ABSTRACT
EF24, a curcumin analog, exerts a potent antitumor effect on various cancers. However, whether EF24 retards the progression of triple-negative breast cancer (TNBC) remains unclear. In this study, we explored the role of EF24 in TNBC and clarified the underlying mechanism. In a mouse model of TNBC xenograft, EF24 administration reduced the tumor volume, suppressed cell proliferation, promoted cell apoptosis, and downregulated long noncoding RNA human leukocyte antigen complex group 11 (HCG11) expression. In TNBC cell lines, EF24 administration reduced cell viability, suppressed cell invasion, and downregulated HCG11 expression. HCG11 overexpression reenhanced the proliferation and invasion of TNBC cell lines suppressed by EF24. The following mechanism research revealed that HCG11 overexpression elevated Sp1 transcription factor (Sp1) expression by reducing its ubiquitination, thereby enhanced Sp1-mediated cell survival and invasion in the TNBC cell line. Finally, the in vivo study showed that HCG11-overexpressed TNBC xenografts exhibited lower responsiveness in response to EF24 treatment. In conclusion, EF24 treatment reduced HCG11 expression, resulting in the degradation of Sp1 expression, thereby inhibiting the proliferation and invasion of TNBC cells.
KEYWORDS: EF24, lncRNA HCG11, proliferation, invasion, triple-negative breast cancer
INTRODUCTION
Triple-negative breast cancer (TNBC) is a heterogeneous breast cancer subtype characterized by the absence of progesterone receptor, estrogen receptor (ER), and human epidermal growth factor receptor 2 (1). The lack of ER renders TNBC patients do not respond to the targeted therapeutics for hormone receptor-positive breast cancer subtypes (2). In addition, TNBC usually shows an aggressive pattern of progression with a higher possibility of metastasis than other subtypes of breast cancer (3). The paucity of targeted therapy and the relatively aggressive biologic behavior place huge burdens on the clinical management of TNBC. Therefore, there is an urgent need to screen and develop effective therapeutic drugs for the treatment of TNBC.
Curcumin, a natural product derived from Curcuma longa, demonstrates a strong antitumor activity in a variety of cancers, including TNBC (4). However, the clinical application of curcumin is greatly limited by its low solubility and poor bioavailability (5). To overcome these challenges, attempts are being made to develop curcumin derivatives and analogs. EF24, an analog of curcumin, displays an increased bioavailability compared to curcumin (6). Meanwhile, the potent antitumor effect of EF24 has been widely confirmed. For instance, Lin et al. (7) reported that EF24 induced cell death in osteosarcoma cells by promoting ferroptosis. Yin et al. (8) found that EF24 suppressed the metastatic phenotype of cholangiocarcinoma in a mouse model of spontaneous tumor metastasis by inhibiting the nuclear factor kappa B-dependent pathway. Of note, Adams et al. (9) found that EF24 induced the apoptosis of MDA-MB-231 cells (a human TNBC cell line) by causing a cell cycle arrest at the G2/M phase, suggesting the potential regulatory effect on the progression of TNBC.
Long noncoding RNAs (lncRNAs), a type of noncoding RNAs (ncRNAs) with more than 200 nucleotides, are closely associated with the pathophysiological progression by modulating target gene expression (10). LncRNA human leukocyte antigen complex group 11 (HCG11) is a 5,688-nucleotide lncRNA encoded on human chromosome 6 (11) and plays a contradictory role in cancers. For instance, the research of Zhang et al. (12) showed that HCG11 was highly expressed in gastric cancer (GC) tissues and facilitated the proliferation of GC cells by targeting catenin beta 1. While the research of Wang et al. (13) clarified that HCG1 increased the expressions of caspase-3 and cleaved caspase-3, thereby enhancing the apoptosis of non-small-cell lung cancer cells. In breast cancer, high expression of HCG11 is related to poor overall survival (11) and the tumor specimens with high expression of HCG11 are always ER-negative (14). However, little is known about the specific biological function of HCG11 in TNBC.
In this study, we evaluated the effect of EF24 on the proliferation and migration of TNBC cells, investigated whether HCG11 was involved in the regulatory effect of EF24, hoping to provide evidence for the antitumor effect of EF24 on TNBC, and offered a novel insight into the pathological progression of TNBC.
RESULTS
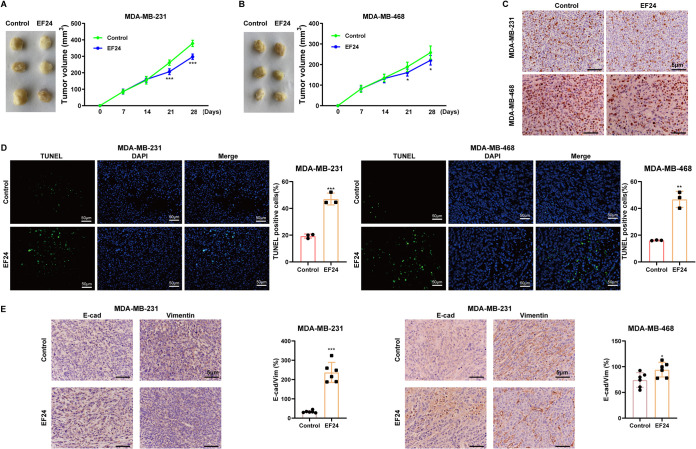
EF24 attenuated malignant phenotypes of TNBC cells in vivo.
The xenograft tumors of MDA-MB-231/MDA-MB-468 cells in nude mice were established. EF24 administration obviously reduced tumor volume in the mice xenograft model (Fig. 1A and B). Then, the proliferation and apoptosis of MDA-MB-231/MDA-MB-468 cells in vivo were evaluated using immunohistochemical staining for Ki-67 and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay, respectively. The results showed that the EF24-treated xenograft tumors exhibited fewer Ki-67 positive cells (Fig. 1C) and more TUNEL-positive cells (Fig. 1D) compared with vehicle-treated xenograft tumors. Meanwhile, the immunohistochemical staining for E-cadherin (E-cad; epithelial marker (15)) and vimentin (a mesenchymal marker (15)) demonstrated that EF24 administration increased E-cad-positive cell numbers while decreased vimentin-positive cell numbers in xenograft tumors (Fig. 1E). This suggested that EF24 retarded the epithelial-mesenchymal transition (EMT) of MDA-MB-231/MDA-MB-468 cells in vivo.
FIG 1.
EF24 attenuated malignant phenotypes of triple-negative breast cancer (TNBC) cells and decreased HCG11 expression in vivo. BALBC/c nude mice were divided into MDA-MB-231+control, MDA-MB-231+EF24, MDA-MB-468+control, and MDA-MB-468+EF24 groups (n = 6 for each group). On day 0, 1 × 106 MDA-MB-231/MDA-MB-468 were subcutaneously injected into mice. On days 14 to 28, EF24 (3 mg/kg/d) was intraperitoneally injected into mice in MDA-MB-231+EF24 and MDA-MB-468+EF24 groups. The xenograft tumor volumes were calculated at days 0, 7, 14, 21, and 28 in (A) MDA-MB-231+control and MDA-MB-231+EF24 groups, and (B) MDA-MB-468+control and MDA-MB-468+EF24 groups. On day 28, all mice were euthanized and the xenograft tumors were collected. (C) Representative images of immunohistochemical staining for Ki-67 (scale bars = 5 μm). (D) Representative images of TUNEL assay (scale bars = 50 μm). (E) Representative images of immunohistochemical staining for E-cadherin (E-cad) and vimentin (scale bars = 5 μm). Shapiro-Wilk test was used to check whether the data follow a normal distribution and F-test was used to verify if the variances were significantly different. F-test P < 0.05: a heteroskedasticity t test. F-test P ≥ 0.05: a homoscedastic t test. ***, P < 0.001 versus MDA-MB-231+control; *, P < 0.05; **, P < 0.01 versus MDA-MB-468+control.
