Abstract
Antimicrobial resistance (AMR) is a global health issue that plays a significant role in morbidity and mortality, especially in immunocompromised patients. It also becomes a serious threat to the successful treatment of many bacterial infections. The widespread and irrelevant use of antibiotics in hospitals and local clinics is the leading cause of AMR. Under this scenario, the study was conducted in a tertiary care hospital in Lahore, Pakistan, from 2 August 2021 to 31 October 2021 to discover the prevalence of bacterial infections and AMR rates in COVID-19 patients admitted in surgical intensive care units (SICUs). Clinical samples were collected from the patients and we proceeded to identify bacterial isolates, followed by antibiotic susceptibility testing (AST) using the Kirby Bauer disk diffusion method and minimum inhibitory concentration (MIC). The data of other comorbidities were also collected from the patient’s medical record. The current study showed that the most common pathogens were E. coli (32%) and Klebsiella pneumoniae (17%). Most E. coli were resistant to ciprofloxacin (16.8%) and ampicillin (19.8%). Klebsiella pneumoniae were more resistant to ampicillin (13.3%) and amoxycillin (12.0%). The most common comorbidity was chronic kidney disease (CKD) and urinary tract infections (UTIs). Around 17 different types of antibiotic, the carbapenem, fluoroquinolones, aminoglycoside, and quinolones, were highly prevalent in ICU patients. The current study provides valuable data on the clinical implication of antibiotics consumed by COVID-19 patients in SICUs and the AMR rates, especially with different comorbidities.
Keywords: antibiotic susceptibility, antimicrobial resistance pattern, antimicrobial stewardship, comorbidity, COVID-19
1. Introduction
The pandemic of coronavirus disease 2019 (COVID-19) and antimicrobial resistance (AMR) are two simultaneous and interacting health concerns that provide significant learning opportunities. They may interact because, given the lack of particular therapies, there is a desire to employ current antimicrobials to treat critically ill COVID-19 patients [1]. The COVID-19 pandemic helps to illustrate the possible long-term effects of AMR, which is less severe but not less critical since their measurements and outcomes are comparable. Understanding how COVID-19 influences AMR trends and what to assume if they remain the same or increase in AMR will help us plan the next steps in addressing AMR [2].
COVID-19 infections have far exceeded bacterial co-infection and mortality rates compared to other common respiratory viral infections [2]. The co-infection of SARS-CoV-2 with other microbes, mainly bacteria and fungus, is a determining factor in COVID-19 development, making diagnosis, treatment, and prognosis more complicated. In individuals with COVID-19, bacterial co-infection has been linked to disease progression and prognosis. This scenario increases the need for critical care units, antibiotic therapy, and mortality [3]. Unfortunately, due to their widespread use, we may face the emergence of multi-drug resistant (MDR) pathogens leading to reduced efficacy of most potent antimicrobials [3,4]. AMR is a global problem that poses a severe threat to the success of treating a wide range of bacterial infections and affects many hospitalised patients, and most probably becomes a serious threat to the patients who are admitted to the SICUs [5,6].
Overall, the selection and development of highly drug-resistant bacteria due to the increased use of antibiotics and disinfectants may impact the clinical prognosis of severe COVID-19 patients receiving emergency hospital care, resulting in poor patient outcomes [7,8]. In this context, highly and extensively drug-resistant organisms have been documented to cause significant co-infections in COVID-19 patients, and mortality has recently been recorded in situations when bacterial co-infections were reported in COVID-19 patients [9,10].
The bacterial co-infections that arise during SARS-CoV-2 infection must be identified and characterised in a timely fashion [11,12]. Several studies have looked into the prevalence of bacterial co-infections in COVID-19 patients, finding highly heterogeneous distributions (with differences of >50%) that can be attributed to clinical and epidemiological characteristics of each geographical location, as well as diagnostic methods and criteria, used [13,14,15]. As a result, a study of hospitalised patients might increase our understanding of how viruses and bacteria interact during severe disease and give detailed information on COVID-19 in our environment. Similarly, identifying the main sociodemographic and clinical factors linked to bacterial co-infection in COVID-19 patients is critical to prioritising potential risk groups and institutional clinical and epidemiological surveillance programs to guide future etiological studies. Keeping in view the threat of high AMR rates in the COVID-19 pandemic, the current study was conducted to discover the prevalence of bacterial infections and AMR rates and other comorbidities with mortality rates in COVID-19 patients who were admitted to SICUs.
2. Materials and Methods
2.1. Ethical Consideration
Before starting the research, ethical approval was obtained from the Human Research Ethics Committee, University of Central Punjab, Lahore, Pakistan. Before proceeding with the sample collection, written informed consent was obtained from the patient (or from the dependent if the patient was unstable) before proceeding with the sample collection. Studied patients have been followed up for the consumption of antibiotics from the day of admission to the day of discharge (recovery or death). The demographic data (age, gender), comorbidities, recommended antibacterial drugs, the total number of given antibacterial drugs, and the brand names were recorded in a pre-defined data collection sheet.
