ABSTRACT
The activity of AMP-activated protein kinase α (AMPKα) is reduced in type 2 diabetes, and type 2 diabetes is associated with muscular atrophy. To date, there is little known about the mechanism by which free fatty acid (FFA) participates in muscular impairment. The purpose of the present study was to explore whether FFA damages myogenesis through the AMPKα-histone deacetylase 4 (HDAC4)-microRNA 206 (miR-206) pathway. The results showed that 1 mM FFA produced lipid accumulation, significantly impaired the insulin signaling pathway, and decreased the myogenic differentiation of C2C12 myoblast cells. FFA reduced the LKB1-AMPKα pathway, and the activation of AMPKα rescued the myogenic impairment caused by FFA (P < 0.05). AMPKα promoted myogenesis by regulating the expression of miR-206 through HDAC4 (P < 0.05) and affected the cell cycle and cell proliferation to promote myogenesis by regulating miR-206 and miR-206’s target cyclin D1 gene. In addition, AICAR (5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside) and HDAC4 small interfering RNA (siRNA) promoted myogenic differentiation compared with the FFA group; however, this positive effect was significantly downregulated after transfection with the miR-206 inhibitor. In summary, AMPKα plays positive roles in myogenic differentiation and myogenesis, and FFA decreased myogenic differentiation and myotube formation through the AMPKα–HDAC4–miR-206 pathway.
KEYWORDS: AMPKα, miR-206, cell cycle, FFA, myogenic differentiation
INTRODUCTION
Skeletal muscle makes up over 40% of body weight (1). In livestock, skeletal muscle is the main source of meat, determining the quality and taste of meat products (2, 3). It also plays a critical role in metabolism (4). One study found that it accounts for approximately 80% of insulin-mediated glucose uptake (5). During the past decades, skeletal muscle has been identified as a secretory organ (6), and the peptides secreted from skeletal muscle not only participate in muscle development but also interact with other organs, such as the liver, adipose tissue, and the immune system. Impairment of skeletal muscle is connected with the pathological processes of other organs (7, 8). Therefore, it has become increasingly recognized that skeletal muscle dysfunction is an important feature in many diseases, such as chronic obstructive pulmonary disease, amyotrophic lateral sclerosis (ALS), and type 2 diabetes (5, 9, 10).
Type 2 diabetes is associated with muscular atrophy, resulting in greater declines in muscle mass and muscle strength (11, 12), which deteriorates type 2 diabetes in turn. Elevated serum free fatty acid (FFA) levels and the occurrence of insulin resistance were found in type 2 diabetes patients; the accumulation of FFA is associated with impaired glucose metabolism and insulin sensitivity (13–16). AMP-activated protein kinase (AMPK) is a crucial cellular energy sensor (17). Once activated by energetic stress, it switches on catabolic pathways that produce ATP and switches off biosynthetic pathways that consume ATP (18). The phosphorylation of AMPKα reflects the activity of AMPK; the activity of AMPKα is reduced in type 2 diabetes, and the activation of AMPKα by drugs can significantly enhance glucose uptake and GLUT4 translocation, indicating its important role in the occurrence and treatment of type 2 diabetes (19–21). However, there is little known about whether AMPKα participates in muscular impairment associated with type 2 diabetes (22). MicroRNA (miRNA) 206 (miR-206), a skeletal muscle-specific miRNA, is involved in cell proliferation and differentiation of skeletal muscle by downregulating a large number of target genes, such as the genes for cyclin D1, Cx43, and glucose-6-phosphate dehydrogenase (G6PD) (23–25). miR-206 is decreased after FFA treatment, and Wu et al. found that the expression of miR-206 is reduced in high-fat diet (HFD)-fed mice, consistent with the activity of AMPKα (26). In addition, it is well documented that AMPKα modulates histone deacetylase 4 (HDAC4) (27); HDAC4 deacetylates miR-206’s promoter and decreases the expression of miR-206 (28), which makes us speculate that AMPKα regulates myogenesis by the HDAC4–miR-206 pathway.
In the current study, we investigated whether AMPKα participates in myogenic damage caused by FFA and explored whether AMPKα regulates myogenic differentiation through the HDAC4–miR-206 pathway in FFA-treated C2C12 cells. Our results demonstrate the positive role of AMPKα in myogenic differentiation and elucidate the regulatory mechanisms of FFA damaging skeletal muscle development through AMPKα–HDAC4–miR-206, which provides new insight to treat type 2 diabetes-associated skeletal muscle disease.
RESULTS
Myogenic differentiation was suppressed by 1 mM FFA.
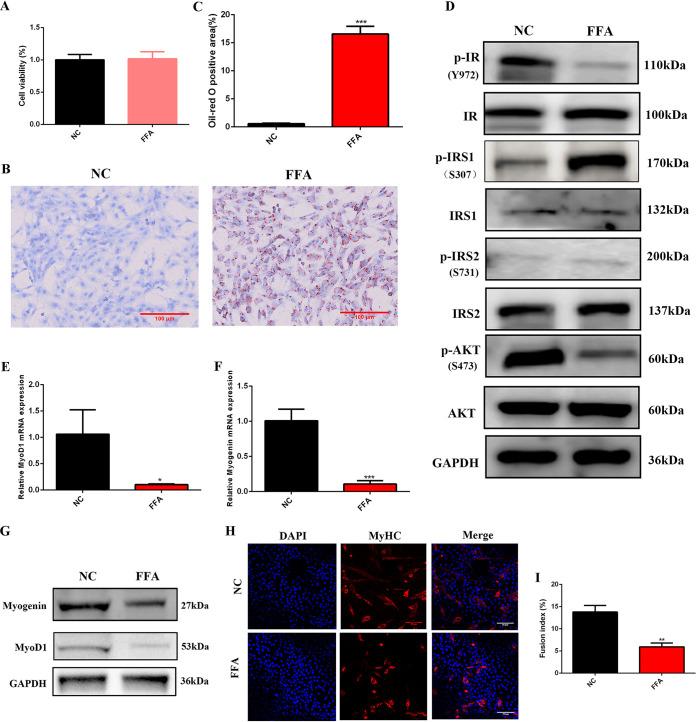
One micromolar FFA was used to treat C2C12 myoblasts. The cytotoxic effect of 1 mM FFA on C2C12 myoblast cells was determined by a cell counting kit 8 (CCK-8) assay. As shown in Fig. 1A, cell viability was not affected by 1 mM FFA. Oil red O staining indicated that 1 mM FFA significantly increased intracellular lipid accumulation (Fig. 1B and C). Insulin signaling was tested by Western blotting, and the results showed that 1 mM FFA significantly decreased the phosphorylation of insulin receptor (pY972-IR) and AKT (pS473-AKT) but increased the phosphorylation of insulin receptor substrate 1 (pS307-IRS1), a negative mediator of insulin signaling. There was no significant difference in the phosphorylation of insulin receptor substrate 2 (pS731-IRS2) (Fig. 1D). After FFA treatment, C2C12 myoblast cells were induced to differentiate for 3 days to explore whether FFA affects myogenic differentiation. The results showed that the MyoD1 and myogenin genes, two important myogenic differentiation-related genes, were significantly decreased in C2C12 myotubes after 1 mM FFA treatment (Fig. 1E to G). Myosin heavy chain (MyHC) is the marker of mature myotubes, and the fusion index was determined as the ratio of the number of nuclei in multinucleated myotubes (myotubes have two or more nuclei) to the total number of nuclei; the results showed that the myotubes’ infusion index was significantly suppressed after FFA treatment (Fig. 1H and I). All these results demonstrated that FFA impaired normal myogenic differentiation of skeletal muscle cells.