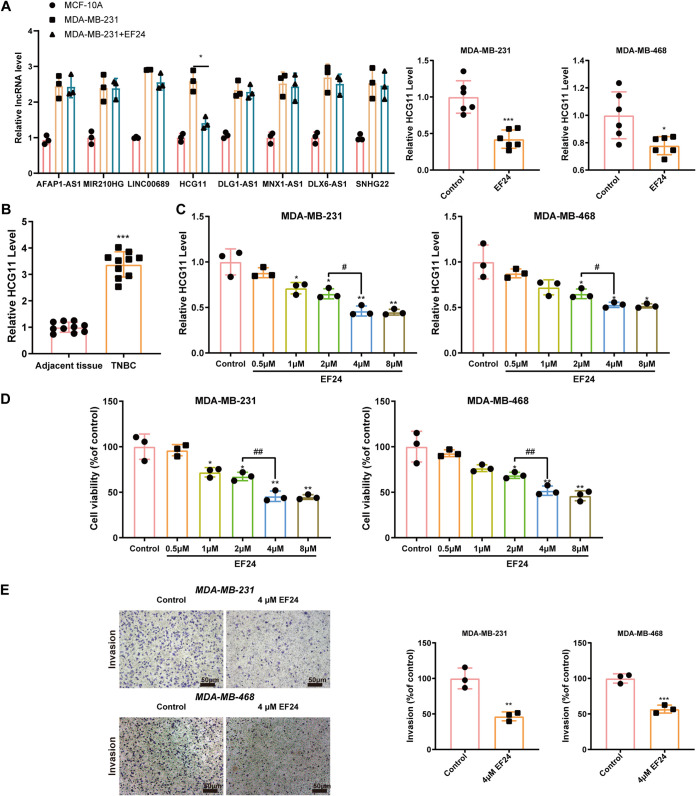
EF24 downregulated HCG11 expression, reduced cell viability, and suppressed cell invasion in TNBC cells in vitro.
To determine the downstream regulator of the EF24 effect in TNBC cells, the expression levels of 8 lncRNAs that have been reported to be dysregulated in breast cancer or TNBC (lncRNA AFAP1-AS1 (16), MIR210HG (17), linc00689 (18), HCG11 (14), DLG1-AS1 (19), MNX1-AS1 (20), DLX6-AS1 (21), SNHG22 (22)) were measured in MCF-10A, MDA-MB-231, and EF24-treated MDA-MB-231 cells. The results showed that the HCG11 level exhibited the greatest changes in response to EF24 treatment in MDA-MB-231 cells (Fig. 2A). Similarly, EF24 treatment could also lessen the HCG11 level in xenograft tumors (Fig. 2A). What’s more, HCG11 expression in cancerous tissues of TNBC patients was significantly higher than that in adjacent tissues of TNBC patients (Fig. 2B), suggesting a potential role of HCG11 in the progression of TNBC. EF24 treatment downregulated HCG11 level in a dose-dependent manner in both MDA-MB-231 and MDA-MB-468 cells and reached the threshold of HCG11 expression inhibition at 4 μM (Fig. 2C). Similarly, 4 μM EF24 showed the best suppressive effect on cell viability in MDA-MB-231 and MDA-MB-468 cells (Fig. 2D). Therefore, 4 μM EF24 was adopted in the following experiments. Except for reducing HCG11 expression and cell viability, EF24 treatment also lessened cell invasion in MDA-MB-231 and MDA-MB-468 cells (Fig. 2E).
FIG 2.
EF24 downregulated HCG11 expression, reduced cell viability, and suppressed cell invasion in TNBC cells in vitro. (A) Left: qRT-PCR analysis of candidate lncRNA expressions in MCD-10A, MDA-MB-231, and MDA-MB-231+EF24 (4 μm) cells (*, P < 0.05). Right: qRT-PCR analysis of HCG11 expression in xenograft tumors of mice from MDA-MB-231+control, MDA-MB-231+EF24, MDA-MB-468+control, and MDA-MB-468+EF24 groups (n = 6 for each group). (B) qRT-PCR analysis of HCG11 expression in the cancerous tissues and adjacent tissues of TNBC patients (n = 10). (C and D) MDA-MB-231 and MDA-MB-468 cells were incubated with EF24 (0, 0.5, 1, 2, 4, or 8 μm) for 48 h. (C) qRT-PCR analysis of HCG11 expression. (D) Methyl thiazolyl tetrazolium (MTT) assay of cell viability. (E) Transwell assay of cell invasion in MDA-MB-231 and MDA-MB-468 cells incubated with EF24 (0 or 4 μm) for 48 h (scale bars = 50 μm). Shapiro-Wilk test was used to check whether the data follow a normal distribution and F-test was used to verify if the variances were significantly different. F-test P < 0.05: a heteroskedasticity t test (two groups) and Brown-Forsythe and Welch ANOVA test (more than two groups). F-test P ≥ 0.05: a homoscedastic t test (two groups) and ordinary ANOVA test (more than two groups). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus adjacent tissues or control (MDA-MB-231/MDA-MB-468 cells treated with 0 μm EF24); #, P < 0.05; ##, P < 0.01.
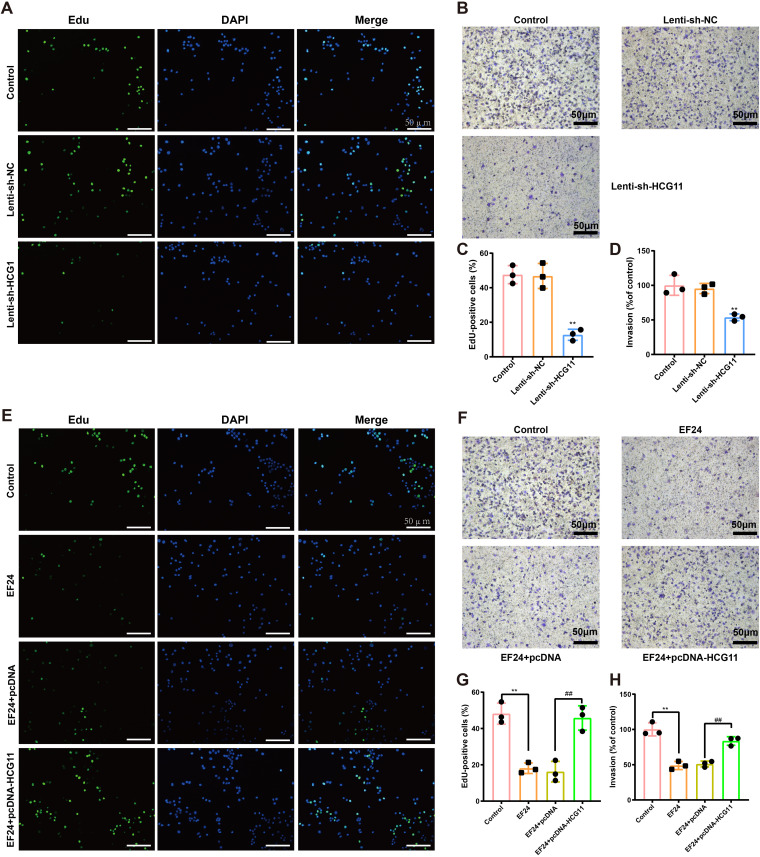
HCG11 mediated the inhibitory effect of EF24 on cell proliferation and invasion in TNBC cells.