2.2. Sample Collection
The current study was conducted by the Department of Microbiology, Faculty of Life Sciences, University of Central Punjab, Lahore, collaborating with a tertiary care hospital in Lahore, Pakistan, from 2 August 2021 to 31 October 2021. A total of 856 COVID-19 positive patients (admitted in SICUs) were recruited for the current study. To include more patients, only one type of sample was collected from each patient. The respiratory samples including sputum (n = 165), tracheal aspirate (n = 156), bronchoalveolar lavage (n = 117), and pleural fluid (n = 3) were collected with other samples including urine (n = 238), wound swab (n = 102), blood (n = 60), Foley’s catheter tip (n = 6), pus swab (n = 6), and abscess (n = 3) to proceed further for bacterial cultures. The samples were collected under sterile conditions and strict standard operating procedures (SOPs). After sample collection, these were immediately transported to the Microbiology laboratory at the University of Central Punjab, Lahore, for further processing.
2.3. Isolation and Identification of Bacterial Isolates
All samples except urine were proceeded for Gram staining first to see the microscopic characteristics of bacteria and then, based on the sample type, inoculated (including urine samples) on blood, MacConkey, chocolate and cysteine electrolyte deficient (CLED) agar media. After culture inoculation, the agar plates were incubated at 37 °C initially for 18 to 24 h. After first incubation, the agar plates were observed for the appearance of bacterial colonies. In case if there were no bacterial colonies, the plates were re-incubated for the second and third reading, respectively, except for blood cultures which were reinoculated and rechecked until the seventh day of sample collection. The negative plates were reported as “No bacterial growth” while the positive bacterial cultures further proceeded for bacterial identification and AST.
The final bacterial identification was undertaken using Gram staining, the appearance of bacterial colonies on the agar plates and biochemical identification. The colonies of Gram-positive bacteria proceeded to the catalase test first, and if the catalase test was positive, the colonies proceeded further for coagulase, DNAs, and Optochin disk tests, accordingly. However, the Gram-negative bacteria were first examined in terms of their appearance as lactose fermenters or non-fermenter and then proceeded for indole, citrate, and oxidase tests and the analytical profile indexing (API) biochemical identification kit.
2.4. Antibiotic Susceptibility Testing (AST)
After the isolation and identification of the organisms, the AST was performed to check their antibiotic susceptibility patterns using a Kirby Bauer disk diffusion assay and MIC (where applicable) [16]. The bacterial colonies were first mixed in a pre-prepared 0.5% MacFarland standard solution to dilute colonies to check the susceptibility patterns. After the inoculum of the test organism, these were inoculated three-dimensionally into a Muller Hinton (MH) agar (MH) agar plate with the help of a sterile cotton swab.
The antibiotics were checked according to the clinical laboratory standards institute (CLSI) guidelines (2017) for each microorganism [16]. The antibiotics were dispensed on inoculated MH agar plates and incubated for 18 to 24 h at 37 °C. After the incubation period, the zone of inhibition was measured using a labelled measuring scale. The CLSI guidelines followed the zone of inhibition (measured in mm) for each of the antibiotic v/s organisms. The final AST results were noted as resistant, sensitive or intermediate.
According to the CLSI guidelines (2017), the zone of inhibition for fosfomycin was only available to be reported in E. coli and Enterococcus faecalis isolates using the disk diffusion method. The sensitivity patterns were based on minimum inhibitory concentrations (MICs) for all other recommended bacterial isolates.
2.5. Statistical Analysis
The final data was recorded in Microsoft Excel and transferred to the SPSS version 26.0 (IBM, New York, NY, USA). The Frequencies and percentages were calculated from the recorded data. The association between the comorbidities and treatment outcome, type of bacterial infection, and severity of COVID-19 were analysed using the Pearson Chi-square (X2) test where p-value of <0.05 was considered statistically significant.
3. Results
3.1. Distribution of Clinical Bacterial Isolates among Coronavirus Disease 2019 (COVID-19) Patients
From the total 856 patient samples, n = 506 (59.11%) were male and n = 350 (40.88%) were females as shown in Table 1. A total of 342 (39.95%) samples were found to be positive for different bacterial isolates. Most of the bacterial cultures were found to be positive in urine (n = 136, 39.76%) and tracheal aspirate cultures (n = 51, 14.91%) followed by broncho-alveolar lavage (n = 48, 14.03%), wound swab (n = 37, 10.81%), blood (n = 32, 9.35%), sputum (n = 24, 7.01%), pus swab (n = 5, 1.46%), Foley’s catheter tip (n = 4, 1.16%), abscess (n = 3, 0.87%), and pleural fluid (n = 2, 0.58%).
Table 1.