FIG 1.
Myogenic differentiation was suppressed by 1 mM FFA. (A) CCK-8 assay 24 h after 1 mM FFA treatment (n = 6). (B and C) Oil red staining 24 h after 1 mM FFA treatment (n = 3). (D) Western blotting of insulin signaling 24 h after 1 mM FFA treatment (n = 3). GAPDH was used as a loading control in all Western blot experiments. (E and F) Relative mRNA expression of MyoD1 and myogenin at 3 days of differentiation after 1 mM FFA treatment (n = 3). (G) Western blotting of MyoD1 and myogenin at 3 days of differentiation after 1 mM FFA treatment (n = 3). (H and I) Immunofluorescence staining of MyHC-positive cells at 3 days of differentiation after 1 mM FFA treatment (n = 3). *, P < 0.05 compared with NC; **, P < 0.01 compared with NC; ***, P < 0.001 compared with NC. Error bars indicate SD.
FFA impaired myogenic differentiation by inhibiting the activity of AMPKα.
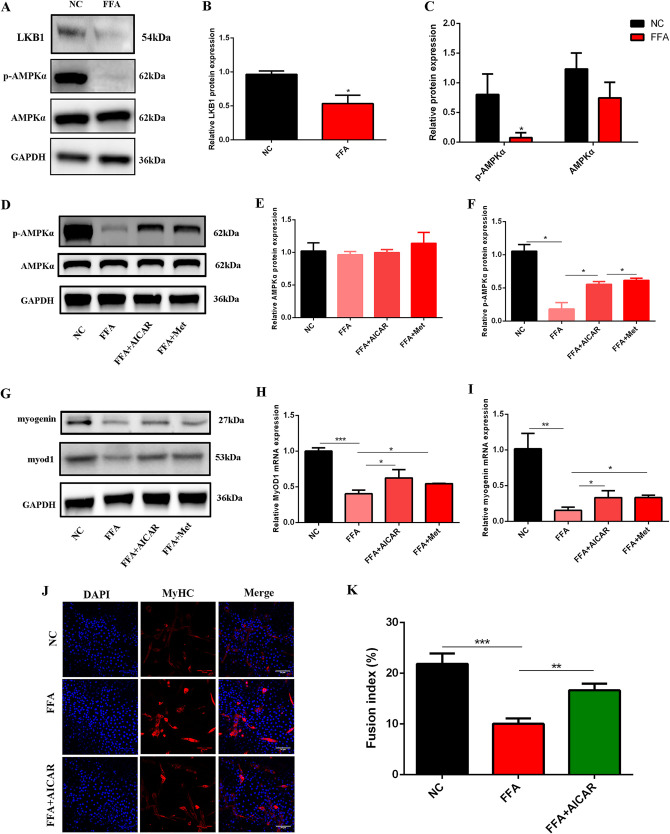
It is well documented that the LKB1-AMPKα signaling pathway was impaired after FFA treatment (29), so we next explored whether AMPKα is involved in myogenic damage induced by FFA. First, we detected the protein expression of LKB1, p-AMPKα, and AMPKα in FFA-treated C2C12 myoblast cells. The results showed that FFA significantly decreased LKB1 and p-AMPKα (Fig. 2A to C); the expression of AMPKα was constant (Fig. 2A and C). Next, to investigate whether decreased p-AMPKα is associated with myogenic damage, AICAR (5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside) and metformin, two activators of AMPKα, were used to rescue p-AMPKα. The results showed that both AICAR and metformin significantly increased the expression of p-AMPKα compared to FFA treatment (Fig. 2D to F). Myogenin and MyoD1 were increased after AICAR or metformin treatment compared to FFA (Fig. 2G to I). In addition, AICAR significantly promoted myogenic infusion compared to FFA (Fig. 2J and K). All these results illustrated that FFA inhibited myogenic differentiation by suppressing the activity of AMPKα; the activation of AMPKα rescued myogenic impairment caused by FFA, suggesting that AMPKα plays a positive role in myogenesis.
FIG 2.
FFA impaired myogenic differentiation by inhibiting the activity of AMPKα. (A to C) Western blotting of LKB1, p-AMPKα, and AMPKα after 1 mM FFA treatment (n = 3). *, P < 0.05 compared with NC. (D to F) Western blotting of p-AMPKα and AMPKα after FFA and AICAR-metformin (Met) cotreatment (n = 3). (G) Western blotting of myogenin and MyoD1 after FFA and AICAR-metformin cotreatment at 3 days of differentiation (n = 3). (H and I) Relative mRNA expression of MyoD1 and myogenin after FFA and AICAR-metformin cotreatment at 3 days of differentiation (n = 3). (J and K) Immunofluorescence assay for MyHC-positive cells at 3 days of differentiation after FFA and AICAR cotreatment (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate SD.
AMPKα promoted myogenesis during skeletal muscle development.
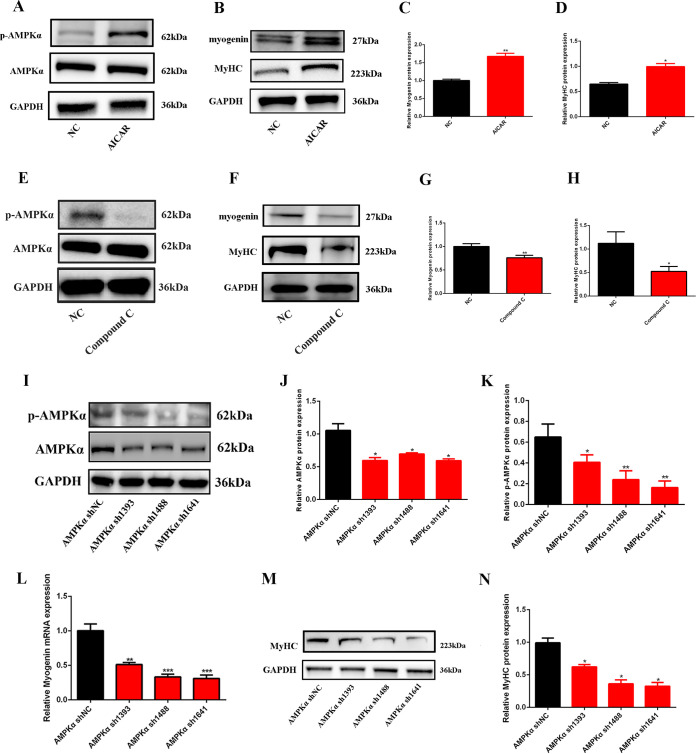
To further confirm the role of AMPKα in myogenesis, C2C12 cells were treated with AICAR, compound C, or AMPK short hairpin RNAs (shRNAs) to test the expression of myogenin and MyHC. The results showed that AICAR significantly increased the expression of p-AMPKα without changing total AMPKα (Fig. 3A); myogenin and MyHC were upregulated after AICAR treatment (Fig. 3B to D). Compound C, an AMPKα inhibitor, significantly inhibited the expression of p-AMPKα (P < 0.05) without changing the expression of AMPKα (P > 0.05) (Fig. 3E); myogenin and MyHC were significantly decreased after compound C treatment (P < 0.05) (Fig. 3F to H). In accordance with compound C, three AMPKα shRNAs were used to inhibit the expression of AMPKα. As shown in Fig. 3I to K, AMPKα shRNA significantly decreased the expression of AMPKα and p-AMPKα; meanwhile, the expression of myogenin and MyHC was also suppressed (Fig. 3L to N). All these results illustrated that AMPKα plays positive roles in myogenic differentiation and myotube formation.