Since an obvious elevation of HCG11 level was observed in cancerous tissues of TNBC patients (Fig. 2A), we then explored the role of HCG11 in TNBC. In response to Lenti-sh-HCG11 infection, the EdU-positive cell numbers were reduced in MDA-MB-231 cells (Fig. 3A and C), suggesting that HCG11 knockdown repressed the proliferation of TNBC cells. Meanwhile, the results of the Transwell assay showed that HCG11 knockdown decreased cell invasion in MDA-MB-231 cells (Fig. 3B and D). Considering EF24 treatment reduced HCG11 level and suppressed cell viability and invasion in TNBC cells (Fig. 2B to D), we hypothesized that the antitumor effect of EF24 was involved in its regulation of HCG11 expression. To verify our suspicion, MDA-MB-231 cells were transfected with pcDNA-HCG11, followed by incubation of EF24, and the proliferation and invasion of cells were evaluated. The results showed that HCG11 overexpression effectively reversed the inhibitory effect of EF24 on cell proliferation and invasion in MDA-MB-231 cells (Fig. 3E to H), indicating that EF24 inhibited proliferation and invasion of TNBC cells by suppressing HCG11 expression.
FIG 3.
HCG11 mediated the inhibitory effect of EF24 on cell proliferation and invasion in TNBC cells. (A to D) MDA-MB-231 cells were transfected with lentivirus (Lenti)-sh-HCG11 or its negative control (Lenti-sh-NC). Cell proliferation was measured using 5-ethynyl-2’-deoxyuridine (EdU) staining. Cell nuclei were counterstained with DAPI. Representative images (scale bar = 50 μm) were shown in panel A, and quantitative results were shown in panel C. Cell invasion was measured using Transwell assay. Representative images (scale bar = 50 μm) were shown in panel B and quantitative results were shown in panel D. (E to H) MDA-MB-231 cells were transfected with pcDNA-HCG11 or its negative control (pcDNA), followed by incubation of EF24. (E and G) Representative images (scale bar = 50 μm) (E) quantitative results (G) of EdU staining. (F and H) Representative images (scale bar = 50 μm) (F) quantitative results (H) of Transwell assay. Shapiro-Wilk test was used to check whether the data follow a normal distribution and F-test was used to verify if the variances were significantly different. F-test P < 0.05: Brown-Forsythe and Welch ANOVA test. F-test P ≥ 0.05: ordinary ANOVA test. (C and D) **, P < 0.01 versus Lenti-sh-NC; (G and H) **, P < 0.01; ##, P < 0.01.
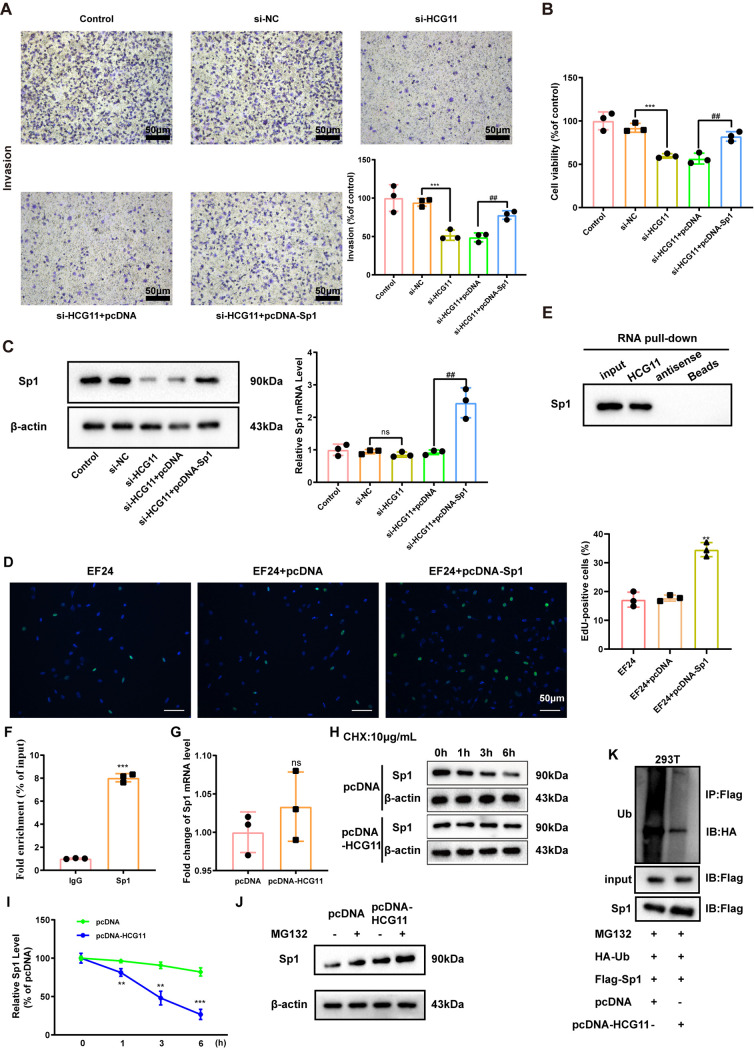
HCG11 modulate viability and invasion of TNBC cells by targeting Sp1.
Next, the mechanism by which HCG11 regulating proliferation and invasion of TNBC cells was investigated. Through a bioinformatics database RAID (http://www.rna-society.org/raid2/index.html), we found there was a possible combination between HCG11 and Sp1. As reported, SP1 is a transcription factor and facilitates TNBC progression by modulating the cell cycle and metastasis (23). Therefore, we then explored whether Sp1 was involved in the modulatory effect of HCG11 on the migration and viability of TNBC cells. Results showed that interferences of HCG11 suppressed migration and viability of MDA-MB-231 cells, which were then reversed by Sp1 overexpression (Fig. 4A and B). Meanwhile, interferences of HCG11 lessened Sp1 protein level without affecting its mRNA level in MDA-MB-231 cells (Fig. 4C). The results of the EdU assay showed that Sp1 overexpression reversed the inhibitory effect of EF24 on cell proliferation in MDA-MB-231 cells (Fig. 4D). The above data suggested that HCG11 exerted its modulatory effect on TNBC cells by increasing the Sp1 protein level.
FIG 4.
HCG11 modulate viability and invasion of TNBC cells by targeting Sp1 transcription factor (Sp1). (A to C) MDA-MB-231 cells were transfected with si-NC, si-HCG11, si-HCG11+pcDNA-Sp1, and si-HCG11+the negative control of pcDNA-Sp1 (pcDNA). (A) Cell invasion was measured using Transwell assay (scale bars = 50 μm). (B) Cell viability was measured using an MTT assay. (C) The protein and mRNA levels of Sp1 were measured using Western blot and qRT-PCR, respectively. β-Actin was served as the internal control. (D) Cell proliferation was measured by EdU staining in MDA-MB-231 cells treated with EF24, EF24+pcDNA, or EF24+pcDNA-Sp1. (E) Western blot analysis of Sp1 level in the complexes pulled down by biotinylated HCG11/antisense probe in MDA-MB-231 cells. (F) qRT-PCR analysis of HCG11 level in the immunoprecipitates of anti-Sp1/IgG antibody in MDA-MB-231 cells. (G) qRT-PCR analysis of Sp1 level in MDA-MB-231 cells transfected with pcDNA or pcDNA-HCG11. (H and I) MDA-MB-231 cells were transfected with pcDNA or pcDNA-HCG11, followed by incubating with 10 μg/ml cyclohexane (CHX) for 0, 1, 3, and 6 h. Representative bands were shown in panel H, and quantitative results were shown in panel I. (J) Western blot analysis of Sp1 in MDA-MB-231 cells treated with pcDNA, pcDNA+MG132 (10 μM, 2 h), pcDNA-HCG11, and pcDNA-HCG11+ MG132 (10 μM, 2 h). (K) Coimmunoprecipitation (Co-IP) analysis was performed to evaluate the combination between HA-ubiquitin (Ub) and Flag-Sp1 in 293T cells treated with MG132+HA-Ub+Flag-Sp1+pcDNA/pcDNA-HCG11. Shapiro-Wilk test was used to check whether the data follow a normal distribution and F-test was used to verify if the variances were significantly different. F-test P < 0.05: a heteroskedasticity t test (two groups) and Brown-Forsythe and Welch ANOVA test (more than two groups). F-test P ≥ 0.05: a homoscedastic t test (two groups) and ordinary ANOVA test (more than two groups). (A, B, C) ***, P < 0.001; ##, P < 0.01; ns, no significance; (D) **, P < 0.01 versus EF24+pcDNA; (F) ***, P < 0.001 versus IgG; (G) ns, no significance; and (I) **, P < 0.01; ***, P < 0.001 versus pcDNA.