Gender-wise and age group-wise distribution of coronavirus disease 2019 (COVID-19) patients in intensive care unit (ICU).
| Age Group (Years) | Gender | Total | |
|---|---|---|---|
| Male | Female | ||
| 01–20 | 72 | 74 | 146 |
| 21–40 | 99 | 102 | 201 |
| 41–60 | 132 | 76 | 208 |
| 61–80 | 181 | 86 | 267 |
| >80 | 22 | 12 | 34 |
| Total | 506 | 350 | 856 |
3.2. Clinical Isolates from Various Infections
The most common diagnosed comorbidity was chronic kidney disease (CKD), along with urinary tract infection (UTI) (21.4%), which was caused mainly by E. coli (50%), followed by the Klebsiella pneumoniae (20%). The second most diagnosed comorbidity was sepsis and hematuria (19.1%), also caused mainly by E. coli (52.9%), followed by Klebsiella pneumoniae (11.8%) and Proteus spp. (11.7%), respectively. The least common diagnosis was respiratory tract infections (1.11%), caused mainly by Klebsiella pneumoniae (100%). The BSI followed by the lung abscesses (1.07%) was caused mainly by the Klebsiella pneumoniae. (100%). The prevalence of bacterial isolates is shown in Table 2. Supplementary Materials: Figure S1 shows the beta hemolysis by Streptococcus spp. On blood agar after 24 h of incubation at 37 °C. Figure S2 shows the appearance of Gram-negative rods under observation at the 100× lens of the microscope. At the same time, Figure S3 shows the appearance of Gram-positive cocci. Figure S4 shows the positive bile esculin test, which is for Enterococcus faecalis. Figure S5 shows the positive results for Escherichia coli using the API kit.
Table 2.
The general prevalence of bacterial co-infections in COVID-19 patients admitted in ICU.
| Bacterial Isolates | n | % | |
|---|---|---|---|
| Gram-Positive bacteria (n = 39, 11.40%) | |||
| Staphylococcus aureus | MRSA | 4 | 1.16 |