FIG 3.
AMPKα promoted myogenesis during skeletal muscle development. (A) Western blotting of p-AMPKα and AMPKα after AICAR treatment (n = 3). (B to D) Western blotting of myogenin and MyHC after AICAR treatment (n = 3). (E) Western blotting of p-AMPKα and AMPKα after compound C treatment (n = 3). (F to H) Western blotting of myogenin and MyHC after compound C treatment (n = 3). (I to K) Western blotting of p-AMPKα and AMPKα after AMPKα shRNA transfection (n = 3). (L) Relative mRNA expression of myogenin after AMPKα shRNA transfection (n = 3). (M and N) Western blotting of MyHC after AMPKα shRNA transfection (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate SD.
AMPKα regulated the expression of miR-206.
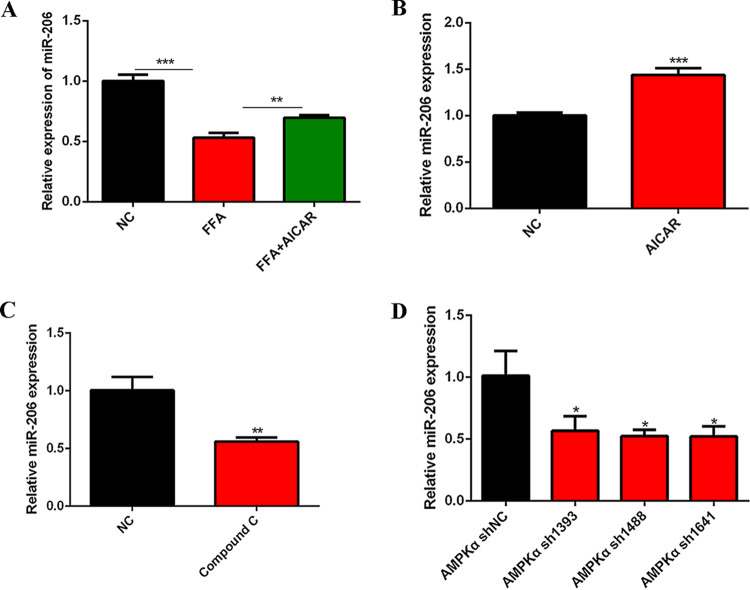
To investigate whether AMPKα participates in myogenesis through miR-206, we tested miR-206 expression after AICAR treatment in FFA-treated C2C12 cells. The results showed that FFA significantly reduced the expression of miR-206, and AICAR rescued the downregulation of miR-206 in FFA-treated C2C12 cells (Fig. 4A). Consistently, miR-206 was upregulated after AICAR treatment (Fig. 4B) and decreased after compound C or AMPKα shRNA treatment (Fig. 4C and D). All these results illustrate that AMPKα regulates the expression of miR-206. Considering the vital role of miR-206 in myogenesis, we next explored whether AMPKα regulates myogenesis through miR-206.
FIG 4.
AMPKα regulated the expression of miR-206. (A) Relative miR-206 expression after FFA and AICAR cotreatment for 24 h (n = 3). (B) Relative miR-206 expression after AICAR treatment (n = 3). (C) Relative miR-206 expression after compound C treatment (n = 3). (D) Relative miR-206 expression after AMPKα shRNA transfection (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate SD.
AMPKα promoted myogenesis by arresting the cell cycle and decreasing cell proliferation via miR-206.
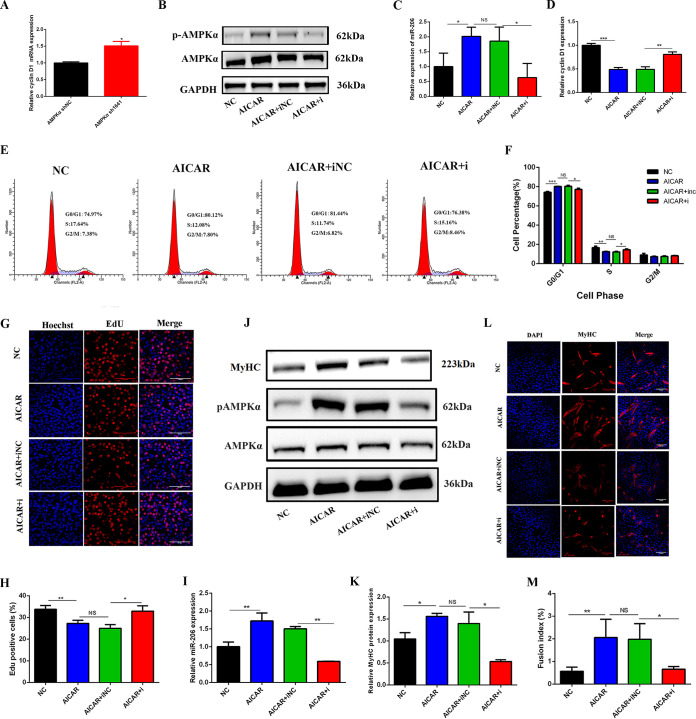
Cell cycle arrest and decreased cell proliferation are the two key events during myogenesis (23). miR-206 blocks the cell cycle and decreases cell proliferation to promote myogenesis by downregulating its target genes, such as cyclin D1 (30). Therefore, we explored whether AMPKα promoted myogenesis by arresting the cell cycle and decreasing cell proliferation through miR-206–cyclin D1. First, we found that the expression of cyclin D1 was increased after AMPKα shRNA transfection (Fig. 5A). Next, we confirmed that the activity of AMPKα regulates cyclin D1 through miR-206 by cotreatment with AICAR and an miR-206 inhibitor; p-AMPKα and miR-206 were significantly increased after AICAR treatment but decreased after cotreatment with AICAR and the miR-206 inhibitor, and the expression of AMPKα was not changed (Fig. 5B and C). On the contrary, cyclin D1 was significantly decreased after AICAR treatment; cotreatment of AICAR with the miR-206 inhibitor increased its expression (Fig. 5D).
FIG 5.
AMPKα promoted myogenesis by arresting the cell cycle and decreasing cell proliferation through miR-206. (A) Relative mRNA expression of cyclin D1 after AMPKα shRNA transfection (n = 3). (B) Western blotting of AMPKα and p-AMPKα in C2C12 myoblasts 24 h after cotreatment with AICAR and the miR-206 inhibitor (AICAR+i) (n = 3). (C) Relative expression of miR-206 in C2C12 myoblasts 24 h after cotreatment with AICAR and the miR-206 inhibitor (n = 3). (D) Relative expression of cyclin D1 in C2C12 myoblasts 24 h after cotreatment with AICAR and the miR-206 inhibitor (n = 3). (E and F) Effect of cotreatment with AICAR and the miR-206 inhibitor on the cell cycle (n = 3). C2C12 cells were treated with AICAR and the miR-206 inhibitor, and the cell cycle was then analyzed by flow cytometry 24 h later. The percentage of cells in each phase of the cell cycle was calculated using Modfit32 software. (G and H) Effect of cotreatment with AICAR and the miR-206 inhibitor on cell proliferation (n = 3). C2C12 cells were cotreated with AICAR and the miR-206 inhibitor, and proliferating cells were then stained with EdU 24 h later. The nuclei were stained blue, and EdU-positive cells were stained red. The percentage of EdU-positive cells was calculated with IPP software. (I) Relative expression of miR-206 in C2C12 myotubes at 3 days of differentiation after cotreatment with AICAR and the miR-206 inhibitor (n = 3). (J and K) Western blotting of MyHC, AMPKα, and p-AMPKα in C2C12 myotubes at 3 days of differentiation after cotreatment with AICAR and the miR-206 inhibitor (n = 3). (L and M) Immunofluorescence staining of MyHC-positive cells at 3 days of differentiation after cotreatment with AICAR and the miR-206 inhibitor (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. Error bars indicate SD.