Following, we explored the specific regulatory mechanism of HCG11 upregulating the Sp1 protein level. The results of RNA pulldown showed that biotinylated HCG11, rather than biotinylated antisense, was able to pull down abundant Sp1 in MDA-MB-231 cells (Fig. 4E). In the RIP experiment performed on MDA-MB-231 cells, the mixtures immunoprecipitated by anti-Sp1 antibody exhibited an obvious enrichment of HCG11 (Fig. 4F). Besides, HCG11 overexpression did not significantly change the Sp1 mRNA level (Fig. 4G) while significantly retarded by Sp1 protein degradation in the presence of CHX (a protein synthesis inhibitor) (Fig. 4H and I). When blocking the proteasome degradation pathway using MG132, the promoting effect of HCG11 overexpression on the Sp1 protein level disappeared (Fig. 4J), indicating that HCG11 mediated Sp1 protein degradation through the ubiquitin-proteasome system. Be consistent with this indication, the following coimmunoprecipitation (Co-IP) assay showed that HCG11 overexpression decreased the ubiquitination of Sp1 (Fig. 4L). To sum up, HCG11 directly combined with Sp1 and increased Sp1 protein level by decreasing its ubiquitination, and HCG11 knockdown suppressed invasion and viability of TNBC cells through suppressing Sp1 protein level.
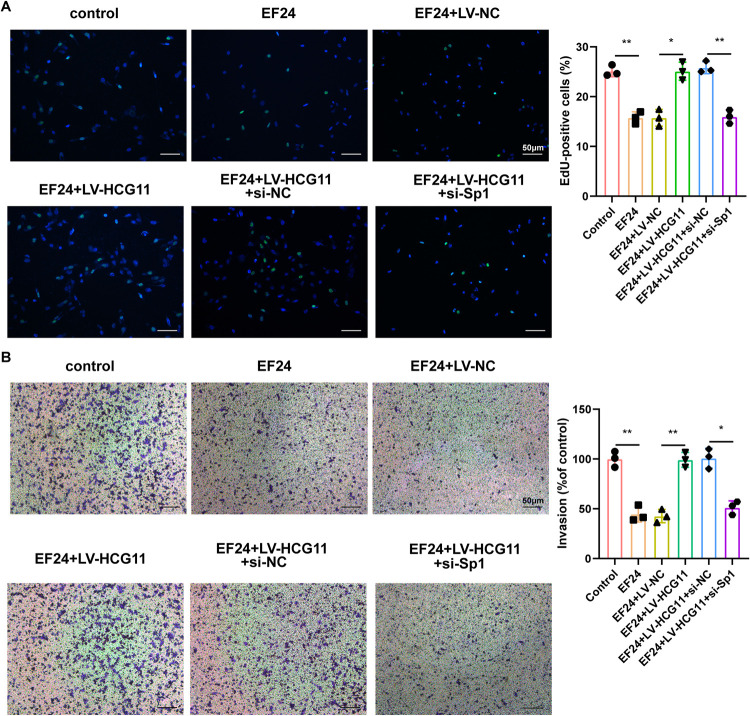
Sp1 mediated the regulatory effect of HCG11 on cell proliferation and invasion in EF24-treated TNBC.
The role of the EF24/HCG11/Sp1 axis on TNBC cell proliferation and migration was assessed in the other TNBC cell line (BT-549). EF24 treatment suppressed proliferation and invasion capacities of BT-549 cells. In response to HCG11 overexpression, the EdU-positive cells and invasion ratio were increased in BT-549 cells, while the promoting effects of HCG11 overexpression on the proliferation and invasion capacities of EF-24-treated BT-549 cells were removed by Sp1 knockdown (Fig. 5A and B).
FIG 5.
Sp1 mediated the regulatory effect of HCG11 on cell proliferation and invasion in EF24-treated TNBC cells. BT-549 cells were divided into control, EF24, EF24+lentivirus (LV)-NC, EF24+LV-HCG11, EF24+LV-HCG11+si-NC, EF24+LV-HCG11+si-Sp1. Cell (A) proliferation and (B) invasion were measured by EdU staining and Transwell assay. Shapiro-Wilk test was used to check whether the data follow a normal distribution and F-test was used to verify if the variances were significantly different. F-test P < 0.05: Brown-Forsythe and Welch ANOVA test. F-test P ≥ 0.05: ordinary ANOVA test.*, P < 0.05; **, P < 0.01.
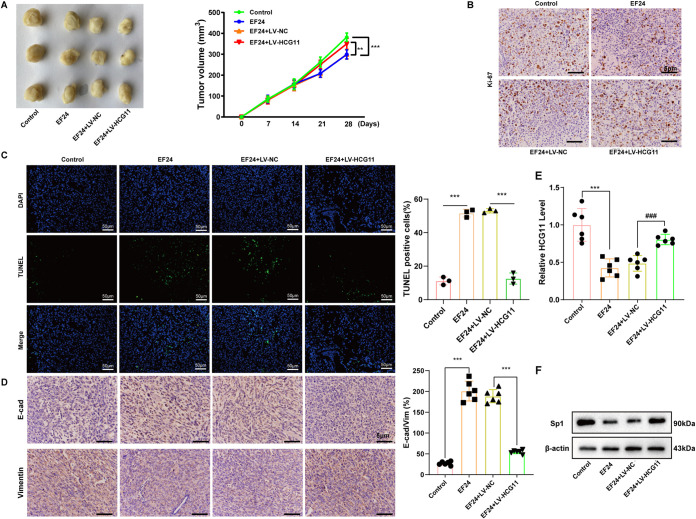
HCG11 overexpression eliminated the antitumor effect of EF24 on TNBC in vivo.
In the end, we evaluated whether HCG11 mediated the antitumor effect of EF24 on TNBC in vivo. Nude mice underwent a subcutaneous injection of MDA-MB-231, LV-NC-infected MDA-MB-231, or LV-HCG11-infected MDA-MB-231, and then treated with EF24 or vehicle. The results showed that compared with xenograft tumors of mice treated with LV-NC-infected MDA-MB-231+EF24 (EF24+LV-NC group), xenograft tumors of mice treated with LV-HCG11-infected MDA-MB-231+EF24 (EF24+LV-HCG11 group) exhibited a bigger tumor volume (Fig. 6A), increased Ki-67-positive cell numbers (Fig. 6B), decreased TUNEL-positive cell numbers (Fig. 6C), reduced E-cad-positive cell numbers (Fig. 6D), elevated vimentin-positive cell numbers (Fig. 6D), and upregulated HCG11 level (Fig. 6E). These data suggested that HCG11 overexpression abrogated the inhibitory effect of EF24 on the proliferation and EMT of TNBC cells and the promoting effect of EF24 on the apoptosis of TNBC cells. In addition, the Sp1 protein level in the xenograft tumors of mice treated with MDA-MB-231+EF24 (EF24 group) was lessened compared to xenograft tumors of mice treated with MDA-MB-231 (control group) (Fig. 6F). However, the Sp1 protein level in the xenograft tumors of mice treated with EF24+LV-HCG11-infected MDA-MB-231 (EF24+ LV-HCG11 group) was upregulated compared to xenograft tumors of mice treated with EF24+LV-NC -infected MDA-MB-231(EF24+ LV-NC group) (Fig. 6F). These data suggested that HCG11 overexpression reversed the antitumor effect of EF24 through increasing Sp1 expression.