| MSSA | 20 | 5.84 | |
| Enterococcus faecalis | Non-VRE | 8 | 2.33 |
| VRE | 1 | 0.29 | |
| Streptococcus agalactiae | 6 | 1.75 | |
| Gram-Negative bacteria (n = 303, 88.59%) | |||
| Escherichia coli | Non-CRE | 93 | 27.19 |
| CRE | 5 | 1.46 | |
| Klebsiella pneumoniae | Non-CRE | 68 | 19.88 |
| CRE | 16 | 4.67 | |
| Pseudomonas aeruginosa | 51 | 14.91 | |
| Acinetobacter baumannii | 49 | 14.32 | |
| Stenotrophomonas maltophilia | 5 | 1.46 | |
| Klebsiella oxytoca | 5 | 1.46 | |
| Citrobacter freundii | 4 | 1.16 | |
| Serratia liquefaciens | 4 | 1.16 | |
| Proteus vulgaris | 3 | 0.87 | |
MRSA: Methicillin-resistant S. aureus. MSSA: Methicillin sensitive S. aureus. VRE: Vancomycin resistant Enterobacteriaceae. CRE: Carbapenem resistant Enterobacteriaceae.
3.3. Antibiotic Susceptibility Patterns of Clinical Isolates
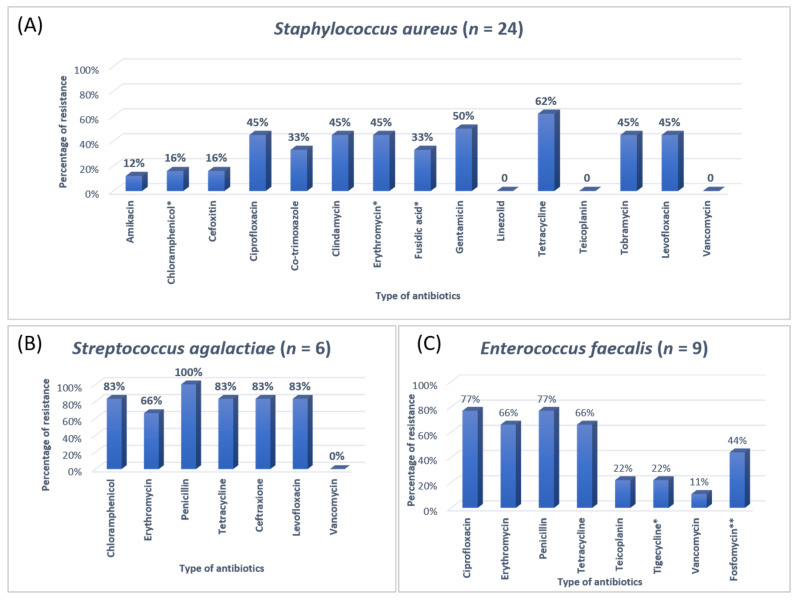
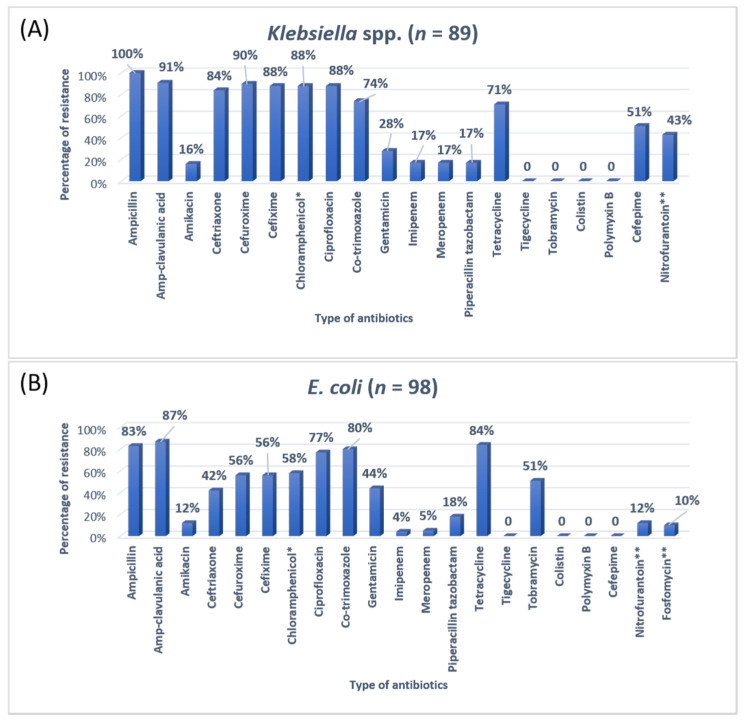
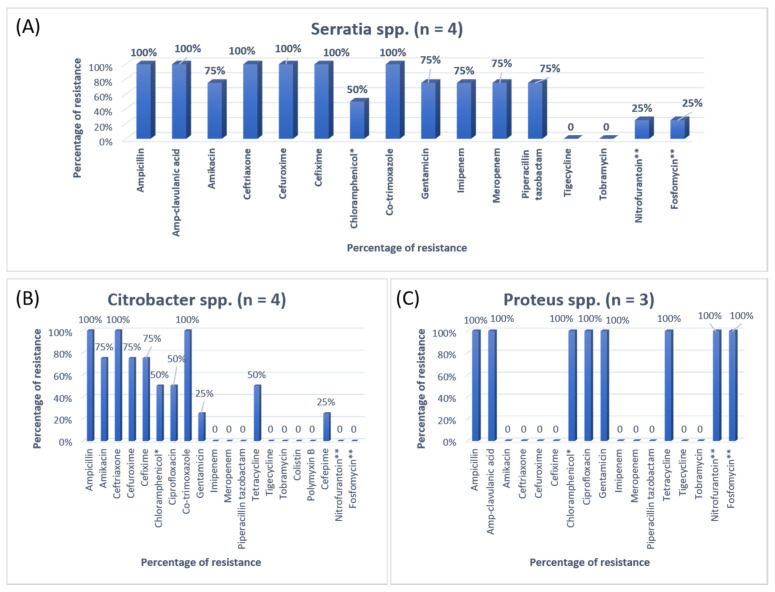
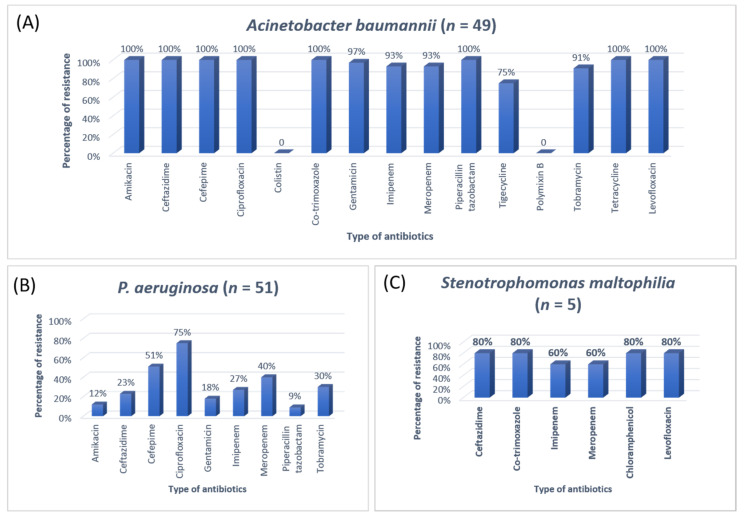
The antibiotic resistance patterns of individual bacterial isolates are shown in Figure 1, Figure 2, Figure 3 and Figure 4.
Figure 1.
The antibiotic resistance patterns in (A) Staphylococcus aureus, (B) Streptococcus agalactiae, (C) Enterococcus faecalis. * Not reported in urinary isolates. ** Reported in urinary isolates only.
Figure 2.
The antibiotic resistance patterns in (A) Klebsiella spp, (B) Escherichia coli * Not reported in urinary isolates. ** Only reported in urinary isolates.
Figure 3.
The antibiotic resistance patterns in (A) Serratia spp., (B) Citrobacter spp., (C) Proteus spp. * Not reported in urinary isolates. ** Only reported in urinary isolates.
Figure 4.
The antibiotic resistance patterns in (A) Acinetobacter baumannii, (B) Pseudomonas aeruginosa, (C) Stenotrophomonas maltophilia.