Cell cycle analysis showed that AICAR significantly increased the percentage of cells in G0/G1 phase (P < 0.05) and decreased the percentage of cells in S phase (P < 0.05), whereas cotreatment with AICAR and the miR-206 inhibitor significantly decreased the percentage of cells in G0/G1 phase and increased the percentage of cells in S phase compared to the AICAR and miR-206 inhibitor negative control (NC) cotreatment group (P < 0.05) (Fig. 5E and F). Moreover, AICAR significantly decreased the percentage of 5-ethynyl-2′-deoxyuridine (EdU)-positive cells (P < 0.05), and cotreatment with AICAR and the miR-206 inhibitor significantly increased the percentage of EdU-positive cells compared with the AICAR and miR-206 inhibitor NC cotreatment group (P < 0.05) (Fig. 5G and H), indicating that AICAR caused cell cycle arrested in G0/G1 phase and inhibited the proliferation of skeletal muscle cells by regulating miR-206–cyclin D1.
C2C12 myoblasts were induced to differentiate for 3 days after AICAR and miR-206 inhibitor cotreatment. miR-206 and p-AMPKα were significantly increased in the AICAR group but decreased in the AICAR and miR-206 inhibitor cotreatment group (Fig. 5I and J). In addition, the protein expression of MyHC (Fig. 5J and K) and the fusion index of mature myotubes (Fig. 5L and M) were significantly increased in the AICAR treatment group but decreased in the AICAR and miR-206 inhibitor cotreatment group (Fig. 5J to M). All these results illustrated that the activation of AMPKα promoted myogenesis by arresting the cell cycle and decreasing cell proliferation through miR-206–cyclin D1.
AMPKα controlled the expression of miR-206 by regulating HDAC4.
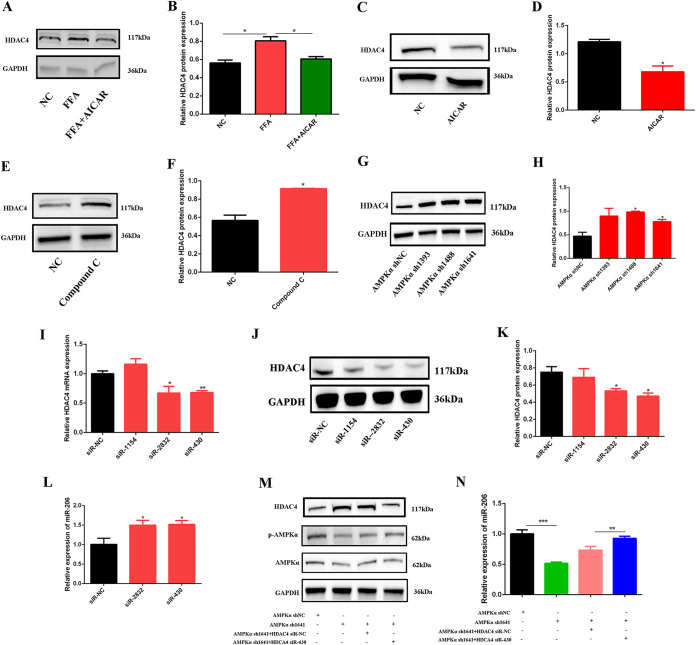
Next, to explore whether AMPKα regulates miR-206 through HDAC4, we tested the expression of HDAC4 after AICAR treatment in FFA-treated cells. As shown in Fig. 6A and B, HDAC4 was significantly increased after FFA treatment, and AICAR decreased HDAC4 compared to the FFA group. In addition, AICAR significantly decreased HDAC4 (Fig. 6C and D), and compound C and AMPKα shRNAs significantly increased HDAC4 (Fig. 6E to H).
FIG 6.
AMPKα controlled the expression of miR-206 by regulating HDAC4. (A and B) Western blotting of HDAC4 expression after FFA and AICAR cotreatment (n = 3). (C and D) Western blotting of HDAC4 expression after AICAR treatment (n = 3). (E and F) Western blotting of HDAC4 expression after compound C treatment (n = 3). (G and H) Western blotting of HDAC4 expression after AMPKα shRNA transfection (n = 3). (I) Relative mRNA expression of HDAC4 after HDAC4 siRNA transfection (n = 3). (J and K) Western blotting of HDAC4 expression after HDAC4 siRNA transfection (n = 3). (L) Relative miR-206 expression after HDAC4 siRNA transfection (n = 3). U6 was used as a control. (M) Western blotting of HDAC4, p-AMPKα, and AMPKα after cotreatment with AMPKα shRNA and HDAC4 siRNA (n = 3). (N) Relative miR-206 expression after cotreatment with AMPKα shRNA and HDAC4 siRNA (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate SD.
Next, HDAC4 small interfering RNAs (siRNAs) were used to knock down HDAC4. The results showed that HDAC4 siRNAs significantly decreased the expression of HDAC4 (Fig. 6I to K) and increased the expression of miR-206 (Fig. 6L). After cotreating C2C12 cells with AMPKα shRNA and HDAC4 siRNA, we found that AMPKα shRNA significantly decreased miR-206 expression, and HDAC4 siRNA rescued the downregulation of miR-206 caused by AMPKα shRNA (Fig. 6M and N). All these results illustrated that AMPKα controls the expression of miR-206 by regulating HDAC4.
FFA damaged myogenesis through the AMPKα–HDAC4–miR-206 pathway.
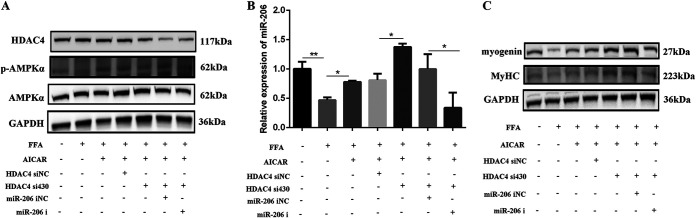
Next, we explored whether AMPKα regulates myogenesis through the HDAC4–miR-206 pathway in FFA-treated C2C12 cells by the coadministration of AICAR with HDAC4 siRNA and miR-206 mimics. The results showed that FFA decreased p-AMPKα (Fig. 7A) and miR-206 (Fig. 7B) and increased HDAC4 (Fig. 7A); meanwhile, myogenin and MyHC were decreased after FFA treatment (Fig. 7C). Cotreatment of AICAR with FFA increased the expression of p-AMPKα, miR-206, myogenin, and MyHC (Fig. 7A to C) and decreased HDAC4 (Fig. 7A) compared to the FFA group; transfection with HDAC4 siRNA further decreased HDAC4 expression (Fig. 7A) and increased the expression of miR-206, myogenin, and MyHC (Fig. 7B and C). The miR-206 inhibitor abolished the upregulation of miR-206, myogenin, and MyHC caused by AICAR and HDAC4 siRNA (Fig. 7B and C). All these results illustrated that FFA decreased the activity of AMPKα and then increased HDAC4, thereby decreasing the expression of miR-206 and causing the impairment of skeletal muscle cell differentiation.