FIG 6.
HCG11 overexpression eliminated the antitumor effect of EF24 on TNBC in vivo. BALBC/c nude mice were divided into control, EF24, EF24+LV-HCG11, and EF24+the negative control of LV-HCG11 (LV-NC) groups (n = 6 for each group). On day 0, 1 × 106 control/LV-HCG11-infected/LV-NC-infected MDA-MB-231 cells were subcutaneously injected into mice. On days 14 to 28, mice in EF24, EF24+LV-NC, and EF24+LV-HCG11 groups were treated with EF24 (3 mg/kg/d) and mice in the control group were treated with the same volume of vehicle. (A) Left: representative images of xenograft tumor in each group on day 28. Right: the xenograft tumor volumes were calculated at days 0, 7, 14, 21, and 28 in each group. (B) Representative images of immunohistochemical staining for Ki-67 performed on xenograft tumors (scale bar = 5 μm). (C) Representative images of TUNEL assay performed on xenograft tumors (scale bars = 50 μm). (D) Representative images of immunohistochemical staining for E-cadherin and vimentin performed on xenograft tumors (scale bars = 5 μm). (E) qRT-PCR analysis of HCG11 expression in xenograft tumors. (F) Western blot analysis of Sp1 protein level performed in xenograft tumors. Shapiro-Wilk test was used to check whether the data follow a normal distribution and F-test was used to verify if the variances were significantly different. F-test P < 0.05: Brown-Forsythe and Welch ANOVA test. F-test P ≥ 0.05: ordinary ANOVA test.**, P < 0.01; ***, P < 0.001; ###, P < 0.001.
DISCUSSION
Drugs derived from plants have been widely used as an adjunctive treatment strategy against cancer due to their potent bioactivity, lower toxicity, and limited adverse effects (24). In this study, we showed that EF24, an analog of curcumin, reduced HCG11 expression, resulting in the downregulation of Sp1 expression, thereby inhibiting the proliferation and invasion of TNBC cells. Our study may open up new avenues for the exploration of TNBC treatment.
EF24 is one of the top candidate curcumin analogs, and its IC50 value was 10 times lower than that of curcumin in the solid tumor cells, such as cervical and breast cancer cells (25). Recently, researchers found that EF24 exerts its antitumor effect by targeting ncRNAs. Zhang et al. (26) found that EF24 increased microRNA 33b (miR-33b) level and concomitantly reduced high mobility group AT-hook 2 expression, thereby suppressing EMT of melanoma cell lines at a noncytotoxic concentration. The research of Yang et al. (27) showed that the EF24 administration effectively repressed the growth of prostate cancer xenografts by decreasing miR-21 expression. However, no study has reported whether the antitumor effect of EF24 is involved in lnRNAs till now. In the present study, HCG11 possessed a low expression in EF24-treated TNBC xenografts and EF24-treated TNBC cell lines (Fig. 2). The subsequent in vitro study demonstrated that HCG11 overexpression reenhanced the cell proliferation and invasion suppressed by EF24 in TNBC cell lines (Fig. 3). Meanwhile, the in vivo study showed that HCG11-overexpressed TNBC xenografts exhibited lower responsiveness in response to EF24 treatment compared to control TNBC xenografts, manifesting as larger tumor xenografts size, upregulated cell proliferation, reduced cell apoptosis, and enhanced EMT (Fig. 6). Thus, our study provided evidence for the association of lncRNAs with the bioactivity of EF24 for the first time.
N6-methyladenosine (m6A) is the most common RNA modification in eukaryotic cells and has the ability to modulate RNA (including lncRNAs) splicing, stability, translation, and so on (28). As reported, the m6A level is closely related to the development of various cancers (29). The research of Shi et al. (30) showed that methyltransferase-like 3 (METTL3), a methyltransferase of m6A, was downregulated in TNBC tissues and the low expression of METTL3 increased collagen type III alpha 1 chain expression by decreasing the abundance of m6A, resulting in increased migration ability in TNBC cells, indicating that the progression of TNBC is associated with the downregulation of m6A level. Of note, it has been reported that curcumin could modulate downstream genes’ expressions through m6A modification. The research of Lu et al. (31) showed that curcumin inhibited stearoyl-coenzyme A desaturase 1 expression by increasing the m6A level in the liver of piglets. Therefore, we speculated that, as the analog of curcumin, EF24 might also have the ability to modulate the disordered m6A level, thereby downregulating the HCG11 level in TNBC cells. In the following research, we will validate this speculation to elucidate the specific mechanism by which EF24 downregulates HCG11 expression.
With the in-depth study of HCG11 in various cancers, researchers find that the role of HCG11 in cancer seems to be contradictory. In vestibular schwannoma cells, HCG11 was downregulated and HCG11 overexpression facilitated cell apoptosis via targeting the miR-620/ETS transcription factor ELK4 axis (32). Nevertheless, HCG11 possessed a high expression in ovarian cancer tissues and HCG11 knockdown reduced cell viability of ovarian cancer cells via regulating the miR-144-3p/PBX homeobox 3 axis (33). In our study, a nearly 3.5-fold elevation in HCG11 expression was observed in TNBC cancerous tissues compared to the adjacent tissues (Fig. 2). HCG11 silencing obviously reduced cell proliferative and invasive properties in the TNBC cell lines (Fig. 3). Meanwhile, HCG11 expression removed the suppressive effect of EF24 on cell proliferation and invasion in the TNBC cell line (Fig. 3). These data suggested the promoting effect of HCG11 on the progression of TNBC. So, what leads to the contradictory role of HCG11 in different cancers? As reported, lncRNAs can modulate target gene expression at epigenetic, transcriptional, and posttranscriptional levels, thereby participating in multiple biological processes (34). Therefore, the different roles of HCG11 in different types of cancers may be due to its specific target genes. To further explore the mechanism by which HCG11 modulates the proliferative and invasive properties of TNBC cells, we then determined the downstream gene of HCG11 in TNBC cells.
Through bioinformatics database prediction and experimental verification (RNA pulldown and RIP assays), we verified that HCG11 could directly bind to Sp1 in TNBC cells. SP1 is a member of the Sp transcription factor family and is responsible for the transcriptions of a huge number of genes that play important roles in the onset and development of various cancers (35). In patients with nasopharyngeal or gastric cancer, the high expression of Sp1 is considered to be an independent poor prognostic factor (36, 37). A gene-selective Sp1 inhibitor (mithramycin A) suppresses the growth of TNBC xenografts in vivo through decreasing Sp1-enhanced matrix metalloproteinase 2 transcription (38). Meanwhile, mithramycin A inhibits TNBC cell survival by suppressing the transcription of Krüppel-like factor 5 induced by Sp1 (23), suggesting the suppression of Sp1 is a potential intervention strategy for slowing down the progression of TNBC. In our study, we found that HCG11 reduced the Sp1 ubiquitination, thereby increasing the Sp1 protein level in TNBCs. The ubiquitin-proteasome system is a principal protein degradation pathway, which consists of two main steps: the ubiquitination of protein and the degradation of ubiquitinated protein by 26S proteasome (39). Like most proteins in the eukaryotic cells, the expression level of SP1 is also regulated by the ubiquitin-proteasome system. Cui et al. (40) reported that NEDD4 like E3 ubiquitin-protein ligase promoted the SP1 degradation by ubiquitinating SP1 at the K685 site. Dong et al. (41) reported that ubiquitin-specific peptidase 39, a deubiquitinating enzyme, stabilized SP1 through the deubiquitylation pathway. The results of our study, for the first time, suggested that the ubiquitination of Sp1 can be regulated by lncRNA. Consistent with our study, several studies confirmed the ability of lncRNAs to modulate the ubiquitination of downstream genes at the post-translation level (42, 43). For example, the research of Zhu et al. (44) showed that lncRNA GAS5 enhanced itchy E3 ubiquitin-protein ligase-mediated thioredoxin-interacting protein ubiquitination, thereby reducing its expression. In the follow-up research work, we will further explore whether HCG11 suppressed SP1 ubiquitination by weakening the combination between ubiquitin-protein ligase and Sp1 or enhancing the combination between deubiquitinating enzyme and Sp1.