In SICUs, different antibiotics were prescribed and given to patients with a different diagnosis, as shown in Table 3.
Table 3.
Clinical isolates from various samples and their prescribed antibiotics.
| Variables | Drug Administration | Number (n) | Percentage (%) | |
|---|---|---|---|---|
| Type of antibiotic used | Vancomycin | IV | 23 | 6.72 |
| Teicoplanin | IV | 2 | 0.58 | |
| Linezolid | IV | 4 | 1.16 | |
| Imipenem | IV | 29 | 8.47 | |
| Meropenem | IV | 27 | 7.89 | |
| Amikacin | IV | 54 | 15.78 | |
| Gentamicin | IV/Oral | 36 | 10.52 | |
| Tobramycin | IV/Oral | 23 | 6.72 | |
| Nitrofurantoin | IV/Oral | 76 | 22.22 | |
| Fosfomycin | IV/Oral | 84 | 24.56 | |
| Piperacillin-tazobactam | IV | 62 | 18.12 | |
| Colistin | IV | 21 | 6.14 | |
| Polymyxin B | IV | 16 | 1.75 | |
| Ciprofloxacin | IV/Oral | 73 | 21.34 | |
| Ceftriaxone | IV | 34 | 9.94 | |
| Cefepime | IV | 23 | 6.72 | |
| Combination of antibiotics | One | 229 | 66.95 | |
| Multiple (two or more) | 113 | 33.04 | ||
| Days of antibiotic treatment | 1 to 7 days | 204 | 59.64 | |
| 8 to 14 days | 91 | 26.60 | ||
| >15 days | 47 | 18.71 | ||
IV: Intravenous.
3.4. Distribution of COVID-19 Patients with Major Comorbidities
Apart from COVID-19, 36.09% (n = 309) of patients also suffered from bacterial pneumoniae, UTIs, meningoencephalitis and sepsis. The patients with pneumoniae were also diagnosed with aspiration, diabetes mellitus (DM), chronic obstructive pulmonary disease (COPD), ischemic heart disease (IHD) and hypertension (HTN). The patients with UTIs, meningoencephalitis and sepsis were also diagnosed with other comorbidities, which were the main reason for mortality, as shown in Table 4.
Table 4.
Distribution of comorbidities among COVID-19 patients admitted in the intensive care unit and their outcome.
| Sr. No | Comorbidities | Total Number of Patients | Outcome | p-Value |
|---|---|---|---|---|
| 1. | Pneumonia, Aspiration, DM, HTN, IHD, COPD, CLD | 135 | 64 Recovered 71 Died |
<0.001 |
| 2. | UTIs, Dementia, DM, HTN, CKD | 140 | 121 Recovered 19 Died |
|
| 3. | Meningoencephalitis, Parkinsonism, HTN, Stroke | 2 | 2 Recovered | |
| 4. | Sepsis, DM, GI disorders | 32 | 29 Recovered 3 Died |
CKD: Chronic kidney diseases. CLD: Chronic liver disease. GI: Gastrointestinal.
A significant association (p < 0.001) was found between the COVID-19 patients, comorbidities and the mortality rate. The correlation between co-infections and COVID-19 severity has been shown in Table 5.
Table 5.
Distribution of co-infections among COVID-19 patients admitted in SICUs.
| Co-Infections | COVID-19 Severity | p-Value | ||
|---|---|---|---|---|
| Mild | Moderate | Severe | ||
| Upper respiratory tract infections | 4 | 5 | 15 | 0.028 |
| Lower respiratory tract infections | 13 | 21 | 67 | |
| Bacteremia | 3 | 8 | 21 | |
| Gastrointestinal infections | 4 | 23 | 18 | |
| Urinary tract infections | 11 | 41 | 88 | |
3.5. Mortality and Recovery Rate of the Patient
The recovery and death rate among the COVID-19 patients was recorded during their stay in the hospital. From the total 856 COVID-19 patients, the recovery rate was 76.16% (n = 652). The death rate was 23.83% (n = 204). The most common death rate among COVID-19 patients with different comorbidities was the CLD cases, followed by pneumoniae, UTIs, sepsis, cellulitis, pancreatic cancer, ascites, CKD, pyelonephritis rigours, cystitis and hematuria.
4. Discussion
Antibiotics are the most commonly prescribed drugs among hospitalised patients, especially in SICUs. It is imperative to use the appropriate antibiotics in intensive care units with few prescriptions as an acceptable quality of care, infection control, cost reduction, and length of hospital stay [17,18,19]. Patients admitted in the SICU are critically ill requiring prescribed the medicine without waiting for the culture reports that give information about the antimicrobial resistance pattern of the suspected organism for a specific cause [20,21]. In terms of the culture reports, there is no possibility to wait for reports because these take a minimum of 48 h, and as a result antibiotic resistance occurs in patients admitted to the SICUs [22]. The current study was conducted among COVID-19 patients admitted in SICUs of tertiary care hospitals, requiring monitoring and special care to analyse the antibiotics utilisation pattern and determine the prevalence of AMR. Our analysis included each organism’s antibiotics sensitivity and resistance pattern isolated from patients admitted in SICUs.