FIG 7.
FFA damaged myogenesis through the AMPKα–HDAC4–miR-206 pathway. (A) Western blotting of AMPKα, p-AMPKα, and HDAC4 after cotreatment of AICAR with HDAC4 siRNA and the miR-206 inhibitor in FFA-treated C2C12 cells for 24 h (n = 3). (B) Relative miR-206 expression after cotreatment of AICAR with HDAC4 siRNA and the miR-206 inhibitor in FFA-treated C2C12 cells for 24 h (n = 3). (C) Western blotting of MyHC and myogenin after cotreatment of AICAR with HDAC4 siRNA and the miR-206 inhibitor in FFA-treated C2C12 cells for 24 h (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate SD.
DISCUSSION
IRS1 and IRS2 are the most important IRS proteins in insulin signaling (31). Studies found that IRS1 is crucial for the normal growth of myofibers and insulin-dependent glucose uptake in skeletal muscle, whereas the role of IRS2 in skeletal muscle is limited (32, 33). Consistently, we found that the phosphorylation of IRS1 at Ser307 was significantly increased after FFA treatment, which demonstrates that IRS1, but not IRS2, plays an important role in skeletal muscle cells. The decreased phosphorylation of AKT at Ser473 in the IRS1–phosphatidylinositol 3-kinase (PI3K) pathway suggests that the insulin signaling pathway is impaired (34). In this study, FFA treatment led to increased phosphorylation of IRS1 at serine sites and decreased phosphorylation of AKT at Ser473, indicating that the insulin signaling pathway was damaged. Lipid overload decreases skeletal muscle mass and inhibits myogenesis (35). In animals with insulin resistance induced by a high-fat diet (HFD), the proliferation and differentiation of skeletal muscle were impaired (36). Our study also indicates that FFA increased the accumulation of lipid and impaired skeletal muscle differentiation.
LKB1 is an important upstream kinase for AMPKα (37), and the expression of LKB1 is necessary for AMPKα activation (38, 39) in skeletal muscle. To date, the function of the LKB1-AMPKα pathway in FFA-treated cells or HFD-fed mice has been well documented (29, 40, 41). We confirmed that the LKB1-AMPKα signaling pathway was decreased after FFA treatment. Next, we speculated that the decreased activity of AMPKα may be connected with skeletal muscle damage. As expected, our results showed that both AICAR and metformin rescued the decreased expression of myogenin and promoted impaired myotube fusion under the conditions of FFA treatment, indicating that the decreased activity of AMPKα is one of the reasons for skeletal muscle impairment after FFA treatment, and this also provides new insight to explore the role of AMPKα in skeletal muscle development.
Recently, researchers found that AMPKα knockout (KO) mice are associated with reduced muscle mass (42), and Okamoto et al. found that the knockdown of AMPKα leads to a reduction of the myotube diameter (43). However, in dominant negative AMPKα (AMPK-DN) transgenic mice, muscles tend to be larger than those in wild-type (WT) mice, suggesting that AMPKα might negatively regulate basal muscle mass (44). In this study, we identified the positive role of AMPKα in myogenic differentiation by treating C2C12 cells with AMPKα shRNAs, AICAR, and compound C; myogenic differentiation was significantly reduced when the activity of AMPKα was inhibited by compound C or AMPKα shRNA but increased after AICAR treatment. These results indicate that the activity of AMPKα is critical for skeletal muscle differentiation.
The proliferation and differentiation of myogenic cells in embryonic and postnatal muscle play major roles in determining the rate and efficiency of muscle growth (45), and cell differentiation always requires an irreversible exit from the cell cycle (46). Our results found that AICAR increased miR-206 and decreased cyclin D1, leading to an arrest of the cell cycle in G0/G1 phase and a decrease of C2C12 cell proliferation. Interestingly, we found that that activation of AMPKα increased the expression of miR-206; meanwhile, the activity of AMPKα was suppressed when miR-206 was inhibited, which illustrates that bidirectional regulation may exist between AMPKα and miR-206; however, how miR-206 affects AMPKα activity needs to be further studied.
Finally, AMPKα shRNA decreased miR-206, and HDAC4 siRNA increased miR-206 expression compared to AMPKα shRNA, suggesting that AMPKα regulates miR-206 by HDAC4. FFA induced myogenic impairment, and this effect was decreased by AICAR and HDAC4 siRNA; however, the miR-206 inhibitor abolished the positive role of AICAR and HDAC4 siRNA in myogenesis. All these results confirmed that FFA impairs myogenic differentiation by inhibiting the AMPKα–HDAC4–miR-206 pathway.
In summary, this study demonstrates that AMPKα activity plays a positive role in myogenic differentiation and myogenesis. Mechanically, the activation of AMPKα reduces the expression of HDAC4, which thereby increases miR-206 and then decreases the expression of cyclin D1. AMPKα causes cell cycle arrest in G0/G1 phase and inhibits cell proliferation to promote myogenesis through miR-206–cyclin D1. FFA induces myogenic impairment by inhibiting the AMPKα–HDAC4–miR-206 pathway.
MATERIALS AND METHODS
Cell culture.
The mouse myoblast cell line (C2C12) was purchased from the Stem Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and seeded into T25 flasks. These cells were maintained in growth medium (GM), which comprised Dulbecco’s modified Eagle’s medium (DMEM; HyClone Laboratories, Logan, UT, USA) containing 10% fetal bovine serum (Millipore Sigma, Burlington, MA, USA), and differentiated in differentiation medium (DM), which comprised DMEM containing 2% horse serum (Millipore Sigma). Free fatty acid (FFA) was dissolved in 5% bovine serum albumin (BSA; Millipore Sigma), and its final concentration was 1 mM. 5-Aminoimidazole-4-carboxamide 1-β-d-ribofuranoside (AICAR) (catalog number S1802; Selleck) and compound C (catalog number S7306; Selleck) were dissolved in dimethyl sulfoxide (DMSO); the final concentration of compound C in the culture medium was 10 μM, and that of AICAR was 250 μM. Metformin (catalog number S5958; Selleck) was dissolved in diethyl pyrocarbonate (DEPC), and the final concentration was 1 mM. All cells were incubated at 37°C in an atmosphere containing 5% CO2 and 95% air.
CCK-8 analysis.
The cell viability of C2C12 myoblast cells after FFA treatment was evaluated by examining the conversion of 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium sodium salt (WST-8) to formazan through dehydrogenase in viable cells according to the manufacturer’s directions (Dojindo Laboratories, Kumamoto, Japan). Briefly, cells were seeded into 48-well plates and cultured for 24 h. After FFA treatments, 20 μL of cell counting kit 8 (CCK-8) solution was added to each well containing 200 μL of medium and incubated at 37°C for 2 h. Cell viability was then measured by reading the optical density at 450 nm under a microplate spectrophotometer (Tecan).
Oil red staining.