To sum up, the present study confirms the therapeutic effect of EF24 on TNBC and clarifies its specific mechanism. Meanwhile, the results of our study emphasize the potential of HCG11 as a novel intervention target for TNBC treatment.
MATERIALS AND METHODS
Cell culture and transfection.
Human TNBC cell lines MDA-MB-231 and MDA-MB-468 (Procell Life Science & Technology, Wuhan, China) were maintained in Leibovitz's L-15 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution. Human TNBC BT-549 cell lines (Procell Life Science & Technology) were maintained in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin solution.
si-HCG11, si-SP1, pcDNA-Sp1, and the negative controls (si-NC and pcDNA) were synthesized by Ribobio Biotechnology (Guangzhou, China). According to the desired experimental protocol, si-HCG11/si-NC/pcDNA-Sp1/pcDNA were transfected into cells utilizing Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) under the manufacturer’s protocol. The lentivirus (Lenti)-sh-HCG11, lentivirus (LV)-HCG11, and the negative control (Lenti-sh-NC and LV-NC) were synthesized by YunZhou Biotechnology (Guangzhou, China). For infection, Lenti-sh-HCG11/Lenti-sh-NC/LV-HCG11/LV-NC were infected into well-grown MDA-MB-231 or BT-549 cells (multiplicity of infection = 10) using Polybrene (6 μg/ml) (Mito Biotechnology, Shanghai, China).
Mouse xenograft model.
Female BALBC/c nude mice were purchased from Cyagen Biosciences (Guangzhou, China). On day 0, mice were randomly allocated to MDA-MB-231 and MDA-MB-468 groups (n = 12 for each group) and received a subcutaneous injection of MDA-MB-231 or MDA-MB-468 cells (1 × 106, resuspended in 100 μl PBS). On days 14 to 28, six mice in each group were received an intraperitoneal injection of EF24 (3 mg/kg, dissolved in a PBS solution containing 6% castor oil) once a day, and the other mice in each group were injected intraperitoneally with the same volume of vehicle (served as the control group). The tumor volumes were calculated (0.5 × length × width2) at days 0, 7, 14, 21, and 28. After the last measurement of tumor volume, mice in MDA-MB-231+control (n = 6), MDA-MB-231+EF24 (n = 6), MDA-MB-468+control (n = 6), and MDA-MB-468+EF24 (n = 6) were euthanized and the xenograft tumors were collected.
To evaluate whether HCG11 mediated the effect of EF24 on TNBC in vivo, the LV-HCG11 and its negative control (LV-NC) were infected into MDA-MB-231 cells using Polybrene (Mito Biotechnology). BALBC/c nude mice were divided into control, EF24, EF24+LV-NC, and EF24+LV-HCG11 groups (n = 6 for each group). On day 0, 1 × 106 control/LV-NC-infected/LV-HCG11-infected MDA-MB-231 cells were subcutaneously injected into mice. On days 14 to 28, mice were treated with EF24 or vehicle as mentioned above. The tumor volumes were calculated at days 0, 7, 14, 21, and 28. After the last measurement of tumor volume, all mice were euthanized and the xenograft tumors were collected. All animal experiments were approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medical University.
Immunohistochemical staining.
The expressions of Ki-67, E-cadherin (E-cad), and vimentin were evaluated using immunohistochemical staining. Xenograft tumor tissues were prepared into 4 μm thick sections, followed by dewaxing and rehydrating. The sections were treated with 3% H2O2 for 10 min, sodium citrate buffer solution (pH = 6.0) for 20 min, and 5% bovine serum albumin for 30 min. Subsequently, sections were incubated with the primary antibody against Li-67 (1:250; Biobryt, Cambridge, UK), E-cad (1:100; Biorbyt), and vimentin (1:50; Biorbyt) overnight at 4°C. On the next day, sections were incubated with anti-rabbit IgG antibody (1:100; Boster Biological Technology, Wuhan, China) for 30 min and SABC (Boster Biological Technology) for 30 min. After being counterstained with hematoxylin for 4 min, the sections were photographed under a microscope (Leica, Wetzlar, Germany).
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay.
TUNEL assay was conducted using a TUNEL Apoptosis Detection Kit (Beyotime Biotechnology. Shanghai, China). For paraffin sections of xenograft tumor tissues, sections were dewaxed, rehydrated, and incubated with proteinase K for 20 min. After washing with PBS, the sections were incubated with TUNEL detection solution for 60 min in the dark. Then, cell nuclei were stained with DAPI. The sections were photographed under a microscope (Leica).
For MDA-MB-231 and MDA-MB-468 cells, cells were seeded on 6-well plates and subjected to the desired experimental protocol. Then, cells were fixed using 4% paraformaldehyde, treated with 0.3% Triton X-100 (in PBS) for 5 min, and incubated with 50 μl TUNEL detection solution for 60 min in the dark. Then, cell nuclei were stained with DAPI. The stained cells were photographed under a microscope (Leica).
qRT-PCR.
Tissue RNA purification kit and total RNA purification kit (all from Norgen Biotek, Thorold, Canada) were employed to extract total RNA samples from xenograft tumors/cancerous tissues of TNBC patients and MDA-MB-231/MDA-MB-468 cells, respectively. Samples of cDNA were synthesized from purified RNA samples using RevertAid H Minus Transcriptase (Thermo Fisher Scientific). qRT-PCR analysis was performed using PowerUp SYBR Green (Thermo Fisher Scientific). The expressions of HCG11 and Sp1 transcription factor (Sp1) were measured by the 2-ΔΔCt method. GAPDH was used for loading control.
Clinical samples.
Ten pairs of cancerous tissues and adjacent tissues from 10 patients were obtained from The First Affiliated Hospital of Zhejiang Chinese Medical University. All of them were newly diagnosed and had not undergone any medication. This study was approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medical University. The HCG11 expression in cancerous tissues and adjacent tissues was determined using qRT-PCR.
Cell proliferation, viability, and invasion.
Cell proliferation was examined using BeyoClick EdU-488 assay kit (Beyotime Biotechnology). MDA-MB-231 cells were seeded on 6-well plates and subjected to the desired experimental protocol. Then, cells were incubated with 10 μM EdU working solution (in Leibovitz's L-15 medium) for 2 h, 4% paraformaldehyde for 10 min, 0.3% Triton X-100 (in PBS) for 10 min, and 500 μl reaction mixture for 30 min. Cell nuclei were stained with DAPI. The stained cells were photographed under a microscope (Leica).
The viability of MDA-MB-231 and MDA-MB-468 cells was examined using MTT assay kit (Beyotime Biotechnology) as described previously (45).
The invasion of MDA-MB-231, MDA-MB-468, and BT-549 cells was examined using Tranwell assay. Briefly, 2 × 104 cells subjected to the desired experimental protocol were harvested and transferred into the upper chamber precoated with Matrigel matrix (Corning, Corning, NY, USA) in a 24-well plate. Cells in the upper chamber were cultured in Leibovitz's L-15 medium. The lower chamber was added with 700 μl Leibovitz's L-15/1640 medium containing 10% FBS. After 48 h normal culture, crystal violet was used to stain the cells that invaded the reverse side of the membrane. In the end, cells were observed under a microscope (Leica).