A previous study on microbial infection and antibiotic resistance patterns in COVID-19 patients admitted in SICUs of tertiary care hospitals showed that Pseudomonas was the most common organism identified in the medical ICU, followed by Klebsiella pneumonia [23]. A study on the prevalence of microorganisms and bacterial resistance in the SICU of the Bangabandhu Sheikh Mujib Medical University of Bangladesh showed that the maximum identified organism was Acetobacter. (45.4%), of P. aeruginosa (32.2%), Proteus (11%), Klebsiella pneumoniae 10%, and E. coli (3%) were identified [24]. A study by Mehta et al., 2015 on ICU patients revealed that the Pseudomonas spp. (29.1%) was the most common organism, followed by Acinetobacter spp. (27.5%) [18]. Another previous analysis of AST and bacteriology profile on patients at tertiary care hospitals in Ahmadabad showed that Acinetobacter spp. [30.9%] was the most common organism, after coming Klebsiella spp. (29.8%) and P. aeruginosa (22.9%) [19]. However, in the current study, the most common isolated organism was E. coli (38%), followed by Klebsiella pneumoniae (24%), P. aeruginosa (14%). While Streptococcus agalactiae, Citrobacter freundii, Serratia liqeuficiens, and Stenotrophomonas maltophilla were 1.7%, 1.1%, 1.1% and 1.4%, respectively.
In Jordan, a study was conducted to see the AMR rates, the prevalence of bacterial infections, and misuse of antibiotics. They conducted a study to resolve the resistance rate of Gram-negative bacteria in patients admitted to the ICU of Prince Hasen hospital. It revealed that P. aeruginosa was the most resistant pathogen and broad-spectrum antibiotic-resistant bacteria [15]. A similar previous study on the uropathogens of ICU showed that E. coli was highly susceptible to imipenem, meropenem, and nitrofurantoin [25]. The second most common organism in the current study was Klebsiella pneumoniae, 100% resistant to ampicillin and 91% to Amp-clavulanic acid. For Klebsiella pneumoniae, the treatment option was amikacin and gentamicin. Work on the AMR pattern of the bacterial pathogen in the ICU of Fatimawati hospital showed high resistance to cephalexin (96.3%), cefotaxime (64.3%), and ceftriaxone (61.0%) shown by P. aeruginosa. The most effective antibiotic against P. aeruginosa was amikacin after imipenem (81.3%) and meropenem (75.2%). Klebsiella pneumoniae showed resistance to cephalexin, ceftriaxone, ceftazidime (86.6%) (75.9%) (73.4%) respectively [26]. In the current study, the most common organism, E. coli, showed ampicillin resistance (83%) and Amp-clavulanic acid (87%). In that case, the most effective antibiotic was imipenem and meropenem. Most E. coli showed high sensitivity to the imipenem and meropenem (96%). Imipenem and meropenem are usually used against both GPIs and GNIs. However, a study conducted in India showed that Klebsiella was more susceptible to imipenem and nitrofurantoin and showed more resistance to penicillin and gentamicin [18].
Most of the Gram-positive organisms in the current study were resistant to penicillin and tetracycline. For the treatment of the Gram-positive bacteria, linezolid, gentamicin, and vancomycin were used. Most Gram-negative organisms were resistant to two or more alarming antibiotics and shortly caused high mortality and morbidity. This will also affect the management of Gram-negative bacteria. Around 17 different types of antibiotics were used in the ICU. Carbapenem, fluoroquinolones, aminoglycoside, and quinolones were highly consumed among all the antibiotics. In a study in India, the high consumption of beta-lactam, carbapenem, metronidazole was observed. Another study in Nepal showed that ampicillin, metronidazole, and amoxicillin were the most common antibiotics [27]. Another research study showed that beta-lactam, nitroimidazoles, and fluoroquinolones were commonly used in an ICU [12]. The prevalence of MRSA in the current study was 16%, VRE was 11%, and CRE 17%, showing a high rate of AMR against most effective drugs. These high rates of AMR in SICU patients are an alarming situation for all of the clinicians and researchers and the time to take some actions to ensure the correct usage of antibiotics.
Results of the current study showed that approximately 36% of COVID-19 patients had pneumonia followed by aspiration, diabetes mellitus (DM), chronic obstructive pulmonary disease (COPD), ischemic heart disease (IHD) and hypertension. UTIs, meningoencephalitis and sepsis followed by different comorbidities were also diagnosed. COVID-19 with bacterial pneumonia were the most important cause of distress and mortality. DM, HTN, CKD were significant comorbidities among ICU patients. Many patients had DM and HTN at the same time. Females experienced more complications and comorbidities than males. However, a previous systematic review and meta-analysis by Yang et al. (2020) reported that HTN and diabetes were the most common comorbidities, followed by cardiovascular diseases (CVD) and pneumonia. The pooled odds ratio of HTN, pneumonia and CVD in severe patients was lower than in non-severe patients [28]. We looked at the incidence of comorbidities in COVID-19 infection patients and discovered that underlying conditions, such as HTN, pneumonia, DM, CKD and COPD, might be a risk factor for high AMR rates among COVID-19 patients.
