. C.F. Meyer is an employee/paid consultant for Bayer, and reports receiving speakers bureau honoraria from Novartis. D.J. McConkey is an employee/paid consultant for Janssen, Rainier, and H3 Biomedicine, and reports receiving commercial research grants from Astra-Zeneca and Rainier. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Burningham Z, Hashibe M, Spector L, Schiffman JD. The epidemiology of sarcoma. Clin Sarcoma Res 2012;2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ries LAG, Smith MA, Gurney JG, Linet M, Tamra T, Young JL, et al. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995, NCI. Available from: https://seer.cancer.gov/archive/publications/childhood/childhood-monograph.pdf. [Google Scholar]
- 3.Linabery AM, Ross JA. Childhood and adolescent cancer survival in the U.S. by race and ethnicity (diagnostic period 1975–1999). Cancer 2008;113:2575–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng VY, Scharschmidt TJ, Mayerson JL, Fisher JL. Incidence and survival in sarcoma in the United States: a focus on musculoskeletal lesions. Anticancer Res 2013;33:2597–604. [PubMed] [Google Scholar]
- 5.Malempati S, Hawkins DS. Rhabdomyosarcoma: review of the Children's Oncology Group (COG) Soft-Tissue Sarcoma Committee experience and rationale for current COG studies. Pediatr Blood Cancer 2012;59:5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwinger W, Klass V, Benesch M, Lackner H, Dornbusch HJ, Sovinz P, et al. Feasibility of high-dose interleukin-2 in heavily pretreated pediatric cancer patients. Ann Oncol 2005;16:1199–206. [DOI] [PubMed] [Google Scholar]
- 8.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol 1999;17:2105–16. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg SA, Lotze MT, Yang JC, Aebersold PM, Linehan WM, Seipp CA, et al. Experience with the use of high-dose interleukin-2 in the treatment of 652 cancer patients. Ann Surg 1989;210:474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielack SS, Smeland S, Whelan JS, Marina N, Jovic G, Hook JM, et al. Methotrexate, doxorubicin, and cisplatin (MAP) plus maintenance pegylated interferon alfa-2b versus map alone in patients with resectable high-grade osteosarcoma and good histologic response to preoperative MAP: first results of the EURAMOS-1 good response randomized controlled trial. J Clin Oncol 2015;33:2279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinerman ES, Jia SF, Griffin J, Seibel NL, Benjamin RS, Jaffe N. Phase II study of liposomal muramyl tripeptide in osteosarcoma: the cytokine cascade and monocyte activation following administration. J Clin Oncol 1992;10:1310–6. [DOI] [PubMed] [Google Scholar]
- 12.Chou AJ, Kleinerman ES, Krailo MD, Chen Z, Betcher DL, Healey JH, et al. Addition of muramyl tripeptide to chemotherapy for patients with newly diagnosed metastatic osteosarcoma: a report from the Children's Oncology Group. Cancer 2009;115:5339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagher R, Long LM, Read EJ, Leitman SF, Carter CS, Tsokos M, et al. Pilot trial of tumor-specific peptide vaccination and continuous infusion interleukin-2 in patients with recurrent Ewing sarcoma and alveolar rhabdomyosarcoma: an inter-institute NIH study. Med Pediatr Oncol 2002;38:158–64. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi S, Tsukahara T, Ida K, Kimura S, Murase M, Kano M, et al. SYT-SSX breakpoint peptide vaccines in patients with synovial sarcoma: a study from the Japanese Musculoskeletal Oncology Group. Cancer Sci 2012;103:1625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi S, Wada T, Ida K, Sato Y, Nagoya S, Tsukahara T, et al. Phase I vaccination trial of SYT-SSX junction peptide in patients with disseminated synovial sarcoma. J Transl Med 2005;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins PF, Kassim SH, Tran TLN, Crystal JS, Morgan RA, Feldman SA, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res 2015;21:1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Angelo SP, Melchiori L, Merchant MS, Bernstein D, Glod J, Kaplan R, et al. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 c259T cells in synovial sarcoma. Cancer Discov 2018;8:944–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paoluzzi L, Cacavio A, Ghesani M, Karambelkar A, Rapkiewicz A, Weber J, et al. Response to anti-PD1 therapy with nivolumab in metastatic sarcomas. Clin Sarcoma Res 2016;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Angelo SP, Mahoney MR, Van Tine BA, Atkins J, Milhem MM, Jahagirdar BN, et al. Nivolumab with or without ipilimumab treatment for metastatic sarcoma (Alliance A091401): two open-label, non-comparative, randomised, phase 2 trials. Lancet Oncol 2018;19:416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tawbi HA, Burgess M, Bolejack V, Van Tine BA, Schuetze SM, Hu J, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol 2017;18:1493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. [DOI] [PubMed] [Google Scholar]
- 22.Pardoll D. Cancer and the immune system: basic concepts and targets for intervention. Semin Oncol 2015;42:523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petitprez F, de Reyniès A, Keung EZ, Chen TW, Sun C, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577:556–60. [DOI] [PubMed] [Google Scholar]