RNA pulldown assay.
Ribobio Biotechnology provided the biotin-labeled HCG11 probe and the biotin-labeled antisense probe used in this study. Cell lysates of MDA-MB-231 cells were prepared using the NP40 lysis buffer (Serviebio Biotechnology. Wuhan, China) and then reacted with HCG11/antisense probe-conjugated streptavidin magnetic beads (Thermo Fisher) at 4°C overnight. On the next day, the proteins attached to the probe-bead complex were extracted and subjected to Western blot analysis.
RNA binding protein immunoprecipitation (RIP) assay.
Before assay, anti-Sp1 (Abcam), and anti-human IgG antibodies (Abcam) were conjugated with protein A/G magnetic beads (Thermo Fisher). Then, cell lysates of MDA-MB-231 cells were incubated with antibody-bead complexes at 4°C overnight. On the next day, the RNA attached to the antibody-bead complex was extracted and subjected to qRT-PCR.
Co-IP assay.
The ubiquitination of Sp1 was evaluated using the Co-IP assay. Briefly, MDA-MB-231 cells were transfected with hemagglutinin (HA)-ubiquitin (Ub), Flag-Sp1, and pcDNA-HCG11/pcDNA. Forty-eight hours later, cells were treated with 10 μM MG132 for 2 h and then lysed using NP40 lysis buffer. Cell lysates were reacted with anti-Flag antibody (Abcam)-conjugated protein A/G magnetic beads at 4°C overnight. On the next day, the proteins attached to the antibody-bead complex were extracted and subjected to Western blot analysis.
Western blot.
Protein samples obtained from MDA-MB-231 cells and xenograft tumors were separated by 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. Nonfat powdered milk (5%; Bio Channel Biotechnology Co., Ltd., Nanjing, China) was used to block the membranes. Then, the membranes were incubated with an anti-Sp1 antibody (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) 4°C overnight and goat anti-mouse IgG–horseradish peroxidase (1:1,000; Santa Cruz Biotechnology) for 2 h at room temperature. Membranes were visualized using a ChemiDoc XRS+ System (Bio-Rad, Hercules, CA, USA).
Statistical analysis.
Data were expressed as the means ± standard deviations . All statistical analyses were conducted using SPSS Statistics 20.0 (IBM, Armonk, NY, USA). A value of P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Nature Science Fund of China (grant no. 81904032).
We declare that we have no conflicts of interest.
Footnotes
[This article was published on 20 January 2022 with Min Ling’s name misspelled as “Ming Lin” in the byline. The byline was updated in the current version, posted on 26 January 2022.]
REFERENCES
- 1.Naik A, Decock J. 2020. Lactate metabolism and immune modulation in breast cancer: a focused review on triple negative breast tumors. Front Oncol 10:598626. 10.3389/fonc.2020.598626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren D, Cheng H, Wang X, Vishnoi M, Teh BS, Rostomily R, Chang J, Wong ST, Zhao H. 2020. Emerging treatment strategies for breast cancer brain metastasis: from translational therapeutics to real-world experience. Ther Adv Med Oncol 12:1758835920936151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Guan X, Tang J. 2021. The long non-coding RNA landscape in triple-negative breast cancer. Cell Prolif 54:e12966. 10.1111/cpr.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou X, Jiao D, Dou M, Zhang W, Lv L, Chen J, Li L, Wang L, Han X. 2020. Curcumin inhibits the growth of triple-negative breast cancer cells by silencing EZH2 and restoring DLC1 expression. J Cell Mol Med 24:10648–10662. 10.1111/jcmm.15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. 2007. Bioavailability of curcumin: problems and promises. Mol Pharm 4:807–818. 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 6.Bertazza L, Barollo S, Mari ME, Faccio I, Zorzan M, Redaelli M, Rubin B, Armanini D, Mian C, Pezzani R. 2019. Biological effects of EF24, a curcumin derivative, alone or combined with mitotane in adrenocortical tumor cell lines. Molecules 24:2202. 10.3390/molecules24122202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin H, Chen X, Zhang C, Yang T, Deng Z, Song Y, Huang L, Li F, Li Q, Lin S, Jin D. 2021. EF24 induces ferroptosis in osteosarcoma cells through HMOX1. Biomed Pharmacother 136:111202. 10.1016/j.biopha.2020.111202. [DOI] [PubMed] [Google Scholar]
- 8.Yin DL, Liang YJ, Zheng TS, Song RP, Wang JB, Sun BS, Pan SH, Qu LD, Liu JR, Jiang HC, Liu LX. 2016. EF24 inhibits tumor growth and metastasis via suppressing NF-kappaB dependent pathways in human cholangiocarcinoma. Sci Rep 6:32167. 10.1038/srep32167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams BK, Cai J, Armstrong J, Herold M, Lu YJ, Sun A, Snyder JP, Liotta DC, Jones DP, Shoji M. 2005. EF24, a novel synthetic curcumin analog, induces apoptosis in cancer cells via a redox-dependent mechanism. Anticancer Drugs 16:263–275. 10.1097/00001813-200503000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Xu K-l. 2018. LncRNA NEAT1 is involved in temozolomide resistance by regulating MGMT in glioblastoma multiforme. Clin Surg Res Commun 2. 10.31491/CSRC.2018.3.011. [DOI] [Google Scholar]
- 11.Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, Wang Z, Wang C, Zhang X, Mo YY. 2016. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget 7:20584–20596. 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Huang H, Xu X, Wang H, Wang J, Yao Z, Xu X, Wu Q, Xu F. 2019. LncRNA HCG11 promotes proliferation and migration in gastric cancer via targeting miR-1276/CTNNB1 and activating Wnt signaling pathway. Cancer Cell Int 19:350. 10.1186/s12935-019-1046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Liu L, Zhang J, Huang C, Chen Y, Bai W, Wang Y, Zhao K, Li S. 2020. LncRNA HCG11 suppresses cell proliferation and promotes apoptosis via sponging miR-224-3p in non-small-cell lung cancer cells. Onco Targets Ther 13:6553–6563. 10.2147/OTT.S244181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dashti S, Taherian-Esfahani Z, Kholghi-Oskooei V, Noroozi R, Arsang-Jang S, Ghafouri-Fard S, Taheri M. 2020. In silico identification of MAPK14-related lncRNAs and assessment of their expression in breast cancer samples. Sci Rep 10:8316. 10.1038/s41598-020-65421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko JH, Yang MH, Baek SH, Nam D, Jung SH, Ahn KS. 2019. Theacrine attenuates epithelial mesenchymal transition in human breast cancer MDA-MB-231 cells. Phytother Res 33:1934–1942. 10.1002/ptr.6389. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Zhou Y, Mao F, Lin Y, Shen S, Sun Q. 2020. lncRNA AFAP1-AS1 promotes triple negative breast cancer cell proliferation and invasion via targeting miR-145 to regulate MTH1 expression. Sci Rep 10:7662. 10.1038/s41598-020-64713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y, Wei N, Ma R, Jiang SH, Song D. 2020. Long noncoding RNA MIR210HG promotes the Warburg effect and tumor growth by enhancing HIF-1α translation in triple-negative breast cancer. Front Oncol 10:580176. 10.3389/fonc.2020.580176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma T, Liu H, Liu Y, Liu T, Wang H, Qiao F, Song L, Zhang L. 2020. USP6NL mediated by LINC00689/miR-142-3p promotes the development of triple-negative breast cancer. BMC Cancer 20:998. 