The current study was performed in a single hospital, and only those patients recruited for the current study were admitted in the SICUs. The sample size was smaller due to the single centre/unit study. Furthermore, multicentral studies with a larger sample size are recommended.
5. Conclusions
The current study points to a significant prevalence of bacterial infections and high AMR rates in COVID-19 patients, especially with specific comorbidities and complications, making it challenging to identify the priority of treatment groups and improve the care of these infections. These high rates of AMR may lead to high mortality rates; that is why a specific antibacterial usage policy of institutions and SICUs are needed to control this problem. Periodic audits are required to monitor compliance with these guidelines. In a hospital setup, antibiotic sensitivity and AMR patterns must be considered to direct the infection control expert or clinician to begin empirical antibiotics in severe cases. An intensive and systematic effort is needed to quickly classify high-risk patients and minimise the irrelevant use of antibiotics, one of the primary reasons for increased AMR rates in developing countries.
Acknowledgments
This research was supported by Taif University Researchers support project Number (TURSP-2020/62), Taif University, P.O. Box-11099, Taif-21944, Saudi Arabia.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11010035/s1, Figure S1: Clear beta hemolysis by Streptococcus. Figure S2: Gram stain showing Gram-negative bacteria. Figure S3: Gram stain showing Gram-positive bacteria. Figure S4: A positive bile asculine reaction (blacking of media). Figure S5: Result of E. coli on API.
Author Contributions
Conceptualisation, B.Z., M.I.K. and N.A. (Naveed Ahmed); methodology, B.Z.; software, N.A. (Nadia Afzal); validation, S.N.B., S.W.P. and N.A. (Naveed Ahmed); formal analysis, S.B., A.S. and S.W.P.; investigation, B.Z. and N.A. (Nadia Afzal) resources, B.Z.; data curation, A.S. and N.A. (Naveed Ahmed); writing—original draft preparation, B.Z., M.I.K., C.Y.Y. and S.B.; writing—review and editing, M.I.K., S.N.B., M.M., T.Y.N. and N.H.A.D.; visualisation, M.M., N.H.A.D., C.Y.Y. and T.Y.N.; supervision, B.Z. and M.I.K.; project administration, N.A.; funding acquisition, S.B. and A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (Human Research Ethics Committee) of the University of Central Punjab.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data will be shared upon a reasonable request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clancy C.J., Buehrle D.J., Nguyen M.H. PRO: The COVID-19 pandemic will result in increased antimicrobial resistance rates. JAC-Antimicrob. Resist. 2020;2:dlaa049. doi: 10.1093/jacamr/dlaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mondal M.K., Roy B.R., Yeasmeen S., Haque F., Huda A.Q., Banik D. Prevalence of microorganism and emergence of bacterial resistance in ICU of Bangabandhu Sheikh Mujib Medical University of Bangladesh. J. Bangladesh Soc. Anaesthesiol. 2013;26:20–26. doi: 10.3329/jbsa.v26i1.19811. [DOI] [Google Scholar]
- 3.Parveen S., Saqib S., Ahmed A., Shahzad A., Ahmed N. Prevalence of MRSA colonisation among healthcare-workers and effectiveness of decolonisation regimen in ICU of a Tertiary care Hospital, Lahore, Pakistan. Adv. Life Sci. 2020;8:38–41. [Google Scholar]
- 4.Ventola C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. Pharm. Ther. 2015;40:344. [PMC free article] [PubMed] [Google Scholar]
- 5.Pickens C.I., Wunderink R.G. Principles and practice of antibiotic stewardship in the ICU. Chest. 2019;156:163–171. doi: 10.1016/j.chest.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laxminarayan R., Duse A., Wattal C., Zaidi A.K., Wertheim H.F., Sumpradit N., Vlieghe E., Hara G.L., Gould I.M., Goossens H., et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith R., Coast J. The true cost of antimicrobial resistance. BMJ. 2013;346:1–5. doi: 10.1136/bmj.f1493. [DOI] [PubMed] [Google Scholar]
- 8.Dyar O.J., Castro-Sánchez E., Holmes A.H. What makes people talk about antibiotics on social media? A retrospective analysis of Twitter use. J. Antimicrob. Chemother. 2014;69:2568–2572. doi: 10.1093/jac/dku165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight G.M., Glover R.E., McQuaid C.F., Olaru I.D., Gallandat K., Leclerc Q.J., Fuller N.M., Willcocks S.J., Hasan R., van Kleef E., et al. Antimicrobial resistance and COVID-19: Intersections and implications. Elife. 2021;10:e64139. doi: 10.7554/eLife.64139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monnet D.L., Harbarth S. Will coronavirus disease (COVID-19) have an impact on antimicrobial resistance? Eurosurveillance. 2020;25:2001886. doi: 10.2807/1560-7917.ES.2020.25.45.2001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali Z., Jatoi M.A., Al-Wraikat M., Ahmed N., Li J. Time to Enhance Immunity via Functional Foods and Supplements: Hope for SARS-CoV-2 Outbreak. Altern. Ther. Health Med. 2020;27:30–44. [PubMed] [Google Scholar]