Oli red staining was conducted to test lipid accumulation in C2C12 myoblast cells after 1 mM FFA treatment for 24 h. Briefly, C2C12 myoblast cells were washed with phosphate-buffered saline (PBS) and then fixed in 4% paraformaldehyde for 30 min at ambient temperature. After that, the cells were stained using oil red for 10 min. After washing with PBS, the cell nuclei were stained using hematoxylin for 3 min. The morphology of the cells was observed under a light microscope (Olympus Corporation, Tokyo, Japan).
RNA oligonucleotides and cell transfection.
The miR-206 inhibitor is a single-strand RNA whose function is to reduce miR-206 expression; the miR-206 mimics are double-strand RNAs whose function is to promote miR-206’s function. Both the miR-206 inhibitor and the mimics were purchased from GenePharma (Shanghai, China), and their sequences are shown in Table 1. The sequence of HDAC4 siRNA and the target sequence of AMPKα shRNA are shown in Table 1. Transfection was performed using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
TABLE 1.
Sequences of RNA oligonucleotides
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| miR-206 inhibitor | CCACACACUUCCUUACAUUCCA |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
| miR-206 mimics | UGGAAUGUAAGGAAGUGUGUGG |
| ACACACUUCCUUACAUUCCAUU | |
| Negative control | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT | |
| HDAC4-siR1154 | CACCAUCCUUACCCAACAUTT |
| AUGUUGGGUAAGGAUGGUGTT | |
| HDAC4-siR2832 | GGUUAUGCCUAUCGCAAAUTT |
| AUUUGCGAUAGGCAUAACCTT | |
| HDAC4-siR430 | CACAGUUGCAUGAACAUAUTT |
| AUAUGUUCAUGCAACUGUGTT | |
| AMPKα shNC | TTCTCCGAACGTGTCACGT |
| AMPKα shRNA1393 | GGTAGTGAATGCATACCATCT |
| AMPKα shRNA1488 | GGAGVTATCTTCTGGACTTCA |
| AMPKα shRNA1641 | GCTCTCTCACTGGCTCTTTGA |
RNA extraction and real-time quantitative PCR.
Total RNA was extracted from cultured cells using TRIzol reagent (Thermo Fisher Scientific). mRNAs and miRNA were reverse transcribed into cDNA using PrimeScript RT master mix (TaKaRa Bio, Kusatsu, Japan) and a Mir-X miRNA real-time quantitative PCR (RT-qPCR) SYBR kit (TaKaRa Bio), respectively. RT-qPCR was performed on a Step-One Plus real-time PCR system using AceQ qPCR SYBR green master mix (TaKaRa Bio). Relative expression levels were calculated using the 2−ΔΔCT method. mRNA and miRNA expression levels were normalized against those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6, respectively. All primers were synthesized by Genewiz (Suzhou, China). The sequences of the primers are listed in Table 2.
TABLE 2.
Primer sequences for RT-qPCR
| Primer or gene name | Primer directiona | Primer sequence (5′–3′) |
|---|---|---|
| miR-206-specific primer | TCGGCAGGTGGAATGTAAGG | |
| Myogenin | F | TGGAGCTGTATGAGACATCCC |
| R | TGGACAATGCTCAGGGGTCCC | |
| MyOD1 | F | GCCCGCGCTCCAACTGCTCTGAT |
| R | TCTTTTGGGCGTGAAGAACCAG | |
| GAPDH | F | ATCACTGCCACCCAGAAGACT |
| R | CATGCCAGTGAGCTTCCCGTT | |
| HDAC4 | F | CTGCAAGTGGCCCCTACAG |
| R | CTGCTCATGTTGACGCTGGA | |
| Cyclin D1 | F | CACCGGCCTCTGGCTAAAC |
| R | CGCAGGCTTGACTCCAGAAG | |
F, forward; R, reverse.
Protein extraction and Western blot analysis.
Total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer (Applygen, Beijing, China). The protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Beyotime, Shanghai, China). Western blotting was performed to investigate protein expression. Briefly, samples (each containing 30 μg protein) were electrophoresed on a 4 to 20% ExpressPlus PAGE gel (GeneScript, Nanjing, Jiangsu, China) and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore Sigma). The membrane was subsequently blocked with 5% BSA (Millipore Sigma) prepared in Tris-buffered saline containing Tween 20 for 2 h at room temperature. Thereafter, the membrane was incubated with an anti-AMPKα rabbit polyclonal antibody (pAb) (1:1,000) (catalog number 5832; Cell Signaling Technology [CST], Danvers, MA, USA), an anti-pAMPKα rabbit pAb (1:1,000) (catalog number 2535; CST), an anti-MyHC mouse pAb (0.2 μg/mL) (catalog number MF20; DSHB, IA, USA), an anti-MyoD1 rabbit pAb (1:1,000) (catalog number 18943-1-AP; Proteintech, Wuhan, China), an antimyogenin rabbit pAb (1:1,000) (catalog number A6664; ABclonal, Woburn, MA, USA), an anti-pIRS1 rabbit pAb (2 μg/mL) (catalog number ab5599; Abcam, Cambridge, UK), an anti-pIR rabbit pAb (0.2 μg/mL) (catalog number ab5678), an anti-IRS1 rabbit pAb (1:1,000) (catalog number ab52167), anti-IR (1:1,000) (catalog number ab137747), anti-pIRS2 (0.5 μg/mL) (catalog number ab3690), anti-IRS2 (1:1,000) (catalog number ab134101), an anti-pAKT rabbit pAb (1:1,000) (catalog number 4060), an anti-AKT rabbit monoclonal antibody (mAb) (1:1,000) (catalog number 4691), an anti-HDAC4 rabbit pAb (1:1,000) (catalog number 17449-1-AP; Proteintech), or an anti-LKB1 rabbit pAb (1:1,000) (catalog number 10746-1-AP; Proteintech) overnight at 4°C. The membrane was then incubated with an anti-GAPDH rabbit pAb (1:2,000) (catalog number ab9485; Abcam) as an internal control. Next, the membrane was incubated with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (1:2,000) (catalog number 7074; Cell Signaling Technology, Danvers, MA, USA) for 2 h at room temperature. Signals were visualized using the WesternBright ECL chemiluminescent HRP substrate (Advansta, San Jose, CA, USA) and an ImageQuant LAS-4000 system (Fujifilm, Tokyo, Japan). Signal intensities were analyzed using ImageJ software.
Cell cycle analysis.
Twenty-four hours after transfection, cells grown in GM were trypsinized, washed with PBS, and fixed in precooled 75% (vol/vol) ethanol overnight at 4°C. The following day, ethanol was removed after centrifugation, and cells were resuspended in 400 μL PBS and then incubated with 20 μL of an RNase A solution for 30 min at 37°C. Finally, cells were incubated with 400 μL of a propidium iodide (PI) staining solution for 60 min at 4°C in the dark. The RNase A and PI staining solutions were obtained from a cell cycle assay kit (Vazyme, Nanjing, China). Samples were assessed on a FACSCalibur flow cytometer (Becton, Dickinson, San Diego, CA, USA). Data were analyzed using ModFit32 software (Verity Software House, Topsham, ME, USA). A total of 20,000 cells were analyzed per sample.
Cell proliferation assay.
Twenty-four hours after transfection, C2C12 cells were cultured in fresh GM containing 50 μM 5-ethynyl-2′-deoxyuridine (EdU) obtained from a Cell-Light EdU Apollo567 in vitro kit (RiboBio, Guangzhou, China) for 2 h. Next, cells were stained according to the manufacturer’s instructions. Cells were observed using a confocal microscope (LSM 700; Carl Zeiss, Oberkochen, Germany). Nuclei were stained blue with Hoechst 33342, and proliferating cells were stained red with EdU.