10.1186/s12885-020-07394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S. 2020. LncRNA DLG1-AS1 promotes cancer cell proliferation in triple negative breast cancer by downregulating miR-203. J Breast Cancer 23:343–354. 10.4048/jbc.2020.23.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Pan Y, Pan Y, Wang O. 2019. MNX1-AS1 is a functional oncogene that induces EMT and activates the AKT/mTOR pathway and MNX1 in breast cancer. Cancer Manag Res 11:803–812. 10.2147/CMAR.S188007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du C, Wang Y, Zhang Y, Zhang J, Zhang L, Li J. 2020. LncRNA DLX6-AS1 contributes to epithelial-mesenchymal transition and cisplatin resistance in triple-negative breast cancer via modulating mir-199b-5p/paxillin axis. Cell Transplant 29:963689720929983. 10.1177/0963689720929983. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Fang X, Zhang J, Li C, Liu J, Shi Z, Zhou P. 2020. Long non-coding RNA SNHG22 facilitates the malignant phenotypes in triple-negative breast cancer via sponging miR-324-3p and upregulating SUDS3. Cancer Cell Int 20:252. 10.1186/s12935-020-01321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Zhi X, Zhou Z, Zhang H, Yang R, Zou T, Chen C. 2018. Mithramycin A suppresses basal triple-negative breast cancer cell survival partially via down-regulating Krüppel-like factor 5 transcription by Sp1. Sci Rep 8:1138. 10.1038/s41598-018-19489-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park CR, Lee JS, Son CG, Lee NH. 2021. A survey of herbal medicines as tumor microenvironment-modulating agents. Phytother Res 35:78–94. 10.1002/ptr.6784. [DOI] [PubMed] [Google Scholar]
- 25.Kasinski AL, Du Y, Thomas SL, Zhao J, Sun SY, Khuri FR, Wang CY, Shoji M, Sun A, Snyder JP, Liotta D, Fu H. 2008. Inhibition of IkappaB kinase-nuclear factor-kappaB signaling pathway by 3,5-bis(2-flurobenzylidene)piperidin-4-one (EF24), a novel monoketone analog of curcumin. Mol Pharmacol 74:654–661. 10.1124/mol.108.046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Bai H, Liu G, Wang H, Chen F, Zhang B, Zeng P, Wu C, Peng C, Huang C, Song Y, Song E. 2015. MicroRNA-33b, upregulated by EF24, a curcumin analog, suppresses the epithelial-to-mesenchymal transition (EMT) and migratory potential of melanoma cells by targeting HMGA2. Toxicol Lett 234:151–161. 10.1016/j.toxlet.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Yang CH, Yue J, Sims M, Pfeffer LM. 2013. The curcumin analog EF24 targets NF-κB and miRNA-21, and has potent anticancer activity in vitro and in vivo. PLoS One 8:e71130. 10.1371/journal.pone.0071130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan Y, Liu B, Guo H. 2021. The role of M(6)A modification in the regulation of tumor-related lncRNAs. Mol Ther Nucleic Acids 24:768–779. 10.1016/j.omtn.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Chen TQ, Fang K, Zeng ZC, Ye H, Chen YQ. 2021. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J Hematol Oncol 14:117. 10.1186/s13045-021-01129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y, Zheng C, Jin Y, Bao B, Wang D, Hou K, Feng J, Tang S, Qu X, Liu Y, Che X, Teng Y. 2020. Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6A methylation-mediated COL3A1 up-regulation. Front Oncol 10:1126. 10.3389/fonc.2020.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu N, Li X, Yu J, Li Y, Wang C, Zhang L, Wang T, Zhong X. 2018. Curcumin attenuates lipopolysaccharide-induced hepatic lipid metabolism disorder by modification of m6A RNA methylation in piglets. Lipids 53:53–63. 10.1002/lipd.12023. [DOI] [PubMed] [Google Scholar]
- 32.Long R, Liu Z, Li J, Zhang Y, Yu H. 2021. HCG11 up-regulation induced by ELK4 suppressed proliferation in vestibular schwannoma by targeting miR-620/ELK4. Cancer Cell Int 21:5. 10.1186/s12935-020-01691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li XF, Hu DM, Zhao YX, Zhang L, Jin Y. 2020. Knockdown of lncRNA HCG11 suppresses cell progression in ovarian cancer by modulating miR-144-3p/PBX3. Eur Rev Med Pharmacol Sci 24:11032–11040. 10.26355/eurrev_202011_23588. [DOI] [PubMed] [Google Scholar]
- 34.Hu C, Yan S, Yang X. 2020. Pioglitazone up-regulates MALAT1 and promotes the proliferation of endothelial progenitor cells through increasing c-Myc expression in type 2 diabetes mellitus. Apt 2:38–44. 10.31491/APT.2020.03.011. [DOI] [Google Scholar]
- 35.Vizcaíno C, Mansilla S, Portugal J. 2015. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther 152:111–124. 10.1016/j.pharmthera.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Zhu ZG, Ji J, Yuan F, Yu YY, Liu BY, Lin YZ. 2005. Transcription factor Sp1 expression in gastric cancer and its relationship to long-term prognosis. WJG 11:2213–2217. 10.3748/wjg.v11.i15.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Kang M, Qin YT, Wei ZX, Xiao JJ, Wang RS. 2015. Sp1 is over-expressed in nasopharyngeal cancer and is a poor prognostic indicator for patients receiving radiotherapy. Int J Clin Exp Pathol 8:6936–6943. [PMC free article] [PubMed] [Google Scholar]
- 38.Luo CW, Hou MF, Huang CW, Wu CC, Ou-Yang F, Li QL, Wu CC, Pan MR. 2020. The CDK6-c-Jun-Sp1-MMP-2 axis as a biomarker and therapeutic target for triple-negative breast cancer. Am J Cancer Res 10:4325–4341. [PMC free article] [PubMed] [Google Scholar]
- 39.Hariri H, St-Arnaud R. 2021. Expression and role of ubiquitin-specific peptidases in osteoblasts. Int J Mol Sci 22:7746. 10.3390/ijms2214774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui J, Shu C, Xu J, Chen D, Li J, Ding K, Chen M, Li A, He J, Shu Y, Yang L, Zhang R, Zhou J. 2020. JP1 suppresses proliferation and metastasis of melanoma through MEK1/2 mediated NEDD4L-SP1-Integrin αvβ3 signaling. Theranostics 10:8036–8050. 10.7150/thno.45843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong X, Liu Z, Zhang E, Zhang P, Wang Y, Hang J, Li Q. 2021. USP39 promotes tumorigenesis by stabilizing and deubiquitinating SP1 protein in hepatocellular carcinoma. Cell Signal 85:110068. 10.1016/j.cellsig.2021.110068. [DOI] [PubMed] [Google Scholar]
- 42.Hull R, Mbita Z, Dlamini Z. 2021. Long non-coding RNAs (LncRNAs), viral oncogenomics, and aberrant splicing events: therapeutics implications. Am J Cancer Res 11:866–883. [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T, Beeharry MK, Wang Z, Zhu Z, Li J, Li C. 2021. YY1-modulated long non-coding RNA SNHG12 promotes gastric cancer metastasis by activating the miR-218-5p/YWHAZ axis. Int J Biol Sci 17:1629–1643. 10.7150/ijbs.58921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu QQ, Lai MC, Chen TC, Wang X, Tian L, Li DL, Wu ZH, Wang XH, He YY, He YY, Shang T, Xiang YL, Zhang HK. 2021. LncRNA SNHG15 relieves hyperglycemia-induced endothelial dysfunction via increased ubiquitination of thioredoxin-interacting protein. Lab Invest 101:1142–1152. 10.1038/s41374-021-00614-5. [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Yuhua H, Jianquan H, Jinxing L, Xiang D. 2020. MiR-181c inhibits prostatic epithelial cell proliferation caused by chronic non-bacterial prostatitis through downregulating COX-2. Apt 2:210–218. 10.31491/APT.2020.12.042. [DOI] [Google Scholar]