- 12.Adiga M.S., Alwar M., Pai M., Adiga U.S. Pattern of antimicrobial agents use in hospital deliveries: A prospective comparative study. Online J. Health Allied Sci. 2010;8:10. [Google Scholar]
- 13.Vincent J.-L., Rello J., Marshall J., Silva E., Anzueto A., Martin C.D., Moreno R., Lipman J., Gomersall C., Sakr Y., et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 14.Zahra N., Zeshan B., Qadri M.M.A., Ishaq M., Afzal M., Ahmed N. Phenotypic and Genotypic Evaluation of Antibiotic Resistance of Acinetobacter baumannii Bacteria Isolated from Surgical Intensive Care Unit Patients in Pakistan. Jundishapur J. Microbiol. 2021;14:e113008. doi: 10.5812/jjm.113008. [DOI] [Google Scholar]
- 15.Bataineh H.A., Alrashed K.M. Resistant gram-negative bacilli and antibiotic consumption in Zarqa, Jordan. Pak. J. Med. Sci. 2007;23:59–63. [Google Scholar]
- 16.Clinical Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical Laboratory Standards Institute (CLSI) Guidelines; CLSI; Wayne, PA, USA: 2017. CLSI Supplement M100. [Google Scholar]
- 17.Ahmed N., Ali Z., Riaz M., Zeshan B., Wattoo J.I., Aslam M.N. Evaluation of Antibiotic Resistance and Virulence Genes among Clinical Isolates of Pseudomonas aeruginosa from Cancer Patients. Asian Pac. J. Cancer Prev. APJCP. 2020;21:1333–1338. doi: 10.31557/APJCP.2020.21.5.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta T., Chauhan B., Rathod S., Pethani J., Shah P.D. Bacterilogical profile and drug resistance pattern of isolates of the patients admitted in medical intensive care unit of a tertiary care hospital in Ahmedabad. Med. Sci. 2015;4:222–225. [Google Scholar]
- 19.Barai L., Fatema K., Haq J.A., Faruq M.O., Ahsan A.A., Morshed M.A.H.G., Hossain M.B. Bacterial profile and their antimicrobial resistance pattern in an intensive care unit of a tertiary care hospital of Dhaka. Ibrahim Med. Coll. J. 2010;4:66–69. doi: 10.3329/imcj.v4i2.6499. [DOI] [Google Scholar]
- 20.Lockhart S.R., Abramson M.A., Beekmann S.E., Gallagher G., Riedel S., Diekema D.J., Quinn J.P., Doern G.V. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J. Clin. Microbiol. 2007;45:3352–3359. doi: 10.1128/JCM.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang S.Y., Kumar A. Empiric antimicrobial therapy in severe sepsis and septic shock: Optimising pathogen clearance. Curr. Infect. Dis. Rep. 2015;17:36. doi: 10.1007/s11908-015-0493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed N., Zeshan B., Naveed M., Afzal M., Mohamed M. Antibiotic resistance profile in relation to virulence genes fimH, hlyA and usp of uropathogenic E. coli isolates in Lahore, Pakistan. Trop. Biomed. 2019;36:559–568. [PubMed] [Google Scholar]
- 23.Sanjana R., Shah R., Chaudhary N., Singh Y. Prevalence and antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus (MRSA) in CMS-teaching hospital: A preliminary report. J. Coll. Med. Sci.-Nepal. 2010;6:1–6. doi: 10.3126/jcmsn.v6i1.3595. [DOI] [Google Scholar]
- 24.Radji M., Fauziah S., Aribinuko N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pac. J. Trop. Biomed. 2011;1:39–42. doi: 10.1016/S2221-1691(11)60065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuem K.B., Desta R., Bitew H., Ibrahim S., Hishe H.Z. Antimicrobial resistance patterns of uropathogens isolated between 2012 and 2017 from a tertiary hospital in Northern Ethiopia. J. Glob. Antimicrob. Resist. 2019;18:109–114. doi: 10.1016/j.jgar.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Mohamad N.A., Jusoh N.A., Htike Z.Z., Win S.L. Bacteria identification from microscopic morphology: A survey. Int. J. Soft Comput. Artif. Intell. Appl. (IJSCAI) 2014;3:1–12. doi: 10.5121/ijscai.2014.3201. [DOI] [Google Scholar]
- 27.Phillips-Jones M.K., Harding S.E. Antimicrobial resistance (AMR) nanomachines—Mechanisms for fluoroquinolone and glycopeptide recognition, efflux and/or deactivation. Biophys. Rev. 2018;10:347–362. doi: 10.1007/s12551-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020;10:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be shared upon a reasonable request to the corresponding author.