- 24.D'Angelo SP, Shoushtari AN, Agaram NP, Kuk D, Qin L, Carvajal RD, et al. Prevalence of tumor-infiltrating lymphocytes and PD-L1 expression in the soft tissue sarcoma microenvironment. Hum Pathol 2015;46:357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oike N, Kawashima H, Ogose A, Hotta T, Hatano H, Ariizumi T, et al. Prognostic impact of the tumor immune microenvironment in synovial sarcoma. Cancer Sci 2018;109:3043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Erp AEM, Versleijen-Jonkers YMH, Hillebrandt-Roeffen MHS, van Houdt L, Gorris MAJ, van Dam LS, et al. Expression and clinical association of programmed cell death-1, programmed death-ligand-1 and CD8+lymphocytes in primary sarcomas is subtype dependent. Oncotarget 2017;8:71371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majzner RG, Simon JS, Grosso JF, Martinez D, Pawel BR, Santi M, et al. Assessment of programmed death-ligand 1 expression and tumor-associated immune cells in pediatric cancer tissues. Cancer 2017;123:3807–15. [DOI] [PubMed] [Google Scholar]
- 28.Dancsok AR, Setsu N, Gao D, Blay J, Thomas D, Maki RG, et al. Expression of lymphocyte immunoregulatory biomarkers in bone and soft-tissue sarcomas. Mod Pathol 2019;32:1772–85. [DOI] [PubMed] [Google Scholar]
- 29.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 30.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck AH, Espinosa I, Edris B, Li R, Montgomery K, Zhu S, et al. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res 2009;15:778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppola D, Nebozhyn M, Khalil F, Dai H, Yeatman T, Loboda A, et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol 2011;179:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T, et al. The immune landscape of cancer. Immunity 2018;48:812–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delespaul L, Lesluyes T, Pérot G, Brulard C, Lartigue L, Baud J, et al. Recurrent TRIO fusion in nontranslocation-related sarcomas. Clin Cancer Res 2017;23:857–67. [DOI] [PubMed] [Google Scholar]
- 38.Cannarile MA, Weisser M, Jacob W, Jegg A, Ries CH, Rüttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 2017;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley ER, Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol 2014;6:a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colbeck EJ, Ager A, Gallimore A, Jones GW. Tertiary Lymphoid structures in cancer: drivers of antitumor immunity, immunosuppression, or bystander sentinels in disease?. Front Immunol 2017;8:1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano C, George S. Leiomyosarcoma. Hematol Oncol Clin North Am 2013;27:957–74. [DOI] [PubMed] [Google Scholar]
- 42.Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, et al. Multidimensional, quantitative assessment of PD-1/PD-L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. J Immunother Cancer 2018;6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeong J, Lim JCT, Lee B, Li H, Ong CCH, Thike AA, et al. Prognostic value of CD8 + PD-1 + immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J Immunother Cancer 2019;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan KW, Chacko A, Chew V. PD-1 expression and its significance in tumour microenvironment of hepatocellular carcinoma. Transl Gastroenterol Hepatol 2019;4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shigeoka M, Urakawa N, Nakamura T, Nishio M, Watajima T, Kuroda D, et al. Tumor associated macrophage expressing CD204 is associated with tumor aggressiveness of esophageal squamous cell carcinoma. Cancer Sci 2013;104:1112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012;12:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boström MM, Irjala H, Mirtti T, Taimen P, Kauko T, Ålgars A, et al. Tumor-associated macrophages provide significant prognostic information in urothelial bladder cancer. PLoS One 2015;10:e0133552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang Z, Wen C, Chen X, Yin R, Zhang C, Wang X, et al. Myeloid-derived suppressor cell and macrophage exert distinct angiogenic and immunosuppressive effects in breast cancer. Oncotarget 2017;8:54173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espinosa I, Edris B, Lee C, Cheng HW, Gilks CB, Wang Y, et al. CSF1 expression in nongynecological leiomyosarcoma is associated with increased tumor angiogenesis. Am J Pathol 2011;179:2100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmid MC, Varner JA. Myeloid cells in the tumor microenvironment: modulation of tumor angiogenesis and tumor inflammation. J Oncol 2010;2010:201026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis CE, Harney AS, Pollard JW. The multifaceted role of perivascular macrophages in tumors. Cancer Cell 2016;30:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espinosa I, Beck AH, Lee C, Zhu S, Montgomery KD, Marinelli RJ, et al. Coordinate expression of colony-stimulating factor-1 and colony-stimulating factor-1-related proteins is associated with poor prognosis in gynecological and nongynecological leiomyosarcoma. Am J Pathol 2009;174:2347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forget MA, Voorhees JL, Cole SL, Dakhlallah D, Patterson IL, Gross AC, et al. Macrophage colony-stimulating factor augments Tie2-expressing monocyte differentiation, angiogenic function, and recruitment in a mouse model of breast cancer. PLoS One 2014;9:e98623. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Eubank TD, Galloway M, Montague CM, Waldman WJ, Marsh CB. M-CSF induces vascular endothelial growth factor production and angiogenic activity from human monocytes. J Immunol 2003;171:2637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019;576:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561–5. [DOI] [PubMed] [Google Scholar]
- 57.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sautès-Fridman C, Petitprez F, Calderaro J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer 2019;19:307–25. [DOI] [PubMed] [Google Scholar]
- 59.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.