Immunofluorescence assay.
An immunofluorescence assay was performed to determine the MyHC-positive cells in C2C12 cells. C2C12 cells were induced to differentiate for 3 days. After that, C2C12 cells were washed 3 times with PBS (HyClone, Logan, UT) and then fixed in 4% paraformaldehyde at ambient temperature for 1 h. Cells were permeabilized with 0.5% Triton X-100 at 4°C for 10 min. Cells were immunostained with rabbit antibody against MyHC (1:100 dilution) overnight at 4°C, followed by blocking in 1% BSA for 1 h. Next, cells were incubated for 1 h with rhodamine (tetramethyl rhodamine isocyanate [TRITC])-conjugated goat anti-rabbit IgG (1:100 dilution) (ZSGB-Bio). Cell nuclei were stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) for 15 min. Fluorescence images were captured using a Zeiss LSM 710 Meta confocal microscope. The fusion index was determined as the ratio of the number of nuclei in multinucleated myotubes (myotubes have two or more nuclei) to the total number of nuclei.
Statistical analysis.
Statistical analyses were performed using Prism 6 software (GraphPad Software, La Jolla, CA, USA). Results were expressed as means ± standard deviations (SD), and error bars represent the SD from 3 replicates unless stated otherwise. Data were compared using two-tailed unpaired Student’s t test and one-way analysis of variance (ANOVA). A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This work was supported by Breeding and Reproduction in The Major Agricultural Plan of New Variety Innovation of Jiangsu Province (PZCZ201734), the Natural Science Foundation of Jiangsu Province (BK20130693), and The National Major Project of Breeding for Transgenic Pigs (2016ZX08006001-003).
We have no conflict of interest to declare.
Funding acquisition, Wangjun Wu and Honglin Liu; Investigation, Aiwen Jiang and Wangjun Wu; Methodology, Liangliang Zhang and Xiaoyu Jiang; Project administration, Wangjun Wu; Visualization, Xiying Zhang; Writing – original draft, Aiwen Jiang and Hongyun Guo; Writing – review & editing, Honglin Liu.
Contributor Information
Wangjun Wu, Email: wuwangjun321@163.com.
Honglin Liu, Email: liuhonglin@njau.edu.cn.
REFERENCES
- 1.Janssen I, Heymsfield SB, Wang Z, Ross R. 2000. Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. J Appl Physiol 89:81–88. 10.1152/jappl.2000.89.1.81. [DOI] [PubMed] [Google Scholar]
- 2.Ozawa S, Mitsuhashi T, Mitsumoto M, Matsumoto S, Itoh N, Itagaki K, Kohno Y, Dohgo T. 2000. The characteristics of muscle fiber types of longissimus thoracis muscle and their influences on the quantity and quality of meat from Japanese Black steers. Meat Sci 54:65–70. 10.1016/S0309-1740(99)00072-8. [DOI] [PubMed] [Google Scholar]
- 3.Plastow GS, Carrión D, Gil M, García-Regueiro JA, I Furnols MF, Gispert M, Oliver MA, Velarde A, Guàrdia MD, Hortós M, Rius MA, Sárraga C, Díaz I, Valero A, Sosnicki A, Klont R, Dornan S, Wilkinson JM, Evans G, Sargent C, Davey G, Connolly D, Houeix B, Maltin CM, Hayes HE, Anandavijayan V, Foury A, Geverink N, Cairns M, Tilley RE, Mormède P, Blott SC. 2005. Quality pork genes and meat production. Meat Sci 70:409–421. 10.1016/j.meatsci.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 4.Archer AE, Von Schulze AT, Geiger PC. 2018. Exercise, heat shock proteins and insulin resistance. Philos Trans R Soc Lond B Biol Sci 373:20160529. 10.1098/rstb.2016.0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFronzo RA, Tripathy D. 2009. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 32(Suppl 2):S157–S163. 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen BK, Febbraio MA. 2012. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol 8:457–465. 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 7.Giudice J, Taylor JM. 2017. Muscle as a paracrine and endocrine organ. Curr Opin Pharmacol 34:49–55. 10.1016/j.coph.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen BK. 2013. Muscle as a secretory organ. Compr Physiol 3:1337–1362. 10.1002/cphy.c120033. [DOI] [PubMed] [Google Scholar]
- 9.Jaitovich A, Barreiro E. 2018. Skeletal muscle dysfunction in chronic obstructive pulmonary disease. What we know and can do for our patients. Am J Respir Crit Care Med 198:175–186. 10.1164/rccm.201710-2140CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. 2009. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326:1549–1554. 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leenders M, Verdijk LB, van der Hoeven L, Adam JJ, van Kranenburg J, Nilwik R, van Loon LJ. 2013. Patients with type 2 diabetes show a greater decline in muscle mass, muscle strength, and functional capacity with aging. J Am Med Dir Assoc 14:585–592. 10.1016/j.jamda.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Bianchi L, Volpato S. 2016. Muscle dysfunction in type 2 diabetes: a major threat to patient’s mobility and independence. Acta Diabetol 53:879–889. 10.1007/s00592-016-0880-y. [DOI] [PubMed] [Google Scholar]
- 13.Boden G, Jadali F, White J, Liang Y, Mozzoli M, Chen X, Coleman E, Smith C. 1991. Effects of fat on insulin-stimulated carbohydrate metabolism in normal men. J Clin Invest 88:960–966. 10.1172/JCI115399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boden G, Cheung P, Stein TP, Kresge K, Mozzoli M. 2002. FFA cause hepatic insulin resistance by inhibiting insulin suppression of glycogenolysis. Am J Physiol Endocrinol Metab 283:E12–E19. 10.1152/ajpendo.00429.2001. [DOI] [PubMed] [Google Scholar]
- 15.Santomauro AT, Boden G, Silva ME, Rocha DM, Santos RF, Ursich MJ, Strassmann PG, Wajchenberg BL. 1999. Overnight lowering of free fatty acids with acipimox improves insulin resistance and glucose tolerance in obese diabetic and nondiabetic subjects. Diabetes 48:1836–1841. 10.2337/diabetes.48.9.1836. [DOI] [PubMed] [Google Scholar]
- 16.Randle PJ, Garland PB, Hales CN, Newsholme EA. 1963. The glucose-fatty acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet i:785–789. 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 17.Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13:251–262. 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Zhang L, Li B, Jiang H, Duan Y, Xie Z, Shuai L, Li J, Li J. 2018. AMP-activated protein kinase (AMPK) regulates energy metabolism through modulating thermogenesis in adipose tissue. Front Physiol 21:122. 10.3389/fphys.2018.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takaguri A, Inoue S, Kubo T, Satoh K. 2016. AMPK activation by prolonged stimulation with interleukin-1β contributes to the promotion of GLUT4 translocation in skeletal muscle cells. Cell Biol Int 40:1204–1211. 10.1002/cbin.10673. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y, Du X, Yao X, Zhao Y. 2019. Vildagliptin inhibits high free fatty acid (FFA)-induced NLRP3 inflammasome activation in endothelial cells. Artif Cells Nanomed Biotechnol 47:1067–1074. 10.1080/21691401.2019.1578783. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, Sim HA, Jung DY, Lim EY, Kim YT, Kim BJ, Jung MH. 2019. Poria cocus wolf extract ameliorates hepatic steatosis through regulation of lipid metabolism, inhibition of ER stress, and activation of autophagy via AMPK activation. Int J Mol Sci 20:4801. 10.3390/ijms20194801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X, Zhu M, Zhang S, Foretz M, Viollet B, Du M. 2016. Obesity impairs skeletal muscle regeneration through inhibition of AMPK. Diabetes 65:188–200. 10.2337/db15-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. 2006. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174:677–687. 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Fu Y, Liu K, Hou L, Zhang W. 2019. miR-206 regulates alveolar type II epithelial cell Cx43 expression in sepsis-induced acute lung injury. Exp Ther Med 18:296–304. 10.3892/etm.2019.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang A, Dong C, Li B, Zhang Z, Chen Y, Ning C, Wu W, Liu H. 2019. MicroRNA-206 regulates cell proliferation by targeting G6PD in skeletal muscle. FASEB J 33:14083–14094. 10.1096/fj.201900502RRRR. [DOI] [PubMed] [Google Scholar]
- 26.Wu H, Zhang T, Pan F, Steer CJ, Li Z, Chen X, Song G. 2017. MicroRNA-206 prevents hepatosteatosis and hyperglycemia by facilitating insulin signaling and impairing lipogenesis. J Hepatol 66:816–824. 10.1016/j.jhep.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng S, Zhang L, Liu X, Li G, Zhang B, Wang Z, Zhang H, Ma H. 2020. Low levels of AMPK promote epithelial-mesenchymal transition in lung cancer primarily through HDAC4- and HDAC5-mediated metabolic reprogramming. J Cell Mol Med 24:7789–7801. 10.1111/jcmm.15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW, Wakabayashi N, Dewi R, Boros LG, Gonzalez FJ, Gabrielson E, Wong KK, Girnun G, Biswal S. 2013. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest 123:2921–2934. 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo MS, Kim JH, Kim HJ, Chang KC, Park SW. 2015. Honokiol activates the LKB1-AMPK signaling pathway and attenuates the lipid accumulation in hepatocytes. Toxicol Appl Pharmacol 284:113–124. 10.1016/j.taap.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Alteri A, De Vito F, Messina G, Pompili M, Calconi A, Visca P, Mottolese M, Presutti C, Grossi M. 2013. Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle 12:3781–3790. 10.4161/cc.26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckstein SS, Weigert C, Lehmann R. 2017. Divergent roles of IRS (insulin receptor substrate) 1 and 2 in liver and skeletal muscle. Curr Med Chem 24:1827–1852. 10.2174/0929867324666170426142826. [DOI] [PubMed] [Google Scholar]
- 32.Previs SF, Withers DJ, Ren JM, White MF, Shulman GI. 2000. Contrasting effects of IRS-1 versus IRS-2 gene disruption on carbohydrate and lipid metabolism in vivo. J Biol Chem 275:38990–38994. 10.1074/jbc.M006490200. [DOI] [PubMed] [Google Scholar]
- 33.Higaki Y, Wojtaszewski JF, Hirshman MF, Withers DJ, Towery H, White MF, Goodyear LJ. 1999. Insulin receptor substrate-2 is not necessary for insulin- and exercise-stimulated glucose transport in skeletal muscle. J Biol Chem 274:20791–20795. 10.1074/jbc.274.30.20791. [DOI] [PubMed] [Google Scholar]
- 34.Smadja-Lamère N, Shum M, Déléris P, Roux PP, Abe J, Marette A. 2013. Insulin activates RSK (p90 ribosomal S6 kinase) to trigger a new negative feedback loop that regulates insulin signaling for glucose metabolism. J Biol Chem 288:31165–31176. 10.1074/jbc.M113.474148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamilarasan KP, Temmel H, Das SK, Al Zoughbi W, Schauer S, Vesely PW, Hoefler G. 2012. Skeletal muscle damage and impaired regeneration due to LPL-mediated lipotoxicity. Cell Death Dis 3:e354. 10.1038/cddis.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ussher JR, Koves TR, Cadete VJ, Zhang L, Jaswal JS, Swyrd SJ, Lopaschuk DG, Proctor SD, Keung W, Muoio DM, Lopaschuk GD. 2010. Inhibition of de novo ceramide synthesis reverses diet-induced insulin resistance and enhances whole-body oxygen consumption. Diabetes 59:2453–2464. 10.2337/db09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh HJ. 2016. Regulation of exercise-stimulated glucose uptake in skeletal muscle. Ann Pediatr Endocrinol Metab 21:61–65. 10.6065/apem.2016.21.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen T, Hill JT, Moore TM, Cheung ECK, Olsen ZE, Piorczynski TB, Marriott TD, Tessem JS, Walton CM, Bikman BT, Hansen JM, Thomson DM. 2020. Lack of skeletal muscle liver kinase B1 alters gene expression, mitochondrial content, inflammation and oxidative stress without affecting high-fat diet-induced obesity or insulin resistance. Biochim Biophys Acta 1866:165805. 10.1016/j.bbadis.2020.165805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan YY, Chen Y, Zhang Q, Zhuang JJ, Tian M, Chen HZ, Zhang LR, Zhang HK, He JP, Wang WJ, Wu R, Wang Y, Shi C, Yang K, Li AZ, Xin YZ, Li TY, Yang JY, Zheng ZH, Yu CD, Lin SC, Chang C, Huang PQ, Lin T, Wu Q. 2012. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat Chem Biol 8:897–904. 10.1038/nchembio.1069. [DOI] [PubMed] [Google Scholar]
- 40.Fei-Wang, Tian D-R, Tso P, Han J-S. 2012. Diet-induced obese rats exhibit impaired LKB1-AMPK signaling in hypothalamus and adipose tissue. Peptides 35:23–30. 10.1016/j.peptides.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Long J-K, Dai W, Zheng Y-W, Zhao S-P. 2019. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol Med 25:26. 10.1186/s10020-019-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu X, Zhao JX, Liang J, Zhu MJ, Foretz M, Viollet B, Du M. 2013. AMP-activated protein kinase mediates myogenin expression and myogenesis via histone deacetylase 5. Am J Physiol Cell Physiol 305:C887–C895. 10.1152/ajpcell.00124.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto S, Asgar NF, Yokota S, Saito K, Minokoshi Y. 2019. Role of the α2 subunit of AMP-activated protein kinase and its nuclear localization in mitochondria and energy metabolism-related gene expressions in C2C12 cells. Metabolism 90:52–68. 10.1016/j.metabol.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Mu J, Barton ER, Birnbaum MJ. 2003. Selective suppression of AMP-activated protein kinase in skeletal muscle: update on ‘lazy mice’. Biochem Soc Trans 31:236–241. 10.1042/bst0310236. [DOI] [PubMed] [Google Scholar]
- 45.Dayton WR, Hathaway MR. 1991. Myogenic cell proliferation and differentiation. Poult Sci 70:1815–1822. 10.3382/ps.0701815. [DOI] [PubMed] [Google Scholar]
- 46.Russo S, Tatò F, Grossi M. 1997. Transcriptional down-regulation of myogenin expression is associated with v-ras-induced block of differentiation in unestablished quail muscle cells. Oncogene 14:63–73. 10.1038/sj.onc.1200805. [DOI] [PubMed] [Google Scholar]